Published online Jun 15, 2025. doi: 10.4239/wjd.v16.i6.105709
Revised: March 27, 2025
Accepted: May 16, 2025
Published online: June 15, 2025
Processing time: 129 Days and 10.9 Hours
Diabetic peripheral neuropathy (DPN) is a common complication of diabetes and is characterized by sensory and motor impairments resulting from neural injury. Schwann cells (SCs), which are important for peripheral nerve function, are compromised under hyperglycemic conditions, leading to impaired axonal re
Core Tip: This article underscores the critical role of autophagy in Schwann cells in the progression of diabetic peripheral neuropathy progression, highlight its potential as a pharmacotherapeutic target. It explored autophagy mechanisms (mammalian target of rapamycin, adenosine monophosphate-activated protein kinase, and phosphatase and tensin homolog-induced putative kinase/parkin pathways), their dysregulation in diabetic peripheral neuropathy, and various autophagy-modulating agents (natural products and chemical drugs) that promote nerve regeneration and recovery of Schwann cell function.
- Citation: Xing QC, Chen J, Liu Z, Li WC, Liu X, Li W. Autophagy in Schwann cells: A potential pharmacotherapeutic target in diabetic peripheral neuropathy. World J Diabetes 2025; 16(6): 105709
- URL: https://www.wjgnet.com/1948-9358/full/v16/i6/105709.htm
- DOI: https://dx.doi.org/10.4239/wjd.v16.i6.105709
Diabetic peripheral neuropathy (DPN) is one of the most prevalent chronic complications of diabetes, encountered in clinical practice. Surveys indicate that DPN accounts for approximately 50% of all diabetic complications[1]. Patients with DPN experience abnormal temperature or pain sensations, as well as diminished conduction function in both sensory and motor neurons during clinical evaluations. The symptoms included numbness, pain, sensory disturbances, and limb muscle weakness. These manifestations significantly compromise the quality of life of patients with diabetes and represent risk factors for disability and mortality in this population[2]. Research has shown that DPN development is linked to various types of injury, including axonal atrophy, demyelination, loss of nerve fiber function, and delayed regeneration under hyperglycemic conditions[3]. The essential process of nerve repair and regeneration after injury involves several key steps: The formation of axonal regeneration pathways and microcirculation, extension and growth of axonal sprouts, myelination of newly regenerated axons, and reinnervation of target cells. This process transitioned from structural to functional recovery[4]. Schwann cells (SCs), the primary glial cells in the peripheral nerves, are responsible for myelin sheath formation. SCs are essential for maintaining the stability of the axonal microenvironment, providing support and protection to axons, facilitating nutrient metabolism, and improving signal conduction[5]. Following peripheral nerve injury (PNI), SCs contribute significantly to nerve repair and regeneration through various mechanisms.
Hyperglycemia has been shown to increases aldose reductase activity and polyol pathway metabolism in SCs. This leads to the accumulation of abnormal metabolites, which in turn causes organelle damage and morphological changes, including swelling and vacuolization[6]. Furthermore, autophagy serves as a crucial protective mechanism in nerve tissues that helps eliminate harmful substances by degrading protein aggregates and removing damaged organelles, thereby mitigating their detrimental effects on nerve cells. Impairment of the autophagy pathway is a well-established contributing factor in DPN progression[7]. Consequently, restoration of autophagy is vital for the effective treatment of DPN. In recent years, the use of medications that affect autophagy has increased significantly. Recent studies have highlighted the potential of various agents to improve PNI and DPN outcomes by altering autophagic processes. This article discusses the modern studies on autophagy in SCs and investigates the mechanisms by which these agents enhance the efficacy of DPN treatment by modulating autophagy modulation. This study highlights the potential of autophagy-modulating agents as viable therapeutic agents for the management of DPN.
Autophagy is a crucial physiological process that functions as the body’s primary defense mechanism in response to stressors, such as nutrient or energy deprivation. Autophagy can be typically divided into three distinct forms: Macroautophagy, microautophagy, and chaperone-mediated autophagy. Macroautophagy, simply referred to as autophagy, represents the most significant pathway, and entails a complex self-degradative process characterized by multiple vesicle fusion events[8]. When a cell detects an autophagy signal, it initiates formation of a membrane-like structure within the cytoplasm. This structure continues to expand and, gradually develops into a crescent-shaped lipid bilayer known as the autophagic vacuole. The membrane extends further, enveloping surrounding organelles, such as the endoplasmic reticulum and mitochondria, ultimately forming an autophagosome with a circular double-membrane architecture. Subsequently, autophagosomes fuse with lysosomes to form autolysosomes, in which lysosomal enzymes degrade the encapsulated contents, yielding amino and fatty acids for cellular utilization[9,10]. Under normal phy
SCs originate from neural crest cells as precursors, undergo a series of developmental stages, and eventually reach maturity. In the peripheral nervous system, mature SCs are classified into two main types: Myelinated SCs, which form myelin sheaths, and non-myelinated SCs, which do not[12,13]. The myelinated SCs encase individual axons in a multilayered myelin sheath, whereas non-myelinated SCs associate with multiple axons, particularly C-fibers, to form a fascicle. Both types of SCs work together to sheathe the axons of peripheral nerves, offering protection, facilitating efficient nerve signal conduction, and delivering crucial nutritional support[14,15]. Under normal physiological conditions, SCs exhibit high autophagic activity, which is essential for the removal of damaged myelin fragments and the promotion of nerve regeneration and repair[16]. Research has demonstrated that SCs can eliminate up to 60% of damaged fragments within 72 hours of injury[17]. Knockdown of autophagy-related genes, such as autophagy related 7 (ATG7) exacerbates the fragmentation of the myelin sheath surrounding nerve fibers and impedes axonal regeneration following injury[18]. Another study found that SCs activate autophagy to degrade myelin debris, which create a favorable en
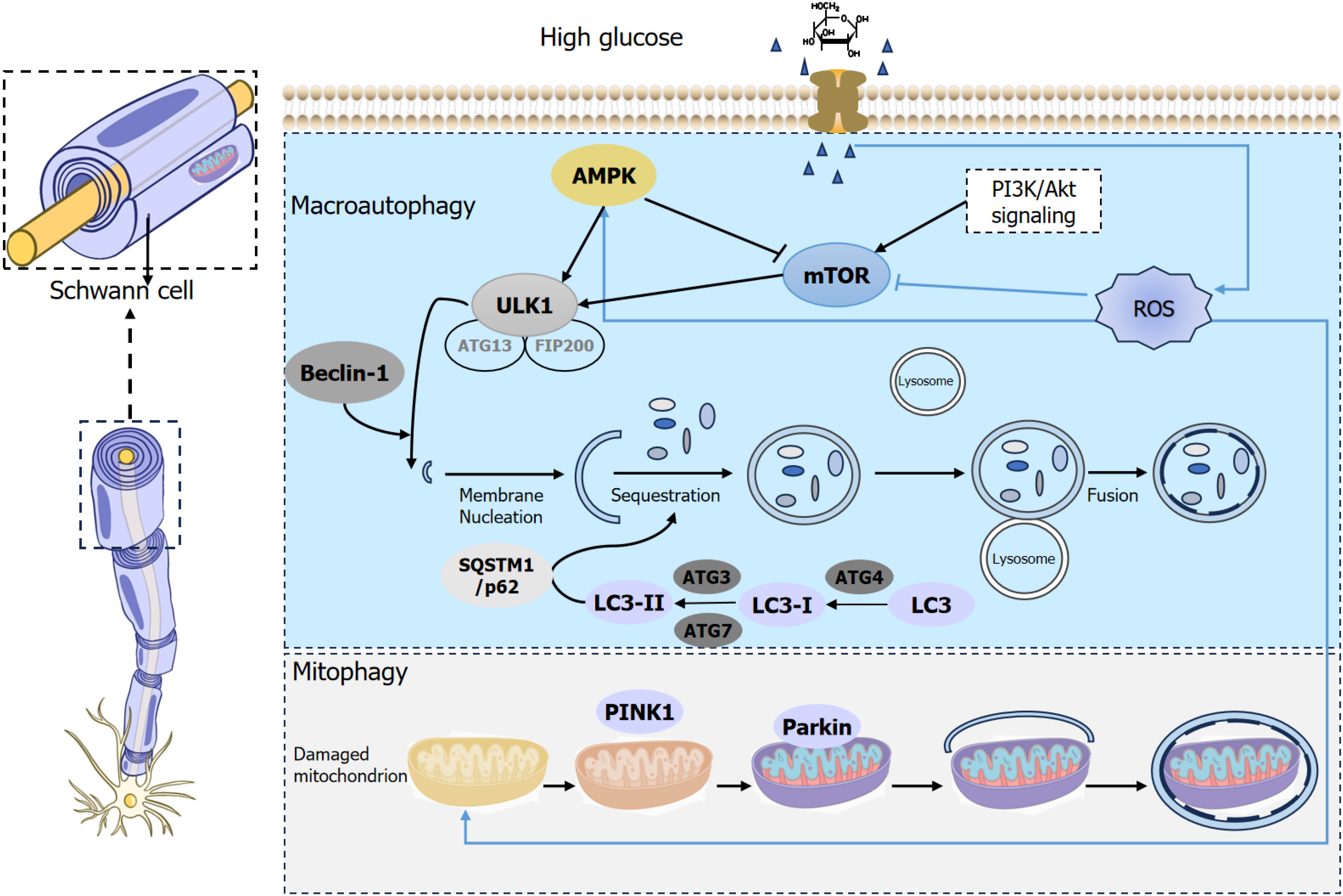
Autophagy plays a crucial role in maturation and structural plasticity of SCs and oligodendrocytes[21]. The major signaling cascades regulating autophagy include the mTOR, AMPK, and phosphatase and tensin homolog-induced putative kinase 1 (PINK1) pathways. In DPN, autophagy is influenced by autophagy-related factors, noncoding RNAs, and oxidative stress (Figure 1). Autophagy supports SC maturation and myelin integrity by regulating lipid metabolism
As a negative regulator of autophagy, mTOR plays a crucial role in neural cell autophagy and mediates multiple signaling pathways involved in this process[22]. MTOR is a 289-kDa serine/threonine protein kinase that comprises two distinct protein complexes: MTORC1 and mTORC2. Notably, mTORC1 is highly sensitive to rapamycin and inhibits the autophagy initiation kinase ATG1. In addition, the inhibition of mTORC2 by rapamycin was caused by the blockage of mTORC2 assembly. Inhibition of unc-51-like kinase 1 (ULK1), a key inducer of autophagy at the ser757 site, effectively suppresses autophagy onset[23]. Studies have shown that high glucose levels upregulate mTOR expression, leading to phosphorylation of its downstream effector, p70 ribosomal S6 kinase. This process enhances mRNA translation while inhibiting autophagosome formation by preventing the detachment of endoplasmic reticulum membranes[24]. MTOR phosphorylation is closely associated with the phosphatidylinositol 3-kinase (PI3K)/protein kinase B (Akt) signaling pathway. High glucose-induced oxidative stress triggers the release of substantial reactive oxygen species (ROS), which stimulates cell proliferation. This process leads to Akt phosphorylation via PI3K activation, inhibits the activation of the downstream tuberous sclerosis complex, and promotes the phosphorylation of downstream factors, including mTOR. This cascade effectively regulates the PI3K/Akt/mTOR signaling pathway. Furthermore, it mitigates excessive autophagy in SCs induced by high glucose conditions, slows cellular apoptosis, and ultimately improves DPN[25].
In contrast to the inhibitory effect of the mTOR signaling pathway on autophagy, the activation of AMPK signaling promotes autophagy. AMPK serves as an intracellular sensor of adenosine nucleotide levels, maintaining cellular energy homeostasis by promoting catabolic pathways that generate adenosine triphosphate, while reducing adenosine triphosphate consumption. Prolonged exposure to high glucose environments causes lysosomal damage, triggering the binding of transforming growth factor-β-activated protein kinase 1 with galectin-9, which subsequently phosphorylates AMPK. AMPK activates ULK1 to induce autophagy[26]. Abdelkader et al[27] have demonstrated a reduction in AMPK phosphorylation in a model of streptozotocin-induced DPN. The sodium-dependent glucose transporter 2 inhibitor empagliflozin enhances AMPK phosphorylation and upregulates the expression of autophagy proteins, specifically beclin-1 and LC3-II. This intervention significantly alleviates high glucose-induced injury in peripheral nerve cells[27].
