Published online Sep 26, 2025. doi: 10.4330/wjc.v17.i9.110278
Revised: June 25, 2025
Accepted: August 11, 2025
Published online: September 26, 2025
Processing time: 99 Days and 5 Hours
Coronary heart disease (CHD) is a prominent cause of mortality and disability worldwide. Like most complex diseases, the risk of CHD in individuals is regulated by the interaction between genetic factors and lifestyle. APOE and SLCO1B1 genetic polymorphisms and LPA KIV-2 copy number variation may influence the development and progression of CHD. Clarifying gene polymor
To investigate the influence of APOE and SLCO1B1 gene polymorphisms, as well as LPA KIV-2 copy number variation on CHD in the Teochew population.
A total of 324 patients with CHD and 143 control participants were involved in this study. Single nucleotide polymorphisms rs429358 and rs7412 in the APOE gene, and rs2306283 and rs4149056 in the SLCO1B1 gene were analyzed via high-resolution melting curve analysis. Additionally, PCR was performed to detect KIV-2 copy number variations. Clinical risk factors and potential effects on CHD patients were subsequently assessed.
In the CHD group, the frequencies of APOE allele ε2, ε3, ε4 were 8.02%, 82.97%, and 9.10%, respectively. Compared to the control groups (13.29%, 79.37%, and 7.34%, respectively), the ε2 allele frequency showed a significant difference (8.02% vs 13.29%, P = 0.012). SLCO1B1 allele frequencies in the CHD group were not significantly different from those in the control group (*1a: 26.69% vs 25.52%, *1b: 61.17% vs 65.38%, *5: 0.15% vs 0.35%, *15: 11.83% vs 8.74%). The number of copies of the KIV-2 gene was significantly lower in the CHD group when compared to controls (23.35 ± 8.78 vs 27.21 ± 9.48; P < 0.01). Logistic regression analysis revealed that sex, age, hypertension, diabetes, smoking, the ε2 allele and KIV-2 copy number were factors influencing the presence of CHD.
In the Teochew population, the APOE ε2 allele and a higher KIV-2 copy number were associated with a reduced risk of CHD. In contrast, the APOE ε4 allele and SLCO1B1 gene were not associated with CHD.
Core Tip: In this study of 324 patients with coronary heart disease (CHD) and 143 control participants, we analysed polymorphisms in the APOE and SLCO1B1 genes, as well as LPA KIV-2 copy number variation in the Teochew population of southern China. We confirmed that both APOE ε2 allele and higher KIV-2 copy number are associated with a reduced risk of CHD in the Teochew population, whereas neither the APOE ε4 allele nor SLCO1B1 gene showed any association.
- Citation: Xu JX, Wu Y, Zhang L, Wu YH, Li CL, Lin F. Correlation of APOE, SLCO1B1 and LPA KIV-2 gene polymorphisms with coronary heart disease in the Teochew population. World J Cardiol 2025; 17(9): 110278
- URL: https://www.wjgnet.com/1949-8462/full/v17/i9/110278.htm
- DOI: https://dx.doi.org/10.4330/wjc.v17.i9.110278
Coronary heart disease (CHD) is a prominent cause of mortality and disability worldwide; it arises from the constriction or obstruction of coronary arteries caused by atherosclerosis, ultimately resulting in decreased blood flow and insufficient oxygen supply to the heart muscle[1]. The mortality and morbidity of CHD continue to rise worldwide, particularly in low- and middle-income countries as well as in the elderly population[2]. Like most complex diseases, the risk of CHD in individuals is regulated by the interaction between genetic factors and lifestyle. The impact of genetic polymorphisms on the occurrence and progression of CHD has been well documented; for example, genome-wide association studies have identified more than 300 genetic loci associated with CHD[3]. Owing to differences in gene distribution, the risk of disease may vary across geographic areas and populations. Clarifying gene polymorphisms can guide clinical precision and prevention, thereby improving treatment outcomes.
Apolipoprotein E (ApoE) is a ubiquitous lipid transport protein that binds a variety of lipid species, including cholesterol, phospholipids, and triglycerides (TG), in lipoprotein particles[4]. ApoE plays an important role in lipid metabolism. The APOE gene encodes APOE and contains two single nucleotide polymorphisms (SNPs), rs429358 (388T>C, Cys130Arg) and rs7412 (526C>T, Arg176Cys). These SNPs can generate three haplotypes, namely, ε2 (388T-526T), ε3 (388T-526C) and ε4 (388C-526C), resulting in six genotypes (ε2/ε2, ε2/ε3, ε2/ε4, ε3/ε3, ε3/ε4 and ε4/ε4)[5]. Previous studies suggested that three isoforms, ApoE2 (Cys112/Cys158), ApoE3 (Cys112/Arg158) and ApoE4 (Arg112/Arg158) can lead to changes in lipid and lipoprotein levels. These single amino acid substitutions drastically alter the functionality of ApoE, resulting in isoform-specific variations in its structure[6].
The SLCO1B1 gene encodes OATP1B1, a drug transporter that contributes to the hepatic uptake of statins[7]. Statins are commonly used for the primary and secondary prevention of CHD. SLCO1B1 gene polymorphisms are the main cause of individual differences in statin efficacy and adverse effects. Rs2306283 (388A>G) and rs4149056 (521T>C) are two common SNPs in the SLCO1B1 gene. Among them, the 521T>C SNP induces in a substitution of alanine for valine at amino acid position 174, whereas the 388A>G variant causes an Asn130Asp mutation within the extracellular region of the transporter[8]. These two SNPs can form four haplotypes, including SLCO1B1*1a (388A-521T), SLCO1B1*1b (388G-521T), SLCO1B1*5 (388A-521C), and SLCO1B1*15 (388G-521C).
