Published online Aug 27, 2022. doi: 10.4240/wjgs.v14.i8.833
Peer-review started: April 12, 2022
First decision: May 11, 2022
Revised: May 14, 2022
Accepted: August 6, 2022
Article in press: August 6, 2022
Published online: August 27, 2022
Processing time: 134 Days and 1.5 Hours
Colorectal cancer (CRC) is the third most common cancer worldwide, and it is the second leading cause of death from cancer in the world, accounting for approximately 9% of all cancer deaths. Early detection of CRC is urgently needed in clinical practice.
To build a multi-parameter diagnostic model for early detection of CRC.
Total 59 colorectal polyps (CRP) groups, and 101 CRC patients (38 early-stage CRC and 63 advanced CRC) for model establishment. In addition, 30 CRP groups, and 62 CRC patients (30 early-stage CRC and 32 advanced CRC) were separately included to validate the model. 51 commonly used clinical detection indicators and the 4 extrachromosomal circular DNA markers NDUFB7, CAMK1D, PIK3CD and PSEN2 that we screened earlier. Four multi-parameter joint analysis methods: binary logistic regression analysis, discriminant analysis, classification tree and neural network to establish a multi-parameter joint diagnosis model.
Neural network included carcinoembryonic antigen (CEA), ischemia-modified albumin (IMA), sialic acid (SA), PIK3CD and lipoprotein a (LPa) was chosen as the optimal multi-parameter combined auxiliary diagnosis model to distinguish CRP and CRC group, when it differentiated 59 CRP and 101 CRC, its overall accuracy was 90.8%, its area under the curve (AUC) was 0.959 (0.934, 0.985), and the sensitivity and specificity were 91.5% and 82.2%, respectively. After validation, when distinguishing based on 30 CRP and 62 CRC patients, the AUC was 0.965 (0.930-1.000), and its sensitivity and specificity were 66.1% and 70.0%. When distinguishing based on 30 CRP and 32 early-stage CRC patients, the AUC was 0.960 (0.916-1.000), with a sensitivity and specificity of 87.5% and 90.0%, distinguishing based on 30 CRP and 30 advanced CRC patients, the AUC was 0.970 (0.936-1.000), with a sensitivity and specificity of 96.7% and 86.7%.
We built a multi-parameter neural network diagnostic model included CEA, IMA, SA, PIK3CD and LPa for early detection of CRC, compared to the conventional CEA, it showed significant improvement.
Core Tip: Most patients with colorectal cancer (CRC) are diagnosed at an advanced stage. The high morbidity and mortality of advanced CRC indicates an urgent need for clinical improvements in early CRC detection and individualized management. Compared with free linear DNA, extrachromosomal circular DNA is not easily degraded by nucleases, and its structure is more stable. In this study, we aimed to build a multi-parameter diagnostic model for early detection of CRC.
- Citation: Li J, Jiang T, Ren ZC, Wang ZL, Zhang PJ, Xiang GA. Early detection of colorectal cancer based on circular DNA and common clinical detection indicators. World J Gastrointest Surg 2022; 14(8): 833-848
- URL: https://www.wjgnet.com/1948-9366/full/v14/i8/833.htm
- DOI: https://dx.doi.org/10.4240/wjgs.v14.i8.833
Colorectal cancer (CRC) is the third most common cancer worldwide, and it is the second leading cause of death from cancer in the world, accounting for approximately 9% of all cancer deaths. Currently, surgery is the most common treatment for nonmetastatic CRC[1]. Most patients with CRC are diagnosed at an advanced stage. The high morbidity and mortality of advanced CRC indicates an urgent need for clinical improvements in early CRC detection and individualized management[2].
In the era of precision oncology, liquid biopsy has become the primary method for characterizing circulating tumor components present in body fluids[3]. This noninvasive tool can identify relevant molecular alterations in CRC patients, including some that indicate disruption of epigenetic mechanisms. Epigenetic alterations found in solid and liquid biopsies have shown great utility as biomarkers for the early detection, prognosis, monitoring, and assessment of the treatment response in CRC patients[4]. Therefore, the term “liquid biopsy” includes blood, the most commonly used human fluid sample, as well as other fluids, such as urine, ascites, pleural effusion, cerebrospinal fluid, and saliva[5,6]. Both primary tumors and metastases can release tumor material into these body fluids, mainly comprised of circulating tumor cells (CTCs), nucleic acids (cNA), and extracellular vesicles (cEVs)[7]. These circulating elements constitute a valuable source of noninvasive biomarkers[8-11].
At present, single-stranded or double-stranded DNA is detected based on ctDNA. With the development of high-throughput sequencing technology and single-cell gene amplification technology, new types of circular cell-free DNA have been discovered such as extrachromosomal circular DNA (eccDNA)[12,13]. eccDNA refers to a closed circular DNA located outside the chromosome in the form of single-stranded or double-stranded DNA, which is widely found in eukaryotes, including humans[14,15]. Compared with free linear DNA, eccDNA is not easily degraded by nucleases, and its structure is more stable.
In our study, we aimed to build a multi-parameter diagnostic model based on the commonly used clinical detection indicators and the 4 eccDNA markers for early detection of CRC which is urgently needed in clinical practice.
After approval by the ethics committee, the research subjects signed informed consent forms. This project included 59 patients with colorectal polyps (CRP) and 101 CRC patients (38 early-stage CRC and 63 advanced CRC) for building the model. An additional 30 CRP and 62 CRC patients (30 early-stage CRC and 32 advanced CRC) were used to validate the model (Table 1).
| Clinical features | Model building | Model validation | ||
| CRC (n = 101) | CRP (n = 59) | CRC (n = 62) | CRP (n = 30) | |
| Age | ||||
| Average | 58 | 56 | 57 | 57 |
| Range | 29-81 | 31-76 | 33-74 | 35-69 |
| Sex | ||||
| Male | 60 | 34 | 37 | 19 |
| Female | 41 | 25 | 25 | 11 |
| Location | ||||
| Ascending colon | 21 | 17 | ||
| Descending colon | 15 | 12 | ||
| Transverse colon | 3 | 4 | ||
| Sigmoid colon | 59 | 28 | ||
| Rectal | 3 | 1 | ||
| Differentiation | ||||
| Well | 21 | 15 | ||
| Moderate | 57 | 33 | ||
| Poorly | 23 | 14 | ||
| TNM stage | ||||
| T1 | 11 | 11 | ||
| T2 | 27 | 21 | ||
| T3 | 44 | 7 | ||
| T4 | 19 | 23 | ||
The inclusion criteria for the CRP group were those with villous/tubular adenoma, with or without mild-to-moderate hyperplasia, confirmed by colonoscopy and pathologically confirmed after adenoma removal, or confirmed by pathology and immunohistochemistry as focal high-grade neoplasia of villous tubular adenoma. All biochemical examinations and auxiliary examinations showed no abnormality, no complaints of gastrointestinal discomfort, no signs of a tumor, adenoma with a diameter less than 1 cm, no villous adenoma or mixed adenoma, and no adenoma with moderate to severe dysplasia.
In the early CRC group, it was confirmed by tumor surgery that the adenocarcinoma of the intestinal wall was confined to the mucosa or submucosa without lymphatic metastasis, that is, stage 1 or 2, and it was pathologically confirmed villous tubular adenoma with focal high-grade neoplasia or intestinal wall glands.
For the advanced CRC group based on tumor staging according to the American Joint Committee on Cancer tumor node metastasis staging, we defined colorectal cancer stages 3 and 4 as advanced stage with pathologically confirmed colorectal cancer; no treatment was performed before sample collection, including surgery, chemotherapy, radiotherapy, or other treatments; and no blood transfusion had occurred within the past 3 mo.
All enrolled patients provided colorectal cancer or polyp specimens and the corresponding clinical examination data. None of the patients received chemotherapy, radiotherapy or immunotherapy before surgery, and other tumors and gastrointestinal diseases were excluded by examination at the time of admission.
