©The Author(s) 2025.
World J Gastrointest Surg. Jul 27, 2025; 17(7): 107610
Published online Jul 27, 2025. doi: 10.4240/wjgs.v17.i7.107610
Published online Jul 27, 2025. doi: 10.4240/wjgs.v17.i7.107610
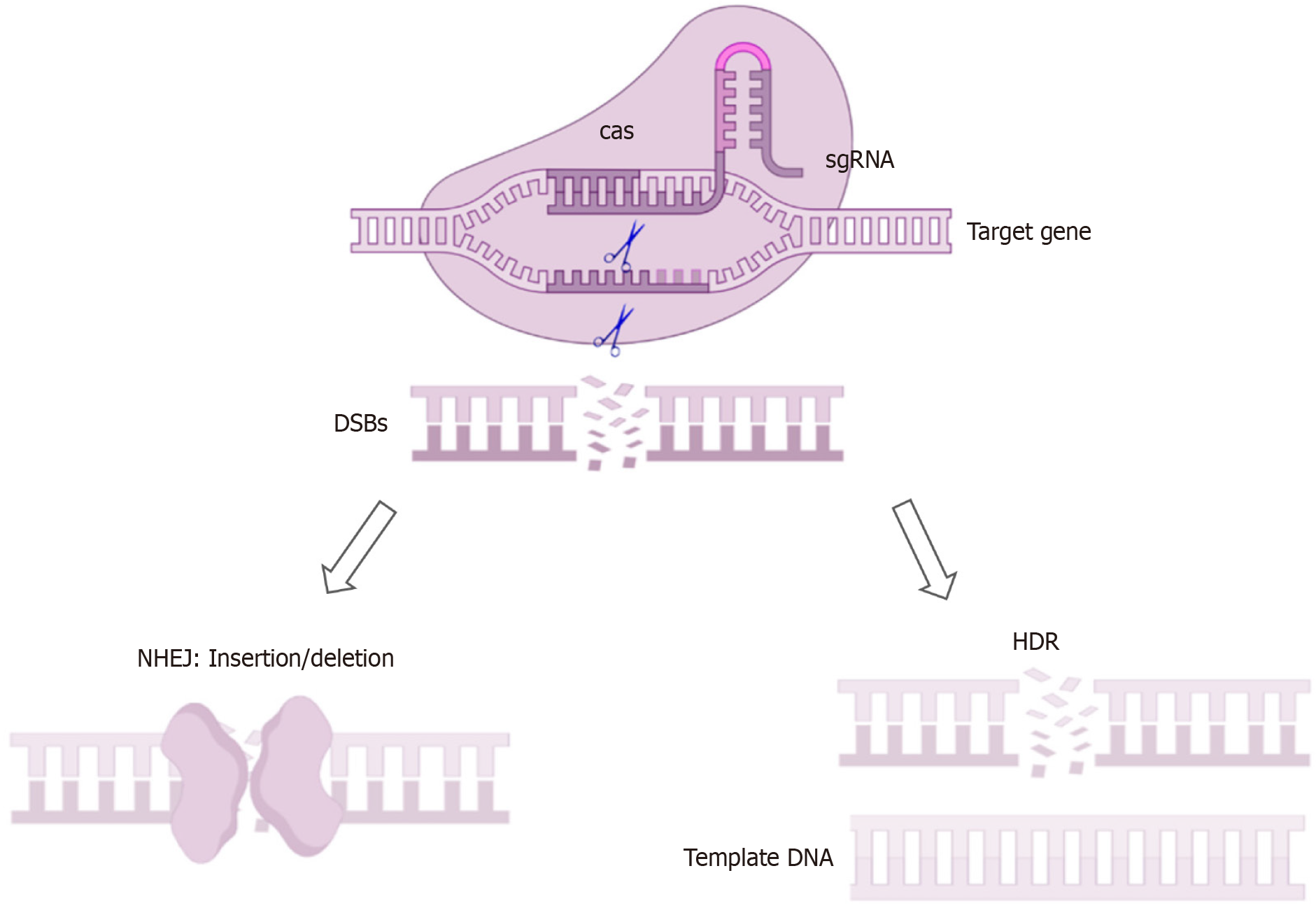
Figure 1 Working principle of clustered regularly interspaced short palindromic repeats/clustered regularly interspaced short palindromic repeats-associated protein 9.
The figures were created using the Figdraw platform. Non-homologous end joining: It directly ligates the broken DNA ends together, often resulting in small insertions or deletions. Homology-directed repair: Provide template DNA containing the sequence to be inserted to achieve gene knock-in, tagging or mutation correction. HDR: Homology-directed repair; NHEJ: Non-homologous end joining; sgDNA: Single-guide DNA; cas: Clustered regularly interspaced short palindromic repeats-associated protein; DSBs: DNA double-strand breaks.
