Copyright
©The Author(s) 2025.
World J Biol Chem. Dec 5, 2025; 16(4): 112768
Published online Dec 5, 2025. doi: 10.4331/wjbc.v16.i4.112768
Published online Dec 5, 2025. doi: 10.4331/wjbc.v16.i4.112768
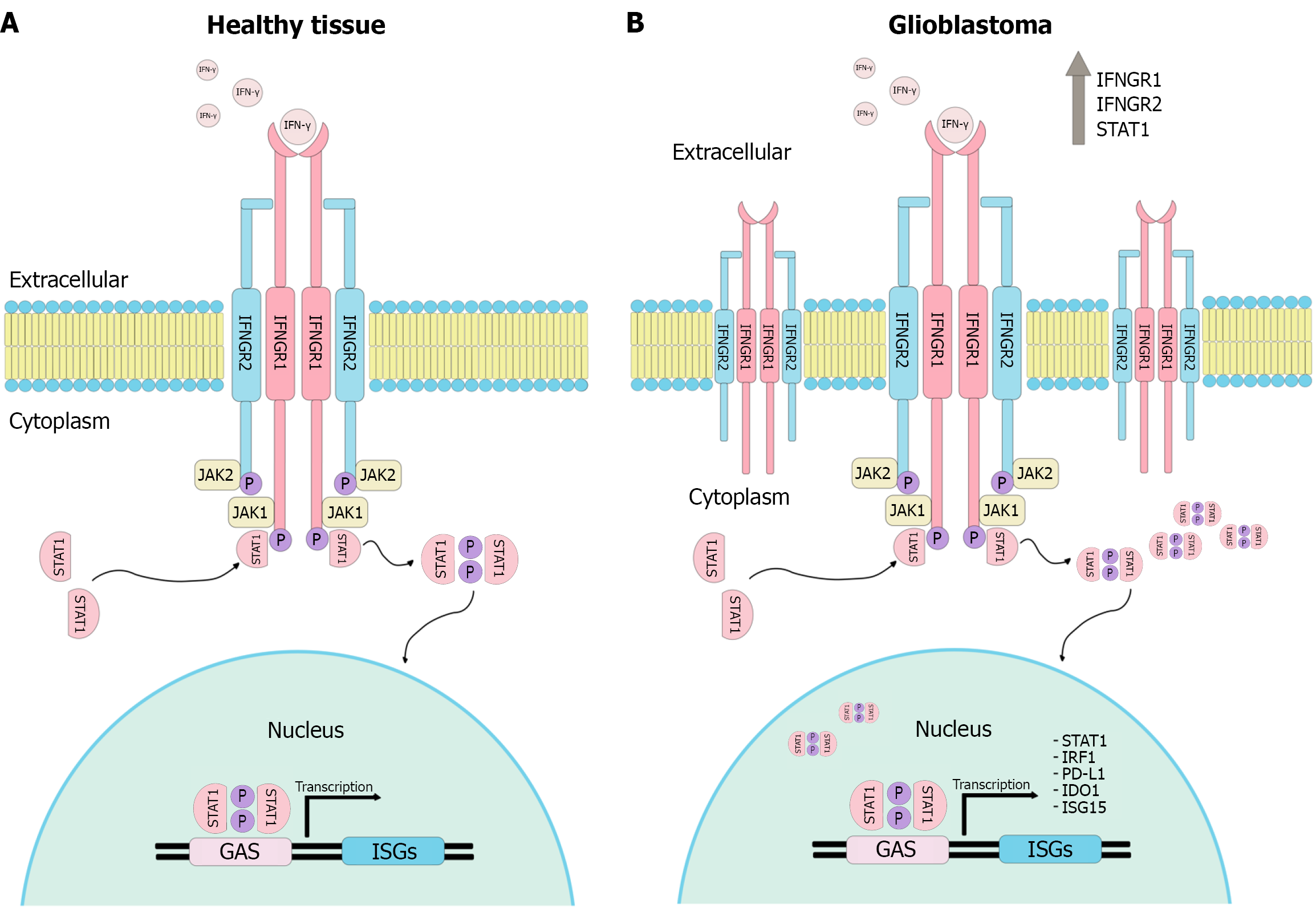
Figure 1 Interferon-gamma pathway in normal tissue and glioblastoma.