Peripheral blood was collected from all subjects included in this study on an empty stomach in the morning. The anticoagulant in the plasma collection tube was EDTA and after collection, the blood was centrifuged at 3000 rpm for 10 min, and the plasma was placed into a new sterile Eppendorf tube. Serum samples were early morning fasting peripheral blood samples collected in tubes containing separation gel and a clot activator. The samples were centrifuged at 3000 rpm for 10 min, and the serum was transferred to new sterile Eppendorf tubes and stored at -80 °C until assayed. The plasma was also stored at -80 °C. During the sample collection process, hemolyzed and chyle blood samples were removed to avoid repeated freezing and thawing. When testing was conducted, normal temperature recovery was performed.
There were 51 commonly used clinical detection indicators, including 13 common tumor-related markers and 38 clinical biochemical indicators. Among them, 13 tumor-related indicators included carcinoembryonic antigen (CEA), alpha fetoprotein (AFP), carbohydrate antigen 125 (CA125), CA199, CA153, CA724, cytokeratin fragment 211 (Cyfra211), ferritin (Ferr), neuron-specific enolase (NSE), squamous cell carcinoma (SCC), pepsinogen (PG) I, PG II and PGI/II. The 38 clinical biochemical indicators included alanine aminotransferase (ALT), aspartate aminotransferase (AST), total protein (TP), albumin (ALB), total bilirubin (TB), direct bilirubin (DB), total bile acid (TBA), alkaline pho
Among the 51 detection indicators, CEA, AFP, CA199, CA724, CA125, CA153, Cyfra211, Ferr, NSE, ALT, AST, TP, ALB, ALP, GGT, Glu, UN, CR, UA, CHO, TG, CK, Ca, P, Mg, K, Na, CL, CO2, HDL, LDL, CRP, AMY, and LPS standards and controls and detection kits were purchased from Roche Diagnostics Ltd. ApoA1, ApoB, CYS, LPa, and CKMB standards and controls and detection kits were purchased from Beijing Leadman Biochemical Co., Ltd. SCC, PG I and PG II standards and controls and test kits were purchased from Abbott Diagnostics. TBA and HCY standards and quality controls and detection kits were purchased from Beijing Jiuqiang Biotechnology Co., Ltd. TB and DB standards and controls and assay kits were purchased from Hitachi Diagnostics Co., Ltd. IMA standards, quality control products, and detection kits were purchased from Changsha Yikang Technology Development Co., Ltd. SA standards, quality control products, and detection kits were purchased from Zhejiang Dongou Diagnostic Products Co., Ltd. SOD standards, quality control products and detection kits were purchased from Fujian Fuyuan Biotechnology Co., Ltd. A modular 7600 automatic biochemical analyzer, Roche E170 immunoassay analyzer and Architect i2000 immunoassay system were used to complete the pre-assay quality control and calibration. After the analysis, the experimental data of each instrument were exported for statistical analysis.
Cell-free DNA was extracted from plasma samples using the QIAamp DNA Blood Kit (Qiagen, 51192) according to the ddPCR detection method established in the second part of this study. ATP-dependent DNase (Epicenter, E310K) was added to the free DNA and digested at 37 °C for 1.5 h to a final concentration of 0.4 U/μL to remove linear double-stranded DNA. The reaction was continued at 70 °C for 30 min to inactivate ATP-dependent DNase activity, and the product was then stored until analysis.
Based on the eccDNA sequence incorporated into the model, primers were designed using Primer3 software. After a homology search was performed with BLAST, the primers were synthesized by Invitrogen. The 5' ends of the primers were modified with a FAM fluorophore, and the 3' ends were modified with a BHQ1 quenching group. (1) NDUFB7. Forward sequence: TACCGTCAGC
Primers and probes were diluted with deionized water to the storage concentration of 200 μmol/L, and the working concentration was 10 μmol/L. The total PCR volume was 20 μL, including 2-fold ddPCRTTM Super mix 10 μL, forward and reverse primers 1.8 μL each (final concentration 900 nmol/L), probe 0.5 μL (final concentration 250 nmol/L), template DNA 4 μg, and ddH2O to make it up to 20 μL. Then, 20 μL of the reaction system mixture was added to the droplet generation card for droplet generation. All of the resulting droplets were transferred to a 96-well plate for PCR amplification. The PCR conditions were: 95 °C/10 min; 94 °C/30 s, 60 °C/1 min, 40 cycles; 98 °C/10 min. Finally, Quanta Soft 1.6 software (Bio-Rad, USA) was used to analyze the results and the Flush System was used before each experiment. After the setup is complete, the sample droplets are analyzed. We analyzed the results of the run and view channels, scatterplots, concentration data, ratio data, and the number of events.
Second, we compared the 51 common clinical indicators and 4 kinds of eccDNA between the CRP group and CRC group based on the difference indicator, tested by the area under the curve (AUC) and the P value, for potential markers to evaluate their diagnostic value for distinguishing the CRP and CRC groups, CRP and early CRC groups, colon polyps and advanced CRC groups.
Based on the differential diagnostic value (CRP group vs CRC group), we established a multiparameter combined auxiliary diagnostic model. The models are binary logistic regression analysis, discriminant analysis, classification tree and neural network. Binary logistic regression analysis was used for the Forward: Conditional method. Discriminant analysis applied the Bayes discriminant method, and stepwise discriminant analysis was used in the fitting function process. A classification tree was the CHAID classification tree method, and a cross-validation evaluation was conducted to establish the classification tree model. An artificial neural network was the neural network's multilayer perceptron used to build the model.
After comparing the diagnostic value of the binary logistic regression analysis, the discriminant analysis, classification tree and neural network with the diagnostic value of a single index were conducted. The optimal multiparameter auxiliary diagnosis model was selected, and 30 CRP groups and 62 CRC patients (30 early-stage CRC patients and 32 advanced CRC patients) were enrolled to validate the multiparameter model. Then, the stability of the model was evaluated. Finally, the validated model was compared with the commonly used clinical detection index CEA, and its clinical application value was evaluated by comparing the sensitivity, specificity, and AUC.
SPSS 22.0 was used for statistical analysis. Measurement data were expressed as medians (25%, 75%). If the data were normally distributed, they were compared by two independent samples t-tests. If nonnormally distributed, comparisons were made by the rank-sum test. The AUC was used to assess the diagnostic value of the index. Four multiparameter analysis methods (binary logistic regression analysis, discriminant analysis, classification tree and neural network) were used to establish a multiparameter joint diagnosis model. The binary logistic regression model used the forward conditional method. The discriminant analysis used the Bayes discriminant method. The classification tree used the CHAID classification tree method, and the established classification tree model was evaluated by cross-validation. Artificial neural networks used multilayer perceptrons of neural networks to build the models. Univariate and multivariate logistic regression were used to analyze Exp (B) of the index. The Z score test was used to compare the AUC of the different groups. P < 0.05 indicates that the difference is statistically significant.
Thirteen tumor markers (CEA, AFP, CA125, CA199, CA153, CA724, CY211, Ferr, NSE, SCC, PG I/II, PG II, and PG I) and 38 blood biochemical indices (ALT, AST, TP, ALB, TB, DB, TBA, ALP, GGT, GLu, UN, Cr, UA, CHO, TG, CK, LDH, CKMB, Ca, P, Mg, K, Na, Cl, CO2, LPa, HDL, LDL, ApoA1, ApoB, CYS, SA, HCY, CRP, AMY, LPS, SOD, and IMA) were compared between the 59 CRP patients and the 101 CRC patients. Among the 51 commonly used clinical indicators, 22 indicators, including IMA, CEA, SA, LPa, CK, TB, HDL, NSE, ALT, Ferr, DB, CA125, LDH, AMY, CY211, CA724, HCY, CHO, P, LDL, Cl and CKMB, were significantly different between the CRP and CRC groups (P < 0.05). The remaining 29 indicators were not significantly different. By comparison, among the four eccDNA indices, two indices, CAMK1D and PIK3CD, showed significant differences between the CRP and CRC groups (P < 0.05). The other two indicators were not significantly different, as shown in Table 2.
