Published online Jan 26, 2025. doi: 10.4330/wjc.v17.i1.99595
Revised: October 23, 2024
Accepted: December 27, 2024
Published online: January 26, 2025
Processing time: 179 Days and 17.9 Hours
Hypertrophic cardiomyopathy (HCM) is an autosomal dominant inherited cardio
Core Tip: Hypertrophic cardiomyopathy (HCM) is an autosomal dominant inherited cardiomyopathy with a highly variable clinical phenotype. This paper provides a detailed reference on the pathogenic genes of HCM and their phenotypic prognosis. These findings hold significant potential for guiding clinical decision-making, particularly in personalizing management strategies for HCM patients.
- Citation: Hong Y, Xi HT, Yang XY, Su WW, Li XP. Pathogenic genes and clinical prognosis in hypertrophic cardiomyopathy. World J Cardiol 2025; 17(1): 99595
- URL: https://www.wjgnet.com/1949-8462/full/v17/i1/99595.htm
- DOI: https://dx.doi.org/10.4330/wjc.v17.i1.99595
Hypertrophic cardiomyopathy (HCM) is a common genetic cardiomyopathy inherited in an autosomal dominant pattern, with offspring having a 50% chance of inheriting the same pathogenic genetic variant[1]. The prevalence of HCM in young adults has been estimated to be 1:200-1:500, though when considering asymptomatic HCM in the general popu
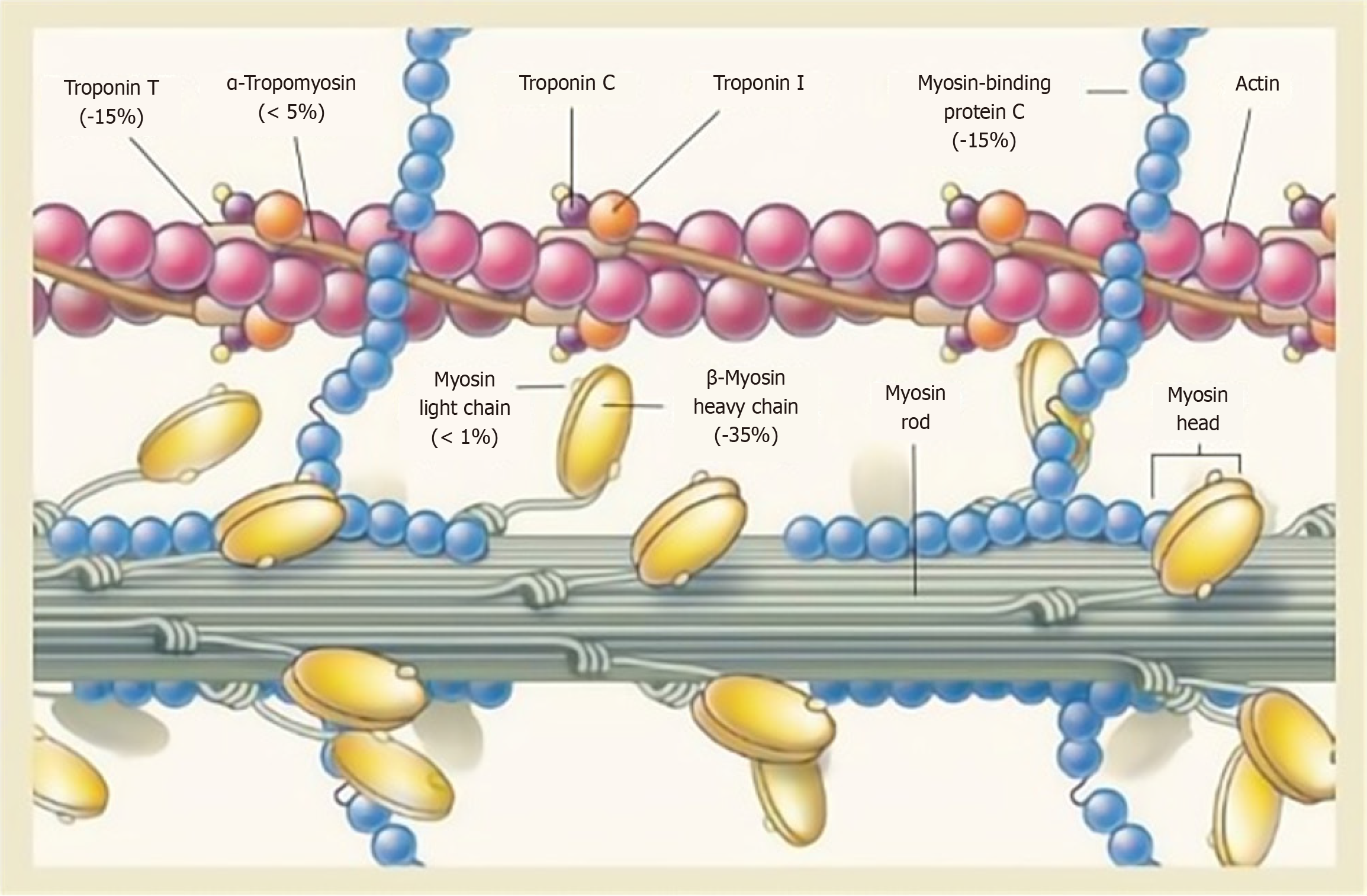
Among patients with HCM, 60% have an identifiable pathogenic genetic variant[10,11]. Several encode sarcomere proteins [myosin binding protein C3 (MYBPC3), myosin heavy chain 7 (MYH7), myosin light chain 2 (MYL2), MYL3, troponin T2 (TNNT2), TNNI3, tropomyosin 1 (TPM1), and actin alpha cardiac muscle 1 (ACTC1)], the two most com
MYBPC3 contains more than 21 kbp in 35 exons. CMyBP-C, encoded by MYBPC3, is an important structural protein component of the sarcomere, and it participates in sarcomere assembly, stabilizes sarcomere structure, and regulates the contraction and relaxation of cardiac muscle through phosphorylation. MYBPC3 mutations contribute to 50% of HCM, and more than 350 individuals have been identified in MYBPC3, making it the most frequently mutated gene in HCM. Heterozygous truncation mutations in MYBPC3 are the principal cause of HCM[17-19]. Sixty percent of the known MYBPC3 mutations associated with HCM are heterozygous truncation mutations, which include nonsense mutations, insertions or deletions, and splicing mutations[19-21]. The main molecular pathogenic mechanism for heterozygous truncating mutations is haploinsufficiency[22]. Haploinsufficiency is caused by nonsense-mediated mRNA decay and/or the ubiquitin-proteasome system[23,24]. These mutations lead to a premature termination codon in the transcribed mRNA and are expected to produce C-terminal truncated cMyBP-C that lacks the necessary myosin-binding and/or titin-binding domains, which results in a reduction in cMyBP-C content that enhances maximal thin myofilament sliding velocities in the thick myofilament C-zone[13,17,19]. The decreased content or complete absence of cMyBP-C leads to enhanced Ca2+ sensitivity of myofilaments, which leads to increased combination of cardiac troponin and Ca2+ and thus abnormal myocardial contraction[25,26], but its mechanism has not been fully clarified.