The PINK/parkin signaling pathway plays a key role in diabetic neuropathic pain by mediating mitophagy[28]. PINK1 is predominantly expressed in mitochondria, where it undergoes degradation and cleavage in the inner mitochondrial membrane. PINK1 is imported into the inner membrane and cleaved in healthy mitochondria, whereas in the event of a potential loss in the inner mitochondrial membrane, PINK1 is unable to enter the inner membrane and thus accumulates in the outer mitochondrial membrane, which in turn recruits parkin. Under sustained high glucose conditions, mi
Autophagy is a complex and dynamic process tightly regulated by various autophagy-related factors. Beclin-1, a gene homologous to the yeast autophagy gene ATG/vps30, plays a crucial role in autophagosome and autophagic vesicle formation, and its expression levels are positively correlated with the extent of autophagy[31]. LC3 serves as a marker protein for autophagy and undergoes mutual transformation between the two forms. Outside of the autophagic membrane, LC3 covalently binds to phosphatidylethanolamine on the surface of autophagosome catalysis, modification, and processing by various autophagy-related enzymes, resulting in the formation of LC3-I and LC3-II. The conversion of LC3-I to LC3-II signifies the formation of autophagosomes, and is an important marker of autophagic activity. p62 protein (also known as sequestosome 1) acts as an autophagy receptor and is characterized by its ubiquitin-binding structural domain. It interacts with LC3-II through specific amino acid sequences to form a complex, which is subsequently degraded during lysosomal digestion and is associated with autophagosome formation. Consequently, p62 levels were inversely correlated with the degree of autophagic activity[32]. Numerous studies involving animal models and cell cultures have demonstrated that beclin-1 expression is significantly downregulated in DPN models. This decline correlated with reduced levels of LC3-II and increased p62 levels.
ROS induced by glycolipid toxicity serve as critical signaling molecules involved in autophagy induction. ROS produced during starvation inactivates Atg4 by oxidizing a critical, catalytic cysteine residue (C81). This prevents Atg4-mediated cleavage of LC3-II, which leads to the accumulation of LC3-II on the phagophore membrane and the formation of autophagosomes[33]. Conversely, ROS induce poly (adenosine diphosphate-ribose) polymerase 1 and activate AMPK through AMP activation, promoting autophagy[22]. The ULK-ATG13-focal adhesion kinase family kinase-interacting protein 200kD complex plays a vital role in the formation of autophagosomes and double-membrane autophagic vacuoles[34]. Furthermore, ROS can inhibit the mTOR signaling pathway, leading to ATG13 dephosphorylation, ULK activation, and focal adhesion kinase family kinase-interacting protein 200kD modulation, thereby regulating autophagy. Zhou et al[35] demonstrated that ROS-induced c-Jun N-terminal kinase activation leads to both autophagy and apoptosis, and is associated with caspase-independent cell death via autophagy.
The field of peripheral neuropathy research has witnessed remarkable advancements in recent years, particularly concerning nerve regeneration and recovery of SCs function. Autophagy, a fundamental cellular mechanism, has gained increasing attention because of its pivotal role in nerve repair following injury. Recent studies have focused on elucidating the mechanism by which various compounds and molecules enhance nerve regeneration by modulating autophagy (Table 1).
| Ref. | Agents | Target | Functions | Dose in vitro |
| [37] | Lycorine | AMPK/MMP9 | Decrease MMP9 expression and induce autophagy (increase) | 2.87 μg/mL |
| [38] | Resveratrol | N/A | Promote autophagy (increase) | 10 μM |
| [25] | Muscone | Akt/mTOR | Inhibit p-AKT/p-mTOR expression and mitigates autophagy (decrease) | 10 μm |
| [40] | Astragaloside IV | MicroRNA-155 | Reduce apoptosis by promoting autophagy (increase) | 15 μg/mL |
| [42] | Salvianolic acid B | JNK | Prevent autophagic cell death by activating the JNK pathway (decrease) | 0.1-10 μM |
| [43] | Quercetin | N/A | Alleviate high glucose-induced damage by autophagy (decrease) | 25 μM |
| [45] | Curcumin | Erk1/2 and Akt | Accelerate cell autophagy and prevent cell apoptosis (increase) | 30 μM |
| [46] | Lentinan | AMPK/mTOR | Promote autophagic flux (increase) | 100 μg/mL |
| [48] | Docosahexaenoic acid | AMPK | DHA attenuates excessive autophagy (decrease) | 10 μM |
| [50] | Imidazole propionate | Erk/mTOR | The reduction of imidazole propionate promotes migration of Schwan cells through enhancing autophagy (increase) | 100 μM |
| [51] | Epothilone B | PI3K | Enhance migration and autophagy (increase) | 1 nM |
| [52] | 5-azacytidine | TXNIP | Reduce TXNIP protein expression, improve autophagy and inhibite apoptosis (increase) | 10 μM |
| [53] | AG490 | STAT3 | Enhance P62 expression in high glucose-stimulated RSC96 cells (increase) | N/A |
| [54] | 4’-chlorodiazepam (Ro5- 4864) | Translocator protein | Improve Schwann cells function by activating autophagy (increase) | 10-5 M |
| [57] | Healthy plasma-derived exosomes (miR-20b-3p) | Stat3 | Alleviate autophagy impairment (increase) | 20 μg/mL |
| [58] | Exosomes produced by adipose-derived stem cells (miRNA-26b) | Kpna2 | Promote the regeneration of the myelin sheath by moderately reducing autophagy (decrease) | 50 μg/mL |
| [60] | Long non-coding RNA XIST | MiR-30d-5p/SIRT1 | Induce autophagy (increase) | N/A |
| [62] | LV-lipin1 overexpressing virus | N/A | Decreased the autophagic hyperactivity and apoptosis (decrease) | 1.0 × 109 TU/mL |
| [64] | Plasmid-UNC5B | AMPK/ULK1 | Facilitates autophagic flux in SCs (increase) | N/A |
| [66] | Methyltransferase-like protein 3 | MiR-192-5p/ATG7 | Stimulated miR-192-5p maturation to depress ATG7 and SCs autophagy (decrease) | N/A |
| [70] | Fibroblast growth factor 21 | ERK/Nrf-2 | Suppresses autophagy-induced cell death (decrease) | 250 ng/mL |
| [71] | Basic fibroblast growth factor | TEFB | Activate autophagy (decrease) | 100 ng/mL |
| [73] | protein inhibitor of activated STAT1 | MiR-124 | Inhibit apoptosis and promote autophagy of Schwann cells (increase) | N/A |
| [75] | Triggering receptor expressed on myeloid cells-2 | N/A | TREM2 deficiency induces mitochondrial damage and autophagy of Schwann cells (increase) | N/A |
| [77] | Rat p66shc siRNA | N/A | Recovers the high glucose-induced impairment of autophagy and ER stress in SCs (increase) | N/A |
| [79] | Mst1 siRNA | N/A | Promoted parkin recruitment to mitochondria in SCs (increase) | N/A |
Lycorine is an alkaloid derived from species of the Amaryllidaceae family that exhibits a range of biological activities, including antitumor, antiviral, and anti-inflammatory effects[36]. Yuan et al[37] indicated that lycorine enhances SCs autophagy by activating the AMPK pathway and downregulating MMP9, leading to LC3-II transformation in DPN. Resveratrol, a natural compound extracted from various food sources, exhibits antioxidant properties. Zhang et al[38] suggested that resveratrol facilitates recovery from sciatic nerve crush injuries by promoting SC autophagy, thereby accelerating myelin clearance. Muscone, a primary active compound extracted from traditional Chinese herbs, has been reported to exhibit several beneficial effects. Muscones exert anti-apoptotic effects during myocardial remodeling post-myocardial infarction in murine models[39]. Dong et al[25] found that muscone ameliorates high glucose-induced apoptosis and autophagy in RSC96 cells by modulating the Akt/mTOR signaling pathway. Astragaloside IV is an important component of Astragalus membranaceus and is known for its efficacy in the treatment of DPN. One study indicated that AS-IV reduced apoptosis in RSC96 cells and promoted autophagy through the regulation of the miR-155-mediated PI3K/Akt/mTOR signaling pathways[40]. Salvianolic acid B, extracted from the roots of “Salviae miltiorrhizae (Danshen)”, represent a new class of natural water-soluble bioactive compounds[41]. Recent findings have demonstrated that salvianolic acid B effectively inhibits c-Jun N-terminal kinase activation, thereby reducing apoptosis and excessive autophagy in SCs by downregulating the caspase-dependent pathway and enhancing B cell leukemia/lymphoma 2 expression[42]. Quercetin, a flavonoid widely found in traditional Chinese medicine and various foods, was recently shown to enhance the proliferation of neurons and glial cells, which are typically suppressed in high glucose en
Docosahexaenoic acid, an n-3 polyunsaturated fatty acid, induces the expression of antioxidant enzymes, thereby protecting immortalized adult mouse SCs (IMS32) from oxidative stress[47]. Research has demonstrated that docosahexaenoic acid can suppress excessive autophagy resulting from oxidative stress in SCs, suggesting that it may be beneficial for preventing or minimizing cell death in vitro[48]. Imidazole propionate, a microbial metabolite derived from histidine, is elevated in the portal and peripheral blood of individuals with type 2 diabetes[49]. A previous study demonstrated that a reduction in imidazole propionate promotes SC migration by enhancing autophagy mediated through the MAPK/Erk/mTOR signaling pathway[50]. Epothilone B, an anti-tumor agent approved by the Food and Drug Administration, is well-known for promoting alpha-tubulin polymerization and enhancing the stability of microtubules. Studies have shown that epothilone B significantly improves axonal regeneration and functional recovery and augments the migratory and autophagic capabilities of SCs[51]. 5-azacytidine is a cytosine analog that functions as a DNA methyltransferase inhibitor (DNMT inhibitor). It can be incorporated into DNA/RNA, where it interacts with DNA methyltransferase, inhibiting its activity and subsequent DNA demethylation within cells. A previous study indicated that 5-azacytidine enhances thioredoxin interacting protein expression by downregulating DNMT1 and DNMT3a, thereby influencing apoptosis and autophagy in RSC96 cells[52]. AG490 is identified as a tyrosine kinase inhibitor that targets epidermal growth factor receptor, signal transducer and activator of transcription 3 (Stat3), and Janus kinase 2/3. Research conducted by Du et al[53] demonstrated that AG490 effectively inhibited STAT3 phosphorylation, which promoted autophagy by modulating both the histone deacetylase 1-Atg3-LC3-II axis and P62. 4’-chlorodiazepam (Ro5-4864), a derivative of diazepam, functions as an effective ligand for the mitochondrial translocation protein 18 kDa, exhibiting notable neuroprotective effects. Research has demonstrated that Ro5-4864 can alleviate allodynia and enhances SC function and regeneration in sciatic nerves via autophagy activation and the nuclear factor-erythroid 2 related factor 2-driven antioxidant system[54].
Exosomes are extracellular vesicles with a lipid bilayer and sizes ranging from 30 to 200 nm that, are produced by virtually all cell types in both physiological and pathological contexts[55]. MiRNAs within these vesicles can be transported to adjacent or distant cells, facilitating subsequent signaling[56]. Li et al[57] revealed that miR-20b-3p is enriched in plasma-derived exosomes from healthy rats and can regulate SC autophagy under pathological conditions by targeting Stat3, thereby inhibiting DPN progression. Distinct stressors or treatment durations alter autophagic responses. Another study suggested that exosomes isolated from adipose-derived stem cells support myelin sheath regeneration by inhibiting nerve injury-induced autophagy in injured SCs via the action of miRNA-26b, which targets karyopherin subunit alpha 2[58]. Long non-coding RNA, defined as transcripts longer than 200 nucleotides, can directly target autophagy-related genes and enhance post-transcription[59]. Liu et al[60] reported that the long non-coding RNA X-inactive specific transcript promoted autophagy in SCs cultured under high-glucose conditions via the miR-30d-5p/sirtuin 1 axis, thereby preventing cell apoptosis and reducing DPN progression.