| Index | CRP (n = 59) | CRC (n = 101) | F value | Sig | P value |
| CEA | 1.86 (1.17, 2.43) | 3.9 (1.67, 13.87) | 11.39 | < 0.01 | < 0.01 |
| AFP | 2.58 (1.87, 3.59) | 2.41 (1.75, 3.36) | 0.02 | 0.90 | 0.41 |
| CA125 | 9.78 (6.77, 13.55) | 11.63 (7.98, 19.9) | 4.80 | 0.03 | 0.04 |
| CA199 | 8.57 (5.44, 14.38) | 13.43 (7.22, 26.48) | 3.62 | 0.06 | 0.22 |
| CA153 | 9.5 (7.08, 13.09) | 9.25 (6.6, 13) | 1.53 | 0.22 | 0.49 |
| CA724 | 1.63 (1.16, 4.39) | 2.55 (1.36, 7.33) | 5.54 | 0.02 | 0.07 |
| CY211 | 1.82 (1.4, 2.89) | 2.3 (1.63, 3.58) | 9.29 | < 0.01 | < 0.01 |
| Ferr | 150.9 (85.62, 269.5) | 72.12 (17.02, 161.5) | 0.11 | 0.74 | 0.01 |
| NSE | 8.06 (6.52, 9.16) | 10 (7.71, 12.63) | 4.58 | 0.03 | < 0.01 |
| SCC | 1.1 (0.7, 1.5) | 0.8 (0.6, 1.2) | 2.96 | 0.09 | 0.19 |
| PG I/II | 4.576 (2.835, 5.914) | 5.12 (3.7, 6.53) | 0.10 | 0.76 | 0.08 |
| PG II | 15.9 (9, 28.3) | 14.6 (9.7, 24.2) | 1.08 | 0.30 | 0.64 |
| PG I | 75.5 (38.5, 101.3) | 71.7 (51.45, 96.3) | 0.49 | 0.49 | 0.82 |
| ALT | 16.8 (12.1, 25) | 12.7 (9.3, 17.75) | 0.59 | 0.44 | 0.03 |
| AST | 17.1 (14.1, 20.6) | 16.6 (12.25, 19.3) | 0.87 | 0.35 | 0.43 |
| TP | 68.3 (64.1, 71.9) | 67.3 (63.15, 70.65) | 0.03 | 0.86 | 0.29 |
| ALB | 41.8 (39.6, 44.4) | 39.5 (36.95, 41.45) | 0.63 | 0.43 | 0.07 |
| TB | 12.5 (10, 16.4) | 9.7 (7.4, 12.8) | 0.75 | 0.39 | < 0.01 |
| DB | 4.1 (3.2, 5.2) | 3.6 (2.3, 4.2) | 0.05 | 0.82 | 0.01 |
| TBA | 4.2 (2.5, 7.2) | 3.5 (2.2, 5.7) | 2.15 | 0.14 | 0.11 |
| ALP | 61.4 (54.8, 73.6) | 67 (56.4, 80.05) | 2.38 | 0.13 | 0.59 |
| GGT | 23.6 (13.5, 37.6) | 22.3 (14.75, 33.95) | 0.01 | 0.95 | 0.98 |
| GLu | 5.02 (4.79, 5.51) | 5.12 (4.74, 5.81) | 0.00 | 0.97 | 0.97 |
| UN | 5.49 (4.64, 6.08) | 5.23 (4.08, 6.29) | 5.94 | 0.02 | 0.43 |
| Cr | 70.2 (61.6, 78.6) | 65.2 (56.35, 75.6) | 0.22 | 0.64 | 0.06 |
| UA | 312.3 (257.9, 386.9) | 292.8 (224, 339.4) | 0.19 | 0.67 | 0.06 |
| CHO | 4.5 (4.04, 5.27) | 4.36 (3.88, 5.09) | 2.31 | 0.13 | 0.02 |
| TG | 1.43 (1.01, 2.01) | 1.25 (0.93, 1.62) | 7.94 | 0.01 | 0.45 |
| CK | 69.8 (55.5, 118.9) | 54.4 (35.2, 71.05) | 15.60 | < 0.01 | 0.04 |
| LDH | 137 (122.2, 153.3) | 148.4 (129.75, 177.75) | 4.13 | 0.04 | < 0.01 |
| CKMB | 6.6 (4, 9.8) | 6.14 (4.05, 9.6) | 1.81 | 0.18 | 0.02 |
| Ca | 2.26 (2.19, 2.31) | 2.21 (2.14, 2.27) | 0.10 | 0.75 | 0.47 |
| P | 1.27 (1.14, 1.39) | 1.25 (1.07, 1.38) | 0.01 | 0.93 | 0.01 |
| Mg | 0.93 (0.85, 1.01) | 0.91 (0.84, 0.97) | 0.01 | 0.94 | 0.29 |
| K | 4.03 (3.78, 4.18) | 4.09 (3.87, 4.33) | 4.98 | 0.03 | 0.53 |
| Na | 143.8 (141.6, 145.4) | 143.1 (141.45, 144.7) | 0.17 | 0.68 | 0.12 |
| Cl | 105.6 (103.4, 107.2) | 105.3 (103.5, 107.4) | 2.08 | 0.15 | 0.04 |
| CO2 | 22.6 (20.7, 26.1) | 24.9 (22.9, 26.65) | 2.31 | 0.13 | 0.40 |
| LPa | 7.83 (3.01, 12.74) | 15.65 (7.82, 31.65) | 13.29 | < 0.01 | 0.01 |
| HDL | 1.27 (1.03, 1.41) | 1.02 (0.89, 1.23) | 0.10 | 0.76 | < 0.01 |
| LDL | 2.63 (2.26, 3.28) | 2.54 (2.07, 3.27) | 1.33 | 0.25 | < 0.01 |
| ApoA1 | 1.39 (1.17, 1.54) | 1.13 (1.01, 1.34) | 0.66 | 0.42 | 0.55 |
| ApoB | 0.83 (0.72, 1.02) | 0.83 (0.72, 1.01) | 0.09 | 0.76 | 0.62 |
| CYS | 1.07 (0.95, 1.16) | 0.97 (0.84, 1.08) | 0.34 | 0.56 | 0.70 |
| SA | 59.3 (55, 66.5) | 67.1 (60.8, 82.4) | 13.50 | < 0.01 | 0.04 |
| HCY | 15.19 (11.54, 19.68) | 13.92 (11.18, 17.42) | 4.71 | 0.03 | < 0.01 |
| CRP | 0.7 (0.4, 1.5) | 3.9 (1, 10.55) | 30.41 | < 0.01 | 0.11 |
| AMY | 59.5 (50, 73.7) | 51.8 (38.95, 64.7) | 1.18 | 0.28 | < 0.01 |
| LPS | 33.1 (25.1, 42.7) | 32.9 (22.25, 44.25) | 2.87 | 0.09 | 0.06 |
| SOD | 136.1 (125, 147) | 136.5 (115.8, 156.9) | 4.82 | 0.03 | 0.35 |
| IMA | 63.8 (60.1, 66.3) | 62.1 (59.45, 67.5) | 0.11 | 0.74 | < 0.01 |
| NDUFB7 | 1.34 (0.94, 2.42) | 2.10 (1.29, 3.08) | 2.666 | 0.105 | 0.155 |
| CAMK1D | 34.21 (17.82, 103.44) | 70.39 (35.26, 155.57) | 3.045 | 0.083 | 0.030 |
| PIK3CD | 105.90 (36.69, 308.35) | 333.22 (259.40, 417.90) | 3.700 | 0.056 | 0.001 |
| PSEN2 | 6.46 (4.44, 11.03) | 8.69 (6.00, 11.67) | 0.144 | 0.705 | 0.154 |
Based on the 22 commonly used clinical indicators and 2 kinds of eccDNA that showed significant differences between the CRP and CRC groups, receiver operating characteristic (ROC) curves were used to evaluate the diagnostic value, as shown in Table 3. Fifteen commonly used clinical indicators and 2 kinds of eccDNA (IMA, CEA, SA, LPa, CK, TB, HDL, NSE, ALT, Ferr, DB, CA125, LDH, AMY, CY211, CAMK1D and PIK3CD) showed statistically significant differences in the area under the curve (P < 0.05) while the other 7 commonly used clinical indicators (CA724, HCY, CHO, P, LDL, Cl and CKMB) showed no significant difference. Therefore, 15 commonly used clinical indicators and 2 kinds of eccDNA with significant differences between the groups and the areas under the ROC curve were selected for subsequent multiparameter combined auxiliary diagnosis model analysis.