Lipoprotein(a) [Lp(a)] is a unique liver-derived lipoprotein associated with key processes in atherosclerosis[9]. Lp(a) consists of an LDL-like moiety with one plasminogen-like apolipoprotein(a) covalently bound to apolipoprotein B (apo B). More than 90% of the variance in Lp(a) concentration is determined by genetic variation in the apo (a) gene (LPA). LPA has a highly repetitive structure consisting of 10 highly homologous kringle IV (KIV) domains (subtypes 1 to 10), a kringle V domain and a protease domain. The KIV-2 domain is encoded in a 5.6-kilobase large DNA copy number variation, which can be present up to > 40 × per allele[10].
Current studies in China and other regions have investigated the APOE gene, SLCO1B1 gene, and LPA KIV-2 copy number as candidate genes for CHD, revealing significant interpopulation and geographic variability in allele fre
The purpose of this study was to analyse polymorphisms in the APOE and SLCO1B1 genes, as well as LPA KIV-2 copy number variation, in the Teochew population of southern China, providing a theoretical basis for the prevention and treatment of CHD.
The subjects included 467 patients who underwent coronary angiography due to chest tightness, chest pain, and dyspnoea at Chaozhou Central Hospital from December 2023 to November 2024. Among them, 324 patients were diagnosed with CHD, while 143 patients were in the control group without CHD. The diagnostic criteria for CHD were based on the classification of the American College of Cardiology/American Heart Association. Coronary angiography revealed that at least one main branch of the left main artery, left anterior descending artery, left circumflex artery or right coronary artery was stenotic, with a degree of stenosis exceeding 50%. Patients with malignant neoplasms, severe heart or liver disease, kidney dysfunction, or autoimmune diseases were excluded. This study adhered to the ethical guidelines outlined in the Helsinki Declaration and was approved by the Institutional Review Board of Chaozhou Central Hospital (No. 2023014).
The electronic medical records of all patients were thoroughly reviewed. The clinical data collected included demogra
Peripheral blood samples from all the subjects were collected into tubes with EDTA-K2, stored at 4 °C, and then sent to the molecular laboratory for analysis. DNA was extracted from peripheral venous blood via whole blood DNA extraction kits (Chaozhou Hybribio Co., Ltd.) according to the manufacturer’s instructions. The DNA concentration was determined by measuring the UV absorption at 260 nm with a spectrophotometer (NanoDrop One, Thermo Fisher Scientific Co., Ltd.).
Polymorphisms in the APOE and SLCO1B1 genes were identified via reagent kits (Hybribio Company, Chaozhou, China), and the fluorescence PCR melting curve method was used to determine the corresponding mutation types of the target genes. The target fragments were amplified via the SLAN-96P Real-Time PCR System (Shanghai Hongshi Medical Technology Co., Ltd.). The amplification protocol included an initial denaturation step of 95 °C for 5 minutes, followed by 45 cycles of 95 °C for 15 minutes, 56 °C for 20 seconds, and 72 °C for 20 seconds. This step was followed by a final step of 95 °C for 1 minute and 40 °C for 3 minutes. The PCR products were analysed by monitoring the fluorescence from 40 °C to 80 °C at a temperature transition rate of 0.05 °C per second.
The KIV-2 repeat copy number was detected via a real-time fluorescent quantitative PCR detection kit (Shanghai WeHealth Co., Ltd.). The real-time fluorescent quantitative PCR cycle conditions were as follows: Initial denaturation at 98 °C for 2 minutes, followed by 45 cycles of 98 °C for 10 seconds, 60 °C for 30 seconds, and 68 °C for 30 seconds (with fluorescence collection). For the melting curve analysis, the conditions were set at 95 °C for 5 seconds, 65 °C for 1 minute, and a gradual temperature increase from 65 °C to 95 °C at an increment of 0.05 °C. The KIV-2 repeat copy number was calculated via the following formula: ΔCtSample = CtSample KIV-2 - CtSample Reference; ΔCtStandard = CtStandard KIV-2 - CtStandard Reference; KIV-2 repeat copy number = 2-ΔΔCt (ΔΔCt = ΔCtSample - ΔCtStandard).
Analysis was performed via the SPSS statistical package version 21.0 (SPSS Inc., Chicago, IL, United States). Hardy-Weinberg equilibrium was assessed via Pearson’s χ2 test of the observed genetic frequencies. The measurement data are presented as the mean ± SD, and nonnormally distributed data are presented as the medians (interquartile ranges). The t test and nonparametric tests were used for comparisons between groups. Enumeration data are expressed as a percentage (%) and were compared via the χ2 test. The factors influencing CHD incidence were analysed via logistic regression. P < 0.05 was considered statistically significant.