The MYH7 gene contains 40 exons spread over 220 kDa of genomic DNA. The MYH7 gene encodes β-MyHC, which is the predominant myosin heavy chain isoform in human cardiomyocytes. Unlike MYBPC3 with its heterozygous truncation mutations, MYH7 mainly suffers missense mutations[26]. Most of the mutations in β-MyHC cluster in functionally significant regions of myosin S1 and S2[27-29]. The molecular pathogenesis by which MYH7 mutation causes HCM is more complex and still unclear. The currently most recognized mechanisms involve Ca2+ homeostasis, myocardial fibrosis, and energy dysregulation[30,31].
The phenotypic characteristics of HCM patients are diverse. Left ventricular hypertrophy can occur in the free wall of the ventricle, apex, and papillary muscles and can be symmetrical or asymmetrical, and it can have a concentric or free-wall distribution[32]. In histopathology, the cardiac myocytes of HCM are usually hypertrophic, with disarray of myocardial fibers and myocardial fibrosis. Patients with HCM may present with chest pain, dyspnea, palpitations, angina, syncope, and even VA and SCD. SCD is the most devastating manifestation of HCM and mainly affects younger people and athletes. Importantly, many patients with HCM are asymptomatic and often identified incidentally through physical examination. SCD can be the first symptom of HCM occurring in asymptomatic or young patients without any warning. Therefore, analyzing the correlations between the pathogenic genes and phenotypes of HCM will help to identify HCM patients early and improve their prognosis.
The pathogenic variants of HCM mostly occur in genes encoding sarcomere thick and thin filament proteins. Thick filament gene mutations (e.g., MYBPC3, MYH7, MYL2, MYL3, MYH6, etc.) cause HCM of the interventricular septum, while mutations in thin filament genes (e.g., TNNT2, TNNI3, TPM1, and ACTC1) commonly lead to apical HCM and centripetal HCM[33]. A meta-analysis of 7675 HCM individuals by Sedaghat-Hamedani et al[34] observed that patients with sarcomeric mutations are more susceptible to SCD and that mutations in thick filament genes lead to a more severe myocardial hypertrophy phenotype and obstruction than in patients without these mutations. Compared to thick-filament mutations, thin-filament mutations are associated with an increased likelihood of advanced left ventricular dysfunction and heart failure[35].