| Indicator | AUC | SE | P value | 95% CI | |
| Lower | Upper | ||||
| IMA | 0.787 | 0.036 | < 0.001 | 0.716 | 0.859 |
| CEA | 0.734 | 0.038 | < 0.001 | 0.658 | 0.809 |
| SA | 0.728 | 0.039 | < 0.001 | 0.651 | 0.804 |
| LPa | 0.715 | 0.042 | < 0.001 | 0.633 | 0.797 |
| CK | 0.702 | 0.042 | < 0.001 | 0.619 | 0.784 |
| TB | 0.672 | 0.044 | < 0.001 | 0.585 | 0.758 |
| HDL | 0.670 | 0.044 | < 0.001 | 0.583 | 0.758 |
| NSE | 0.668 | 0.044 | < 0.001 | 0.580 | 0.755 |
| ALT | 0.667 | 0.044 | < 0.001 | 0.580 | 0.754 |
| Ferr | 0.663 | 0.045 | 0.001 | 0.575 | 0.751 |
| DB | 0.646 | 0.044 | 0.002 | 0.559 | 0.733 |
| CA125 | 0.642 | 0.044 | 0.003 | 0.557 | 0.728 |
| LDH | 0.621 | 0.045 | 0.011 | 0.534 | 0.709 |
| AMY | 0.611 | 0.045 | 0.019 | 0.522 | 0.700 |
| CY211 | 0.602 | 0.046 | 0.032 | 0.513 | 0.691 |
| CA724 | 0.583 | 0.046 | 0.081 | 0.492 | 0.673 |
| HCY | 0.570 | 0.048 | 0.138 | 0.476 | 0.664 |
| CHO | 0.556 | 0.046 | 0.240 | 0.465 | 0.646 |
| P | 0.543 | 0.047 | 0.361 | 0.451 | 0.636 |
| LDL | 0.536 | 0.046 | 0.453 | 0.445 | 0.626 |
| Cl | 0.525 | 0.047 | 0.603 | 0.432 | 0.618 |
| CKMB | 0.516 | 0.047 | 0.736 | 0.424 | 0.608 |
| CAMK1D | 0.652 | 0.046 | 0.001 | 0.561 | 0.742 |
| PIK3CD | 0.753 | 0.047 | < 0.001 | 0.660 | 0.845 |
Indices with statistically significant differences between the CRP and CRC groups and the ROC included IMA, CEA, SA, LP (a), CK, TB, HDL, NSE, ALT, Ferr, DB, CA125, LDH, AMY, CY211, CAMK1D and PIK3CD (P < 0.05). First, univariate logistic regression analysis was performed, as shown in Table 4. The Exp (B)s of CEA, IMA, SA, E3 and LPa were significantly different (P < 0.05), while that of CK, TB, HDL, NSE, CHO, P, LDL, Cl, CKMB and CAMK1D were not significantly different. Second, multivariate logistic regression analysis was performed on the differences in CEA, IMA, SA, E3 and LPa. As shown in Table 5, the Exp (B)s were significantly different for all of them (P < 0.05). CEA, IMA, SA, PIK3CD and LPa were included in the subsequent multiparameter joint auxiliary diagnosis model.
| Indicator | B | SE | Wals | P value | Exp (B) | 95% CI | |
| Lower | Upper | ||||||
| CEA | 0.335 | 0.138 | 5.864 | 0.015 | 1.398 | 1.066 | 1.834 |
| IMA | -0.138 | 0.048 | 8.352 | 0.004 | 0.871 | 0.793 | 0.956 |
| SA | 0.078 | 0.034 | 5.347 | 0.021 | 1.081 | 1.012 | 1.155 |
| LPa | 0.085 | 0.027 | 9.844 | 0.002 | 1.089 | 1.032 | 1.148 |
| CK | -0.004 | 0.008 | 0.207 | 0.649 | 0.996 | 0.980 | 1.013 |
| TB | -0.065 | 0.054 | 1.463 | 0.226 | 0.937 | 0.843 | 1.041 |
| HDL | -0.949 | 0.822 | 1.331 | 0.249 | 0.387 | 0.077 | 1.941 |
| NSE | 0.160 | 0.084 | 3.656 | 0.056 | 1.174 | 0.996 | 1.383 |
| CHO | -0.004 | 0.017 | 0.053 | 0.817 | 0.996 | 0.964 | 1.029 |
| P | 0.886 | 1.104 | 0.644 | 0.422 | 2.426 | 0.279 | 21.139 |
| LDL | 0-.585 | 0.368 | 2.534 | 0.111 | 0.557 | 0.271 | 1.145 |
| Cl | 0.112 | 0.086 | 1.682 | 0.195 | 1.119 | 0.944 | 1.325 |
| CKMB | -0.025 | 0.057 | 0.202 | 0.653 | 0.975 | 0.872 | 1.089 |
| CAMK1D | 0.003 | 0.003 | 1.189 | 0.275 | 1.003 | 0.998 | 1.009 |
| PIK3CD | 0.003 | 0.001 | 4.429 | 0.035 | 1.003 | 1.000 | 1.005 |
| Indicator | B | SE | Wals | P value | Exp (B) | 95% CI | |
| Lower | Upper | ||||||
| CEA | 0.326 | 0.109 | 8.904 | 0.003 | 1.385 | 1.118 | 1.716 |
| IMA | -0.136 | 0.035 | 14.765 | < 0.001 | 0.873 | 0.815 | 0.936 |
| SA | 0.092 | 0.027 | 11.601 | 0.001 | 1.097 | 1.040 | 1.156 |
| PIK3CD | 0.002 | 0.001 | 5.852 | 0.016 | 1.002 | 1.000 | 1.004 |
| LPa | 0.064 | 0.022 | 8.888 | 0.003 | 1.066 | 1.022 | 1.112 |
Based on CEA, IMA, SA, PIK3CD and LPa, a multiparameter combined auxiliary diagnosis model was built to distinguish the 59 CRP group and 101 CRC group (including 38 cases of early CRC and 63 cases of advanced CRC).
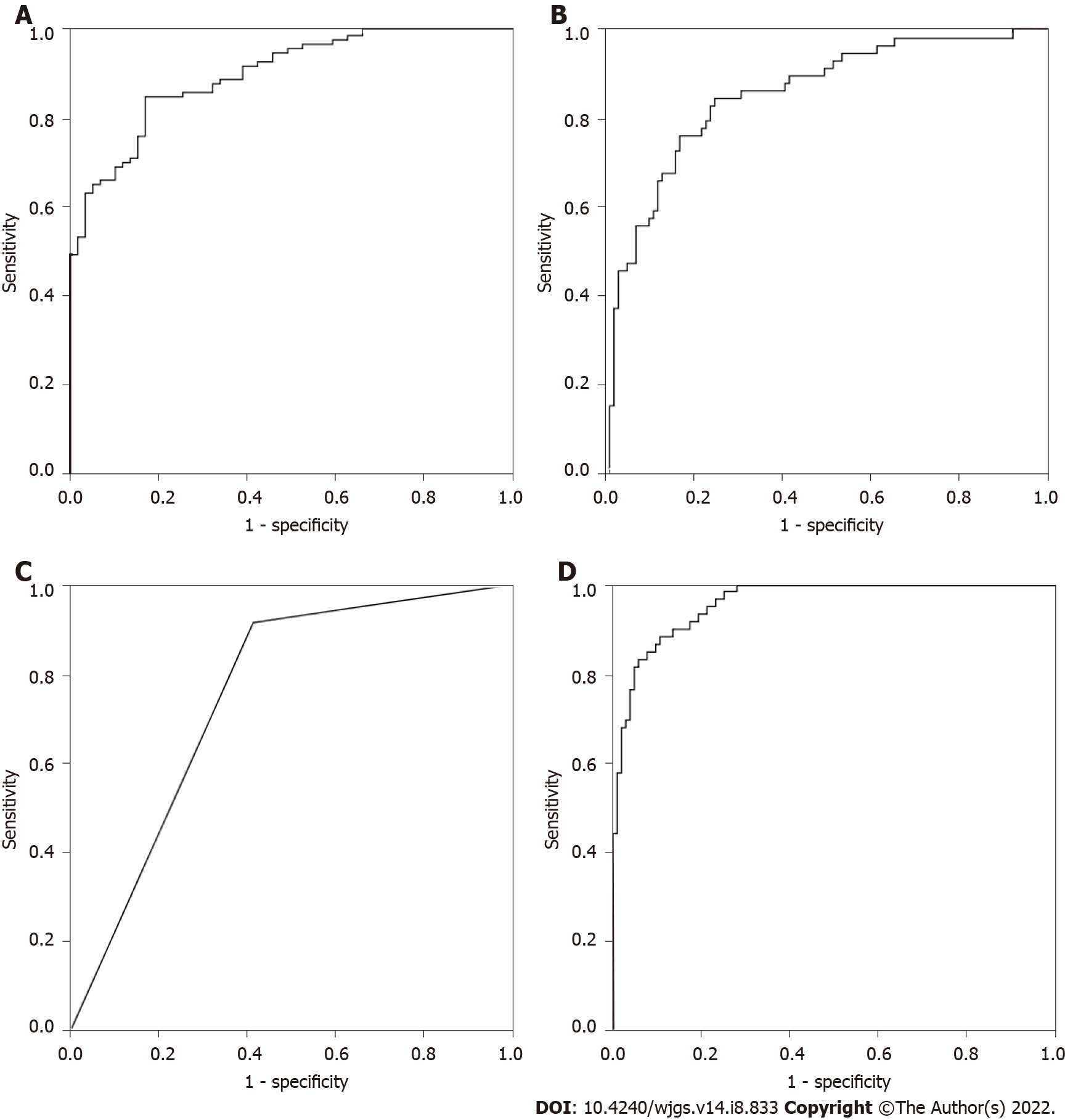
As shown in Table 6, binary logistic regression analysis based on CEA, IMA, SA, PIK3CD and LPa showed that the correct rate of CRP was 76.3%, the correct rate of CRC was 85.1%, and the overall accuracy was 81.9%. The predicted probability of each sample was used as an independent variable, as shown in Figure 1A, and the AUC was 0.900 (0.855-0.946).