Previous studies have indicated that Lipin1, an enzyme essential for glycolipid metabolism encoded by LIPIN, is necessary to ensure normal peripheral nerve conduction[61]. Studies have also revealed that Lipin1 overexpression in RSC96 cells significantly diminishes hyperglycemia-induced excessive autophagy and apoptosis[62]. Uncoordinated gene 5 homolog B, recognized as the netrin-1 receptor, is specifically present in SCs and is involved in neuropathic pain mechanisms[63]. Xiao et al[64] indicated that uncoordinated gene 5 homolog B may preserve myelin by increasing autophagic flux in SCs via the phosphorylation of AMPK and ULK1, a process dependent on its ligand, netrin-1. The modification of m6A, influenced by methyltransferase-like protein 3, is crucial for regulating RNA cleavage, stability, transport, localization, and post-transcriptional translation[65]. Liu et al[66] demonstrated that methyltransferase-like protein 3 enhances the maturation of miR-192-5p via m6A methylation, suppressing ATG7 and SCs autophagy while exacerbating PNI. Molecular therapies utilizing fibroblast growth factors (FGFs) are widely employed to promote peripheral nerve myelination and regeneration[67]. FGFs are a class of signaling molecules that regulate cell differentiation, proliferation, and metabolism[68]. Among them, FGF21 is a key metabolic regulator that modulates blood glucose and lipid homeostasis[69]. Research suggests that FGF21 facilitates remyelination and nerve regeneration following PNI by inhibiting excessive activation of the ERK/nuclear factor-erythroid 2 related factor 2 signaling pathway, which regulates oxidative stress and autophagy-related cell death[70]. Basic FGF, a powerful neurotrophic molecule produced by SCs and diverse neuronal groups, plays a significant role in the initial stages of peripheral nerve regeneration. According to Jiang et al[71], this function is intricately associated with clearance of myelin debris by SCs, potentially triggering SCs-mediated autophagy. Protein inhibitor of activated STAT1 functions as a small ubiquitin-like modifier E3 ligase, significantly influencing multiple cellular processes, including cell growth regulation, DNA damage response, and inflammatory modulation[72]. Moreover, studies by Hou et al[73] offer further evidence that protein inhibitor of activated STAT1 can enhance autophagy and suppress apoptosis in SCs via the peroxisome proliferator-activated receptors-γ-miR-124- enhancer of zeste homolog 2/STAT3 signaling pathway. Triggering receptor expressed on myeloid cells-2 is a membrane protein primarily expressed in immune cells of the central nervous system, particularly microglia. This protein is essential for the regulation of microglial proliferation, migration, and phagocytic activity[74]. Zhang et al[75] showed that the absence of triggering receptor expressed on myeloid cells-2 leads to impaired glycolytic flux and oxidative metabolism in SCs, thereby hindering cell proliferation. The resulting energy deficit triggers the activation of AMPK and disrupts the PI3K/AKT/mTOR signaling pathway, ultimately causing mitochondrial dysfunction and inducing au
Emerging evidence indicates that autophagy is a key regulator of DPN development. The dual roles of SCs autophagy, protective and pathogenic, in diabetes must be discussed. Under stress conditions, such as nerve injury or metabolic dysfunction, AMPK compensates for mTOR suppression to maintain autophagy, whereas the PINK1/parkin pathway ensures mitochondrial integrity. However, dysregulation of these pathways, such as mTOR overactivation in diabetes, impairs autophagy, thus leading to SC dysfunction and neuropathy. Based on available data, it can be posited that a moderate level of constructive autophagy is protective; conversely, hyperactivation of autophagy appears to be detrimental to DPN. Therefore, when considering autophagy as a potential therapeutic target, it is crucial to accurately assess its status in the SCs. In early DPN, moderate autophagy (LC3-II/LC3-I ratio increase by 2-3 folds)[80] helps clear damaged mitochondria and protein aggregates to maintain myelin integrity. This phase is characterized by sustained AMPK activation, partial mTOR inhibition, and coordinated lysosomal biogenesis. However, when chronic hyper
Several pharmacotherapeutic agents have demonstrated their efficacy in alleviating DPN by modulating SCs autophagy via various mechanisms. For instance, numerous growth factors, plant extracts, and compounds have been identified to expedite autophagy after nerve injury, thereby facilitating the reconstruction and functional recovery of the nerve myelin sheath. Moreover, some studies have elucidated how oxidative stress, high-glucose environments, and other factors influence the autophagic function of SCs, and subsequently contribute to neuropathy. Conversely, exosomes exhibit considerable potential for repairing nerve injuries; they are thought to transport essential signaling molecules that regulate both autophagy and other functions within receptor cells. This article integrates the latest findings on autophagy mechanisms in SCs and their potential as pharmacotherapeutic targets in DPN. While previous reviews have touched upon autophagy in SCs or DPN treatment strategies, our study comprehensively explored the intricate interplay between autophagy pathways (such as mTOR, AMPK, and PINK/parkin) and DPN progression. Thus, we provided a novel perspective on how autophagy dysregulation underlies DPN pathogenesis and how it can be targeted therapeutically.
In conclusion, these observations indicate that the regulation of autophagy in SCs may represent a promising therapeutic strategy for DPN. However, autophagy within SCs is a critical factor in determining appropriate therapeutic agents. Numerous pharmacotherapeutic agents that mitigate DPN through mechanisms involving autophagy regulation and target either specific or multiple pathways have been identified in preclinical studies. However, clinical success with a specific autophagy regulator in DPN remains unclear. While some of these agents have demonstrated the potential for advancement in clinical trials against DPN, it is essential to gain a comprehensive understanding of the interplay between the signaling events involved in their autophagy regulatory mechanisms. In the future, it will be essential to conduct a more in-depth exploration of the mechanisms regulating autophagy and the associated signaling pathways, as this may lead to the identification of novel therapeutic targets. Secondly, advancements in technology, including the development of robust biomarkers to monitor autophagy in vivo and the creation of targeted delivery systems to enhance drug efficacy, could offer new insights into the study of neuropathies. Further investigations should focus on personalized medical approaches to tailor therapeutic interventions based on individual patient characteristics and autophagy status. Finally, additional clinical trials are required to validate the therapeutic potential of these agents. By addressing these issues, we can move closer to develop more effective treatments for DPN.
| 1. | Atallah SM, Al-Jaghbir MT, Zayed AA. The prevalence of diabetic peripheral neuropathy among diabetic Palestinian refugees in the Nuzha area, Jordan: a cross-sectional study. Lancet. 2021;398 Suppl 1:S15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 2. | Yu Y. Gold Standard for Diagnosis of DPN. Front Endocrinol (Lausanne). 2021;12:719356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 64] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 3. | Singh R, Kishore L, Kaur N. Diabetic peripheral neuropathy: current perspective and future directions. Pharmacol Res. 2014;80:21-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 271] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 4. | Akram R, Anwar H, Javed MS, Rasul A, Imran A, Malik SA, Raza C, Khan IU, Sajid F, Iman T, Sun T, Han HS, Hussain G. Axonal Regeneration: Underlying Molecular Mechanisms and Potential Therapeutic Targets. Biomedicines. 2022;10:3186. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 67] [Reference Citation Analysis (1)] |
| 5. | Geuna S, Raimondo S, Ronchi G, Di Scipio F, Tos P, Czaja K, Fornaro M. Chapter 3: Histology of the peripheral nerve and changes occurring during nerve regeneration. Int Rev Neurobiol. 2009;87:27-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 188] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 6. | Yang Y, Zhao B, Wang Y, Lan H, Liu X, Hu Y, Cao P. Diabetic neuropathy: cutting-edge research and future directions. Signal Transduct Target Ther. 2025;10:132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 64] [Reference Citation Analysis (0)] |
| 7. | Choi SJ, Kim S, Lee WS, Kim DW, Kim CS, Oh SH. Autophagy Dysfunction in a Diabetic Peripheral Neuropathy Model. Plast Reconstr Surg. 2023;151:355-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (1)] |
| 8. | Zhou Y, Manghwar H, Hu W, Liu F. Degradation Mechanism of Autophagy-Related Proteins and Research Progress. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 31] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 9. | Condello M, Pellegrini E, Caraglia M, Meschini S. Targeting Autophagy to Overcome Human Diseases. Int J Mol Sci. 2019;20:725. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 95] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 10. | Moparthi SB, Wollert T. Reconstruction of destruction - in vitro reconstitution methods in autophagy research. J Cell Sci. 2018;132:jcs223792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 11. | Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861-2873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2692] [Cited by in RCA: 3138] [Article Influence: 165.2] [Reference Citation Analysis (0)] |