The participants of this study included 467 Teochew individuals aged between 20 and 89 years. Among them, 324 were CHD patients (234 men, 85 women) and 143 were non-CHD patients (78 men, 65 women). As shown in Table 1, the age, male proportion, prevalence of hypertension, prevalence of diabetes, history of smoking, TG, homocysteine, and Lp(a) levels in the CHD group were significantly greater than those in the control group, whereas the levels of HDL-C, and ApoA1 were lower than those in the control group. There was no significant difference BMI, history of alcohol con
| CHD group (n = 324) | Control group (n = 143) | P value | |
| Clinical data | |||
| Male | 239 (73.77) | 78 (54.55) | < 0.001 |
| Age, years | 64.93 ± 11.58 | 57.27 ± 13.74 | < 0.001 |
| BMI, kg/m2 | 23.21 ± 3.06 | 23.56 ± 3.89 | 0.746 |
| Comorbidities | |||
| Hypertension | 187 (57.72) | 43 (30.07) | < 0.001 |
| Diabetes | 104 (32.10) | 15 (10.49) | < 0.001 |
| History of smoking | 114 (35.19) | 27 (18.88) | < 0.001 |
| History of drinking | 21 (6.48) | 5 (3.50) | 0.273 |
| Laboratory data | |||
| TG, mmol/L | 1.40 (0.89) | 1.26 (0.72) | 0.024 |
| TC, mmol/L | 4.69 (1.63) | 4.87 (1.40) | 0.580 |
| HCY, μmol/L | 11.87 (5.93) | 9.82 (3.04) | < 0.001 |
| HDL-C, mmol/L | 1.07 (0.37) | 1.19 (0.40) | < 0.001 |
| LDL-C, mmol/L | 2.95 (1.42) | 2.87 (1.17) | 0.422 |
| ApoA1, g/L | 1.05 (0.25) | 1.16 (0.30) | < 0.001 |
| ApoB, g/L | 0.85 (0.35) | 0.83 (0.30) | 0.298 |
| Lp(a), mg/L | 122.50 (266.13) | 100.2 (168.00) | 0.007 |
| ApoE, mg/L | 38.65 (14.40) | 38.95 (15.00) | 0.627 |
In the CHD group, the frequencies of genotypes ε3/ε3, ε3/ε4, ε2/ε3, ε2/ε2, ε2/ε4, and ε4/ε4 were 69.75%, 15.12%, 11.11%, 1.85%, 1.23%, and 0.93%, respectively. The frequencies of alleles ε3, ε4, and ε2 were 82.97%, 9.10%, and 8.02%, respectively. In the control group, the frequencies of genotypes ε3/ε3, ε2/ε3, ε3/ε4, ε2/ε4, ε2/ε2 and ε4/ε4 were 62.94%, 21.68%, 11.19%, 2.10%, 1.40% and 0.70%, respectively. The frequencies of alleles ε3, ε2 and ε4 were 79.37%, 13.29% and 7.34%, respectively. The results indicate that the control group was in Hardy-Weinberg equilibrium (χ2 = 0.243, P = 0.886) for genetic polymorphisms. ε3/ε3 was the most common APOE genotype, and ε3 was also the most frequent allele. Compared with the control group, there was a statistically significant difference in genotype ε2/ε3 among the CHD group (P = 0.003), and the frequency of the allele ε2 was significantly lower in the CHD group than in the control group (P = 0.012). In addition, there was no statistically significant difference in the genotypes of APOE genes (ε2/ε2, ε2/ε4, ε3/ε3, ε3/ε4, and ε4/ε4) or alleles (ε3 and ε4) between the groups (P > 0.05) (Table 2).
| Genotype | CHD group (n = 324) | Control group (n = 143) | Total (n = 467) | P value |
| ε2/ε2 | 6 (1.85) | 2 (1.40) | 8 (1.71) | 1.000 |
| ε2/ε3 | 36 (11.11) | 31 (21.68) | 67 (14.35) | 0.003 |
| ε2/ε4 | 4 (1.23) | 3 (2.10) | 7 (1.50) | 0.442 |
| ε3/ε3 | 226 (69.75) | 90 (62.94) | 316 (67.67) | 0.147 |
| ε3/ε4 | 49 (15.12) | 16 (11.19) | 65 (13.92) | 0.258 |
| ε4/ε4 | 3 (0.93) | 1 (0.70) | 4 (0.86) | 1.000 |
| Allele | CHD group (n = 648) | Control group (n = 286) | Total (n = 934) | |
| ε2 | 52 (8.02) | 38 (13.29) | 90 (9.64) | 0.012 |
| ε3 | 537 (82.87) | 227 (79.37) | 764 (81.80) | 0.201 |
| ε4 | 59 (9.10) | 21 (7.34) | 80 (8.57) | 0.375 |
In the CHD group, the frequencies of genotypes *1b/*1b, *1a/*1b, *1b/*15, *1a/*1a, *1a/*15, *15/*15 and*1a/*5 were 36.72%, 32.10%, 16.98%, 8.02%, 4.94%, 0.93% and 0.31%, respectively. In the control group, the percentages were 42.66%, 31.47%, 13.99%, 7.69%, 3.50%, 0%, and 0.70%, respectively. The *5/*5 and *5/*15 genotypes were not detected in either group. The control group was in Hardy-Weinberg equilibrium (χ2 = 4.931, P = 0.085) for this polymorphism. The *1b allele was the most common in both the CHD group and the control group (61.17% and 65.38%), followed by alleles *1a, *15 and *5. There was no statistically significant difference in the frequencies of SLCO1B1 genotypes and alleles between CHD patients and the control group (P > 0.05) (Table 3).