A: Under physiological conditions, interferon-gamma binds to its receptor and activates the Janus kinases 1 and 2, promoting the phosphorylation and dimerization of the signal transducer and activator of transcription 1. The signal transducer and activator of transcription 1 (STAT1) dimer translocates to the nucleus, where it binds to gamma-activated sequence elements in the promoters of interferon-stimulated genes; B: In glioblastoma cells, the pathway is amplified. This is evidenced by increased interferon gamma receptor density on the plasma membrane and higher levels of STAT1, resulting in enhanced transcription of genes implicated in glioblastoma progression, including STAT1, interferon regulatory factor 1, programmed death-ligand 1, indoleamine 2,3-dioxygenase 1, and interferon-stimulated gene 15. JAK1: Janus kinases 1; JAK2: Janus kinases 2; STAT1: Signal transducer and activator of transcription 1; ISGs: Interferon-stimulated genes; PD-L1: Programmed death-ligand 1; IDO1: Indoleamine 2,3-dioxygenase 1; ISG15: Interferon-stimulated gene 15; GAS: Gamma-activated sequences; IRF1: Interferon regulatory factor 1.
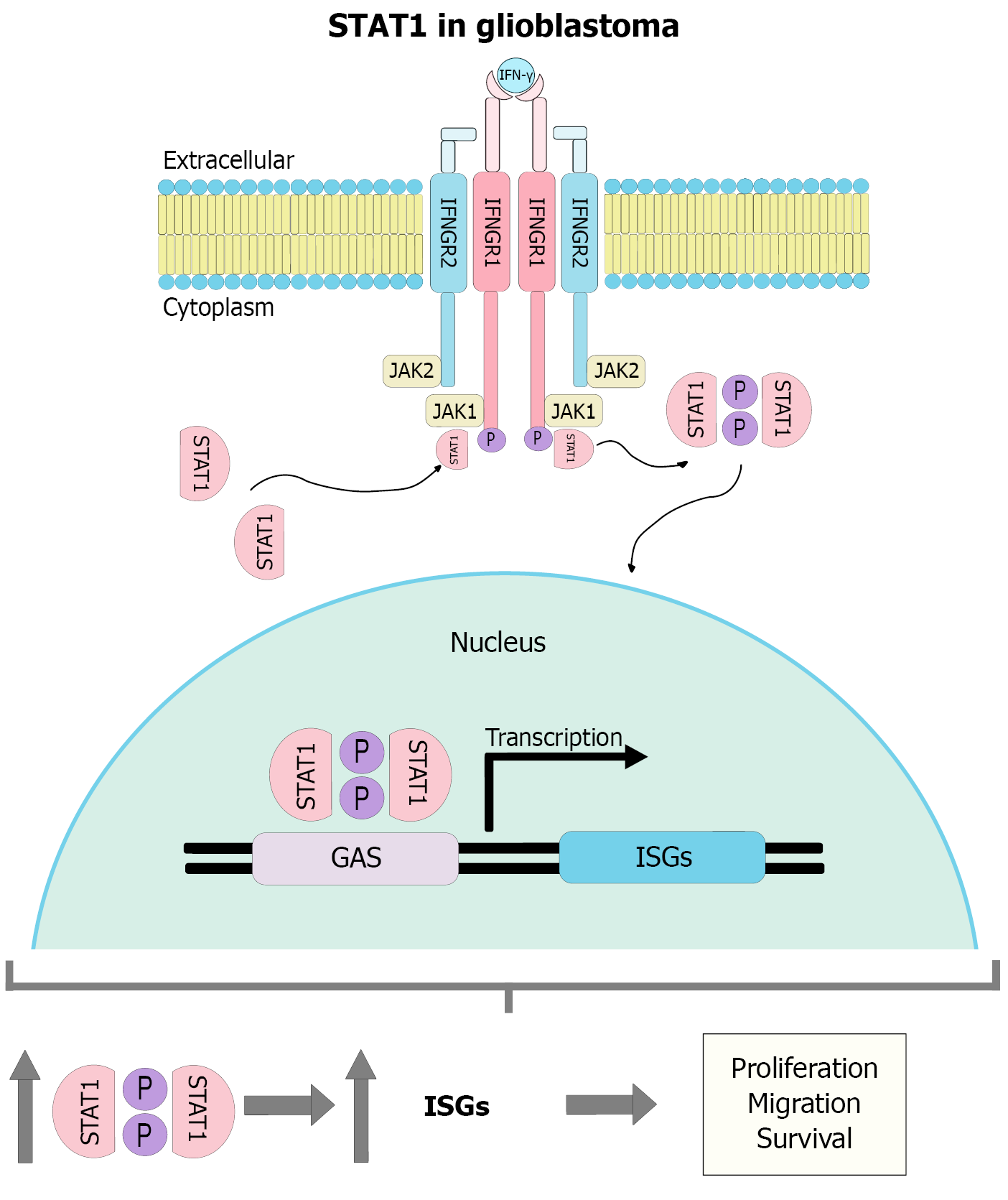
Figure 2 The molecular function of signal transducer and activator of transcription 1 and its relationship with glioblastoma.
The binding of interferon-gamma to its heterotetrameric receptor, composed of two dimers of IFNGR1 and IFNGR2, activates the Janus kinases 1 and 2, which leads to the phosphorylation and activation of the signal transducer and activator of transcription 1 (STAT1). Once phosphorylated, STAT1 forms dimers that translocate to the nucleus, where it binds to gamma-activated sequence elements in the promoters of interferon-stimulated genes, promoting their transcription. Sustained activation of this pathway induces the transcription of genes involved in cell proliferation, migration, and survival, contributing to the aggressive phenotype of glioblastoma. GAS: Gamma-activated sequences; STAT1: Signal transducer and activator of transcription 1; JAK1: Janus kinases 1; JAK2: Janus kinases 2; ISGs: Interferon-stimulated genes.
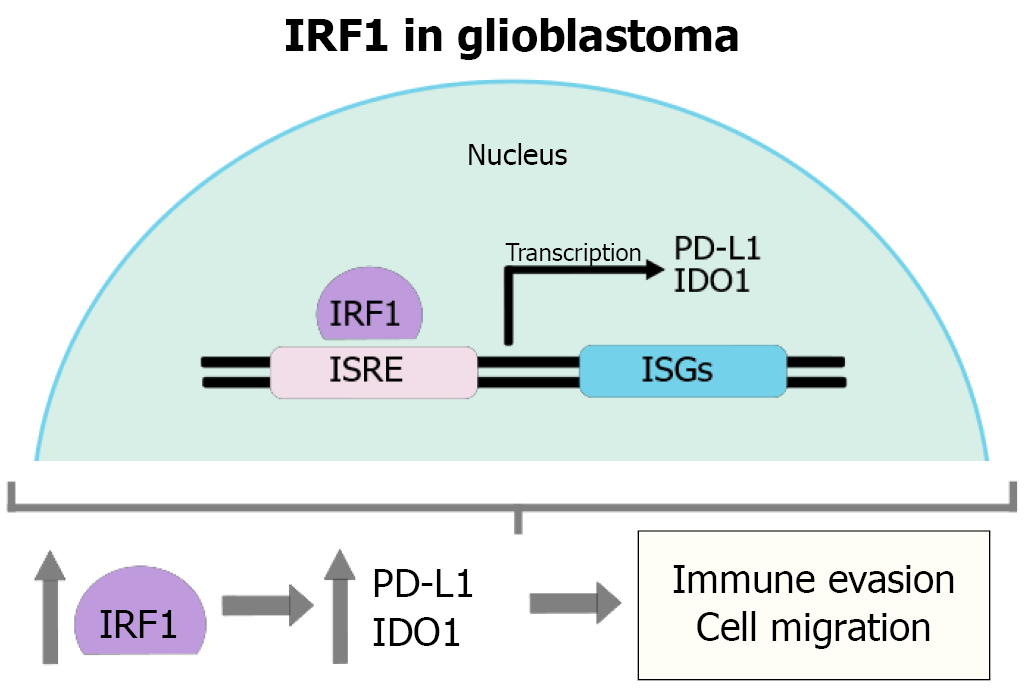
Figure 3 The molecular function of interferon regulatory factor 1 and its relationship with glioblastoma.