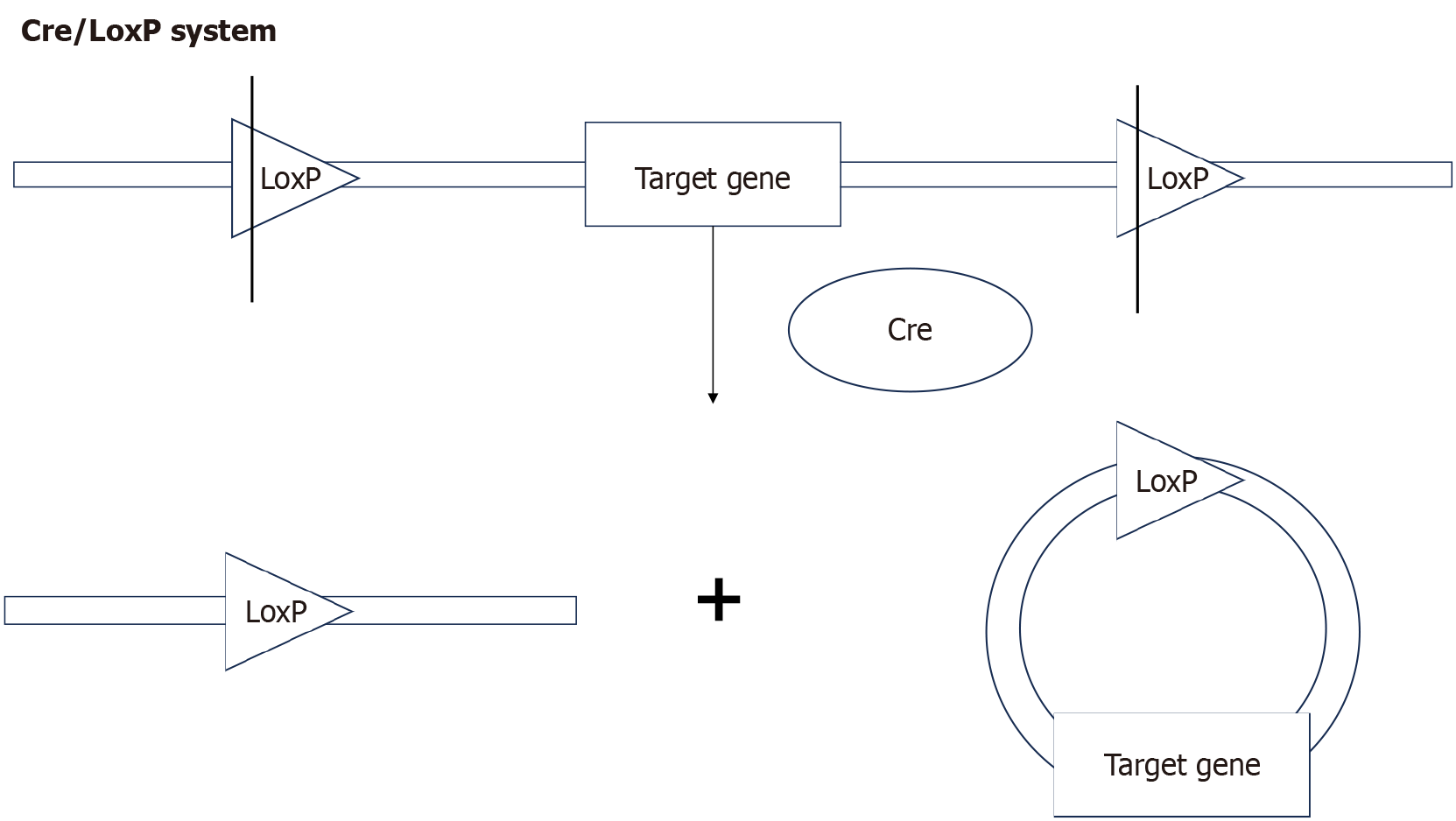
Figure 2 Working principle of cyclization recombinase/locus of X-over P1 system.
The figures were created using the Figdraw platform. Cyclization recombinase recombinase cuts the DNA at the locus of X-over P1 sites and then recombines the remaining DNA, ultimately resulting in the deletion of the target gene between the locus of X-over P1 sites. LoxP: Locus of X-over P1; Cre: Cyclization recombinase.
- Citation: Quan Y, Jia YB, Wu CH, Jia QL, Chen YQ, Gu ZJ, Ling JH. Genetically engineered mouse models in gastric precancerous lesions research. World J Gastrointest Surg 2025; 17(7): 107610
- URL: https://www.wjgnet.com/1948-9366/full/v17/i7/107610.htm
- DOI: https://dx.doi.org/10.4240/wjgs.v17.i7.107610