| Observed | Predicted | ||
| CRP | CRC | Correct percentage | |
| Binary logistic regression analysis building | |||
| CRP | 45 | 14 | 76.30% |
| CRC | 15 | 86 | 85.10% |
| Total percentage | 81.90% | ||
| Discriminant analysis building | |||
| CRP | 51 | 8 | 86.40% |
| CRC | 31 | 70 | 69.30% |
| Total percentage | 75.60% | ||
| Classification tree building | |||
| CRP | 54 | 5 | 91.50% |
| CRC | 42 | 59 | 58.40% |
| Total percentage | 70.60% | ||
| Neural network building | |||
| CRP | 31 | 8 | 79.50% |
| CRC | 2 | 68 | 97.10% |
| Total percentage | 90.80% | ||
| Neural network validation | |||
| CRP | 18 | 2 | 90.00% |
| CRC | 4 | 27 | 87.10% |
| Total percentage | 88.20% | ||
The discriminant analysis based on CEA, IMA, SA, PIK3CD and LPa showed that the correct rate of CRP was 86.4%, the correct rate of CRC was 69.3%, and the overall accuracy was 75.6%. Taking the predicted probability of each sample as an independent variable, as shown in Figure 1B, the AUC was 0.855 (0.794-0.916).
In the classification tree analysis based on CEA, IMA, SA, PIK3CD and LPa, the final independent variables included CEA, IMA, SA, PIK3CD and LPa, the number of nodes was 3, the number of terminal nodes was 2, and the depth was 1. Among them, the correct rate of CRP was 91.5%, the correct rate of CRC was 58.4%, and the overall accuracy rate was 70.6%. Taking the predicted probability of each sample as an independent variable, as shown in Figure 1C, the AUC was 0.750 (0.674-0.826).
The artificial neural network analysis based on CEA, IMA, SA, PIK3CD and LPa, CEA, IMA, SA, PIK3CD and LPa all entered the input layer. The number of hidden layers included 1 Layer, and the output layer included 2 Layers. The training set included 39 cases of CRP and 70 cases of CRC, among which the correct rate of identifying healthy controls was 79.5%, the correct rate of identifying colorectal cancer was 97.1%, and the overall accuracy rate was 90.8%. The test set included 20 cases of CRP and 31 cases of CRC, among which the correct rate of identifying CRP was 90.0%, the correct rate of identifying CRC was 87.1%, and the overall accuracy rate was 88.2%. Taking the predicted probability of each sample as an independent variable, as shown in Figure 1D, the AUC was 0.959 (0.934-0.985).
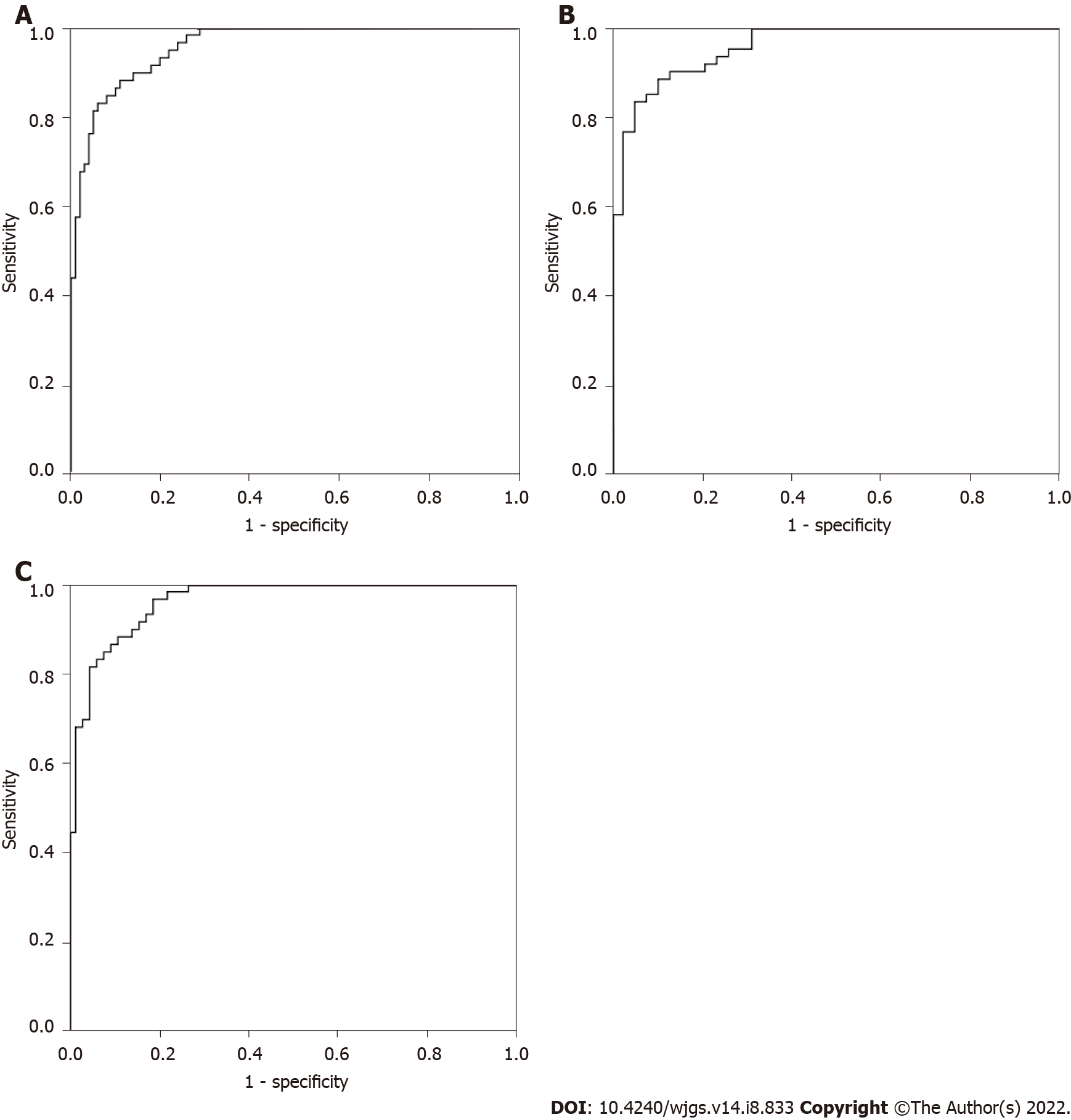
Based on CEA, IMA, SA, PIK3CD and LPa, binary logistic regression analysis, discriminant analysis, classification tree and neural network were used to predict the CRP and CRC groups, and the accuracy rates were 81.9%, 75.6%, 70.6%, and 90.8%, respectively. Therefore, we chose the neural network as the optimal multiparameter joint auxiliary diagnosis model. As shown above, the overall accuracy rate was 90.8%, as shown in Figure 2A. The area under the curve was 0.959 (0.934-0.985), and the sensitivity and specificity were 91.5% and 82.2%, respectively. As shown in Figure 2B, when the CRP and early CRC groups were differentiated, the area under the curve was 0.956 (0.921-0.992), and the sensitivity and specificity were 89.8% and 86.8%, respectively. As shown in Figure 2C, when the CRP and advanced CRC groups were differentiated, the area under the curve was 0.961 (0.932-0.990), and the sensitivity and specificity were 88.1% and 87.3%, respectively.