Interferon regulatory factor 1 protein has the molecular function of a transcription factor; through this mechanism, it can positively regulate the gene expression of programmed death-ligand 1 and indoleamine 2,3-dioxygenase 1, promoting immune evasion and cell migration. IRF1: Interferon regulatory factor 1; PD-L1: Programmed death-ligand 1; IDO1: Indoleamine 2,3-dioxygenase 1; ISRE: Interferon-stimulated response element.
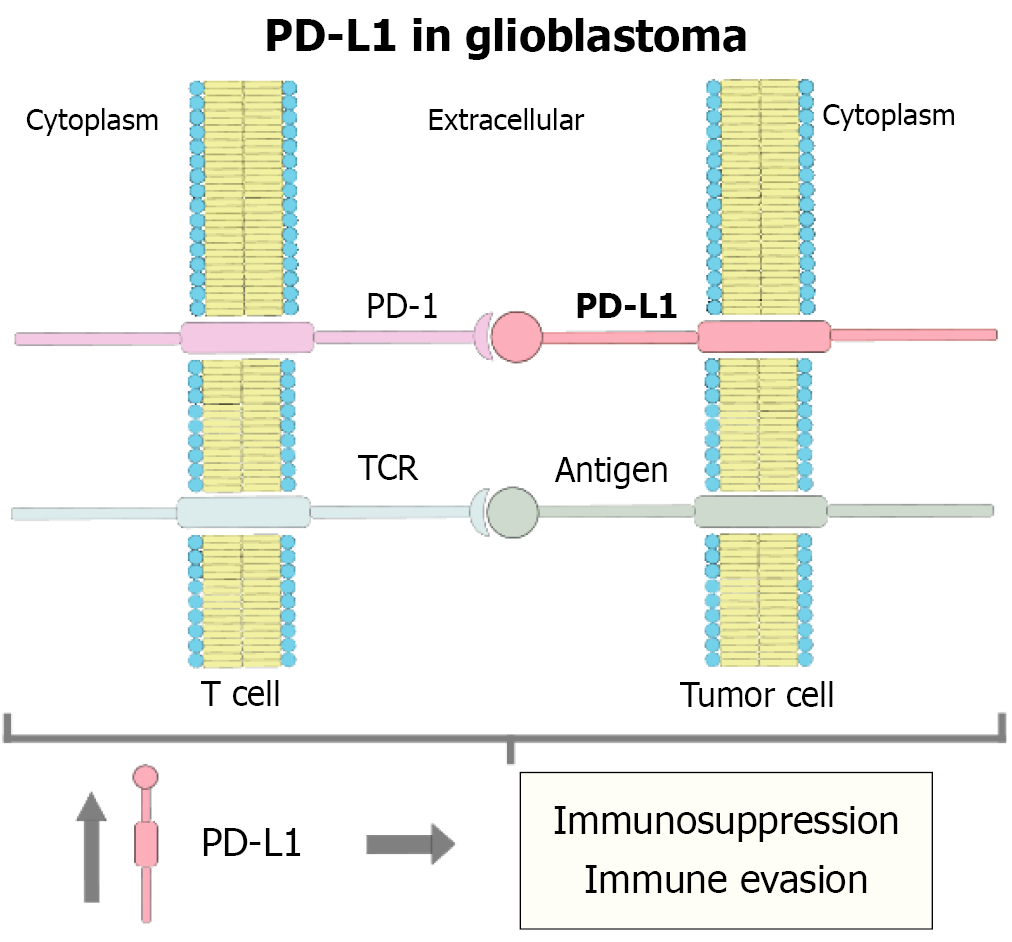
Figure 4 The molecular function of programmed death-ligand 1 and its relationship with glioblastoma.