| Genotype | CHD group (n = 324) | Control group (n = 143) | Total (n = 467) | P value |
| *1a/*1a | 26 (8.02) | 11 (7.69) | 37 (7.92) | 0.902 |
| *1a/*1b | 104 (32.10) | 45 (31.47) | 149 (31.91) | 0.893 |
| *1b/*1b | 119 (36.72) | 61 (42.66) | 180 (38.54) | 0.225 |
| *1a/*5 | 1 (0.31) | 1 (0.70) | 2 (0.43) | 0.519 |
| *1a/*15 | 16 (4.94) | 5 (3.50) | 21 (4.50) | 0.488 |
| *1b/*15 | 55 (16.98) | 20 (13.99) | 75 (16.06) | 0.417 |
| *5/*5 | 0 (0) | 0 (0) | 0 (0) | NA |
| *5/*15 | 0 (0) | 0 (0) | 0 (0) | NA |
| *15/*15 | 3 (0.93) | 0 (0) | 3 (0.64) | 0.556 |
| Allele | CHD group (n = 648) | Control group (n = 286) | Total (n = 934) | |
| *1a | 173 (26.69) | 73 (25.52) | 246 (26.34) | 0.640 |
| *1b | 397 (61.17) | 187 (65.38) | 584 (62.53) | 0.231 |
| *5 | 1 (0.15) | 1 (0.35) | 2 (0.21) | 0.520 |
| *15 | 77 (11.83) | 25 (8.74) | 102 (10.92) | 0.156 |
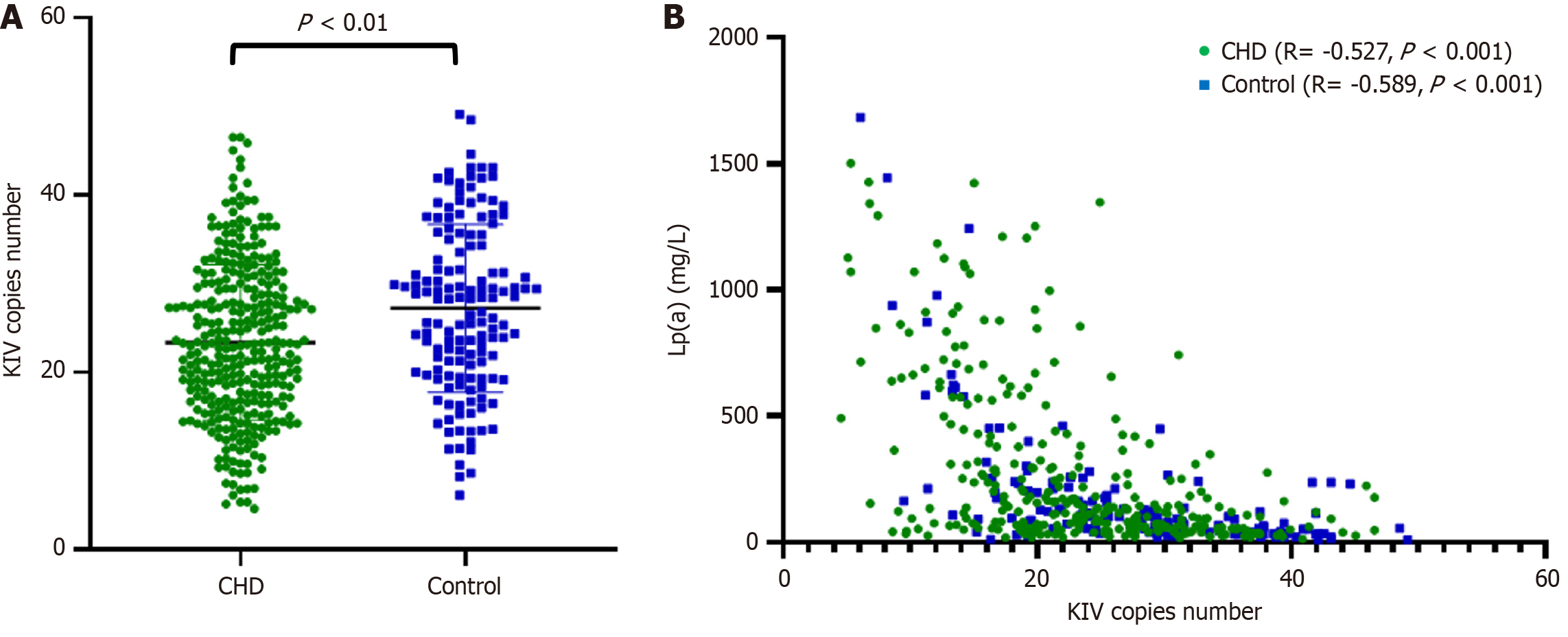
The LPA KIV-2 copy number variation is shown in Figure 1. In the CHD group, the LPA KIV-2 copy number was
The CHD patients were further classified into three groups according to their genotypes: The E2 group (genotypes ε2/ε2 and ε2/ε3), the E3 group (genotypes ε3/ε3), and the E4 group (genotypes ε3/ε4 and ε4/ε4). The relationships between alleles and blood lipid levels were analysed. Owing to the opposite roles of the ε2 and ε4 alleles in lipid metabolism, four patients with ε2/ε4 alleles were excluded. The results revealed that the levels of TC, LDLC, and ApoB in the E4 group were significantly greater than those in the E3 and E2 groups. Similarly, the TG levels in the E4 group were greater than those in the E3 group (Table 4).
| E2 (n = 42) | E3 (n = 226) | E4 (n = 52) | P value | |
| TG, mmol/L | 1.62 ± 0.89 | 1.59 ± 1.10 | 2.09 ± 0.73a | 0.022 |
| TC, mmol/L | 4.57 ± 1.22 | 4.72 ± 1.26 | 5.47 ± 1.70a,b | 0.001 |
| HCY, μmol/L | 14.24 ± 6.73 | 13.33 ± 8.24 | 14.82 ± 8.77 | 0.442 |
| HDL-C, mmol/L | 1.09 ± 0.30 | 1.11 ± 0.29 | 1.08 ± 0.25 | 0.792 |
| LDL-C, mmol/L | 2.69 ± 0.98 | 2.95 ± 1.06 | 3.54 ± 1.36a,b | < 0.001 |
| ApoA1, g/L | 1.05 ± 0.21 | 1.11 ± 0.62 | 1.06 ± 0.21 | 0.730 |
| ApoB, g/L | 0.79 ± 0.28 | 0.86 ± 0.36 | 1.00 ± 0.33a,b | 0.009 |
| LP(a), mg/L | 214.55 ± 290.31 | 275.79 ± 321.48 | 259.50 ± 331.44 | 0.517 |
| ApoE, mg/L | 46.69 ± 15.84 | 39.86 ± 14.11 | 45.43 ± 26.79 | 0.013 |
According to the SLCO1B1 genotype, we divided the CHD group into three categories: Extensive metabolizer I (*1a/*1a, *1a/*1b, *1b/*1b), intermediate metabolizer II (*1a/*5, *1a/*15, *1b/*15), and poor metabolizer III (*5/*5, *5/*15, *15/*15). Due to the relatively small number of patients in the poor metabolizer III group (3 cases), we excluded them from the comparison. We subsequently analysed the relationships between genotypes and blood lipid levels in columns I and II. The results revealed that the levels of LDL-C (3.08 ± 1.16 vs 2.69 ± 0.91, P < 0.016) and ApoB (0.89 ± 0.37 vs 0.79 ± 0.25, P < 0.045) in group I were significantly greater than those in group II (Table 5).