| 12. | Corfas G, Velardez MO, Ko CP, Ratner N, Peles E. Mechanisms and roles of axon-Schwann cell interactions. J Neurosci. 2004;24:9250-9260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 143] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 13. | Liu B, Xin W, Tan JR, Zhu RP, Li T, Wang D, Kan SS, Xiong DK, Li HH, Zhang MM, Sun HH, Wagstaff W, Zhou C, Wang ZJ, Zhang YG, He TC. Myelin sheath structure and regeneration in peripheral nerve injury repair. Proc Natl Acad Sci U S A. 2019;116:22347-22352. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 112] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 14. | Hyung S, Yoon Lee B, Park JC, Kim J, Hur EM, Francis Suh JK. Coculture of Primary Motor Neurons and Schwann Cells as a Model for In Vitro Myelination. Sci Rep. 2015;5:15122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 54] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 15. | Takaku S, Yako H, Niimi N, Akamine T, Kawanami D, Utsunomiya K, Sango K. Establishment of a myelinating co-culture system with a motor neuron-like cell line NSC-34 and an adult rat Schwann cell line IFRS1. Histochem Cell Biol. 2018;149:537-543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 16. | Webber CA, Christie KJ, Cheng C, Martinez JA, Singh B, Singh V, Thomas D, Zochodne DW. Schwann cells direct peripheral nerve regeneration through the Netrin-1 receptors, DCC and Unc5H2. Glia. 2011;59:1503-1517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 80] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 17. | Gomez-Sanchez JA, Carty L, Iruarrizaga-Lejarreta M, Palomo-Irigoyen M, Varela-Rey M, Griffith M, Hantke J, Macias-Camara N, Azkargorta M, Aurrekoetxea I, De Juan VG, Jefferies HB, Aspichueta P, Elortza F, Aransay AM, Martínez-Chantar ML, Baas F, Mato JM, Mirsky R, Woodhoo A, Jessen KR. Schwann cell autophagy, myelinophagy, initiates myelin clearance from injured nerves. J Cell Biol. 2015;210:153-168. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 301] [Cited by in RCA: 364] [Article Influence: 33.1] [Reference Citation Analysis (0)] |
| 18. | Jang SY, Shin YK, Park SY, Park JY, Rha SH, Kim JK, Lee HJ, Park HT. Autophagy is involved in the reduction of myelinating Schwann cell cytoplasm during myelin maturation of the peripheral nerve. PLoS One. 2015;10:e0116624. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 45] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 19. | Li R, Li D, Wu C, Ye L, Wu Y, Yuan Y, Yang S, Xie L, Mao Y, Jiang T, Li Y, Wang J, Zhang H, Li X, Xiao J. Nerve growth factor activates autophagy in Schwann cells to enhance myelin debris clearance and to expedite nerve regeneration. Theranostics. 2020;10:1649-1677. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 195] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 20. | Jessen KR, Mirsky R. The repair Schwann cell and its function in regenerating nerves. J Physiol. 2016;594:3521-3531. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 550] [Cited by in RCA: 873] [Article Influence: 87.3] [Reference Citation Analysis (0)] |
| 21. | Bankston AN, Forston MD, Howard RM, Andres KR, Smith AE, Ohri SS, Bates ML, Bunge MB, Whittemore SR. Autophagy is essential for oligodendrocyte differentiation, survival, and proper myelination. Glia. 2019;67:1745-1759. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 69] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 22. | Zhu Z, Yang C, Iyaswamy A, Krishnamoorthi S, Sreenivasmurthy SG, Liu J, Wang Z, Tong BC, Song J, Lu J, Cheung KH, Li M. Balancing mTOR Signaling and Autophagy in the Treatment of Parkinson's Disease. Int J Mol Sci. 2019;20:728. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 198] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 23. | Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4208] [Cited by in RCA: 5810] [Article Influence: 387.3] [Reference Citation Analysis (0)] |
| 24. | Li Q, Zeng Y, Jiang Q, Wu C, Zhou J. Role of mTOR signaling in the regulation of high glucose-induced podocyte injury. Exp Ther Med. 2019;17:2495-2502. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 25. | Dong J, Li H, Bai Y, Wu C. Muscone ameliorates diabetic peripheral neuropathy through activating AKT/mTOR signalling pathway. J Pharm Pharmacol. 2019;71:1706-1713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 26. | Jia J, Abudu YP, Claude-Taupin A, Gu Y, Kumar S, Choi SW, Peters R, Mudd MH, Allers L, Salemi M, Phinney B, Johansen T, Deretic V. Galectins control MTOR and AMPK in response to lysosomal damage to induce autophagy. Autophagy. 2019;15:169-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 122] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 27. | Abdelkader NF, Elbaset MA, Moustafa PE, Ibrahim SM. Empagliflozin mitigates type 2 diabetes-associated peripheral neuropathy: a glucose-independent effect through AMPK signaling. Arch Pharm Res. 2022;45:475-493. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 28. | Dai CQ, Guo Y, Chu XY. Neuropathic Pain: the Dysfunction of Drp1, Mitochondria, and ROS Homeostasis. Neurotox Res. 2020;38:553-563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 67] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 29. | Ramirez A, Old W, Selwood DL, Liu X. Cannabidiol activates PINK1-Parkin-dependent mitophagy and mitochondrial-derived vesicles. Eur J Cell Biol. 2022;101:151185. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 63] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 30. | He J, Qin Z, Chen X, He W, Li D, Zhang L, Le Y, Xiong Q, Zhang B, Wang H. HIF-1α Ameliorates Diabetic Neuropathic Pain via Parkin-Mediated Mitophagy in a Mouse Model. Biomed Res Int. 2022;2022:5274375. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 31. | Tomaipitinca L, Petrungaro S, D'Acunzo P, Facchiano A, Dubey A, Rizza S, Giulitti F, Gaudio E, Filippini A, Ziparo E, Cecconi F, Giampietri C. c-FLIP regulates autophagy by interacting with Beclin-1 and influencing its stability. Cell Death Dis. 2021;12:686. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 32. | Larsen KB, Lamark T, Øvervatn A, Harneshaug I, Johansen T, Bjørkøy G. A reporter cell system to monitor autophagy based on p62/SQSTM1. Autophagy. 2010;6:784-793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 138] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 33. | Scherz-Shouval R, Shvets E, Elazar Z. Oxidation as a post-translational modification that regulates autophagy. Autophagy. 2007;3:371-373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 142] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 34. | Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, Mizushima N. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981-1991. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1386] [Cited by in RCA: 1640] [Article Influence: 96.5] [Reference Citation Analysis (0)] |
| 35. | Zhou YY, Li Y, Jiang WQ, Zhou LF. MAPK/JNK signalling: a potential autophagy regulation pathway. Biosci Rep. 2015;35:e00199. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 236] [Cited by in RCA: 359] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 36. | Liang Q, Cai W, Zhao Y, Xu H, Tang H, Chen D, Qian F, Sun L. Lycorine ameliorates bleomycin-induced pulmonary fibrosis via inhibiting NLRP3 inflammasome activation and pyroptosis. Pharmacol Res. 2020;158:104884. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 138] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 37. | Yuan Q, Zhang X, Wei W, Zhao J, Wu Y, Zhao S, Zhu L, Wang P, Hao J. Lycorine improves peripheral nerve function by promoting Schwann cell autophagy via AMPK pathway activation and MMP9 downregulation in diabetic peripheral neuropathy. Pharmacol Res. 2022;175:105985. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 48] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 38. | Zhang J, Ren J, Liu Y, Huang D, Lu L. Resveratrol regulates the recovery of rat sciatic nerve crush injury by promoting the autophagy of Schwann cells. Life Sci. 2020;256:117959. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 39. | Wang X, Meng H, Chen P, Yang N, Lu X, Wang ZM, Gao W, Zhou N, Zhang M, Xu Z, Chen B, Tao Z, Wang L, Yang Z, Zhu T. Beneficial effects of muscone on cardiac remodeling in a mouse model of myocardial infarction. Int J Mol Med. 2014;34:103-111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 40. | Yin Y, Qu H, Yang Q, Fang Z, Gao R. Astragaloside IV alleviates Schwann cell injury in diabetic peripheral neuropathy by regulating microRNA-155-mediated autophagy. Phytomedicine. 2021;92:153749. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 41. | Wang SX, Hu LM, Gao XM, Guo H, Fan GW. Anti-inflammatory activity of salvianolic acid B in microglia contributes to its neuroprotective effect. Neurochem Res. 2010;35:1029-1037. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 93] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 42. | Wang QQ, Zhai C, Wahafu A, Zhu YT, Liu YH, Sun LQ. Salvianolic acid B inhibits the development of diabetic peripheral neuropathy by suppressing autophagy and apoptosis. J Pharm Pharmacol. 2019;71:417-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 43. | Qu L, Liang X, Gu B, Liu W. Quercetin alleviates high glucose-induced Schwann cell damage by autophagy. Neural Regen Res. 2014;9:1195-1203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 66] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 44. | Wang YF, Zu JN, Li J, Chen C, Xi CY, Yan JL. Curcumin promotes the spinal cord repair via inhibition of glial scar formation and inflammation. Neurosci Lett. 2014;560:51-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 69] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 45. | Zhao Z, Li X, Li Q. Curcumin accelerates the repair of sciatic nerve injury in rats through reducing Schwann cells apoptosis and promoting myelinization. Biomed Pharmacother. 2017;92:1103-1110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 74] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 46. | Xiao H, Wei C, Liu H, Li Z, Zheng C, Luo J. Lentinan alleviates sciatic nerve injury by promoting autophagy to remove myelin fragments. Phytother Res. 2023;37:4042-4058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 47. | Tatsumi Y, Kato A, Sango K, Himeno T, Kondo M, Kato Y, Kamiya H, Nakamura J, Kato K. Omega-3 polyunsaturated fatty acids exert anti-oxidant effects through the nuclear factor (erythroid-derived 2)-related factor 2 pathway in immortalized mouse Schwann cells. J Diabetes Investig. 2019;10:602-612. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 48. | Tatsumi Y, Kato A, Niimi N, Yako H, Himeno T, Kondo M, Tsunekawa S, Kato Y, Kamiya H, Nakamura J, Higai K, Sango K, Kato K. Docosahexaenoic Acid Suppresses Oxidative Stress-Induced Autophagy and Cell Death via the AMPK-Dependent Signaling Pathway in Immortalized Fischer Rat Schwann Cells 1. Int J Mol Sci. 2022;23:4405. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 49. | Koh A, Molinaro A, Ståhlman M, Khan MT, Schmidt C, Mannerås-Holm L, Wu H, Carreras A, Jeong H, Olofsson LE, Bergh PO, Gerdes V, Hartstra A, de Brauw M, Perkins R, Nieuwdorp M, Bergström G, Bäckhed F. Microbially Produced Imidazole Propionate Impairs Insulin Signaling through mTORC1. Cell. 2018;175:947-961.e17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 322] [Cited by in RCA: 575] [Article Influence: 71.9] [Reference Citation Analysis (0)] |