Among the thick filament genes, MYBPC3 and MYH7 have the most prevalent pathogenic gene mutations. It is now widely believed that MYH7 is more malignant than MYBPC3. The pathogenic mutations of the MYH7 gene are characterized by earlier onset age, larger left ventricular hypertrophy, more severe ventricular systolic and diastolic dysfunction, and a higher risk of complications such as conduction block, atrial fibrillation, VA, and SCD[6,9,36]. MYH7 mutations generally lead to end-stage heart failure with a poor prognosis. We detected three mutations in the MYH7 gene, namely, c.G1346T/p.T449N, c.C1357T/p.R453C and c.5156A>G/p.Q1719R, in three HCM families that exhibited both bradycardia and the dilated-phase HCM (DPHCM) phenotype, which is characterized by gradual left ventricular dilation and left ventricular systolic dysfunction, with an adverse outcome and a higher risk of SCD. Ours was the first report that HCM patients with an MYH7 mutation (c.G1346T/p.T449N) may develop DPHCM and suffer from cardiac block in middle age and beyond (data not published).
By comparison, patients carrying MYBPC3 mutations usually have a delayed disease onset and a milder degree of cardiac hypertrophy with favorable disease progression[37]. Although the prognosis of MYBPC3 mutation is better than that of MYH7 mutation, when MYBPC3 mutation is combined with angiotensin-converting enzyme gene mutation, the phenotype worsens[38]. Patients with the D/D genotype of the angiotensin-converting enzyme gene in the presence of HCM have an increased risk of SCD and an increased severity of hypertrophy[39]. There are relatively few reports on MYL2 and MYL3 mutations. Mutations in the MYL2 gene have been found in fewer than 5% of cases of HCM[40]. According to existing case reports, MYL2 mutations are associated with severe cardiac hypertrophy and SCD[41]. Osborn et al[42] demonstrated that MYL3 loss-of-function mutations can cause cardiomyopathy and SCD. MYH6 encodes the alpha heavy chain subunit of cardiac myosin, mutations in which cause familial HCM, left ventricular dysfunction, and heart failure exacerbation[43]. A new report by Suzuki et al[44] found that the pathogenic MYH6 variant p.Lys364fs might contribute to DPHCM. TNNT2 mutations are the most frequently observed pathogenic thin filament gene variants. TNNT2 gene mutations can manifest as mild left ventricular wall thickness with low penetrance, but they have been associated with a higher incidence of SCD and poor prognosis[6]. The TNNI3 gene encodes a subtype of troponin I and is only expressed in the myocardium. TNNI3 can lead to malignant HCM characterized by a remarkably high rate of early onset SCD[45]. The TPM1 gene accounts for 16% of all variants of thin filament proteins and is not clinically more common[46]. These have higher penetrance, causing a severe disease phenotype and unfavorable prognosis in Vietnamese patients with HCM[47]. The ACTC1 encodes a major component of thin filaments in mature cardiac myocytes and is essential for normal cardiac morphogenesis and muscle contraction. Yang et al[48] found that the ACTC1 D26N amino acid variant had an extremely high penetrance and led to diverse phenotypes. The results are summarized in Table 1.
| Gene | Frequency, % | Change of amino acid | Phenotype | Prognosis |
| MYBPC3 | 50 | p.Glu542Gln | A delayed disease onset and a milder degree of cardiac hypertrophy | Relatively good |
| p.Cys719Arg | ||||
| p.Glu334Lys | ||||
| p.Pro108lafs*9 | ||||
| p.Gly1093Cys | ||||
| p.Arg668His | ||||
| p.Arg502Trp | ||||
| p.F305Pfs*27 | ||||
| P.Lys1209Serfs*28 | ||||
| c.2737+1(IVS26)G>T | ||||
| MYH7 | 30-35 | p.Arg453Cys | An earlier onset age, larger left ventricular hypertrophy, more severe ventricular systolic and diastolic dysfunction, and a higher risk of complications such as conduction block, atrial fibrillation, VA, end-stage heart failure and SCD | Poor |
| p.Arg1045Leu | ||||
| p.Arg 719Trp | ||||
| p.Asn 391Thr | ||||
| p.Gly716Arg | ||||
| p.Arg403Gln | ||||
| p.Arg453Cys | ||||
| p.Glu848Gly | ||||
| p.Asn391Thr | ||||
| p.Thr446Pro | ||||
| p.Phe468Leu | ||||
| MYL2 | < 5 | R58Q | Severe cardiac hypertrophy and SCD | Poor |
| MYL3 | < 5 | c.170C>A | Leading SCD | Poor |
| c. 106G>T | ||||
| c.482-1G>A | ||||
| MYH6 | Not in detail | p.Lys364fs | Causing familial HCM, left ventricular dysfunction, and heart failure exacerbation | Poor |
| TNNT2 | 10 | R92Q | Mild left ventricular wall thickness with low penetrance, a higher incidence of SCD | Poor |