| Extensive metabolizer I (n = 249) | Intermediate metabolizer II (n = 72) | P value | |
| TG, mmol/L | 1.65 ± 1.20 | 1.66 ± 1.05 | 0.992 |
| TC, mmol/L | 4.88 ± 1.22 | 4.56 ± 1.08 | 0.119 |
| HCY, μmol/L | 13.73 ± 8.54 | 13.61 ± 6.54 | 0.801 |
| HDL-C, mmol/L | 1.10 ± 0.29 | 1.11 ± 0.29 | 0.778 |
| LDL-C, mmol/L | 3.08 ± 1.16 | 2.69 ± 0.91 | 0.016 |
| ApoA1, g/L | 1.09 ± 0.59 | 1.09 ± 0.21 | 0.256 |
| ApoB, g/L | 0.89 ± 0.37 | 0.79 ± 0.25 | 0.045 |
| LP(a), mg/L | 248.62 ± 300.96 | 323.28 ± 379.60 | 0.306 |
| ApoE, mg/L | 42.03 ± 18.17 | 39.96 ± 12.88 | 0.419 |
Multivariate logistic regression analysis was used to evaluate the incidence of CHD as the dependent variable. Our results revealed that age, sex, hypertension, diabetes, and smoking were independent risk factors for CHD. Compared with APOE E3 (ε3/ε3), APOE E2 (ε2/ε2, ε2/ε3) was significantly associated with CHD (OR 0.395, 95%CI: 0.209-0.745; P = 0.004). Based on the extensive metabolizer phenotype of the SLCO1B1 gene, poor metabolism and intermediate metabo
| Variables | Unadjusted values | Adjusted values | ||
| OR (95%CI) | P value | OR (95%CI) | P value | |
| Male | 2.343 (1.552-3.537) | < 0.001 | 2.559 (1.395-4.692) | 0.002 |
| Age, years | 1.050 (1.032-1.067) | < 0.001 | 1.063 (1.040-1.085) | < 0.001 |
| Hypertension | 3.174 (2.086-4.831) | < 0.001 | 2.358 (1.420-3.917) | 0.001 |
| Diabetes | 4.034 (2.251-7.230) | < 0.001 | 3.609 (1.813-7.184) | < 0.001 |
| History of smoking | 2.332 (1.448-3.757) | < 0.001 | 2.042 (1.092-3.818) | 0.025 |
| TG, mmol/L | 1.581 (1.009-2.447) | 0.046 | 1.526 (0.855-2.725) | 0.153 |
| HCY, μmol/L | 3.282 (1.824-5.906) | < 0.001 | 1.888 (0.933-3.819) | 0.077 |
| HDL-C, mmol/L | 0.363 (0.227-0.581) | < 0.001 | 0.577 (0.308-1.083) | 0.087 |
| ApoA1, g/L | 2.320 (1.517-3.548) | < 0.001 | 1.402 (0.793-2.478) | 0.245 |
| LP(a), mg/L | 2.358 (1.371-4.056) | 0.002 | 1.577 (0.742-3.350) | 0.236 |
| Genotype | ||||
| E2 | 0.507 (0.302-0.850) | 0.010 | 0.395 (0.209-0.745) | 0.004 |
| E4 | 1.218 (0.669-2.219) | 0.519 | 1.028 (0.500-2.110) | 0.941 |
| SLCO1B1 | 1.355 (0.824-2.229) | 0.231 | 1.666 (0.896-3.096) | 0.107 |
| KIV | 0.954 (0.933-0.976) | < 0.001 | 0.949 (0.919-0.979) | 0.001 |
CHD, distinguished as a widespread chronic ailment and a paramount cause of global mortality, represents a critical public health challenge. Over time, extensive studies have highlighted that genetic predispositions play a crucial role in the aetiology of CHD[13]. The exploration of genetic polymorphisms has become a central theme in investigations into hereditary susceptibility and preventive measures for CHD. Early identification and intervention of modifiable risk factors offer the potential to mitigate the immense toll exacted by this disease on individuals and societies alike. In light of this, our study endeavours to elucidate the interplay between CHD and specific genetic markers within the Teochew population-a region with distinctive genetic and environmental characteristics. Focusing on the APOE and SLCO1B1 gene polymorphisms, alongside the copy number variation of KIV-2, we aim to uncover insights essential for developing personalized and precision medicine approaches tailored to CHD management in the Chaoshan area.
The relationships between several cardiovascular disease risk factors and the incidence of CHD, including age, sex, hypertension, diabetes and smoking, were also verified in this study. These indicators were significantly different between the control and CHD groups. However, there are still several clinically recognized risk factors, such as BMI, alcohol consumption and TC levels, which have not been proven to establish a definitive link with CHD. While the distributions of these factors are similar across both groups, subtle discrepancies exist without statistical significance. Such observations may stem from demographic characteristics intrinsic to the Teochew populations, potentially skewing the results owing to regional biases. To eliminate biases caused by sex, age, diabetes, hypertension, and smoking history, we used logistic regression analysis, including these confounding factors as covariates in the regression model, and calculated the adjusted regression coefficients. We found that APOE E2 (ε2/ε2, ε2/ε3) remained significantly associated with CHD, and a higher copy number of KIV-2 was identified as a protective factor against CHD.
APOE plays a pivotal role in lipoprotein synthesis and metabolism and significantly influences susceptibility to cardiovascular diseases. The APOE gene polymorphism has been shown to be related to atherosclerosis[14]. Regional and ethnic differences result in varied frequencies of APOE genotype distributions. In our study, we found that the ε3 allele had the highest distribution frequency in the Chaoshan area, accounting for 81.80%, followed by ε2 (9.64%) and ε4 (8.57%). This distribution pattern is consistent with those observed in numerous cities across China, such as Guangzhou, Chongqing, Meizhou, Fuzhou, etc.[15-18]. Compared with that in the control group, the frequency of the ε2 allele in the CHD group was significantly lower. Although the proportion of ε4 was greater in the CHD group than in the control group, the difference was not statistically significant. APOE ε3/ε3 is the most common genotype, with ε3 being the predominant allele in most populations and considered the wild-type reference subtype[19]. In comparison, individuals carrying the ε4 allele present elevated levels of TG, TC, LDL-C and APOB; this may be attributed to the enhanced lipid and very low-density lipoprotein (VLDL) binding ability of ApoE4 relative to that of ApoE3[20]. Compared with ApoE3, ApoE4 binds better to the surface of VLDL particles and impairs their lipolytic processing in circulation; thus, ApoE4 is associated with a greater pro-atherogenic lipoprotein-cholesterol distribution (a higher VLDL-cholesterol/high-density lipoprotein-cholesterol ratio). Logistic regression analysis revealed that carrying the E2 allele significantly reduces the risk of developing CHD. A study by Xu et al[21] suggested that the APOE ε4 mutation was associated with an increased risk of CHD, whereas the APOE ε2 allele appeared protective against CHD in Caucasians. Conversely, Zhao et al[22] presented a differing perspective, suggesting that the ε2 allele of APOE is a risk factor for premature coronary artery disease, particularly among Asians. This may be attributed to interactions between genetic factors and environmental influences, especially diet, as well as specific lipid metabolism characteristics. Regional differences in the study populations contribute to the variability in research findings.
In our study, the most common SLCO1B1 genotype was *1b/*1b, followed by *1a/*1b*, 1b/*15 *, 1a/*1a*, and 1a/*15. The *1b allele dominated at 62.53%, followed by *1a (26.34%), *15 (10.92%), and *5 (0.21%). The genetic distribution of the Teochew population is consistent with findings from Asian populations[23,24]. Currently, research concerning SLCO1B1 has focused primarily on how genetic polymorphisms affect drug metabolism, drug efficacy, and related side effects. Mutations in SLCO1B1-encoded OATP1B1 lead to a decrease in protein transport capacity, thereby diminishing the liver's ability to take up medications, causing elevated blood concentrations of statins and increasing the likelihood of conditions such as rhabdomyolysis or myopathy. Our study revealed that there was no significant correlation between SLCO1B1 gene polymorphisms and the incidence of CHD. Liu et al[25] suggested that the SNPs rs2306283 and rs4149056 of the SLCO1B1 gene are not associated with CAD susceptibility in the southern Chinese Hakka population. In contrast, Niu et al[26] reported that the SLCO1B1 *1a/*15 genotype reduces the risk of CHD in the Mongolian population, acting as a protective factor against the disease. Variations in SLCO1B1 gene polymorphisms among CHD populations demonstrate ethnic and regional differences. We can develop personalized treatment plans based on the results derived from SLCO1B1 gene polymorphisms. Personalized treatment strategies can be developed to achieve lipid-level reductions while preventing the adverse effects of statin use.
Currently, the precise mechanism by which elevated Lp(a) levels increase the risk of CHD remains incompletely understood, although high Lp(a) seems to provoke inflammation, atherosclerosis, and thrombosis[27]. Lp(a) concentrations are predominantly driven by the LPA gene. We confirmed that the repeat numbers of KIV-2 in the LPA gene were inversely correlated with serum Lp(a) levels, which aligns with Paultre’s conclusion that fewer KIV copies in apo(a) isoforms are associated with higher Lp(a) concentrations, serving as a predictive risk factor for CHD[28]. We also observed that the CHD group presented a significantly lower KIV-2 copy number than did the control group. Logistic regression analysis using CHD risk factors as dependent variables revealed that for every unit increase in copy number, the risk of CHD decreased by 0.949 times, indicating that a higher KIV-2 copy number corresponds to a lower risk. In a study targeting the Danish general population, it was proposed that high Lp(a) levels correlate with a greater risk of all-cause mortality, possibly attributable to low LPA KIV-2 repeat counts[29]. Additionally, some reports suggest that the causal relationship between LPA mutations and CHD remains uncertain[30]. Further research is needed to clarify the pathogenic mechanisms of Lp(a) and lipid levels during different stages of CHD. Dynamic monitoring of lipid profiles in patients with varying degrees of CHD will be more conducive to guiding lipid-lowering treatment and preventing disease progression.
Since this study included both cases and controls, biases may have been introduced. The limited sample size in certain subgroups may compromise the representativeness. There could be interactions and linkage disequilibrium among genes, and in addition to the three genes investigated, many other genes may also play roles in the pathogenesis of CHD.