| 50. | Tang W, Yin X, Liu K, Shao T, Gao Q, Shen H, Zhong X, Zhang Z. The reduction of imidazole propionate induced by intermittent fasting promotes recovery of peripheral nerve injury by enhancing migration of Schwann cells. Exp Cell Res. 2024;442:114261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 51. | Zhou J, Li S, Gao J, Hu Y, Chen S, Luo X, Zhang H, Luo Z, Huang J. Epothilone B Facilitates Peripheral Nerve Regeneration by Promoting Autophagy and Migration in Schwann Cells. Front Cell Neurosci. 2020;14:143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 52. | Zhang X, Zhao S, Yuan Q, Zhu L, Li F, Wang H, Kong D, Hao J. TXNIP, a novel key factor to cause Schwann cell dysfunction in diabetic peripheral neuropathy, under the regulation of PI3K/Akt pathway inhibition-induced DNMT1 and DNMT3a overexpression. Cell Death Dis. 2021;12:642. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 64] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 53. | Du W, Wang N, Li F, Jia K, An J, Liu Y, Wang Y, Zhu L, Zhao S, Hao J. STAT3 phosphorylation mediates high glucose-impaired cell autophagy in an HDAC1-dependent and -independent manner in Schwann cells of diabetic peripheral neuropathy. FASEB J. 2019;33:8008-8021. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 54. | Gao N, Ma B, Jia H, Hao C, Jin T, Liu X. Translocator protein alleviates allodynia and improves Schwann cell function against diabetic peripheral neuropathy via activation of the Nrf2-dependent antioxidant system and promoting autophagy. Diabet Med. 2023;40:e15090. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 55. | He C, Zheng S, Luo Y, Wang B. Exosome Theranostics: Biology and Translational Medicine. Theranostics. 2018;8:237-255. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 700] [Cited by in RCA: 995] [Article Influence: 124.4] [Reference Citation Analysis (0)] |
| 56. | Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13:17-24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1443] [Cited by in RCA: 1644] [Article Influence: 149.5] [Reference Citation Analysis (1)] |
| 57. | Li J, Wu G, Li W, Zhou X, Li W, Xu X, Xu K, Cao R, Cui S. Plasma exosomes improve peripheral neuropathy via miR-20b-3p/Stat3 in type I diabetic rats. J Nanobiotechnology. 2023;21:447. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 58. | Yin G, Yu B, Liu C, Lin Y, Xie Z, Hu Y, Lin H. Exosomes produced by adipose-derived stem cells inhibit schwann cells autophagy and promote the regeneration of the myelin sheath. Int J Biochem Cell Biol. 2021;132:105921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 59. | Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, Ghaffari SH. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J Cell Physiol. 2019;234:5451-5465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1431] [Cited by in RCA: 1463] [Article Influence: 209.0] [Reference Citation Analysis (0)] |
| 60. | Liu BY, Li L, Bai LW, Xu CS. Long Non-coding RNA XIST Attenuates Diabetic Peripheral Neuropathy by Inducing Autophagy Through MicroRNA-30d-5p/sirtuin1 Axis. Front Mol Biosci. 2021;8:655157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 61. | Mul JD, Nadra K, Jagalur NB, Nijman IJ, Toonen PW, Médard JJ, Grès S, de Bruin A, Han GS, Brouwers JF, Carman GM, Saulnier-Blache JS, Meijer D, Chrast R, Cuppen E. A hypomorphic mutation in Lpin1 induces progressively improving neuropathy and lipodystrophy in the rat. J Biol Chem. 2011;286:26781-26793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 62. | Wang M, Xie M, Yu S, Shang P, Zhang C, Han X, Fan C, Chen L, Zhuang X, Chen S. Lipin1 Alleviates Autophagy Disorder in Sciatic Nerve and Improves Diabetic Peripheral Neuropathy. Mol Neurobiol. 2021;58:6049-6061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 63. | Chen JY, Huang Z, Xiao PY, Yu J, Liao SJ. Local uncoordinated gene 5H2 contributes to nerve injury-induced mechanical allodynia associated to its role in autophagy. Clin Exp Pharmacol Physiol. 2021;48:361-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 64. | Xiao PY, Chen JY, Zeng Q, Huang Z, Huang BX, Yu J, Liao SJ. UNC5B Overexpression Alleviates Peripheral Neuropathic Pain by Stimulating Netrin-1-Dependent Autophagic Flux in Schwann Cells. Mol Neurobiol. 2022;59:5041-5055. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 65. | Huang H, Weng H, Chen J. m(6)A Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell. 2020;37:270-288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 434] [Cited by in RCA: 966] [Article Influence: 161.0] [Reference Citation Analysis (0)] |
| 66. | Liu X, Lv J, Tang W, Hu Y, Wen Y, Shen H. METTL3-mediated maturation of miR-192-5p targets ATG7 to prevent Schwann cell autophagy in peripheral nerve injury. J Neuropathol Exp Neurol. 2023;82:1010-1019. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (1)] |
| 67. | Allodi I, Mecollari V, González-Pérez F, Eggers R, Hoyng S, Verhaagen J, Navarro X, Udina E. Schwann cells transduced with a lentiviral vector encoding Fgf-2 promote motor neuron regeneration following sciatic nerve injury. Glia. 2014;62:1736-1746. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 51] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 68. | Itoh N. Hormone-like (endocrine) Fgfs: their evolutionary history and roles in development, metabolism, and disease. Cell Tissue Res. 2010;342:1-11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 157] [Cited by in RCA: 189] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 69. | Kim KH, Lee MS. FGF21 as a mediator of adaptive responses to stress and metabolic benefits of anti-diabetic drugs. J Endocrinol. 2015;226:R1-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 70. | Lu Y, Li R, Zhu J, Wu Y, Li D, Dong L, Li Y, Wen X, Yu F, Zhang H, Ni X, Du S, Li X, Xiao J, Wang J. Fibroblast growth factor 21 facilitates peripheral nerve regeneration through suppressing oxidative damage and autophagic cell death. J Cell Mol Med. 2019;23:497-511. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 64] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 71. | Jiang Y, Liang J, Li R, Peng Y, Huang J, Huang L. Basic fibroblast growth factor accelerates myelin debris clearance through activating autophagy to facilitate early peripheral nerve regeneration. J Cell Mol Med. 2021;25:2596-2608. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 72. | Liu Y, Zhang YD, Guo L, Huang HY, Zhu H, Huang JX, Liu Y, Zhou SR, Dang YJ, Li X, Tang QQ. Protein inhibitor of activated STAT 1 (PIAS1) is identified as the SUMO E3 ligase of CCAAT/enhancer-binding protein β (C/EBPβ) during adipogenesis. Mol Cell Biol. 2013;33:4606-4617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 51] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 73. | Hou Z, Chen J, Yang H, Hu X, Yang F. PIAS1 alleviates diabetic peripheral neuropathy through SUMOlation of PPAR-γ and miR-124-induced downregulation of EZH2/STAT3. Cell Death Discov. 2021;7:372. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 74. | Ulland TK, Colonna M. TREM2 - a key player in microglial biology and Alzheimer disease. Nat Rev Neurol. 2018;14:667-675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 509] [Article Influence: 63.6] [Reference Citation Analysis (0)] |
| 75. | Zhang N, Ji Q, Chen Y, Wen X, Shan F. TREM2 deficiency impairs the energy metabolism of Schwann cells and exacerbates peripheral neurological deficits. Cell Death Dis. 2024;15:193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 20] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 76. | Carlomosti F, D'Agostino M, Beji S, Torcinaro A, Rizzi R, Zaccagnini G, Maimone B, Di Stefano V, De Santa F, Cordisco S, Antonini A, Ciarapica R, Dellambra E, Martelli F, Avitabile D, Capogrossi MC, Magenta A. Oxidative Stress-Induced miR-200c Disrupts the Regulatory Loop Among SIRT1, FOXO1, and eNOS. Antioxid Redox Signal. 2017;27:328-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 119] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 77. | Choi SJ, Vu GH, Nagar H, Kim S, Lee I, Piao S, Jeon BH, Irani K, Oh SH, Kim CS. p66shc deficiency attenuates high glucose-induced autophagy dysfunction in Schwann cells. Korean J Physiol Pharmacol. 2025;29:57-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 78. | Ma S, Meng Z, Chen R, Guan KL. The Hippo Pathway: Biology and Pathophysiology. Annu Rev Biochem. 2019;88:577-604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 340] [Cited by in RCA: 970] [Article Influence: 138.6] [Reference Citation Analysis (1)] |
