Published online Aug 10, 2016. doi: 10.4239/wjd.v7.i15.316
Peer-review started: March 18, 2016
First decision: May 19, 2016
Revised: May 26, 2016
Accepted: July 11, 2016
Article in press: July 13, 2016
Published online: August 10, 2016
Processing time: 146 Days and 0.1 Hours
AIM: To investigate the presence of total gut viral content in obese mice, and establish correlation with obesity associated metabolic measures and gut microbiome.
METHODS: Fresh fecal samples were collected from normal and obese (Leptin deficient: Lepob/ob) mice. Total viral DNA and RNA was isolated and quantified for establishing the correlation with metabolic measures and composition of gut bacterial communities.
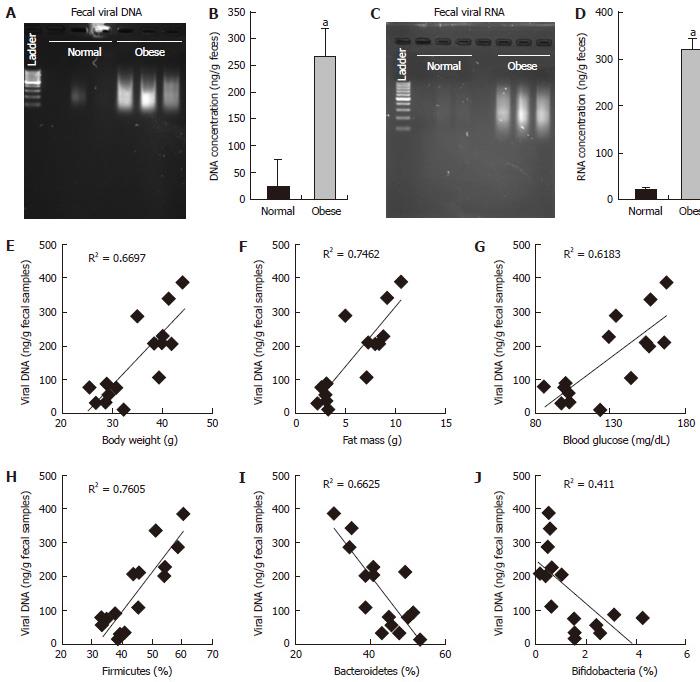
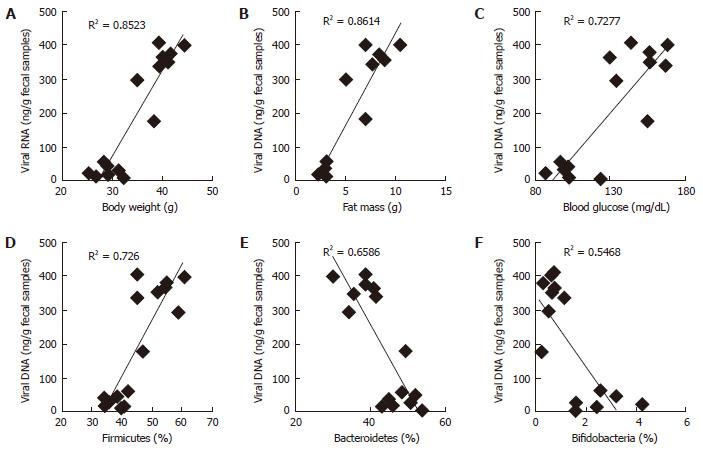
RESULTS: In this report, we found that obese mice feces have higher viral contents in terms of total viral DNA and RNA (P < 0.001). Interestingly, these increased viral DNA and RNA content were tightly correlated with metabolic measures, i.e., body weight, fat mass and fasting blood glucose. Total viral content were positively correlated with firmicutes (R2 > 0.6), whilst negatively correlated with bacteroidetes and bifidobacteria.
CONCLUSION: This study suggests the strong correlation of increased viral population into the gut of obese mice and opens new avenues to explore the role of gut virome in pathophysiology of obesity.