In summary, this study investigated the relationship between APOE, SLCO1B1 gene polymorphisms and LPA KIV-2 copy number variation in the Teochew population with CHD. We confirmed that the APOE ε2 allele and a higher KIV-2 copy number are associated with a reduced risk of CHD. Building on these findings, further research is needed to explore the correlations between genetic polymorphisms and myocardial injury, as well as their prognostic implications, using using multicentre and multiregional samples.
| 1. | Björkegren JLM, Lusis AJ. Atherosclerosis: Recent developments. Cell. 2022;185:1630-1645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 424] [Cited by in RCA: 772] [Article Influence: 193.0] [Reference Citation Analysis (0)] |
| 2. | Martin SS, Aday AW, Almarzooq ZI, Anderson CAM, Arora P, Avery CL, Baker-Smith CM, Barone Gibbs B, Beaton AZ, Boehme AK, Commodore-Mensah Y, Currie ME, Elkind MSV, Evenson KR, Generoso G, Heard DG, Hiremath S, Johansen MC, Kalani R, Kazi DS, Ko D, Liu J, Magnani JW, Michos ED, Mussolino ME, Navaneethan SD, Parikh NI, Perman SM, Poudel R, Rezk-Hanna M, Roth GA, Shah NS, St-Onge MP, Thacker EL, Tsao CW, Urbut SM, Van Spall HGC, Voeks JH, Wang NY, Wong ND, Wong SS, Yaffe K, Palaniappan LP; American Heart Association Council on Epidemiology and Prevention Statistics Committee and Stroke Statistics Subcommittee. 2024 Heart Disease and Stroke Statistics: A Report of US and Global Data From the American Heart Association. Circulation. 2024;149:e347-e913. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1087] [Cited by in RCA: 1591] [Article Influence: 795.5] [Reference Citation Analysis (35)] |
| 3. | Nelson CP, Goel A, Butterworth AS, Kanoni S, Webb TR, Marouli E, Zeng L, Ntalla I, Lai FY, Hopewell JC, Giannakopoulou O, Jiang T, Hamby SE, Di Angelantonio E, Assimes TL, Bottinger EP, Chambers JC, Clarke R, Palmer CNA, Cubbon RM, Ellinor P, Ermel R, Evangelou E, Franks PW, Grace C, Gu D, Hingorani AD, Howson JMM, Ingelsson E, Kastrati A, Kessler T, Kyriakou T, Lehtimäki T, Lu X, Lu Y, März W, McPherson R, Metspalu A, Pujades-Rodriguez M, Ruusalepp A, Schadt EE, Schmidt AF, Sweeting MJ, Zalloua PA, AlGhalayini K, Keavney BD, Kooner JS, Loos RJF, Patel RS, Rutter MK, Tomaszewski M, Tzoulaki I, Zeggini E, Erdmann J, Dedoussis G, Björkegren JLM; EPIC-CVD Consortium; CARDIoGRAMplusC4D; UK Biobank CardioMetabolic Consortium CHD working group, Schunkert H, Farrall M, Danesh J, Samani NJ, Watkins H, Deloukas P. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet. 2017;49:1385-1391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 575] [Cited by in RCA: 519] [Article Influence: 57.7] [Reference Citation Analysis (0)] |
| 4. | Mahley RW, Weisgraber KH, Huang Y. Apolipoprotein E: structure determines function, from atherosclerosis to Alzheimer's disease to AIDS. J Lipid Res. 2009;50 Suppl:S183-S188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 378] [Cited by in RCA: 427] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 5. | Seripa D, D'Onofrio G, Panza F, Cascavilla L, Masullo C, Pilotto A. The genetics of the human APOE polymorphism. Rejuvenation Res. 2011;14:491-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 89] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 6. | Raulin AC, Doss SV, Trottier ZA, Ikezu TC, Bu G, Liu CC. ApoE in Alzheimer's disease: pathophysiology and therapeutic strategies. Mol Neurodegener. 2022;17:72. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 436] [Reference Citation Analysis (0)] |
| 7. | Ramsey LB, Gong L, Lee SB, Wagner JB, Zhou X, Sangkuhl K, Adams SM, Straka RJ, Empey PE, Boone EC, Klein TE, Niemi M, Gaedigk A. PharmVar GeneFocus: SLCO1B1. Clin Pharmacol Ther. 2023;113:782-793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 47] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 8. | Lee HH, Ho RH. Interindividual and interethnic variability in drug disposition: polymorphisms in organic anion transporting polypeptide 1B1 (OATP1B1; SLCO1B1). Br J Clin Pharmacol. 2017;83:1176-1184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 72] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 9. | Kamstrup PR. Lipoprotein(a) and Cardiovascular Disease. Clin Chem. 2021;67:154-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 162] [Article Influence: 32.4] [Reference Citation Analysis (0)] |
| 10. | Schachtl-Riess JF, Kheirkhah A, Grüneis R, Di Maio S, Schoenherr S, Streiter G, Losso JL, Paulweber B, Eckardt KU, Köttgen A, Lamina C, Kronenberg F, Coassin S; GCKD Investigators. Frequent LPA KIV-2 Variants Lower Lipoprotein(a) Concentrations and Protect Against Coronary Artery Disease. J Am Coll Cardiol. 2021;78:437-449. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 55] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 11. | Hu Y, He H, Ou Q, Nai J, Pan L, Chen X, Tu J, Zeng X, Pei G, Wang L, Lin B, Liu Q, Shan G. Prevalence of common chronic disease and multimorbidity patterns in Guangdong province with three typical cultures: analysis of data from the Diverse Life-Course Cohort study. Front Public Health. 2023;11:1163791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 12. | Zheng XY, Yi Q, Xu YJ, Zeng XY, Xu XJ, Chen G, Ma SL, Tang SL, Lin LF. Health transition of the causes of mortality between 2005 and 2015 in Guangdong, China. Postgrad Med J. 2022;98:346-353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 13. | Abraham G, Havulinna AS, Bhalala OG, Byars SG, De Livera AM, Yetukuri L, Tikkanen E, Perola M, Schunkert H, Sijbrands EJ, Palotie A, Samani NJ, Salomaa V, Ripatti S, Inouye M. Genomic prediction of coronary heart disease. Eur Heart J. 2016;37:3267-3278. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 218] [Cited by in RCA: 240] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 14. | Gungor Z, Anuurad E, Enkhmaa B, Zhang W, Kim K, Berglund L. Apo E4 and lipoprotein-associated phospholipase A2 synergistically increase cardiovascular risk. Atherosclerosis. 2012;223:230-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 23] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 15. | Li Z, Yang N, Lei X, Lin C, Li N, Jiang X, Wei X, Xu B. The association between the ApoE polymorphisms and the MRI-defined intracranial lesions in a cohort of southern China population. J Clin Lab Anal. 2019;33:e22950. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 16. | Zheng L, Duan J, Duan X, Zhou W, Chen C, Li Y, Chen J, Zhou W, Wang YJ, Li T, Song W. Association of Apolipoprotein E (ApoE) Polymorphism with Alzheimer's Disease in Chinese Population. Curr Alzheimer Res. 2016;13:912-917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 17. | Zhong Z, Wu H, Li B, Li C, Liu Z, Yang M, Zhang Q, Zhong W, Zhao P. Analysis of SLCO1B1 and APOE genetic polymorphisms in a large ethnic Hakka population in southern China. J Clin Lab Anal. 2018;32:e22408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 18. | Liu X, Lin Q, Fan K, Tang M, Zhang W, Yang B, Ou Q. The effects of genetic polymorphisms of APOE on circulating lipid levels in middle-aged and elderly chinese Fujian Han population: toward age- and sex-personalized management. Lipids Health Dis. 2021;20:158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 19. | Serrano-Pozo A, Das S, Hyman BT. APOE and Alzheimer's disease: advances in genetics, pathophysiology, and therapeutic approaches. Lancet Neurol. 2021;20:68-80. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 474] [Cited by in RCA: 680] [Article Influence: 136.0] [Reference Citation Analysis (0)] |
| 20. | Li H, Dhanasekaran P, Alexander ET, Rader DJ, Phillips MC, Lund-Katz S. Molecular mechanisms responsible for the differential effects of apoE3 and apoE4 on plasma lipoprotein-cholesterol levels. Arterioscler Thromb Vasc Biol. 2013;33:687-693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 51] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 21. | Xu M, Zhao J, Zhang Y, Ma X, Dai Q, Zhi H, Wang B, Wang L. Apolipoprotein E Gene Variants and Risk of Coronary Heart Disease: A Meta-Analysis. Biomed Res Int. 2016;2016:3912175. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 55] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 22. | Zhao QR, Lei YY, Li J, Jiang N, Shi JP. Association between apolipoprotein E polymorphisms and premature coronary artery disease: a meta-analysis. Clin Chem Lab Med. 2017;55:284-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 23. | Na Nakorn C, Waisayarat J, Dejthevaporn C, Srisawasdi P, Wongwaisayawan S, Sukasem C. Genetic Variations and Frequencies of the Two Functional Single Nucleotide Polymorphisms of SLCO1B1 in the Thai Population. Front Pharmacol. 2020;11:728. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 24. | Namgoong S, Cheong HS, Kim JO, Kim LH, Na HS, Koh IS, Chung MW, Shin HD. Comparison of genetic variations of the SLCO1B1, SLCO1B3, and SLCO2B1 genes among five ethnic groups. Environ Toxicol Pharmacol. 2015;40:692-697. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 25. | Liu Q, Wu H, Yu Z, Huang Q, Zhong Z. APOE gene ɛ4 allele (388C-526C) effects on serum lipids and risk of coronary artery disease in southern Chinese Hakka population. J Clin Lab Anal. 2021;35:e23925. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 26. | Niu RB, Dong XX, Guo LP, Pan L, Hai YQ, Chen XX, Duan BS. Study on the relationship between SLCO1B1 and ApoE gene polymorphisms and the risk of coronary heart disease in the Mongolian population. Clin Exp Hypertens. 2021;43:788-792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 27. | Tasdighi E, Adhikari R, Almaadawy O, Leucker TM, Blaha MJ. LP(a): Structure, Genetics, Associated Cardiovascular Risk, and Emerging Therapeutics. Annu Rev Pharmacol Toxicol. 2024;64:135-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 32] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 28. | Paultre F, Pearson TA, Weil HF, Tuck CH, Myerson M, Rubin J, Francis CK, Marx HF, Philbin EF, Reed RG, Berglund L. High levels of Lp(a) with a small apo(a) isoform are associated with coronary artery disease in African American and white men. Arterioscler Thromb Vasc Biol. 2000;20:2619-2624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 128] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 29. | Langsted A, Kamstrup PR, Nordestgaard BG. High lipoprotein(a) and high risk of mortality. Eur Heart J. 2019;40:2760-2770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 204] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 30. | Li ZG, Li G, Zhou YL, Chen ZJ, Yang JQ, Zhang Y, Sun S, Zhong SL. Lack of association between lipoprotein(a) genetic variants and subsequent cardiovascular events in Chinese Han patients with coronary artery disease after percutaneous coronary intervention. Lipids Health Dis. 2013;12:127. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/