| TNNI3 | 8 | p.Arg21Cys | Leading to malignant HCM characterized by a remarkably high rate of early onset SCD | Poor |
In addition to the thick and thin filament protein genes, genes encoding Z-disk structural proteins (e.g., ACTN2, ankyrin repeat domain 1, CSRP3, and FHL1) also regulate the phenotype of HCM. ACTN2 is expressed in cardiac muscle. Mutations of this protein have been implicated in a mild to moderate HCM and may be benign[49,50]. CSRP3 encodes muscle LIM protein, which is mainly expressed in striated muscle tissues. Heterozygous mutations of CSRP3 have been associated with mild late-onset HCM and had an apparently better prognosis, while homozygous individuals with missense mutations in CSRP3 had a more severe phenotype[49,51,52]. FHL1 is involved in sarcomere formation. Giucă et al[53] reported that FHL1 mutation leads to an elevated risk for arrhythmia and SCD and recommended that patients with HCM carrying FHL1 mutations have an implantable cardioverter defibrillator installed in advance. Other genes encoding calcium regulation-related proteins (e.g., JPH2, phospholamban, and calreticulin 3) also have an effect on the phenotype of HCM. JPH2 is the major structural protein in cardiomyocytes and is also a component of junctional mem
Patients with more than one HCM-related mutation often develop a more severe phenotype[56-58] and worse prognosis than patients with only one. Fourteen percent of childhood-onset HCM is caused by compound genetic mutations[59]. This indicates that a gene dosage effect may be responsible for early onset. Significant clinical heterogeneity remains because of different combinations of mutations. Zhang et al[60] analyzed a Chinese HCM pedigree with compound mutations and demonstrated that the combination of Met822Thr and Arg1420Trp in MYH7 was causal but relatively benign.
Collectively, HCM is a widespread inherited disease with a highly variable clinical phenotype. As sequencing technology advances, the pathogenic gene mutation spectrum and phenotypic characteristics of HCM are gradually becoming clearer. However, the precise mechanisms linking known pathogenic gene mutations and the clinical course of this heterogeneous condition remain elusive. Its onset and development are influenced by multiple factors, leading to phe
| 1. | Ho CY, Day SM, Ashley EA, Michels M, Pereira AC, Jacoby D, Cirino AL, Fox JC, Lakdawala NK, Ware JS, Caleshu CA, Helms AS, Colan SD, Girolami F, Cecchi F, Seidman CE, Sajeev G, Signorovitch J, Green EM, Olivotto I. Genotype and Lifetime Burden of Disease in Hypertrophic Cardiomyopathy: Insights from the Sarcomeric Human Cardiomyopathy Registry (SHaRe). Circulation. 2018;138:1387-1398. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 504] [Cited by in RCA: 634] [Article Influence: 79.3] [Reference Citation Analysis (0)] |
| 2. | Ommen SR, Mital S, Burke MA, Day SM, Deswal A, Elliott P, Evanovich LL, Hung J, Joglar JA, Kantor P, Kimmelstiel C, Kittleson M, Link MS, Maron MS, Martinez MW, Miyake CY, Schaff HV, Semsarian C, Sorajja P. 2020 AHA/ACC Guideline for the Diagnosis and Treatment of Patients With Hypertrophic Cardiomyopathy: Executive Summary: A Report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. Circulation. 2020;142:e533-e557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 161] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 3. | Semsarian C, Ingles J, Maron MS, Maron BJ. New perspectives on the prevalence of hypertrophic cardiomyopathy. J Am Coll Cardiol. 2015;65:1249-1254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 633] [Cited by in RCA: 1023] [Article Influence: 93.0] [Reference Citation Analysis (0)] |
| 4. | Ommen SR, Semsarian C. Hypertrophic cardiomyopathy: a practical approach to guideline directed management. Lancet. 2021;398:2102-2108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 55] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 5. | Maron BJ. Clinical Course and Management of Hypertrophic Cardiomyopathy. N Engl J Med. 2018;379:1977. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 6. | Hong Y, Su WW, Li X. Risk factors of sudden cardiac death in hypertrophic cardiomyopathy. Curr Opin Cardiol. 2022;37:15-21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 7. | Mazzarotto F, Girolami F, Boschi B, Barlocco F, Tomberli A, Baldini K, Coppini R, Tanini I, Bardi S, Contini E, Cecchi F, Pelo E, Cook SA, Cerbai E, Poggesi C, Torricelli F, Walsh R, Olivotto I. Defining the diagnostic effectiveness of genes for inclusion in panels: the experience of two decades of genetic testing for hypertrophic cardiomyopathy at a single center. Genet Med. 2019;21:284-292. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 8. | Kraft T, Montag J, Radocaj A, Brenner B. Hypertrophic Cardiomyopathy: Cell-to-Cell Imbalance in Gene Expression and Contraction Force as Trigger for Disease Phenotype Development. Circ Res. 2016;119:992-995. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 9. | Velicki L, Jakovljevic DG, Preveden A, Golubovic M, Bjelobrk M, Ilic A, Stojsic S, Barlocco F, Tafelmeier M, Okwose N, Tesic M, Brennan P, Popovic D, Ristic A, MacGowan GA, Filipovic N, Maier LS, Olivotto I. Genetic determinants of clinical phenotype in hypertrophic cardiomyopathy. BMC Cardiovasc Disord. 2020;20:516. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 49] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 10. | Authors/Task Force members; Elliott PM, Anastasakis A, Borger MA, Borggrefe M, Cecchi F, Charron P, Hagege AA, Lafont A, Limongelli G, Mahrholdt H, McKenna WJ, Mogensen J, Nihoyannopoulos P, Nistri S, Pieper PG, Pieske B, Rapezzi C, Rutten FH, Tillmanns C, Watkins H. 2014 ESC Guidelines on diagnosis and management of hypertrophic cardiomyopathy: the Task Force for the Diagnosis and Management of Hypertrophic Cardiomyopathy of the European Society of Cardiology (ESC). Eur Heart J. 2014;35:2733-2779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2292] [Cited by in RCA: 3163] [Article Influence: 263.6] [Reference Citation Analysis (0)] |
| 11. | Ahluwalia M, Ho CY. Cardiovascular genetics: the role of genetic testing in diagnosis and management of patients with hypertrophic cardiomyopathy. Heart. 2021;107:183-189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 12. | Walsh R, Thomson KL, Ware JS, Funke BH, Woodley J, McGuire KJ, Mazzarotto F, Blair E, Seller A, Taylor JC, Minikel EV; Exome Aggregation Consortium; MacArthur DG, Farrall M, Cook SA, Watkins H. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med. 2017;19:192-203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 412] [Cited by in RCA: 562] [Article Influence: 56.2] [Reference Citation Analysis (1)] |
| 13. | Alfares AA, Kelly MA, McDermott G, Funke BH, Lebo MS, Baxter SB, Shen J, McLaughlin HM, Clark EH, Babb LJ, Cox SW, DePalma SR, Ho CY, Seidman JG, Seidman CE, Rehm HL. Results of clinical genetic testing of 2,912 probands with hypertrophic cardiomyopathy: expanded panels offer limited additional sensitivity. Genet Med. 2015;17:880-888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 356] [Article Influence: 32.4] [Reference Citation Analysis (0)] |
| 14. | Ochoa JP, Sabater-Molina M, García-Pinilla JM, Mogensen J, Restrepo-Córdoba A, Palomino-Doza J, Villacorta E, Martinez-Moreno M, Ramos-Maqueda J, Zorio E, Peña-Peña ML, García-Granja PE, Rodríguez-Palomares JF, Cárdenas-Reyes IJ, de la Torre-Carpente MM, Bautista-Pavés A, Akhtar MM, Cicerchia MN, Bilbao-Quesada R, Mogollón-Jimenez MV, Salazar-Mendiguchía J, Mesa Latorre JM, Arnaez B, Olavarri-Miguel I, Fuentes-Cañamero ME, Lamounier A Jr, Pérez Ruiz JM, Climent-Payá V, Pérez-Sanchez I, Trujillo-Quintero JP, Lopes LR, Repáraz-Andrade A, Marín-Iglesias R, Rodriguez-Vilela A, Sandín-Fuentes M, Garrote JA, Cortel-Fuster A, Lopez-Garrido M, Fontalba-Romero A, Ripoll-Vera T, Llano-Rivas I, Fernandez-Fernandez X, Isidoro-García M, Garcia-Giustiniani D, Barriales-Villa R, Ortiz-Genga M, García-Pavía P, Elliott PM, Gimeno JR, Monserrat L. Formin Homology 2 Domain Containing 3 (FHOD3) Is a Genetic Basis for Hypertrophic Cardiomyopathy. J Am Coll Cardiol. 2018;72:2457-2467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 74] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 15. | Yotti R, Seidman CE, Seidman JG. Advances in the Genetic Basis and Pathogenesis of Sarcomere Cardiomyopathies. Annu Rev Genomics Hum Genet. 2019;20:129-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 137] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 16. | Hong Y. Analysis of clinical characteristics, detection of pathogenic genes and distribution of TCM syndrome types in patients with hypertrophic cardiomyopathy. M.Sc. Thesis, Chengdu University of Traditional Chinese Medicine. 2022. [DOI] [Full Text] |