| 79. | Huang Z, Xiao PY, Chen JY, Zeng Q, Huang BX, Yu J, Liao SJ. Mammalian Sterile 20-Like Kinase 1 Mediates Neuropathic Pain Associated with Its Effects on Regulating Mitophagy in Schwann Cells. Oxid Med Cell Longev. 2022;2022:3458283. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 80. | Klionsky DJ, Abdel-Aziz AK, Abdelfatah S, Abdellatif M, Abdoli A, Abel S, Abeliovich H, Abildgaard MH, Abudu YP, Acevedo-Arozena A, Adamopoulos IE, Adeli K, Adolph TE, Adornetto A, Aflaki E, Agam G, Agarwal A, Aggarwal BB, Agnello M, Agostinis P, Agrewala JN, Agrotis A, Aguilar PV, Ahmad ST, Ahmed ZM, Ahumada-Castro U, Aits S, Aizawa S, Akkoc Y, Akoumianaki T, Akpinar HA, Al-Abd AM, Al-Akra L, Al-Gharaibeh A, Alaoui-Jamali MA, Alberti S, Alcocer-Gómez E, Alessandri C, Ali M, Alim Al-Bari MA, Aliwaini S, Alizadeh J, Almacellas E, Almasan A, Alonso A, Alonso GD, Altan-Bonnet N, Altieri DC, Álvarez ÉMC, Alves S, Alves da Costa C, Alzaharna MM, Amadio M, Amantini C, Amaral C, Ambrosio S, Amer AO, Ammanathan V, An Z, Andersen SU, Andrabi SA, Andrade-Silva M, Andres AM, Angelini S, Ann D, Anozie UC, Ansari MY, Antas P, Antebi A, Antón Z, Anwar T, Apetoh L, Apostolova N, Araki T, Araki Y, Arasaki K, Araújo WL, Araya J, Arden C, Arévalo MA, Arguelles S, Arias E, Arikkath J, Arimoto H, Ariosa AR, Armstrong-James D, Arnauné-Pelloquin L, Aroca A, Arroyo DS, Arsov I, Artero R, Asaro DML, Aschner M, Ashrafizadeh M, Ashur-Fabian O, Atanasov AG, Au AK, Auberger P, Auner HW, Aurelian L, Autelli R, Avagliano L, Ávalos Y, Aveic S, Aveleira CA, Avin-Wittenberg T, Aydin Y, Ayton S, Ayyadevara S, Azzopardi M, Baba M, Backer JM, Backues SK, Bae DH, Bae ON, Bae SH, Baehrecke EH, Baek A, Baek SH, Baek SH, Bagetta G, Bagniewska-Zadworna A, Bai H, Bai J, Bai X, Bai Y, Bairagi N, Baksi S, Balbi T, Baldari CT, Balduini W, Ballabio A, Ballester M, Balazadeh S, Balzan R, Bandopadhyay R, Banerjee S, Banerjee S, Bánréti Á, Bao Y, Baptista MS, Baracca A, Barbati C, Bargiela A, Barilà D, Barlow PG, Barmada SJ, Barreiro E, Barreto GE, Bartek J, Bartel B, Bartolome A, Barve GR, Basagoudanavar SH, Bassham DC, Bast RC Jr, Basu A, Batoko H, Batten I, Baulieu EE, Baumgarner BL, Bayry J, Beale R, Beau I, Beaumatin F, Bechara LRG, Beck GR Jr, Beers MF, Begun J, Behrends C, Behrens GMN, Bei R, Bejarano E, Bel S, Behl C, Belaid A, Belgareh-Touzé N, Bellarosa C, Belleudi F, Belló Pérez M, Bello-Morales R, Beltran JSO, Beltran S, Benbrook DM, Bendorius M, Benitez BA, Benito-Cuesta I, Bensalem J, Berchtold MW, Berezowska S, Bergamaschi D, Bergami M, Bergmann A, Berliocchi L, Berlioz-Torrent C, Bernard A, Berthoux L, Besirli CG, Besteiro S, Betin VM, Beyaert R, Bezbradica JS, Bhaskar K, Bhatia-Kissova I, Bhattacharya R, Bhattacharya S, Bhattacharyya S, Bhuiyan MS, Bhutia SK, Bi L, Bi X, Biden TJ, Bijian K, Billes VA, Binart N, Bincoletto C, Birgisdottir AB, Bjorkoy G, Blanco G, Blas-Garcia A, Blasiak J, Blomgran R, Blomgren K, Blum JS, Boada-Romero E, Boban M, Boesze-Battaglia K, Boeuf P, Boland B, Bomont P, Bonaldo P, Bonam SR, Bonfili L, Bonifacino JS, Boone BA, Bootman MD, Bordi M, Borner C, Bornhauser BC, Borthakur G, Bosch J, Bose S, Botana LM, Botas J, Boulanger CM, Boulton ME, Bourdenx M, Bourgeois B, Bourke NM, Bousquet G, Boya P, Bozhkov PV, Bozi LHM, Bozkurt TO, Brackney DE, Brandts CH, Braun RJ, Braus GH, Bravo-Sagua R, Bravo-San Pedro JM, Brest P, Bringer MA, Briones-Herrera A, Broaddus VC, Brodersen P, Brodsky JL, Brody SL, Bronson PG, Bronstein JM, Brown CN, Brown RE, Brum PC, Brumell JH, Brunetti-Pierri N, Bruno D, Bryson-Richardson RJ, Bucci C, Buchrieser C, Bueno M, Buitrago-Molina LE, Buraschi S, Buch S, Buchan JR, Buckingham EM, Budak H, Budini M, Bultynck G, Burada F, Burgoyne JR, Burón MI, Bustos V, Büttner S, Butturini E, Byrd A, Cabas I, Cabrera-Benitez S, Cadwell K, Cai J, Cai L, Cai Q, Cairó M, Calbet JA, Caldwell GA, Caldwell KA, Call JA, Calvani R, Calvo AC, Calvo-Rubio Barrera M, Camara NO, Camonis JH, Camougrand N, Campanella M, Campbell EM, Campbell-Valois FX, Campello S, Campesi I, Campos JC, Camuzard O, Cancino J, Candido de Almeida D, Canesi L, Caniggia I, Canonico B, Cantí C, Cao B, Caraglia M, Caramés B, Carchman EH, Cardenal-Muñoz E, Cardenas C, Cardenas L, Cardoso SM, Carew JS, Carle GF, Carleton G, Carloni S, Carmona-Gutierrez D, Carneiro LA, Carnevali O, Carosi JM, Carra S, Carrier A, Carrier L, Carroll B, Carter AB, Carvalho AN, Casanova M, Casas C, Casas J, Cassioli C, Castillo EF, Castillo K, Castillo-Lluva S, Castoldi F, Castori M, Castro AF, Castro-Caldas M, Castro-Hernandez J, Castro-Obregon S, Catz SD, Cavadas C, Cavaliere F, Cavallini G, Cavinato M, Cayuela ML, Cebollada Rica P, Cecarini V, Cecconi F, Cechowska-Pasko M, Cenci S, Ceperuelo-Mallafré V, Cerqueira JJ, Cerutti JM, Cervia D, Cetintas VB, Cetrullo S, Chae HJ, Chagin AS, Chai CY, Chakrabarti G, Chakrabarti O, Chakraborty T, Chakraborty T, Chami M, Chamilos G, Chan DW, Chan EYW, Chan ED, Chan HYE, Chan HH, Chan H, Chan MTV, Chan YS, Chandra PK, Chang CP, Chang C, Chang HC, Chang K, Chao J, Chapman T, Charlet-Berguerand N, Chatterjee S, Chaube SK, Chaudhary A, Chauhan S, Chaum E, Checler F, Cheetham ME, Chen CS, Chen GC, Chen JF, Chen LL, Chen L, Chen L, Chen M, Chen MK, Chen N, Chen Q, Chen RH, Chen S, Chen W, Chen W, Chen XM, Chen XW, Chen X, Chen Y, Chen YG, Chen Y, Chen Y, Chen YJ, Chen YQ, Chen ZS, Chen Z, Chen ZH, Chen ZJ, Chen Z, Cheng H, Cheng J, Cheng SY, Cheng W, Cheng X, Cheng XT, Cheng Y, Cheng Z, Chen Z, Cheong H, Cheong JK, Chernyak BV, Cherry S, Cheung CFR, Cheung CHA, Cheung KH, Chevet E, Chi RJ, Chiang AKS, Chiaradonna F, Chiarelli R, Chiariello M, Chica N, Chiocca S, Chiong M, Chiou SH, Chiramel AI, Chiurchiù V, Cho DH, Choe SK, Choi AMK, Choi ME, Choudhury KR, Chow NS, Chu CT, Chua JP, Chua JJE, Chung H, Chung KP, Chung S, Chung SH, Chung YL, Cianfanelli V, Ciechomska IA, Cifuentes M, Cinque L, Cirak S, Cirone M, Clague MJ, Clarke R, Clementi E, Coccia EM, Codogno P, Cohen E, Cohen MM, Colasanti T, Colasuonno F, Colbert RA, Colell A, Čolić M, Coll NS, Collins MO, Colombo MI, Colón-Ramos DA, Combaret L, Comincini S, Cominetti MR, Consiglio A, Conte A, Conti F, Contu VR, Cookson MR, Coombs KM, Coppens I, Corasaniti MT, Corkery DP, Cordes N, Cortese K, Costa MDC, Costantino S, Costelli P, Coto-Montes A, Crack PJ, Crespo JL, Criollo A, Crippa V, Cristofani R, Csizmadia T, Cuadrado A, Cui B, Cui J, Cui Y, Cui Y, Culetto E, Cumino AC, Cybulsky AV, Czaja MJ, Czuczwar SJ, D'Adamo S, D'Amelio M, D'Arcangelo D, D'Lugos AC, D'Orazi G, da Silva JA, Dafsari HS, Dagda RK, Dagdas Y, Daglia M, Dai X, Dai Y, Dai Y, Dal Col J, Dalhaimer P, Dalla Valle L, Dallenga T, Dalmasso G, Damme M, Dando I, Dantuma NP, Darling AL, Das H, Dasarathy S, Dasari SK, Dash S, Daumke O, Dauphinee AN, Davies JS, Dávila VA, Davis RJ, Davis T, Dayalan Naidu S, De Amicis F, De Bosscher K, De Felice F, De Franceschi L, De Leonibus C, de Mattos Barbosa MG, De Meyer GRY, De Milito A, De Nunzio C, De Palma C, De Santi M, De Virgilio C, De Zio D, Debnath J, DeBosch BJ, Decuypere JP, Deehan MA, Deflorian G, DeGregori J, Dehay B, Del Rio G, Delaney JR, Delbridge LMD, Delorme-Axford E, Delpino MV, Demarchi F, Dembitz V, Demers ND, Deng H, Deng Z, Dengjel J, Dent P, Denton D, DePamphilis ML, Der CJ, Deretic V, Descoteaux A, Devis L, Devkota S, Devuyst O, Dewson G, Dharmasivam M, Dhiman R, di Bernardo D, Di Cristina M, Di Domenico F, Di Fazio P, Di Fonzo A, Di Guardo G, Di Guglielmo GM, Di Leo L, Di Malta C, Di Nardo A, Di Rienzo M, Di Sano F, Diallinas G, Diao J, Diaz-Araya G, Díaz-Laviada I, Dickinson JM, Diederich M, Dieudé M, Dikic I, Ding S, Ding WX, Dini L, Dinić J, Dinic M, Dinkova-Kostova AT, Dionne MS, Distler JHW, Diwan A, Dixon IMC, Djavaheri-Mergny M, Dobrinski I, Dobrovinskaya O, Dobrowolski R, Dobson RCJ, Đokić J, Dokmeci Emre S, Donadelli M, Dong B, Dong X, Dong Z, Dorn Ii GW, Dotsch V, Dou H, Dou J, Dowaidar M, Dridi S, Drucker L, Du A, Du C, Du G, Du HN, Du LL, du Toit A, Duan SB, Duan X, Duarte SP, Dubrovska A, Dunlop EA, Dupont N, Durán RV, Dwarakanath BS, Dyshlovoy SA, Ebrahimi-Fakhari D, Eckhart L, Edelstein CL, Efferth T, Eftekharpour E, Eichinger L, Eid N, Eisenberg T, Eissa NT, Eissa S, Ejarque M, El Andaloussi A, El-Hage N, El-Naggar S, Eleuteri AM, El-Shafey ES, Elgendy M, Eliopoulos AG, Elizalde MM, Elks PM, Elsasser HP, Elsherbiny ES, Emerling BM, Emre NCT, Eng CH, Engedal N, Engelbrecht AM, Engelsen AST, Enserink JM, Escalante R, Esclatine A, Escobar-Henriques M, Eskelinen EL, Espert L, Eusebio MO, Fabrias G, Fabrizi C, Facchiano A, Facchiano F, Fadeel B, Fader C, Faesen AC, Fairlie WD, Falcó A, Falkenburger BH, Fan D, Fan J, Fan Y, Fang EF, Fang Y, Fang Y, Fanto M, Farfel-Becker T, Faure M, Fazeli G, Fedele AO, Feldman AM, Feng D, Feng J, Feng L, Feng Y, Feng Y, Feng W, Fenz Araujo T, Ferguson TA, Fernández ÁF, Fernandez-Checa JC, Fernández-Veledo S, Fernie AR, Ferrante AW Jr, Ferraresi A, Ferrari MF, Ferreira JCB, Ferro-Novick S, Figueras A, Filadi R, Filigheddu N, Filippi-Chiela E, Filomeni G, Fimia GM, Fineschi V, Finetti F, Finkbeiner S, Fisher EA, Fisher PB, Flamigni F, Fliesler SJ, Flo TH, Florance I, Florey O, Florio T, Fodor E, Follo C, Fon EA, Forlino A, Fornai F, Fortini P, Fracassi A, Fraldi A, Franco B, Franco R, Franconi F, Frankel LB, Friedman SL, Fröhlich LF, Frühbeck G, Fuentes JM, Fujiki Y, Fujita N, Fujiwara Y, Fukuda M, Fulda S, Furic L, Furuya N, Fusco C, Gack MU, Gaffke L, Galadari S, Galasso A, Galindo MF, Gallolu Kankanamalage S, Galluzzi L, Galy V, Gammoh N, Gan B, Ganley IG, Gao F, Gao H, Gao M, Gao P, Gao SJ, Gao W, Gao X, Garcera A, Garcia MN, Garcia VE, García-Del Portillo F, Garcia-Escudero V, Garcia-Garcia A, Garcia-Macia M, García-Moreno D, Garcia-Ruiz C, García-Sanz P, Garg AD, Gargini R, Garofalo T, Garry RF, Gassen NC, Gatica D, Ge L, Ge W, Geiss-Friedlander R, Gelfi C, Genschik P, Gentle IE, Gerbino V, Gerhardt C, Germain K, Germain M, Gewirtz DA, Ghasemipour Afshar E, Ghavami S, Ghigo A, Ghosh M, Giamas G, Giampietri C, Giatromanolaki A, Gibson GE, Gibson SB, Ginet V, Giniger E, Giorgi C, Girao H, Girardin SE, Giridharan M, Giuliano S, Giulivi C, Giuriato S, Giustiniani J, Gluschko A, Goder V, Goginashvili A, Golab J, Goldstone DC, Golebiewska A, Gomes LR, Gomez R, Gómez-Sánchez R, Gomez-Puerto MC, Gomez-Sintes R, Gong Q, Goni FM, González-Gallego J, Gonzalez-Hernandez T, Gonzalez-Polo RA, Gonzalez-Reyes JA, González-Rodríguez P, Goping IS, Gorbatyuk MS, Gorbunov NV, Görgülü K, Gorojod RM, Gorski SM, Goruppi S, Gotor C, Gottlieb RA, Gozes I, Gozuacik D, Graef M, Gräler MH, Granatiero V, Grasso D, Gray JP, Green DR, Greenhough A, Gregory SL, Griffin EF, Grinstaff MW, Gros F, Grose C, Gross AS, Gruber F, Grumati P, Grune T, Gu X, Guan JL, Guardia CM, Guda K, Guerra F, Guerri C, Guha P, Guillén C, Gujar S, Gukovskaya A, Gukovsky I, Gunst J, Günther A, Guntur AR, Guo C, Guo C, Guo H, Guo LW, Guo M, Gupta P, Gupta SK, Gupta S, Gupta VB, Gupta V, Gustafsson AB, Gutterman DD, H B R, Haapasalo A, Haber JE, Hać A, Hadano S, Hafrén AJ, Haidar M, Hall BS, Halldén G, Hamacher-Brady A, Hamann A, Hamasaki M, Han W, Hansen M, Hanson PI, Hao Z, Harada M, Harhaji-Trajkovic L, Hariharan N, Haroon N, Harris J, Hasegawa T, Hasima Nagoor N, Haspel JA, Haucke V, Hawkins WD, Hay BA, Haynes CM, Hayrabedyan SB, Hays TS, He C, He Q, He RR, He YW, He YY, Heakal Y, Heberle AM, Hejtmancik JF, Helgason GV, Henkel V, Herb M, Hergovich A, Herman-Antosiewicz A, Hernández A, Hernandez C, Hernandez-Diaz S, Hernandez-Gea V, Herpin A, Herreros J, Hervás JH, Hesselson D, Hetz C, Heussler VT, Higuchi Y, Hilfiker S, Hill JA, Hlavacek WS, Ho EA, Ho IHT, Ho PW, Ho SL, Ho WY, Hobbs GA, Hochstrasser M, Hoet PHM, Hofius D, Hofman P, Höhn A, Holmberg CI, Hombrebueno JR, Yi-Ren Hong CH, Hooper LV, Hoppe T, Horos R, Hoshida Y, Hsin IL, Hsu HY, Hu B, Hu D, Hu LF, Hu MC, Hu R, Hu W, Hu YC, Hu ZW, Hua F, Hua J, Hua Y, Huan C, Huang C, Huang C, Huang C, Huang C, Huang H, Huang K, Huang MLH, Huang R, Huang S, Huang T, Huang X, Huang YJ, Huber TB, Hubert