Glioblastoma tumor cells can evade the immune response through the overexpression of programmed death-ligand 1 on their surface. This ligand binds to the programmed cell death protein 1 receptor present on T cells, inhibiting T cell receptor-mediated signaling, even in the presence of tumor antigens. As a result, T cells are not properly activated, promoting an immunosuppressive microenvironment that allows tumor progression. This regulatory axis represents a critical point in glioblastoma immune evasion. PD-L1: Programmed death-ligand 1; TCR: T cell receptor; PD-1: Programmed cell death protein 1.
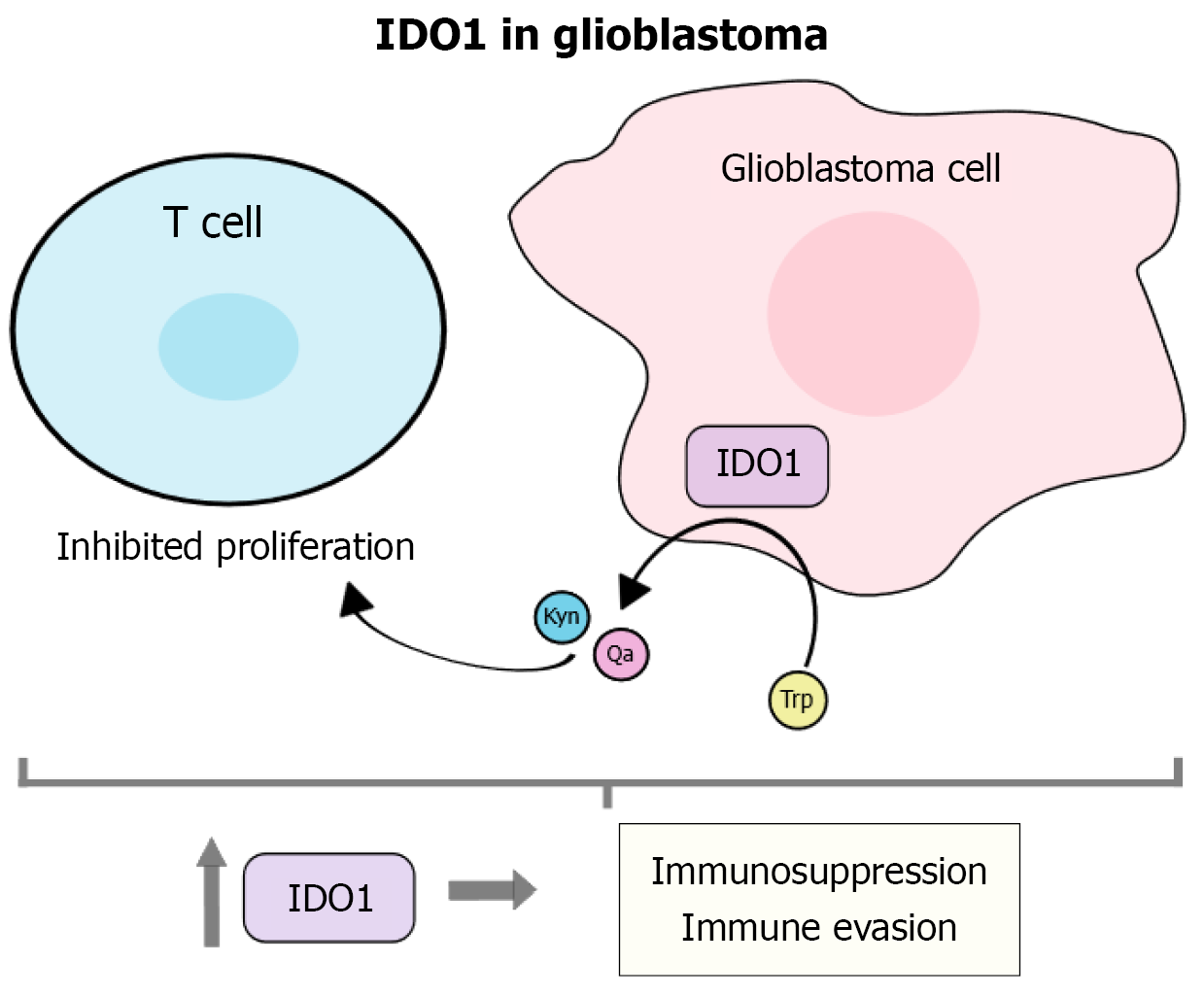
Figure 5 The molecular function of indoleamine 2,3-dioxygenase 1 and its relationship with glioblastoma.