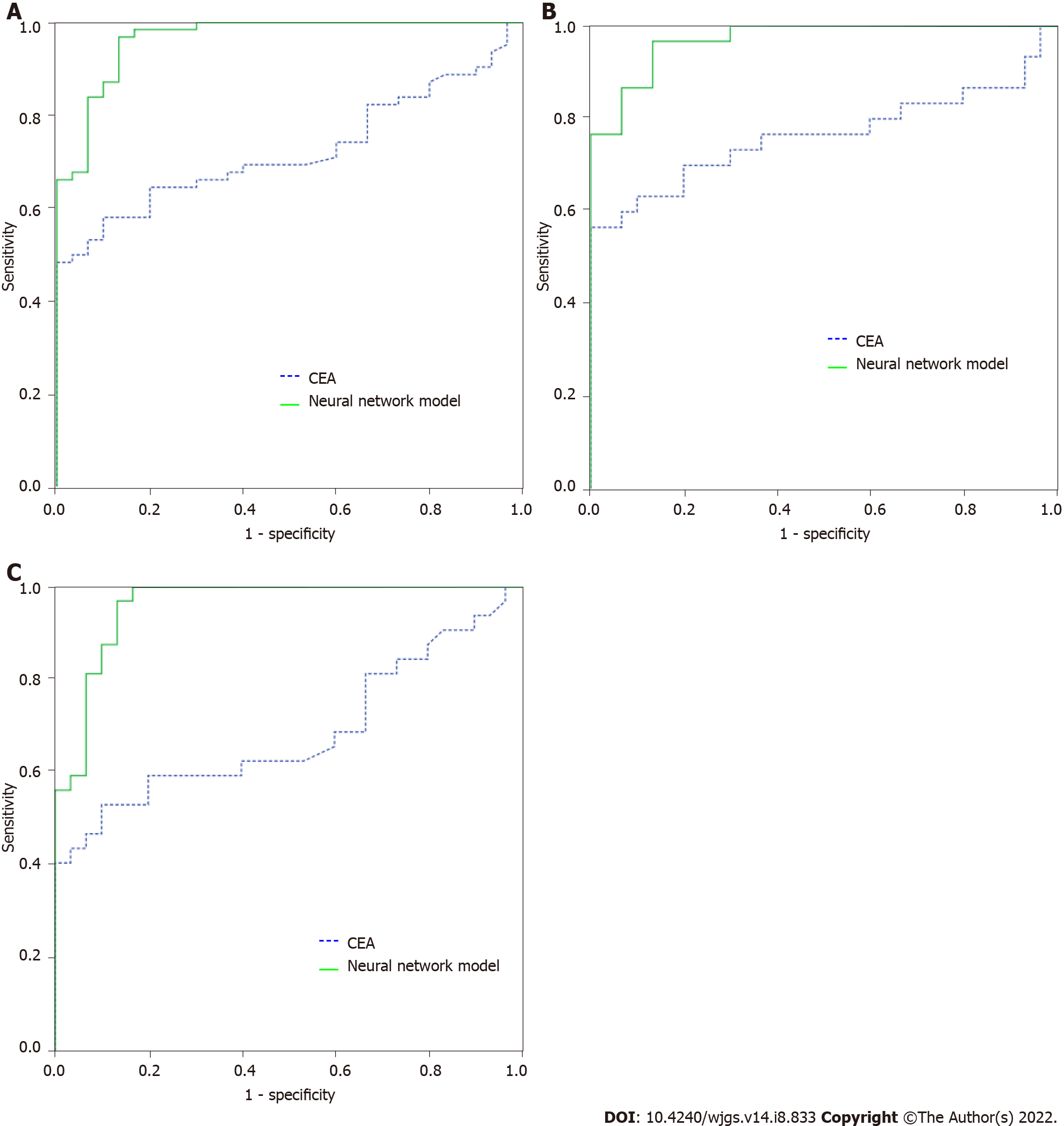
For distinguishing the CRP group from the CRC group, after comparing the multiple multiparameter joint analysis methods, the neural network based on CEA, IMA, SA, PIK3CD and LPa was the optimal multiparameter joint auxiliary diagnosis model. Thirty independent CRP patients and 62 CRC patients (32 in the early-stage CRC group and 30 in the advanced CRC group) were enrolled to validate the model. After validation, as shown in Figure 3A, for distinguishing CRP and CRC, the area under the curve of the neural network for CEA, IMA, SA, PIK3CD and LPa was 0.965 (0.930-1.000), its sensitivity and specificity were 66.1% and 70.0%, the area under the curve of the commonly used clinical indicator CEA was 0.723 (0.622-0.823), and its sensitivity and specificity were 96.8% and 86.7%, respectively. As shown in Figure 3B, for distinguishing CRP and 32 early-stage CRC, the area under the curve of the neural network model was 0.960 (0.916-1.000), with a sensitivity and specificity of 87.5% and 90.0%, the area under the curve of the commonly used clinical indicator CEA was 0.684 (0.548-0.821), and its sensitivity and specificity were 62.5% and 60.0%, respectively. As shown in Figure 3C, for distinguishing CRP and advanced CRC patients, the area under the curve of the neural network model was 0.970 (0.936, 1.000), with a sensitivity and specificity of 96.7% and 86.7%, the area under the curve of the commonly used clinical indicator CEA was 0.763 (0.632-0.895), and its sensitivity and specificity were 76.7% and 63.3%, respectively.
A biomarker is a biological molecule found in blood, other body fluids, or tissues that is a marker of a normal or abnormal process or disease. Biomarkers are primarily based on DNA, RNA, microRNA (miRNA), epigenetic changes, or antibodies. The term tumor marker, considered by some researchers to be synonymous with biomarkers, refers to substances that represent biological structures (most typically proteins, glycolipids) that can be attributed to normal cell development or to different stages of cell development. For example, carcinogenesis-associated antigens (TAAs) are the largest group of clinically meaningful markers. Therefore, the concentration of TAA usually correlates with the quantity (or quality) of specific tumor cells.