Core tip: Gut microbiome is known for major constituents of bacterial population, and their association with obesity, but microbes like viruses are majorly neglected. Our investigation on the basis of hypothesis that viruses are an important part of microbial world, and found in substantial numbers into human gut, we investigated whether viral content have any correlation with obesity in mice models. Interestingly, we found that DNA and RNA viral fecal content was significantly increased in obese mice as compared to normal. This suggests that viral population may have role to regulate host metabolism and might impact obesity prevalence via alteration of gut microbiome composition. Our findings open a new area of research to explore the role of gut virome in obesity.
- Citation: Yadav H, Jain S, Nagpal R, Marotta F. Increased fecal viral content associated with obesity in mice. World J Diabetes 2016; 7(15): 316-320
- URL: https://www.wjgnet.com/1948-9358/full/v7/i15/316.htm
- DOI: https://dx.doi.org/10.4239/wjd.v7.i15.316
Obesity is reaching on higher epidemic around the globe[1]. This is because of lack of successful and effective strategies to prevent and treat this health ailment. The pathophysiology of obesity is highly complex, and involves various factors, i.e., genetics, environment and life style[2]. Abdominal or central obesity (abdominal fat accumulation) is strongly correlated with increased incidence of insulin resistance and metabolic syndrome[3]. Abdominal fat is in close proximity with gastrointestinal tract, and have various gut-adipose communication through various gut hormones and adipokines[4]. Recently, role of gut microbiome in obesity pathophysiology have been well established and known to play significant role in obesity progression[5]. Human and rodent studies suggest that presence of gut microbiome increases the risk of weight gain and insulin resistance[6]. Various mechanisms have been proposed to explain the role of gut-microbiome on obesity progression, i.e., increased energy harvesting capacity, low grade inflammation, endotoxemia and other metabolic interferences[7]. Recently, plethora of literature has been generated to explore the role of gut microbiome (especially bacterial community) in metabolic regulation. Therefore modulation of gut microbiome has been considered one of the important strategies to develop therapies against obesity and diabetes[7].
Most of the studies conducted for exploring the role of gut microbiome in obesity have been focused on bacterial communities and their correlation with host metabolism[8]. Role of viruses present into gut are not investigated in relation with obesity and their impact on host metabolism and associated gut bacterial microbiome. Viruses play a critical role in maintaining bacterial population in specific environment, i.e., gut, where they establish a commensal relation with their partners[9]. Specific viral particles, especially bacteriophages might play an important role in maintaining certain bacterial strains of gut microbiome, that are correlated with obesity occurrence[6]. From our best knowledge, none of the studies investigated the direct link of gut virome with obesity. We hypothesize that changes in gut viral community (gut-virome), might play an important role in maintaining and colonizing gut bacterial species that impact host metabolism. Hence establishing the role of gut-viral community on influencing gut microbiome and host metabolism will open new avenues for development of therapeutic strategies against obesity via targeting gut virome. In this study, we investigated the correlation between gut-virome, obesity associated metabolic measures and gut bacterial communities in mice.
Lepob/ob and C57J/B6 mice (male; age 6-8 wk old) were housed in a light controlled facility by maintaining 12 h light/dark cycle. Mice were maintained in identical conditions and fed with similar diet and water, adlibitum. Body weight was measured using a microscale balance (Cole-Parmer, IL, United States). Total fat mass was measured weighing all the major fat depots, i.e., epididymal, perirenal, mesenteric, supra subscapular, anterior subcutaneous and posterior subcutaneous fat depots). Fasting (12-14 h) blood glucose was measured using Bayer Contour glucometer (Bayers Contour Diabetes Solutions, Thane, India). Fresh fecal samples were collected from each mouse by light abdominal squeezing and immediately stored in a sterile, DNase and RNase free vials at -80 °C till further use. All the animal protocols and procedures were approved by institutional animal ethics committee from University of Punjab and PGIMER, Chandigarh, India.