| 17. | Toepfer CN, Wakimoto H, Garfinkel AC, McDonough B, Liao D, Jiang J, Tai AC, Gorham JM, Lunde IG, Lun M, Lynch TL 4th, McNamara JW, Sadayappan S, Redwood CS, Watkins HC, Seidman JG, Seidman CE. Hypertrophic cardiomyopathy mutations in MYBPC3 dysregulate myosin. Sci Transl Med. 2019;11:eaat1199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 157] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 18. | Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, Bild DE. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA Study. Coronary Artery Risk Development in (Young) Adults. Circulation. 1995;92:785-789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1307] [Cited by in RCA: 1474] [Article Influence: 47.5] [Reference Citation Analysis (0)] |
| 19. | Behrens-Gawlik V, Mearini G, Gedicke-Hornung C, Richard P, Carrier L. MYBPC3 in hypertrophic cardiomyopathy: from mutation identification to RNA-based correction. Pflugers Arch. 2014;466:215-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 20. | Carrier L, Schlossarek S, Willis MS, Eschenhagen T. The ubiquitin-proteasome system and nonsense-mediated mRNA decay in hypertrophic cardiomyopathy. Cardiovasc Res. 2010;85:330-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 70] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 21. | Marian AJ. Hypertrophic cardiomyopathy: from genetics to treatment. Eur J Clin Invest. 2010;40:360-369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 90] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 22. | van der Velden J, Ho CY, Tardiff JC, Olivotto I, Knollmann BC, Carrier L. Research priorities in sarcomeric cardiomyopathies. Cardiovasc Res. 2015;105:449-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 23. | Popp MW, Maquat LE. Leveraging Rules of Nonsense-Mediated mRNA Decay for Genome Engineering and Personalized Medicine. Cell. 2016;165:1319-1322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 235] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 24. | Siwaszek A, Ukleja M, Dziembowski A. Proteins involved in the degradation of cytoplasmic mRNA in the major eukaryotic model systems. RNA Biol. 2014;11:1122-1136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 56] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 25. | O'Leary TS, Snyder J, Sadayappan S, Day SM, Previs MJ. MYBPC3 truncation mutations enhance actomyosin contractile mechanics in human hypertrophic cardiomyopathy. J Mol Cell Cardiol. 2019;127:165-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 26. | Patel BG, Wilder T, Solaro RJ. Novel control of cardiac myofilament response to calcium by S-glutathionylation at specific sites of myosin binding protein C. Front Physiol. 2013;4:336. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 52] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 27. | Colegrave M, Peckham M. Structural implications of β-cardiac myosin heavy chain mutations in human disease. Anat Rec (Hoboken). 2014;297:1670-1680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 90] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 28. | Wang J, Yang F, Liu W, Sun J, Han Y, Li D, Gkoutos GV, Zhu Y, Chen Y. Radiomic Analysis of Native T(1) Mapping Images Discriminates Between MYH7 and MYBPC3-Related Hypertrophic Cardiomyopathy. J Magn Reson Imaging. 2020;52:1714-1721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 29. | Blair E, Redwood C, de Jesus Oliveira M, Moolman-Smook JC, Brink P, Corfield VA, Ostman-Smith I, Watkins H. Mutations of the light meromyosin domain of the beta-myosin heavy chain rod in hypertrophic cardiomyopathy. Circ Res. 2002;90:263-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 63] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 30. | Moore JR, Leinwand L, Warshaw DM. Understanding cardiomyopathy phenotypes based on the functional impact of mutations in the myosin motor. Circ Res. 2012;111:375-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 153] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 31. | Zheng K, Liu L, Zhang YQ. [Recent research on childhood hypertrophic cardiomyopathy caused by MYH7 gene mutations]. Zhongguo Dang Dai Er Ke Za Zhi. 2023;25:425-430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 32. | Naghi JJ, Siegel RJ. Medical management of hypertrophic cardiomyopathy. Rev Cardiovasc Med. 2010;11:202-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 33. | Lafreniere-Roula M, Bolkier Y, Zahavich L, Mathew J, George K, Wilson J, Stephenson EA, Benson LN, Manlhiot C, Mital S. Family screening for hypertrophic cardiomyopathy: Is it time to change practice guidelines? Eur Heart J. 2019;40:3672-3681. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 74] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 34. | Sedaghat-Hamedani F, Kayvanpour E, Tugrul OF, Lai A, Amr A, Haas J, Proctor T, Ehlermann P, Jensen K, Katus HA, Meder B. Clinical outcomes associated with sarcomere mutations in hypertrophic cardiomyopathy: a meta-analysis on 7675 individuals. Clin Res Cardiol. 