V, Hubner CA, Hughes SM, Hughes WE, Humbert M, Hummer G, Hurley JH, Hussain S, Hussain S, Hussey PJ, Hutabarat M, Hwang HY, Hwang S, Ieni A, Ikeda F, Imagawa Y, Imai Y, Imbriano C, Imoto M, Inman DM, Inoki K, Iovanna J, Iozzo RV, Ippolito G, Irazoqui JE, Iribarren P, Ishaq M, Ishikawa M, Ishimwe N, Isidoro C, Ismail N, Issazadeh-Navikas S, Itakura E, Ito D, Ivankovic D, Ivanova S, Iyer AKV, Izquierdo JM, Izumi M, Jäättelä M, Jabir MS, Jackson WT, Jacobo-Herrera N, Jacomin AC, Jacquin E, Jadiya P, Jaeschke H, Jagannath C, Jakobi AJ, Jakobsson J, Janji B, Jansen-Dürr P, Jansson PJ, Jantsch J, Januszewski S, Jassey A, Jean S, Jeltsch-David H, Jendelova P, Jenny A, Jensen TE, Jessen N, Jewell JL, Ji J, Jia L, Jia R, Jiang L, Jiang Q, Jiang R, Jiang T, Jiang X, Jiang Y, Jimenez-Sanchez M, Jin EJ, Jin F, Jin H, Jin L, Jin L, Jin M, Jin S, Jo EK, Joffre C, Johansen T, Johnson GVW, Johnston SA, Jokitalo E, Jolly MK, Joosten LAB, Jordan J, Joseph B, Ju D, Ju JS, Ju J, Juárez E, Judith D, Juhász G, Jun Y, Jung CH, Jung SC, Jung YK, Jungbluth H, Jungverdorben J, Just S, Kaarniranta K, Kaasik A, Kabuta T, Kaganovich D, Kahana A, Kain R, Kajimura S, Kalamvoki M, Kalia M, Kalinowski DS, Kaludercic N, Kalvari I, Kaminska J, Kaminskyy VO, Kanamori H, Kanasaki K, Kang C, Kang R, Kang SS, Kaniyappan S, Kanki T, Kanneganti TD, Kanthasamy AG, Kanthasamy A, Kantorow M, Kapuy O, Karamouzis MV, Karim MR, Karmakar P, Katare RG, Kato M, Kaufmann SHE, Kauppinen A, Kaushal GP, Kaushik S, Kawasaki K, Kazan K, Ke PY, Keating DJ, Keber U, Kehrl JH, Keller KE, Keller CW, Kemper JK, Kenific CM, Kepp O, Kermorgant S, Kern A, Ketteler R, Keulers TG, Khalfin B, Khalil H, Khambu B, Khan SY, Khandelwal VKM, Khandia R, Kho W, Khobrekar NV, Khuansuwan S, Khundadze M, Killackey SA, Kim D, Kim DR, Kim DH, Kim DE, Kim EY, Kim EK, Kim HR, Kim HS, Hyung-Ryong Kim, Kim JH, Kim JK, Kim JH, Kim J, Kim JH, Kim KI, Kim PK, Kim SJ, Kimball SR, Kimchi A, Kimmelman AC, Kimura T, King MA, Kinghorn KJ, Kinsey CG, Kirkin V, Kirshenbaum LA, Kiselev SL, Kishi S, Kitamoto K, Kitaoka Y, Kitazato K, Kitsis RN, Kittler JT, Kjaerulff O, Klein PS, Klopstock T, Klucken J, Knævelsrud H, Knorr RL, Ko BCB, Ko F, Ko JL, Kobayashi H, Kobayashi S, Koch I, Koch JC, Koenig U, Kögel D, Koh YH, Koike M, Kohlwein SD, Kocaturk NM, Komatsu M, König J, Kono T, Kopp BT, Korcsmaros T, Korkmaz G, Korolchuk VI, Korsnes MS, Koskela A, Kota J, Kotake Y, Kotler ML, Kou Y, Koukourakis MI, Koustas E, Kovacs AL, Kovács T, Koya D, Kozako T, Kraft C, Krainc D, Krämer H, Krasnodembskaya AD, Kretz-Remy C, Kroemer G, Ktistakis NT, Kuchitsu K, Kuenen S, Kuerschner L, Kukar T, Kumar A, Kumar A, Kumar D, Kumar D, Kumar S, Kume S, Kumsta C, Kundu CN, Kundu M, Kunnumakkara AB, Kurgan L, Kutateladze TG, Kutlu O, Kwak S, Kwon HJ, Kwon TK, Kwon YT, Kyrmizi I, La Spada A, Labonté P, Ladoire S, Laface I, Lafont F, Lagace DC, Lahiri V, Lai Z, Laird AS, Lakkaraju A, Lamark T, Lan SH, Landajuela A, Lane DJR, Lane JD, Lang CH, Lange C, Langel Ü, Langer R, Lapaquette P, Laporte J, LaRusso NF, Lastres-Becker I, Lau WCY, Laurie GW, Lavandero S, Law BYK, Law HK, Layfield R, Le W, Le Stunff H, Leary AY, Lebrun JJ, Leck LYW, Leduc-Gaudet JP, Lee C, Lee CP, Lee DH, Lee EB, Lee EF, Lee GM, Lee HJ, Lee HK, Lee JM, Lee JS, Lee JA, Lee JY, Lee JH, Lee M, Lee MG, Lee MJ, Lee MS, Lee SY, Lee SJ, Lee SY, Lee SB, Lee WH, Lee YR, Lee YH, Lee Y, Lefebvre C, Legouis R, Lei YL, Lei Y, Leikin S, Leitinger G, Lemus L, Leng S, Lenoir O, Lenz G, Lenz HJ, Lenzi P, León Y, Leopoldino AM, Leschczyk C, Leskelä S, Letellier E, Leung CT, Leung PS, Leventhal JS, Levine B, Lewis PA, Ley K, Li B, Li DQ, Li J, Li J, Li J, Li K, Li L, Li M, Li M, Li M, Li M, Li M, Li PL, Li MQ, Li Q, Li S, Li T, Li W, Li W, Li X, Li YP, Li Y, Li Z, Li Z, Li Z, Lian J, Liang C, Liang Q, Liang W, Liang Y, Liang Y, Liao G, Liao L, Liao M, Liao YF, Librizzi M, Lie PPY, Lilly MA, Lim HJ, Lima TRR, Limana F, Lin C, Lin CW, Lin DS, Lin FC, Lin JD, Lin KM, Lin KH, Lin LT, Lin PH, Lin Q, Lin S, Lin SJ, Lin W, Lin X, Lin YX, Lin YS, Linden R, Lindner P, Ling SC, Lingor P, Linnemann AK, Liou YC, Lipinski MM, Lipovšek S, Lira VA, Lisiak N, Liton PB, Liu C, Liu CH, Liu CF, Liu CH, Liu F, Liu H, Liu HS, Liu HF, Liu H, Liu J, Liu J, Liu J, Liu L, Liu L, Liu M, Liu Q, Liu W, Liu W, Liu XH, Liu X, Liu X, Liu X, Liu X, Liu Y, Liu Y, Liu Y, Liu Y, Liu Y, Livingston JA, Lizard G, Lizcano JM, Ljubojevic-Holzer S, LLeonart ME, Llobet-Navàs D, Llorente A, Lo CH, Lobato-Márquez D, Long Q, Long YC, Loos B, Loos JA, López MG, López-Doménech G, López-Guerrero JA, López-Jiménez AT, López-Pérez Ó, López-Valero I, Lorenowicz MJ, Lorente M, Lorincz P, Lossi L, Lotersztajn S, Lovat PE, Lovell JF, Lovy A, Lőw P, Lu G, Lu H, Lu JH, Lu JJ, Lu M, Lu S, Luciani A, Lucocq JM, Ludovico P, Luftig MA, Luhr M, Luis-Ravelo D, Lum JJ, Luna-Dulcey L, Lund AH, Lund VK, Lünemann JD, Lüningschrör P, Luo H, Luo R, Luo S, Luo Z, Luparello C, Lüscher B, Luu L, Lyakhovich A, Lyamzaev KG, Lystad AH, Lytvynchuk L, Ma AC, Ma C, Ma M, Ma NF, Ma QH, Ma X, Ma Y, Ma Z, MacDougald OA, Macian F, MacIntosh GC, MacKeigan JP, Macleod KF, Maday S, Madeo F, Madesh M, Madl T, Madrigal-Matute J, Maeda A, Maejima Y, Magarinos M, Mahavadi P, Maiani E, Maiese K, Maiti P, Maiuri MC, Majello B, Major MB, Makareeva E, Malik F, Mallilankaraman K, Malorni W, Maloyan A, Mammadova N, Man GCW, Manai F, Mancias JD, Mandelkow EM, Mandell MA, Manfredi AA, Manjili MH, Manjithaya R, Manque P, Manshian BB, Manzano R, Manzoni C, Mao K, Marchese C, Marchetti S, Marconi AM, Marcucci F, Mardente S, Mareninova OA, Margeta M, Mari M, Marinelli S, Marinelli O, Mariño G, Mariotto S, Marshall RS, Marten MR, Martens S, Martin APJ, Martin KR, Martin S, Martin S, Martín-Segura A, Martín-Acebes MA, Martin-Burriel I, Martin-Rincon M, Martin-Sanz P, Martina JA, Martinet W, Martinez A, Martinez A, Martinez J, Martinez Velazquez M, Martinez-Lopez N, Martinez-Vicente M, Martins DO, Martins JO, Martins WK, Martins-Marques T, Marzetti E, Masaldan S, Masclaux-Daubresse C, Mashek DG, Massa V, Massieu L, Masson GR, Masuelli L, Masyuk AI, Masyuk TV, Matarrese P, Matheu A, Matoba S, Matsuzaki S, Mattar P, Matte A, Mattoscio D, Mauriz JL, Mauthe M, Mauvezin C, Maverakis E, Maycotte P, Mayer J, Mazzoccoli G, Mazzoni C, Mazzulli JR, McCarty N, McDonald C, McGill MR, McKenna SL, McLaughlin B, McLoughlin F, McNiven MA, McWilliams TG, Mechta-Grigoriou F, Medeiros TC, Medina DL, Megeney LA, Megyeri K, Mehrpour M, Mehta JL, Meijer AJ, Meijer AH, Mejlvang J, Meléndez A, Melk A, Memisoglu G, Mendes AF, Meng D, Meng F, Meng T, Menna-Barreto R, Menon MB, Mercer C, Mercier AE, Mergny JL, Merighi A, Merkley SD, Merla G, Meske V, Mestre AC, Metur SP, Meyer C, Meyer H, Mi W, Mialet-Perez J, Miao J, Micale L, Miki Y, Milan E, Milczarek M, Miller DL, Miller SI, Miller S, Millward SW, Milosevic I, Minina EA, Mirzaei H, Mirzaei HR, Mirzaei M, Mishra A, Mishra N, Mishra PK, Misirkic Marjanovic M, Misasi R, Misra A, Misso G, Mitchell C, Mitou G, Miura T, Miyamoto S, Miyazaki M, Miyazaki M, Miyazaki T, Miyazawa K, Mizushima N, Mogensen TH, Mograbi B, Mohammadinejad R, Mohamud Y, Mohanty A, Mohapatra S, Möhlmann T, Mohmmed A, Moles A, Moley KH, Molinari M, Mollace V, Møller AB, Mollereau B, Mollinedo F, Montagna C, Monteiro MJ, Montella A, Montes LR, Montico B, Mony VK, Monzio Compagnoni G, Moore MN, Moosavi MA, Mora AL, Mora M, Morales-Alamo D, Moratalla R, Moreira PI, Morelli E, Moreno S, Moreno-Blas D, Moresi V, Morga B, Morgan AH, Morin F, Morishita H, Moritz OL, Moriyama M, Moriyasu Y, Morleo M, Morselli E, Moruno-Manchon JF, Moscat J, Mostowy S, Motori E, Moura AF, Moustaid-Moussa N, Mrakovcic M, Muciño-Hernández G, Mukherjee A, Mukhopadhyay S, Mulcahy Levy JM, Mulero V, Muller S, Münch C, Munjal A, Munoz-Canoves P, Muñoz-Galdeano T, Münz C, Murakawa T, Muratori C, Murphy BM, Murphy JP, Murthy A, Myöhänen TT, Mysorekar IU, Mytych J, Nabavi SM, Nabissi M, Nagy P, Nah J, Nahimana A, Nakagawa I, Nakamura K, Nakatogawa H, Nandi SS, Nanjundan M, Nanni M, Napolitano G, Nardacci R, Narita M, Nassif M, Nathan I, Natsumeda M, Naude RJ, Naumann C, Naveiras O, Navid F, Nawrocki ST, Nazarko TY, Nazio F, Negoita F, Neill T, Neisch AL, Neri LM, Netea MG, Neubert P, Neufeld TP, Neumann D, Neutzner A, Newton PT, Ney PA, Nezis IP, Ng CCW, Ng TB, Nguyen HTT, Nguyen LT, Ni HM, Ní Cheallaigh C, Ni Z, Nicolao MC, Nicoli F, Nieto-Diaz M, Nilsson P, Ning S, Niranjan R, Nishimune H, Niso-Santano M, Nixon RA, Nobili A, Nobrega C, Noda T, Nogueira-Recalde U, Nolan TM, Nombela I, Novak I, Novoa B, Nozawa T, Nukina N, Nussbaum-Krammer C, Nylandsted J, O'Donovan TR, O'Leary SM, O'Rourke EJ, O'Sullivan MP, O'Sullivan TE, Oddo S, Oehme I, Ogawa M, Ogier-Denis E, Ogmundsdottir MH, Ogretmen B, Oh GT, Oh SH, Oh YJ, Ohama T, Ohashi Y, Ohmuraya M, Oikonomou V, Ojha R, Okamoto K, Okazawa H, Oku M, Oliván S, Oliveira JMA, Ollmann M, Olzmann JA, Omari S, Omary MB, Önal G, Ondrej M, Ong SB, Ong SG, Onnis A, Orellana JA, Orellana-Muñoz S, Ortega-Villaizan MDM, Ortiz-Gonzalez XR, Ortona E, Osiewacz HD, Osman AK, Osta R, Otegui MS, Otsu K, Ott C, Ottobrini L, Ou JJ, Outeiro TF, Oynebraten I, Ozturk M, Pagès G, Pahari S, Pajares M, Pajvani UB, Pal R, Paladino S, Pallet N, Palmieri M, Palmisano G, Palumbo C, Pampaloni F, Pan L, Pan Q, Pan W, Pan X, Panasyuk G, Pandey R, Pandey UB, Pandya V, Paneni F, Pang SY, Panzarini E, Papademetrio DL, Papaleo E, Papinski D, Papp D, Park EC, Park HT, Park JM, Park JI, Park JT, Park J, Park SC, Park SY, Parola AH, Parys JB, Pasquier A, Pasquier B, Passos JF, Pastore N, Patel HH, Patschan D, Pattingre S, Pedraza-Alva G, Pedraza-Chaverri J, Pedrozo Z, Pei G, Pei J, Peled-Zehavi H, Pellegrini JM, Pelletier J, Peñalva MA, Peng D, Peng Y, Penna F, Pennuto M, Pentimalli F, Pereira CM, Pereira GJS, Pereira LC, Pereira de Almeida L, Perera ND, Pérez-Lara Á, Perez-Oliva AB, Pérez-Pérez ME, Periyasamy P, Perl A, Perrotta C, Perrotta I, Pestell RG, Petersen M, Petrache I, Petrovski G, Pfirrmann T, Pfister AS, Philips JA, Pi H, Picca A, Pickrell AM, Picot S, Pierantoni GM, Pierdominici M, Pierre P, Pierrefite-Carle V, Pierzynowska K, Pietrocola F, Pietruczuk M, Pignata C, Pimentel-Muiños FX, Pinar M, Pinheiro RO, Pinkas-Kramarski R, Pinton P, Pircs K, Piya S, Pizzo P, Plantinga TS, Platta HW, Plaza-Zabala A, Plomann M, Plotnikov EY, Plun-Favreau H, Pluta R, Pocock R, Pöggeler S, Pohl C, Poirot M, Poletti A, Ponpuak M, Popelka H, Popova B, Porta H, Porte Alcon S, Portilla-Fernandez E, Post M, Potts