Fecal viral DNA and RNA was isolated using Qiagen viral DNA and RNA isolation kits following the manufacturer’s instructions. DNA and RNA quality have been checked using Agilent 2100 Bioanalyzer. Viral DNA and RNA have been quantified using NanoDrop One Spectrophotometer with fluorescent method (Thermo-Fisher Scientific, United States). Viral DNA and RNA quantity has been calculated nanogram per gram of fecal sample.
Mouse total fecal DNA was isolated from separate fecal pallets than viral DNA/RNA isolation, using DNeasy kit (Qiagen). Real time PCR was performed to measure the major obesity associated bacterial community, i.e., Firmicutes, bacteroidetes and Bifidobacteria using microbe specific primers (Table 1). Results are presented here as percent of bacterial DNA abundance normalized by total bacterial DNA.
| Gene Name | Primer sequence (5’→3’) |
| Universal F (Total) | TCCTACGGGAGGCAGCAGT |
| Universal R (Total) | GACTACCAGGGTATCTAATCCTGTT |
| Bifidobacteria F | GCGTGCTTAACACATGCAAGTC |
| Bifidobacteria R | CACCCGTTTCCAGGAGCTATT |
| Bacteroidetes 934F | GGARCATGTGGTTTAATTCGATGAT |
| Bacteroidetes 1060R | AGCTGACGACAACCATGCAG |
| Firmicutes 934F | GGAGYATGTGGTTTAATTCGAAGCA |
| Firmicutes 1060R | AGCTGACGACAACCATGCAC |
All the data expressed is mean and standard error of means. Statistical significance among the groups was calculated using two-tailed t test and/or one way analysis of variance, that followed by post-hoc tests. Data with less than 0.05 P-values considered statistically significance.
Microbiome studies clearly suggest that we are surrounded by microbes, in which viruses makes a significant numbers. Around 1031 viral particles have been estimated on earth, and human feces consist around 109 viral particles/gram[10-12]. Mammalian virome collectively called for viruses that infect eukaryotic cells (eukaryotic virome), bacterial cells (bacterial virome), archeal cells (archeal virome) and virus derived genetic elements in host chromosomes that can change host-gene expression, express proteins, or even generate infectious virus (prophages, endogenous retroviruses, endogenous viral elements)[13]. Viral infections have been associated with prevalence of obesity in animals and humans, and termed as infectobesity[14]. Considering the technological limitations for sequencing and analyzing datasets for viral communities, studies of virome has been lagged behind than bacterial microbiome. In present study, we analyzed total fecal viral content in normal vs obese (leptin deficient Lepob/ob) mice and correlated with obesity related measures. Interestingly, we found that fecal viral DNA and RNA in obese samples was significantly higher than normal mice (Figures 1 and 2), suggesting that total DNA and RNA viral communities have been significantly increased in obese mice. Although, our studies completely lack the types of viruses enriched in obese mice gut as compared to normal, but these very interesting observations indicate that total load of viruses have been increased in obese gut. Therefore, these results provide a strong basis to further explore the role of gut virome in obesity.
We have observed that fecal DNA and RNA viral population positively correlated with firmicutes bacterial communities, which is known to be associated with increased obesity[5]. While viral contents were negatively correlated with bacteroidetes and bifidobacteria, that are known to be associated with lean-ness[5]. These results can be speculated in a way, that gut virome (especially bacteriophages) might have interaction with gut bacterial microbiome to modulate the bacterial species abundance in obese vs normal mice. Although, these studies gives us an intrigued and important preliminary information about the abundance of DNA and RNA viruses in obese and normal mice, but still detailed analysis to find out the types of viruses and their functionality remains completely unknown. Therefore, further studies to explore the types of viruses that are associated with increased viral DNA and RNA contents in normal vs obese mice are highly warranted.