2018;107:30-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 122] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 35. | Coppini R, Ho CY, Ashley E, Day S, Ferrantini C, Girolami F, Tomberli B, Bardi S, Torricelli F, Cecchi F, Mugelli A, Poggesi C, Tardiff J, Olivotto I. Clinical phenotype and outcome of hypertrophic cardiomyopathy associated with thin-filament gene mutations. J Am Coll Cardiol. 2014;64:2589-2600. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 137] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 36. | Lee SP, Ashley EA, Homburger J, Caleshu C, Green EM, Jacoby D, Colan SD, Arteaga-Fernández E, Day SM, Girolami F, Olivotto I, Michels M, Ho CY, Perez MV; SHaRe Investigators. Incident Atrial Fibrillation Is Associated With MYH7 Sarcomeric Gene Variation in Hypertrophic Cardiomyopathy. Circ Heart Fail. 2018;11:e005191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 37. | Carrier L. Targeting the population for gene therapy with MYBPC3. J Mol Cell Cardiol. 2021;150:101-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 38. | Yuan Y, Meng L, Zhou Y, Lu N. Genetic polymorphism of angiotensin-converting enzyme and hypertrophic cardiomyopathy risk: A systematic review and meta-analysis. Medicine (Baltimore). 2017;96:e8639. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 39. | Marian AJ, Yu QT, Workman R, Greve G, Roberts R. Angiotensin-converting enzyme polymorphism in hypertrophic cardiomyopathy and sudden cardiac death. Lancet. 1993;342:1085-1086. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 269] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 40. | Claes GR, van Tienen FH, Lindsey P, Krapels IP, Helderman-van den Enden AT, Hoos MB, Barrois YE, Janssen JW, Paulussen AD, Sels JW, Kuijpers SH, van Tintelen JP, van den Berg MP, Heesen WF, Garcia-Pavia P, Perrot A, Christiaans I, Salemink S, Marcelis CL, Smeets HJ, Brunner HG, Volders PG, van den Wijngaard A. Hypertrophic remodelling in cardiac regulatory myosin light chain (MYL2) founder mutation carriers. Eur Heart J. 2016;37:1815-1822. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 68] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 41. | De Bortoli M, Vio R, Basso C, Calore M, Minervini G, Angelini A, Melacini P, Vitiello L, Vazza G, Thiene G, Tosatto S, Corrado D, Iliceto S, Rampazzo A, Calore C. Novel Missense Variant in MYL2 Gene Associated With Hypertrophic Cardiomyopathy Showing High Incidence of Restrictive Physiology. Circ Genom Precis Med. 2020;13:e002824. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 42. | Osborn DPS, Emrahi L, Clayton J, Tabrizi MT, Wan AYB, Maroofian R, Yazdchi M, Garcia MLE, Galehdari H, Hesse C, Shariati G, Mazaheri N, Sedaghat A, Goullée H, Laing N, Jamshidi Y, Tajsharghi H. Autosomal recessive cardiomyopathy and sudden cardiac death associated with variants in MYL3. Genet Med. 2021;23:787-792. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 43. | Carniel E, Taylor MR, Sinagra G, Di Lenarda A, Ku L, Fain PR, Boucek MM, Cavanaugh J, Miocic S, Slavov D, Graw SL, Feiger J, Zhu XZ, Dao D, Ferguson DA, Bristow MR, Mestroni L. Alpha-myosin heavy chain: a sarcomeric gene associated with dilated and hypertrophic phenotypes of cardiomyopathy. Circulation. 2005;112:54-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 186] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 44. | Suzuki T, Saito K, Yoshikawa T, Hirono K, Hata Y, Nishida N, Yasuda K, Nagashima M. A double heterozygous variant in MYH6 and MYH7 associated with hypertrophic cardiomyopathy in a Japanese Family. J Cardiol Cases. 2022;25:213-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 45. | Marian AJ, Braunwald E. Hypertrophic Cardiomyopathy: Genetics, Pathogenesis, Clinical Manifestations, Diagnosis, and Therapy. Circ Res. 2017;121:749-770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 491] [Cited by in RCA: 1013] [Article Influence: 112.6] [Reference Citation Analysis (0)] |
| 46. | Halder SS, Rynkiewicz MJ, Creso JG, Sewanan LR, Howland L, Moore JR, Lehman W, Campbell SG. Mechanisms of pathogenicity in the hypertrophic cardiomyopathy-associated TPM1 variant S215L. PNAS Nexus. 2023;2:pgad011. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 47. | Tran Vu MT, Nguyen TV, Huynh NV, Nguyen Thai HT, Pham Nguyen V, Ho Huynh TD. Presence of Hypertrophic Cardiomyopathy Related Gene Mutations and Clinical Manifestations in Vietnamese Patients With Hypertrophic Cardiomyopathy. Circ J. 2019;83:1908-1916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 48. | Yang QL, Bian YY, Wang B, Zuo L, Zhou MY, Shao H, Zhang YM, Liu LW. Novel phenotype-genotype correlations of hypertrophic cardiomyopathy caused by mutation in α-actin and myosin-binding protein genes in three unrelated Chinese families. J Cardiol. 2019;73:438-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 49. | Walsh R, Offerhaus JA, Tadros R, Bezzina CR. Minor hypertrophic cardiomyopathy genes, major insights into the genetics of cardiomyopathies. Nat Rev Cardiol. 2022;19:151-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 81] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 50. | Haywood NJ, Wolny M, Rogers B, Trinh CH, Shuping Y, Edwards TA, Peckham M. Hypertrophic cardiomyopathy mutations in the calponin-homology domain of ACTN2 affect actin binding and cardiomyocyte Z-disc incorporation. Biochem J. 2016;473:2485-2493. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 41] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 51. | Huang H, Chen Y, Jin J, Du R, Tang K, Fan L, Xiang R. CSRP3, p.Arg122*, is responsible for hypertrophic cardiomyopathy in a Chinese family. J Gene Med. 2022;24:e3390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 52. | Salazar-Mendiguchía J, Barriales-Villa R, Lopes LR, Ochoa JP, Rodríguez-Vilela A, Palomino-Doza J, Larrañaga-Moreira JM, Cicerchia M, Cárdenas-Reyes I, García-Giustiniani D, Brögger N, Fernández G, García S, Santiago L, Vélez P, Ortiz-Genga M, Elliott PM, Monserrat L. The p.(Cys150Tyr) variant in CSRP3 is associated with late-onset hypertrophic cardiomyopathy in heterozygous individuals. Eur J Med Genet. 2020;63:104079. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 53. | Giucă A, Mitu C, Popescu BO, Bastian AE, Capşa R, Mursă A, Rădoi V, Popescu BA, Jurcuţ R. Novel FHL1 mutation variant identified in a patient with nonobstructive hypertrophic cardiomyopathy and myopathy - a case report. BMC Med Genet. 2020;21:188. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 54. | Vadgama N, Ameen M, Sundaram L, Gaddam S; Genomics England Research Consortium, Gifford C, Nasir J, Karakikes I. De novo and inherited variants in coding and regulatory regions in genetic cardiomyopathies. Hum Genomics. 2022;16:55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 55. | Reynolds JO, Quick AP, Wang Q, Beavers DL, Philippen LE, Showell J, Barreto-Torres G, Thuerauf DJ, Doroudgar S, Glembotski CC, Wehrens XH. Junctophilin-2 gene therapy rescues heart failure by normalizing RyR2-mediated Ca(2+) release. Int J Cardiol. 2016;225:371-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 84] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 56. | Wang J, Wang Y, Zou Y, Sun K, Wang Z, Ding H, Yuan J, Wei W, Hou Q, Wang H, Liu X, Zhang H, Ji Y, Zhou X, Sharma RK, Wang D, Ahmad F, Hui R, Song L. Malignant effects of multiple rare variants in sarcomere genes on the prognosis of patients with hypertrophic cardiomyopathy. Eur J Heart Fail. 2014;16:950-957. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 58] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 57. | Zou Y, Wang J, Liu X, Wang Y, Chen Y, Sun K, Gao S, Zhang C, Wang Z, Zhang Y, Feng X, Song Y, Wu Y, Zhang H, Jia L, Wang H, Wang D, Yan C, Lu M, Zhou X, Song L, Hui R. Multiple gene mutations, not the type of mutation, are the modifier of left ventricle hypertrophy in patients with hypertrophic cardiomyopathy. Mol Biol Rep. 2013;40:3969-3976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 53] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 58. | Burns C, Bagnall RD, Lam L, Semsarian C, Ingles J. Multiple Gene Variants in Hypertrophic Cardiomyopathy in the Era of Next-Generation Sequencing. Circ Cardiovasc Genet. 2017;10:e001666. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 94] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 59. | Morita H, Rehm HL, Menesses A, McDonough B, Roberts AE, Kucherlapati R, Towbin JA, Seidman JG, Seidman CE. Shared genetic causes of cardiac hypertrophy in children and adults. N Engl J Med. 2008;358:1899-1908. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 330] [Cited by in RCA: 311] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 60. | Zhang M, Sun X, Wu G, Wang D, Wang L, Zhang C, Zou Y, Wang J, Song L. Effect of Cis-Compound Variants in MYH7 on Hypertrophic Cardiomyopathy With a Mild Phenotype. Am J Cardiol. 2022;167:104-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/