Indoleamine 2,3-dioxygenase 1 is involved in the degradation of tryptophan into kynurenine and other toxic metabolites such as quinolinic acid; these metabolites function to suppress T-cell proliferation, thereby creating an immunosuppressed environment in glioblastoma. IDO1: Indoleamine 2,3-dioxygenase 1.
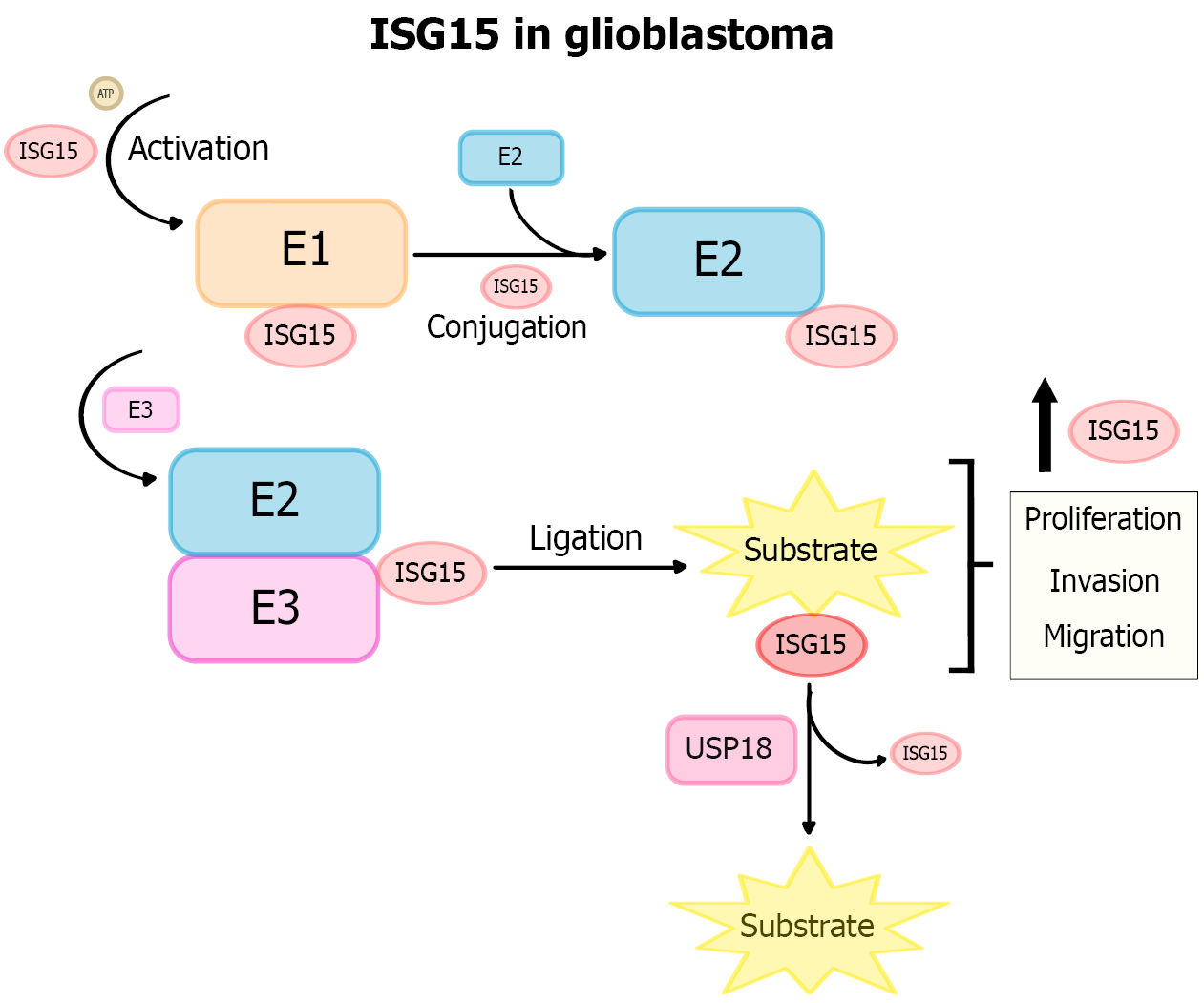
Figure 6 The molecular function of interferon-stimulated gene 15 and its relationship with glioblastoma.
Interferon-stimulated gene 15 (ISG15) carries out its function through a post-translational modification similar to ubiquitination, known as ISGylation, which involves three steps: Activation, conjugation, and ligation of ISG15 to its target. The figure shows that this post-translational modification is reversible by ubiquitin-specific protease 18. ISG15 can be found both free and conjugated to its substrate, and, in the context of glioblastoma, it functions to enhance proliferation, migration, and invasion. ISG15: Interferon-stimulated gene 15; USP18: Ubiquitin-specific protease 18.