Viruses are highly mutagenic and carries individual variations in gut viral communities have been described earlier[9]. Viral proteins can interact with host cells and can induce biological response, i.e., inflammation, receptor based cell signaling or gene expression, to modulate adipose tissue biology[15]. Hence, the results of this study also indicate that increased viral population might be contributing to release of higher amount of certain viral proteins that can interact directly with host cells to modulate metabolism and cause obesity. Therefore it will be very important to establish how these viral species and their end products (i.e., proteins) are playing role in modulation of gut bacterial communities, as well as their impact on host metabolism.
Viruses have been known to infection host, bacterial and other broad array of organisms. Gut bacterial microbiome have been known to play critical role in obesity pathology, but the role of gut virome have not been explored.
The facts that viruses interacts with host cells as well as infects bacterial cells to control bacterial growth, studying virome compositions is one of the important aspects in the microbiome biology and its impact of health. Role of gut virome in obesity and gut bacterial microbiome modulation will open new frontiers of investigations.
The authors first time have reported that total viral population have been changed in obesity mice and correlated with metabolic and gut bacterial microbiome.
This study further open new avenues to find different types of viral populations in obese vs normal population and can develop them as a new drug targets or biomarkers.
Gut virome is considered as collective viral community present in fecal samples.
In the current study, the authors explored the association between fecal viral content and obesity in mice. The results are significant as the fecal viral DNA and RNA content found to be elevated in obese mice model.
Manuscript source: Invited manuscript
Specialty type: Endocrinology and metabolism
Country of origin: India
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Deng WP, Gelaleti RB, Riaz S S- Editor: Qiu S L- Editor: A E- Editor: Wu HL
| 1. | Farag YM, Gaballa MR. Diabesity: an overview of a rising epidemic. Nephrol Dial Transplant. 2011;26:28-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 207] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 2. | Wild SH, Byrne CD. ABC of obesity. Risk factors for diabetes and coronary heart disease. BMJ. 2006;333:1009-1011. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 94] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 3. | Brunzell JD, Hokanson JE. Dyslipidemia of central obesity and insulin resistance. Diabetes Care. 1999;22 Suppl 3:C10-C13. [PubMed] |
| 4. | Yi CX, Tschöp MH. Brain-gut-adipose-tissue communication pathways at a glance. Dis Model Mech. 2012;5:583-587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 60] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 5. | Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022-1023. [PubMed] |
| 6. | Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213-223. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2421] [Cited by in RCA: 2227] [Article Influence: 123.7] [Reference Citation Analysis (0)] |
| 7. | Delzenne NM, Neyrinck AM, Bäckhed F, Cani PD. Targeting gut microbiota in obesity: effects of prebiotics and probiotics. Nat Rev Endocrinol. 2011;7:639-646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 576] [Cited by in RCA: 569] [Article Influence: 37.9] [Reference Citation Analysis (0)] |
| 8. | Cani PD. Gut microbiota and obesity: lessons from the microbiome. Brief Funct Genomics. 2013;12:381-387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 88] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 9. | Sime-Ngando T, Lucas S, Robin A, Tucker KP, Colombet J, Bettarel Y, Desmond E, Gribaldo S, Forterre P, Breitbart M. Diversity of virus-host systems in hypersaline Lake Retba, Senegal. Environ Microbiol. 2011;13:1956-1972. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 67] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 11. | Schoenfeld T, Liles M, Wommack KE, Polson SW, Godiska R, Mead D. Functional viral metagenomics and the next generation of molecular tools. Trends Microbiol. 2010;18:20-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 48] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 13. | Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell. 2009;138:30-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 720] [Cited by in RCA: 811] [Article Influence: 47.7] [Reference Citation Analysis (0)] |
| 14. | Dhurandhar NV. Infectobesity: obesity of infectious origin. J Nutr. 2001;131:2794S-2797S. [PubMed] |
| 15. | Ponterio E, Gnessi L. Adenovirus 36 and Obesity: An Overview. Viruses. 2015;7:3719-3740. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 46] [Article Influence: 4.2] [Reference Citation Analysis (0)] |