Discovered 50 years ago in 1965, CEA is still the only tumor marker with proven efficacy in monitoring treatment in CRC patients. CEA was initially thought to be CRC specific, but elevated CEA levels have since been detected in other tumors, e.g., gastric and pancreatic cancer, and inflammatory states. Rarely, elevated CEA concentrations are found in CRC stage I[16]. Furthermore, CEA cannot differentiate between benign and malignant polyps. Recently, several studies have explored the advantages of mRNA molecules encoding CEA for the detection of CRC, but the results were not superior to CEA[17]. In some studies, high CEA concentrations in patients with CRC stages II and III may be indicative of a more aggressive cancer type. CEA is the marker of choice for monitoring disseminated disease during systemic therapy. Sustained increases in CEA levels are often associated with disease progression, even though radiological examination may prove otherwise. However, chemotherapy may also cause a temporary increase in CEA concentrations, which must be taken into account. Therefore, it is not recommended to measure CEA levels within 2 wk after chemotherapy but only after 4 to 6 wk in oxaliplatin-treated patients. Cancer antigen 19-9 (CA 19-9) is a glycoprotein whose relevance in the diagnosis of CRC remains unclear. Most investigators concluded that the sensitivity of CA 19-9 was much lower than that of CEA and that elevated CA 19-9 Levels indicated a poor prognosis[18]. Other carbohydrate antigens, CA 19-5 and CA 50, have also been investigated with relatively disappointing results. CA 72-4 is a biomarker with poor sensitivity, ranging from 9% to 31%, and good specificity, ranging from 89% to 95%, for screening patients for CRC. The diagnostic information provided by CA 72-4 in recurrent CRC is borderline and far inferior to that of CEA. There is a consensus that CA 72-4 has a rather low sensitivity and incomplete specificity in the screening and follow-up of CRC patients[19]. Tissue polypeptide-specific antigen (TPS) and tissue polypeptide antigen (TPA), which detect cytokeratin 8, 18, and 19 fragments, are not recommended for CRC screening due to their lack of sensitivity and specificity. Most investigators found that elevated levels of TPA and TPS were observed in the metastatic stage of CRC. Further studies showed that the combination of TPA and CEA improved the sensitivity of these biomarkers in identifying patients with CRC recurrence. Other biomarkers, such as thymidine phosphorylase and DNA ploidy, were found to have no utility in the detection, staging or follow-up of CRC patients.
NDUFB is an accessory subunit of NADH dehydrogenase (com-plex I) of the mitochondrial membrane respiratory chain, encoded by nuclear genes[20]. Mutations in NDUFB may promote tumor metastasis[21]. In addition, a SNP (rs7830235) associated with prostate cancer risk is located in the NDUFB gene[22]. In addition to this, most of the other subunits of NADH dehydrogenase (NDUFB1-8/11) family were found to have significant prognostic value (DMFS) in breast cancer patients, and it was the mainstay of MDA-MB-231 breast cancer cell proliferation, inhibition of migration and invasion[23]. Its high expression is positively correlated with the prognosis of gastric cancer, suggesting that these proteins may serve as new candidate diagnostic and prognostic biomarkers for gastric cancer[24]. CAMK1D is a member of the calcium/calmodulin-dependent protein kinase 1 family[25]. It involved in a variety of physiological processes, including activation of CREB-dependent gene transcription, differentiation and activation of neutrophils, and regulation of apoptosis in erythrocytic leukemia[26]. Recent studies have shown that overexpression of CAMK1D can promote the proliferation of breast cancer[27]. Knockdown of CAMK1D in HT-29 and SW480 cells significantly reduced cell proliferation, invasion/migration capacity, and significantly increased apoptosis[28]. Activation of phosphoinositide 3-kinase (PI3K) signaling is one of the most common events in several human cancers, including CRC. PI3K is a family of lipid kinases that phosphorylate phosphatidylinositol 4, 5-bisphosphate to generate phosphatidylinositol-3, 4, 5-triphosphate, which in turn activates serine-threonine[29-31]. PI3Ks are classified into 3 classes according to their substrate specificity and structure in mammals. Of these, class I PI3Ks appear to be most associated with human cancers. Class I PI3Ks are further divided into subclasses IA and IB based on their adapters. Class IA PI3Ks contain a p110 catalytic subunit and a p85 regulatory subunit. The class IA catalytic isoforms p110α, p110β and p110δ are encoded by the genes PIK3CA, PIK3CB and PIK3CD, respectively. PIK3CB and PIK3CD are often overexpressed or amplified in cancer[32,33]. PIK3CD is mainly expressed in leukocytes and plays a key role in some hematological malignancies. Furthermore, PIK3CD has recently been associated with several human solid tumors, including hepatocellular carcinoma, glioma, glioblastoma, neuroblastoma, and breast cancer[33,34]. PIK3CD induces cell growth and invasion in colorectal cancer by activating AKT/GSK-3β/β-catenin signaling[35]. Presenilin 2 (PSEN2) is a protein-coding gene. Diseases associated with PSEN2 include Alzheimer’s disease[36]. Its related pathways include EPH-Ephrin signaling and p75 NTR receptor-mediated signaling. Presenilin (PSEN1 or PSEN2) mutations are generally thought to be present in Alzheimer’s disease patients with inherited disorders[37,38]. Although We have built a multi-parameter neural network diagnostic model for CRC, however, multi-centers and larger sample size still needed in the future study.
In conclusion, we built a multi-parameter neural network diagnostic model included CEA, IMA, SA, PIK3CD and LPa for early detection of CRC, compared to the conventional CEA, it showed significant improvement.
Most patients with colorectal cancer (CRC) are diagnosed at an advanced stage. The high morbidity and mortality of advanced CRC indicates an urgent need for clinical improvements in early CRC detection and individualized management.
Early detection of CRC is urgently needed in clinical practice. Commonly biomarker and extra
This study aimed to build a multi-parameter diagnostic model for early detection of CRC.
Total 59 colorectal polyps (CRP) groups, and 101 CRC patients (38 early-stage CRC and 63 advanced CRC) for model establishment. In addition, 30 CRP groups, and 62 CRC patients (30 early-stage CRC and 32 advanced CRC) were separately included to validate the model. 51 commonly used clinical detection indicators and the 4 eccDNA markers NDUFB7, CAMK1D, PIK3CD and PSEN2 that we screened earlier. Four multi-parameter joint analysis methods: binary logistic regression analysis, discriminant analysis, classification tree and neural network to establish a multi-parameter joint diagnosis model.
Neural network included carcinoembryonic antigen (CEA), ischemia-modified albumin (IMA), sialic acid (SA), PIK3CD and lipoprotein a (LPa) was chosen as the optimal multi-parameter combined auxiliary diagnosis model to distinguish CRP and CRC group, when it differentiated 59 CRP and 101 CRC, its overall accuracy was 90.8%, its area under the curve (AUC) was 0.959 (0.934, 0.985), and the sensitivity and specificity were 91.5% and 82.2%, respectively. After validation, when distinguishing based on 30 CRP and 62 CRC patients, the AUC was 0.965 (0.930, 1.000), and its sensitivity and specificity were 66.1% and 70.0%. When distinguishing based on 30 CRP and 32 early-stage CRC patients, the AUC was 0.960 (0.916, 1.000), with a sensitivity and specificity of 87.5% and 90.0%, distinguishing based on 30 CRP and 30 advanced CRC patients, the AUC was 0.970 (0.936, 1.000), with a sensitivity and specificity of 96.7% and 86.7%.
We built a multi-parameter neural network diagnostic model included CEA, IMA, SA, PIK3CD and LPa for early detection of CRC, compared to the conventional CEA, it showed significant improvement.