MB, Poulton J, Powers T, Prahlad V, Prajsnar TK, Praticò D, Prencipe R, Priault M, Proikas-Cezanne T, Promponas VJ, Proud CG, Puertollano R, Puglielli L, Pulinilkunnil T, Puri D, Puri R, Puyal J, Qi X, Qi Y, Qian W, Qiang L, Qiu Y, Quadrilatero J, Quarleri J, Raben N, Rabinowich H, Ragona D, Ragusa MJ, Rahimi N, Rahmati M, Raia V, Raimundo N, Rajasekaran NS, Ramachandra Rao S, Rami A, Ramírez-Pardo I, Ramsden DB, Randow F, Rangarajan PN, Ranieri D, Rao H, Rao L, Rao R, Rathore S, Ratnayaka JA, Ratovitski EA, Ravanan P, Ravegnini G, Ray SK, Razani B, Rebecca V, Reggiori F, Régnier-Vigouroux A, Reichert AS, Reigada D, Reiling JH, Rein T, Reipert S, Rekha RS, Ren H, Ren J, Ren W, Renault T, Renga G, Reue K, Rewitz K, Ribeiro de Andrade Ramos B, Riazuddin SA, Ribeiro-Rodrigues TM, Ricci JE, Ricci R, Riccio V, Richardson DR, Rikihisa Y, Risbud MV, Risueño RM, Ritis K, Rizza S, Rizzuto R, Roberts HC, Roberts LD, Robinson KJ, Roccheri MC, Rocchi S, Rodney GG, Rodrigues T, Rodrigues Silva VR, Rodriguez A, Rodriguez-Barrueco R, Rodriguez-Henche N, Rodriguez-Rocha H, Roelofs J, Rogers RS, Rogov VV, Rojo AI, Rolka K, Romanello V, Romani L, Romano A, Romano PS, Romeo-Guitart D, Romero LC, Romero M, Roney JC, Rongo C, Roperto S, Rosenfeldt MT, Rosenstiel P, Rosenwald AG, Roth KA, Roth L, Roth S, Rouschop KMA, Roussel BD, Roux S, Rovere-Querini P, Roy A, Rozieres A, Ruano D, Rubinsztein DC, Rubtsova MP, Ruckdeschel K, Ruckenstuhl C, Rudolf E, Rudolf R, Ruggieri A, Ruparelia AA, Rusmini P, Russell RR, Russo GL, Russo M, Russo R, Ryabaya OO, Ryan KM, Ryu KY, Sabater-Arcis M, Sachdev U, Sacher M, Sachse C, Sadhu A, Sadoshima J, Safren N, Saftig P, Sagona AP, Sahay G, Sahebkar A, Sahin M, Sahin O, Sahni S, Saito N, Saito S, Saito T, Sakai R, Sakai Y, Sakamaki JI, Saksela K, Salazar G, Salazar-Degracia A, Salekdeh GH, Saluja AK, Sampaio-Marques B, Sanchez MC, Sanchez-Alcazar JA, Sanchez-Vera V, Sancho-Shimizu V, Sanderson JT, Sandri M, Santaguida S, Santambrogio L, Santana MM, Santoni G, Sanz A, Sanz P, Saran S, Sardiello M, Sargeant TJ, Sarin A, Sarkar C, Sarkar S, Sarrias MR, Sarkar S, Sarmah DT, Sarparanta J, Sathyanarayan A, Sathyanarayanan R, Scaglione KM, Scatozza F, Schaefer L, Schafer ZT, Schaible UE, Schapira AHV, Scharl M, Schatzl HM, Schein CH, Scheper W, Scheuring D, Schiaffino MV, Schiappacassi M, Schindl R, Schlattner U, Schmidt O, Schmitt R, Schmidt SD, Schmitz I, Schmukler E, Schneider A, Schneider BE, Schober R, Schoijet AC, Schott MB, Schramm M, Schröder B, Schuh K, Schüller C, Schulze RJ, Schürmanns L, Schwamborn JC, Schwarten M, Scialo F, Sciarretta S, Scott MJ, Scotto KW, Scovassi AI, Scrima A, Scrivo A, Sebastian D, Sebti S, Sedej S, Segatori L, Segev N, Seglen PO, Seiliez I, Seki E, Selleck SB, Sellke FW, Selsby JT, Sendtner M, Senturk S, Seranova E, Sergi C, Serra-Moreno R, Sesaki H, Settembre C, Setty SRG, Sgarbi G, Sha O, Shacka JJ, Shah JA, Shang D, Shao C, Shao F, Sharbati S, Sharkey LM, Sharma D, Sharma G, Sharma K, Sharma P, Sharma S, Shen HM, Shen H, Shen J, Shen M, Shen W, Shen Z, Sheng R, Sheng Z, Sheng ZH, Shi J, Shi X, Shi YH, Shiba-Fukushima K, Shieh JJ, Shimada Y, Shimizu S, Shimozawa M, Shintani T, Shoemaker CJ, Shojaei S, Shoji I, Shravage BV, Shridhar V, Shu CW, Shu HB, Shui K, Shukla AK, Shutt TE, Sica V, Siddiqui A, Sierra A, Sierra-Torre V, Signorelli S, Sil P, Silva BJA, Silva JD, Silva-Pavez E, Silvente-Poirot S, Simmonds RE, Simon AK, Simon HU, Simons M, Singh A, Singh LP, Singh R, Singh SV, Singh SK, Singh SB, Singh S, Singh SP, Sinha D, Sinha RA, Sinha S, Sirko A, Sirohi K, Sivridis EL, Skendros P, Skirycz A, Slaninová I, Smaili SS, Smertenko A, Smith MD, Soenen SJ, Sohn EJ, Sok SPM, Solaini G, Soldati T, Soleimanpour SA, Soler RM, Solovchenko A, Somarelli JA, Sonawane A, Song F, Song HK, Song JX, Song K, Song Z, Soria LR, Sorice M, Soukas AA, Soukup SF, Sousa D, Sousa N, Spagnuolo PA, Spector SA, Srinivas Bharath MM, St Clair D, Stagni V, Staiano L, Stalnecker CA, Stankov MV, Stathopulos PB, Stefan K, Stefan SM, Stefanis L, Steffan JS, Steinkasserer A, Stenmark H, Sterneckert J, Stevens C, Stoka V, Storch S, Stork B, Strappazzon F, Strohecker AM, Stupack DG, Su H, Su LY, Su L, Suarez-Fontes AM, Subauste CS, Subbian S, Subirada PV, Sudhandiran G, Sue CM, Sui X, Summers C, Sun G, Sun J, Sun K, Sun MX, Sun Q, Sun Y, Sun Z, Sunahara KKS, Sundberg E, Susztak K, Sutovsky P, Suzuki H, Sweeney G, Symons JD, Sze SCW, Szewczyk NJ, Tabęcka-Łonczynska A, Tabolacci C, Tacke F, Taegtmeyer H, Tafani M, Tagaya M, Tai H, Tait SWG, Takahashi Y, Takats S, Talwar P, Tam C, Tam SY, Tampellini D, Tamura A, Tan CT, Tan EK, Tan YQ, Tanaka M, Tanaka M, Tang D, Tang J, Tang TS, Tanida I, Tao Z, Taouis M, Tatenhorst L, Tavernarakis N, Taylor A, Taylor GA, Taylor JM, Tchetina E, Tee AR, Tegeder I, Teis D, Teixeira N, Teixeira-Clerc F, Tekirdag KA, Tencomnao T, Tenreiro S, Tepikin AV, Testillano PS, Tettamanti G, Tharaux PL, Thedieck K, Thekkinghat AA, Thellung S, Thinwa JW, Thirumalaikumar VP, Thomas SM, Thomes PG, Thorburn A, Thukral L, Thum T, Thumm M, Tian L, Tichy A, Till A, Timmerman V, Titorenko VI, Todi SV, Todorova K, Toivonen JM, Tomaipitinca L, Tomar D, Tomas-Zapico C, Tomić S, Tong BC, Tong C, Tong X, Tooze SA, Torgersen ML, Torii S, Torres-López L, Torriglia A, Towers CG, Towns R, Toyokuni S, Trajkovic V, Tramontano D, Tran QG, Travassos LH, Trelford CB, Tremel S, Trougakos IP, Tsao BP, Tschan MP, Tse HF, Tse TF, Tsugawa H, Tsvetkov AS, Tumbarello DA, Tumtas Y, Tuñón MJ, Turcotte S, Turk B, Turk V, Turner BJ, Tuxworth RI, Tyler JK, Tyutereva EV, Uchiyama Y, Ugun-Klusek A, Uhlig HH, Ułamek-Kozioł M, Ulasov IV, Umekawa M, Ungermann C, Unno R, Urbe S, Uribe-Carretero E, Üstün S, Uversky VN, Vaccari T, Vaccaro MI, Vahsen BF, Vakifahmetoglu-Norberg H, Valdor R, Valente MJ, Valko A, Vallee RB, Valverde AM, Van den Berghe G, van der Veen S, Van Kaer L, van Loosdregt J, van Wijk SJL, Vandenberghe W, Vanhorebeek I, Vannier-Santos MA, Vannini N, Vanrell MC, Vantaggiato C, Varano G, Varela-Nieto I, Varga M, Vasconcelos MH, Vats S, Vavvas DG, Vega-Naredo I, Vega-Rubin-de-Celis S, Velasco G, Velázquez AP, Vellai T, Vellenga E, Velotti F, Verdier M, Verginis P, Vergne I, Verkade P, Verma M, Verstreken P, Vervliet T, Vervoorts J, Vessoni AT, Victor VM, Vidal M, Vidoni C, Vieira OV, Vierstra RD, Viganó S, Vihinen H, Vijayan V, Vila M, Vilar M, Villalba JM, Villalobo A, Villarejo-Zori B, Villarroya F, Villarroya J, Vincent O, Vindis C, Viret C, Viscomi MT, Visnjic D, Vitale I, Vocadlo DJ, Voitsekhovskaja OV, Volonté C, Volta M, Vomero M, Von Haefen C, Vooijs MA, Voos W, Vucicevic L, Wade-Martins R, Waguri S, Waite KA, Wakatsuki S, Walker DW, Walker MJ, Walker SA, Walter J, Wandosell FG, Wang B, Wang CY, Wang C, Wang C, Wang C, Wang CY, Wang D, Wang F, Wang F, Wang F, Wang G, Wang H, Wang H, Wang H, Wang HG, Wang J, Wang J, Wang J, Wang J, Wang K, Wang L, Wang L, Wang MH, Wang M, Wang N, Wang P, Wang P, Wang P, Wang P, Wang QJ, Wang Q, Wang QK, Wang QA, Wang WT, Wang W, Wang X, Wang X, Wang Y, Wang Y, Wang Y, Wang YY, Wang Y, Wang Y, Wang Y, Wang Y, Wang Z, Wang Z, Wang Z, Warnes G, Warnsmann V, Watada H, Watanabe E, Watchon M, Wawrzyńska A, Weaver TE, Wegrzyn G, Wehman AM, Wei H, Wei L, Wei T, Wei Y, Weiergräber OH, Weihl CC, Weindl G, Weiskirchen R, Wells A, Wen RH, Wen X, Werner A, Weykopf B, Wheatley SP, Whitton JL, Whitworth AJ, Wiktorska K, Wildenberg ME, Wileman T, Wilkinson S, Willbold D, Williams B, Williams RSB, Williams RL, Williamson PR, Wilson RA, Winner B, Winsor NJ, Witkin SS, Wodrich H, Woehlbier U, Wollert T, Wong E, Wong JH, Wong RW, Wong VKW, Wong WW, Wu AG, Wu C, Wu J, Wu J, Wu KK, Wu M, Wu SY, Wu S, Wu SY, Wu S, Wu WKK, Wu X, Wu X, Wu YW, Wu Y, Xavier RJ, Xia H, Xia L, Xia Z, Xiang G, Xiang J, Xiang M, Xiang W, Xiao B, Xiao G, Xiao H, Xiao HT, Xiao J, Xiao L, Xiao S, Xiao Y, Xie B, Xie CM, Xie M, Xie Y, Xie Z, Xie Z, Xilouri M, Xu C, Xu E, Xu H, Xu J, Xu J, Xu L, Xu WW, Xu X, Xue Y, Yakhine-Diop SMS, Yamaguchi M, Yamaguchi O, Yamamoto A, Yamashina S, Yan S, Yan SJ, Yan Z, Yanagi Y, Yang C, Yang DS, Yang H, Yang HT, Yang H, Yang JM, Yang J, Yang J, Yang L, Yang L, Yang M, Yang PM, Yang Q, Yang S, Yang S, Yang SF, Yang W, Yang WY, Yang X, Yang X, Yang Y, Yang Y, Yao H, Yao S, Yao X, Yao YG, Yao YM, Yasui T, Yazdankhah M, Yen PM, Yi C, Yin XM, Yin Y, Yin Z, Yin Z, Ying M, Ying Z, Yip CK, Yiu SPT, Yoo YH, Yoshida K, Yoshii SR, Yoshimori T, Yousefi B, Yu B, Yu H, Yu J, Yu J, Yu L, Yu ML, Yu SW, Yu VC, Yu WH, Yu Z, Yu Z, Yuan J, Yuan LQ, Yuan S, Yuan SF, Yuan Y, Yuan Z, Yue J, Yue Z, Yun J, Yung RL, Zacks DN, Zaffagnini G, Zambelli VO, Zanella I, Zang QS, Zanivan S, Zappavigna S, Zaragoza P, Zarbalis KS, Zarebkohan A, Zarrouk A, Zeitlin SO, Zeng J, Zeng JD, Žerovnik E, Zhan L, Zhang B, Zhang DD, Zhang H, Zhang H, Zhang H, Zhang H, Zhang H, Zhang H, Zhang H, Zhang HL, Zhang J, Zhang J, Zhang JP, Zhang KYB, Zhang LW, Zhang L, Zhang L, Zhang L, Zhang L, Zhang M, Zhang P, Zhang S, Zhang W, Zhang X, Zhang XW, Zhang X, Zhang X, Zhang X, Zhang X, Zhang XD, Zhang Y, Zhang Y, Zhang Y, Zhang YD, Zhang Y, Zhang YY, Zhang Y, Zhang Z, Zhang Z, Zhang Z, Zhang Z, Zhang Z, Zhang Z, Zhao H, Zhao L, Zhao S, Zhao T, Zhao XF, Zhao Y, Zhao Y, Zhao Y, Zhao Y, Zheng G, Zheng K, Zheng L, Zheng S, Zheng XL, Zheng Y, Zheng ZG, Zhivotovsky B, Zhong Q, Zhou A, Zhou B, Zhou C, Zhou G, Zhou H, Zhou H, Zhou H, Zhou J, Zhou J, Zhou J, Zhou J, Zhou K, Zhou R, Zhou XJ, Zhou Y, Zhou Y, Zhou Y, Zhou ZY, Zhou Z, Zhu B, Zhu C, Zhu GQ, Zhu H, Zhu H, Zhu H, Zhu WG, Zhu Y, Zhu Y, Zhuang H, Zhuang X, Zientara-Rytter K, Zimmermann CM, Ziviani E, Zoladek T, Zong WX, Zorov DB, Zorzano A, Zou W, Zou Z, Zou Z, Zuryn S, Zwerschke W, Brand-Saberi B, Dong XC, Kenchappa CS, Li Z, Lin Y, Oshima S, Rong Y, Sluimer JC, Stallings CL, Tong CK. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)(1). Autophagy. 2021;17:1-382. [PubMed] [DOI] [Full Text] |
| 81. | Vincent AM, Hayes JM, McLean LL, Vivekanandan-Giri A, Pennathur S, Feldman EL. Dyslipidemia-induced neuropathy in mice: the role of oxLDL/LOX-1. Diabetes. 2009;58:2376-2385. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 188] [Cited by in RCA: 208] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 82. | Turner KL, Honasoge A, Robert SM, McFerrin MM, Sontheimer H. A proinvasive role for the Ca(2+) -activated K(+) channel KCa3.1 in malignant glioma. Glia. 2014;62:971-981. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 94] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/