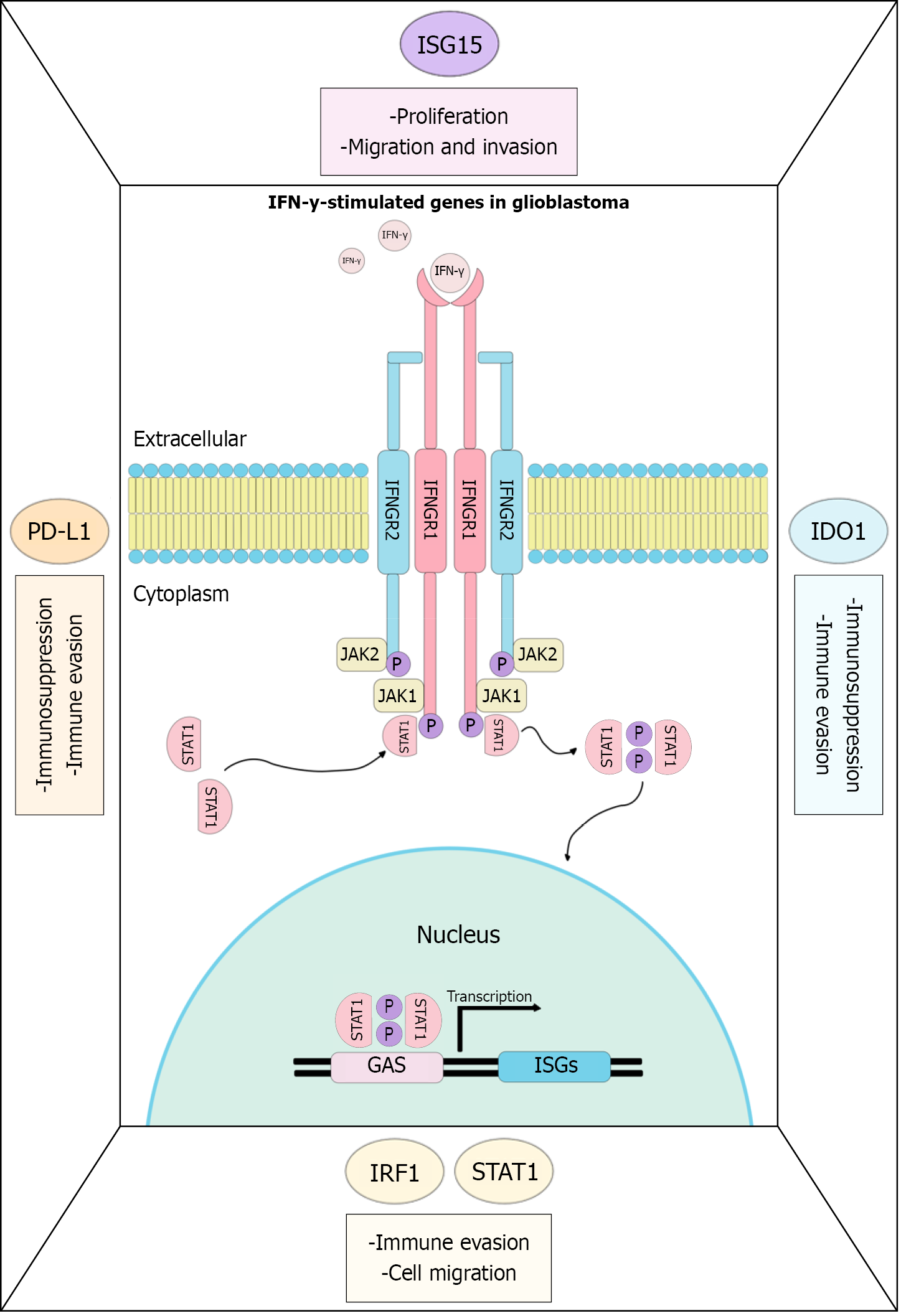
Figure 7 Key interferon-stimulated genes involved in glioblastoma pathology.
The binding of interferon-gamma to its receptor activates the Janus kinase/signal transducer and activator of transcription 1 (STAT1) pathway, promoting the transcription of interferon-stimulated genes. In glioblastoma, this activation is associated with the overexpression of genes such as programmed death-ligand 1 (PD-L1), indoleamine 2,3-dioxygenase 1 (IDO1), interferon regulatory factor 1 (IRF1), interferon-stimulated gene 15 (ISG15), and STAT1, which contribute to the establishment of an immunosuppressive and aggressive tumor microenvironment. PD-L1 and IDO1 promote immune evasion, while IRF1 is associated with poor survival. Meanwhile, STAT1 and ISG15 are linked to processes of cell proliferation, migration, and invasion, all of which are related to an aggressive tumor. PD-L1: Programmed death-ligand 1; ISG15: Interferon-stimulated gene 15; IRF1: Interferon regulatory factor 1; STAT1: Signal transducer and activator of transcription 1; JAK: Janus kinase; IFN-γ: Interferon-gamma; IDO1: Indoleamine 2,3-dioxygenase 1.
- Citation: Oropeza-Martínez E, Palacios Serrato EG, Zamora-Salas SX, Lira-Rodríguez NA, López-Mignon SH, Martinez-Benitez MB, Tecalco-Cruz AC. Interferon-gamma signaling pathway: Modulation of key genes in the progression of glioblastoma. World J Biol Chem 2025; 16(4): 112768
- URL: https://www.wjgnet.com/1949-8454/full/v16/i4/112768.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v16.i4.112768