Larger sample size and multi-center study should be performed to validate the diagnostic model in future studies.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Andersen JB, Denmark; Bartholomeyczik S, Germany; Groome PA, Canada S-Editor: Wang JL L-Editor: A P-Editor: Wang JL
| 1. | Kuipers EJ, Grady WM, Lieberman D, Seufferlein T, Sung JJ, Boelens PG, van de Velde CJ, Watanabe T. Colorectal cancer. Nat Rev Dis Primers. 2015;1:15065. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1014] [Cited by in RCA: 1178] [Article Influence: 107.1] [Reference Citation Analysis (0)] |
| 2. | Cho YA, Lee J, Oh JH, Chang HJ, Sohn DK, Shin A, Kim J. Genetic Risk Score, Combined Lifestyle Factors and Risk of Colorectal Cancer. Cancer Res Treat. 2019;51:1033-1040. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 73] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 3. | Mader S, Pantel K. Liquid Biopsy: Current Status and Future Perspectives. Oncol Res Treat. 2017;40:404-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 169] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 4. | Alix-Panabières C, Pantel K. Liquid Biopsy: From Discovery to Clinical Application. Cancer Discov. 2021;11:858-873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 648] [Article Influence: 129.6] [Reference Citation Analysis (0)] |
| 5. | Poulet G, Massias J, Taly V. Liquid Biopsy: General Concepts. Acta Cytol. 2019;63:449-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 236] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 6. | Mazumder S, Datta S, Ray JG, Chaudhuri K, Chatterjee R. Liquid biopsy: miRNA as a potential biomarker in oral cancer. Cancer Epidemiol. 2019;58:137-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 93] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 7. | Junqueira-Neto S, Batista IA, Costa JL, Melo SA. Liquid Biopsy beyond Circulating Tumor Cells and Cell-Free DNA. Acta Cytol. 2019;63:479-488. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 8. | Tay TKY, Tan PH. Liquid Biopsy in Breast Cancer: A Focused Review. Arch Pathol Lab Med. 2021;145:678-686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 80] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 9. | Ye Q, Ling S, Zheng S, Xu X. Liquid biopsy in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. Mol Cancer. 2019;18:114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 288] [Article Influence: 41.1] [Reference Citation Analysis (0)] |
| 10. | Giannopoulou L, Zavridou M, Kasimir-Bauer S, Lianidou ES. Liquid biopsy in ovarian cancer: the potential of circulating miRNAs and exosomes. Transl Res. 2019;205:77-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 97] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 11. | Chen M, Zhao H. Next-generation sequencing in liquid biopsy: cancer screening and early detection. Hum Genomics. 2019;13:34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 162] [Cited by in RCA: 358] [Article Influence: 51.1] [Reference Citation Analysis (0)] |
| 12. | Kumar P, Kiran S, Saha S, Su Z, Paulsen T, Chatrath A, Shibata Y, Shibata E, Dutta A. ATAC-seq identifies thousands of extrachromosomal circular DNA in cancer and cell lines. Sci Adv. 2020;6:eaba2489. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 126] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 13. | Su Z, Saha S, Paulsen T, Kumar P, Dutta A. ATAC-Seq-based Identification of Extrachromosomal Circular DNA in Mammalian Cells and Its Validation Using Inverse PCR and FISH. Bio Protoc. 2021;11:e4003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 14. | Wang M, Chen X, Yu F, Ding H, Zhang Y, Wang K. Extrachromosomal Circular DNAs: Origin, formation and emerging function in Cancer. Int J Biol Sci. 2021;17:1010-1025. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 15. | Wang T, Zhang H, Zhou Y, Shi J. Extrachromosomal circular DNA: a new potential role in cancer progression. J Transl Med. 2021;19:257. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 16. | Thirunavukarasu P, Sukumar S, Sathaiah M, Mahan M, Pragatheeshwar KD, Pingpank JF, Zeh H 3rd, Bartels CJ, Lee KK, Bartlett DL. C-stage in colon cancer: implications of carcinoembryonic antigen biomarker in staging, prognosis, and management. J Natl Cancer Inst. 2011;103:689-697. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 140] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 17. | Goldstein MJ, Mitchell EP. Carcinoembryonic antigen in the staging and follow-up of patients with colorectal cancer. Cancer Invest. 2005;23:338-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 265] [Cited by in RCA: 297] [Article Influence: 17.5] [Reference Citation Analysis (6)] |
| 18. | Ballehaninna UK, Chamberlain RS. The clinical utility of serum CA 19-9 in the diagnosis, prognosis and management of pancreatic adenocarcinoma: An evidence based appraisal. J Gastrointest Oncol. 2012;3:105-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 369] [Reference Citation Analysis (3)] |
| 19. | Carpelan-Holmström M, Louhimo J, Stenman UH, Alfthan H, Järvinen H, Haglund C. CEA, CA 242, CA 19-9, CA 72-4 and hCGbeta in the diagnosis of recurrent colorectal cancer. Tumour Biol. 2004;25:228-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 47] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 20. | Tan AS, Baty JW, Berridge MV. The role of mitochondrial electron transport in tumorigenesis and metastasis. Biochim Biophys Acta. 2014;1840:1454-1463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 21. | Wang L, McDonnell SK, Hebbring SJ, Cunningham JM, St Sauver J, Cerhan JR, Isaya G, Schaid DJ, Thibodeau SN. Polymorphisms in mitochondrial genes and prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17:3558-3566. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 22. | Lin X, Wells DE, Kimberling WJ, Kumar S. Human NDUFB9 gene: genomic organization and a possible candidate gene associated with deafness disorder mapped to chromosome 8q13. Hum Hered. 1999;49:75-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 23. | Li LD, Sun HF, Liu XX, Gao SP, Jiang HL, Hu X, Jin W. Down-Regulation of NDUFB9 Promotes Breast Cancer Cell Proliferation, Metastasis by Mediating Mitochondrial Metabolism. PLoS One. 2015;10:e0144441. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 54] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 24. | Su F, Zhou FF, Zhang T, Wang DW, Zhao D, Hou XM, Feng MH. Quantitative proteomics identified 3 oxidative phosphorylation genes with clinical prognostic significance in gastric cancer. J Cell Mol Med. 2020;24:10842-10854. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 25. | Yamada T, Suzuki M, Satoh H, Kihara-Negishi F, Nakano H, Oikawa T. Effects of PU.1-induced mouse calcium-calmodulin-dependent kinase I-like kinase (CKLiK) on apoptosis of murine erythroleukemia cells. Exp Cell Res. 2004;294:39-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 26. | Gaines P, Lamoureux J, Marisetty A, Chi J, Berliner N. A cascade of Ca (2+)/calmodulin-dependent protein kinases regulates the differentiation and functional activation of murine neutrophils. Exp Hematol. 2008;36:832-844. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 27. | Bergamaschi A, Kim YH, Kwei KA, La Choi Y, Bocanegra M, Langerød A, Han W, Noh DY, Huntsman DG, Jeffrey SS, Børresen-Dale AL, Pollack JR. CAMK1D amplification implicated in epithelial-mesenchymal transition in basal-like breast cancer. Mol Oncol. 2008;2:327-339. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 53] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 28. | Liu W, Li S. LncRNA ILF3-AS1 Promotes the Progression of Colon Adenocarcinoma Cells Through the miR-619-5p/CAMK1D Axis. Onco Targets Ther. 2021;14:1861-1872. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 29. | Fruman DA, Rommel C. PI3K and cancer: lessons, challenges and opportunities. Nat Rev Drug Discov. 2014;13:140-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1146] [Cited by in RCA: 1386] [Article Influence: 115.5] [Reference Citation Analysis (0)] |
| 30. | Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer. 2015;15:7-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 878] [Cited by in RCA: 1079] [Article Influence: 98.1] [Reference Citation Analysis (10)] |
| 31. | Wang G, Cao X, Lai S, Luo X, Feng Y, Xia X, Yen PM, Gong J, Hu J. PI3K stimulates DNA synthesis and cell-cycle progression via its p55PIK regulatory subunit interaction with PCNA. Mol Cancer Ther. 2013;12:2100-2109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 32. | Singh P, Dar MS, Dar MJ. p110α and p110β isoforms of PI3K signaling: are they two sides of the same coin? FEBS Lett. 2016;590:3071-3082. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 33. | Tzenaki N, Papakonstanti EA. p110δ PI3 kinase pathway: emerging roles in cancer. Front Oncol. 2013;3:40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 49] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 34. | Fang Y, Xue JL, Shen Q, Chen J, Tian L. MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology. 2012;55:1852-1862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 318] [Cited by in RCA: 346] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 35. | Chen JS, Huang JQ, Luo B, Dong SH, Wang RC, Jiang ZK, Xie YK, Yi W, Wen GM, Zhong JF. PIK3CD induces cell growth and invasion by activating AKT/GSK-3β/β-catenin signaling in colorectal cancer. Cancer Sci. 2019;110:997-1011. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 56] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 36. | Gan CL, Zhang T, Lee TH. The Genetics of Alzheimer's Disease in the Chinese Population. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 37. | Xiao X, Liu H, Liu X, Zhang W, Zhang S, Jiao B. APP, PSEN1, and PSEN2 Variants in Alzheimer's Disease: Systematic Re-evaluation According to ACMG Guidelines. Front Aging Neurosci. 2021;13:695808. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 66] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 38. | Lessard CB, Rodriguez E, Ladd TB, Minter LM, Osborne BA, Miele L, Golde TE, Ran Y. γ-Secretase modulators exhibit selectivity for modulation of APP cleavage but inverse γ-secretase modulators do not. Alzheimers Res Ther. 2020;12:61. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |