Published online Feb 15, 2025. doi: 10.4239/wjd.v16.i2.94861
Revised: September 10, 2024
Accepted: November 21, 2024
Published online: February 15, 2025
Processing time: 278 Days and 17.8 Hours
Although numerous single nucleotide polymorphism in multiple genes involve in the risk of type 2 diabetes mellitus (T2D), the single gene defects of T2D with strong family history is not clear yet. SPTLC1 are causative for hereditary sensory and autonomic neuropathy, which is clinical overlapping with diabetic peripheral neuropathy. Mice with adipocyte-specific deletion of SPTLC1 had impaired glucose tolerances and insulin sensitivity. Thus, it is necessary to investigate the SPTLC1 mutations in adult-onset T2D with strong family history.
To analyze the role of SPTLC1 mutation on adult-onset T2D with strong family history.
By whole-exome sequence analysis of a patient with T2D and his family members, an uncertain variant in SPTLC1 was identified. Bioinformation analysis was used to evaluate the influence of mutation, rare variant gene-level associations for SPTLC1 in T2D, and the relationship between SPTLC1 mRNA and T2D in human islets from GSE25724. The effect of G371R of SPTLC1 on the characteristics of inflammatory cytokines and apoptosis was also tested on human embryonic kidney (HEK) 293 cells.
A single nucleotide variation in SPTLC1 (c.1111G>A: p.G371R) was identified in a family with T2D. The deleterious variant was predicted by functional analysis through hidden Markov models and mendelian clinically applicable patho
The study classified SPTLC1 p.G371R mutation as the likely pathogenic mutation from an adult-onset T2D patients with strong family history T2D.
Core Tip: The single gene defects of type 2 diabetes mellitus (T2D) with strong family history is not clear yet. We identified SPTLC1 p.G371R mutation as the likely pathogenic mutation from an adult-onset T2D patients with family history. This mutation induced the expression of tumor necrosis factor-α and the percent of apoptosis in human embryonic kidney 293T cells. Moreover, data from Accelerating Medicines Partnership T2D knowledge showed a positive correlation between rare variant gene-level associations for SPTLC1 and the risk of T2D. This study provides a novel perspective to understand the pathology of T2D.
- Citation: Yi B, Bao Y, Wen ZY. Effect of SPTLC1 on type 2 diabetes mellitus. World J Diabetes 2025; 16(2): 94861
- URL: https://www.wjgnet.com/1948-9358/full/v16/i2/94861.htm
- DOI: https://dx.doi.org/10.4239/wjd.v16.i2.94861
Diabetes is a major global health issue that comes with severe complications, such as diabetic peripheral neuropathy (DPN), and higher mortality rates[1,2]. Type 2 diabetes mellitus (T2D), accounts for 90%-95% of all diabetes, is described as a polygenetic multifactorial and heterogeneous disease[2], while maturity-onset diabetes of the young (MODY) is caused by single gene mutations[2]. A growing number of studies indicated T2D overlaps clinically and genetically with MODY[2,3]. For example, both T2D and MODY present strong family history[2,3]. And some patients with T2D harbor single gene mutations that cause MODY, such as HNF4A p.T131I[2-4]. However, the potential genetic etiology of T2D with strong family history is not clear yet.
Although numerous single nucleotide polymorphism (SNP) in multiple genes involve in the β-cell dysfunction, single gene defects may also influence risk of T2D[2,5-7]. Whole genome sequencing has used to identify mutation in WFS1, PAX6, APPL1 and MAFA gene as novel genetic cause of adult-onset familial diabetes[3]. Interestingly, among those gene, PAX6 mutation have been reported to diminish glucose stimulated insulin release in human and to regulate pancreatic islet development[3]. Additionally, single gene defects were also reported in patients with T2D and some hereditary disease[8,9]. For example, pathogenic mutations in G6PC, which is associated with postprandial hyperglycemia, was found in patients with T2D and glycogen storage disease type 1a[9]. Thus, it is necessary to investigate the single gene mutations in T2D with strong family history.
SPTLC1, the rate-limiting enzyme in sphingolipid biosynthesis, covers serine and palmitoyl-CoA to sphinganine. Pathogenic variants in SPTLC1 are causative for hereditary sensory and autonomic neuropathy[10,11], juvenile amyo
Six participants from a China family with T2D were enrolled and provided written informed consent for genetic analysis according to ethical guidelines of the declaration of Helsinki (1964).
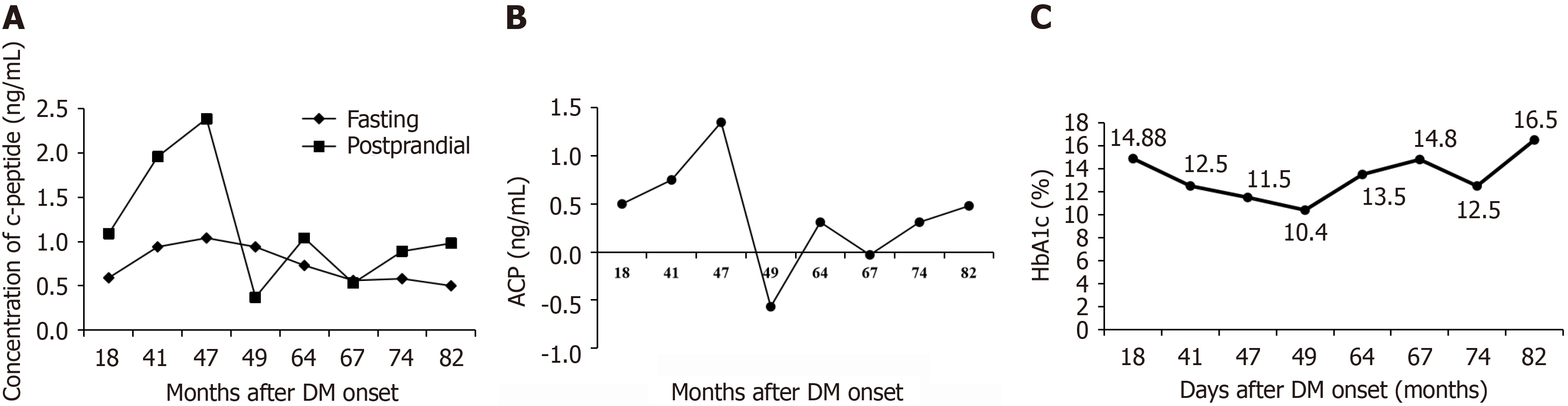
The proband of the family was a 52-year-old man (family member III-1), who presented with a 10-year history of T2D. The initiation symptoms of the index patient were polyuria, excessive drinking and eating, and weight loss. He began to feel fatigue of bilateral lower limb at age 44, and continued to progress to stabbing pain and numbing in a pair of feet. Sensory disturbance of this patients was confirmed through touching by cotton wool, 10-g monofilament, and elec
| Features | Normal range | III-1 (proband) | IV-1 | II-2 |
| Gender | Male | Male | Female | |
| Age at admission, years | 44.00 | 19.00 | 68.00 | |
| Age at DM onset, years | 42.00 | 17.00 | 53.00 | |
| Weight, kg | 50.00 | 70.00 | 51.00 | |
| Height, cm | 170.00 | 174.00 | 156.00 | |
| Waist, cm | 71.00 | 82.00 | 87.00 | |
| BMI, kg/m2 | 17.30 | 23.12 | 20.96 | |
| SBP, mmHg | 118.00 | 130.00 | 129.00 | |
| DBP, mmHg | 90.00 | 85.00 | 68.00 | |
| FCP, ng/mL | 0.81-1.89 | 1.04 | 1.16 | |
| 2hCP, ng/mL | 2.39 | 1.30 | ||
| FINS, μU/L | 4.00-12.00 | 12.65 | ||
| 2hINS, μU/L | 83.15 | |||
| HbA1c, % | 3.60-6.00 | 12.6 | 10.80 | 10.40 |
| TC, mmol/L | 3.10-5.20 | 3.90 | 3.29 | 4.95 |
| TG, mmol/L | 0.56-1.70 | 0.95 | 1.01 | 0.93 |
| HDL, mmol/L | 1.00-1.55 | 1.06 | 0.89 | 1.21 |
| LDL, mmol/L | 1.90-3.10 | 2.45 | 1.84 | 3.03 |
| Hs-CRP, mg/L | 0.00-3.00 | 0.37 | 1.57 | 5.16 |
| GADA | Negative | Negative | Negative | |
| IAA | Negative | Negative | Negative | |
| IA-2A | Negative | Negative | Negative | |
| ICA | Negative | Negative | Negative | |
| ACR, mg/g | 0.00-30.00 | 16.85 | 25.66 | |
| Ketobodies in urine | Negative | Negative | Negative | Negative |
| DR | Negative | No | No | No |
Genomic DNA samples were extracted from peripheral blood leukocytes. WES was analyzed in WeHealth Biomedical Technology Co., Ltd (Shanghai, China) using 150 base pair paired-end sequencing on the Illumina HiSeq platform from the proband and his father, mother, sister. Polymerase chain reaction (PCR) was performed to confirm the identified variant in Tsingke Biotechnology Co., Ltd (Beijing, China) from all 6 participants. Primer sequences are TGGGATACTGAGGTGAGAAGGG (Forward primer) and AGCTGCAATCTGGTCAAACTGA (Reverse primer). Sequences were compared to reference sequence NM006415 of the SPTLC1 gene by SeqMan Pro software.
The raw reads were aligned by Burrows-Wheler Aligner tools, and duplicates were removed from the sorted alignment using Picard. Variants were classified according to the American college of medical genetics guidelines[21]. UniProtKB entry O15269 was used to annotate the STPLC1 protein. The structure of STPLC1 protein was built by using SWISS-MODEL.
The relative expression level of SPTLC1 in tissue specificity was obtained from ProteomicsDB (https://www.proteomicsdb.org/) entry O15269. Rare variant gene-level associations for SPTLC1 in T2D were analyzed in Accelerating Medicines Partnership (AMP) T2D knowledge portal (https://www.type2diabetesgenetics.org), funding by the National Institutes of Health, the United States Food and Drug Administration, biopharmaceutical companies and non-profit organizations[6]. This project seeks to promote understanding and treatment of T2D and its complications from 310 datasets.
The gene expression profile data (GSE25724) were obtained from gene expression omnibus. This array data included 6 type 2 diabetic human islets [mean age and body mass index (BMI) were 71years and 26 kg/m2, and three males] and seven non-diabetic islet samples (mean age: 58years, mean BMI: 24.8 kg/m2, and four males)[22].
The SPTLC1 cDNA (NM006415) was amplified and cloned into the vector FV149 with the use of TTGGTACCGAGCTCG (Forward primer) and GATATCTGCAGAATT (Reverse primer). The SPTLC1 mutation (G371R) was introduced by site-directed mutagenesis with a C-terminally enhanced green fluorescent protein-tagged. All constructs were validated by sequencing. Cell lines expressing wild-type or G371R were created using LipofectamineTM 2000 in human embryonic kidney (HEK) 293T (Invitrogen). The ratio of plasmid and LipofectamineTM 2000 was 5 μL:4 µg.
HEK 293T cells were cultured in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% fetal calf serum and 1% Penicillin-Streptomycin solution at 37 °C and 5% carbon dioxide. Before transfection, the cells were cultivated in serum-free DMEM for 2 hours. After 48 hours incubation time for the transfection, the cells and cultured supernatants were collected.
The concentrations of tumor necrosis factor (TNF)-α and interleukin (IL)-1β in cultured supernatants were measured by commercial enzyme-linked immunosorbent assay kit (Elabscience Biotechnology Co., Ltd, Wuhan, Hubei Province, China).
Real-time quantitative PCR was used to detect the mRNA expression of TNF-α and IL-1β from HEK 293T cells. The forward and reverse primes of TNF-α (Homo) were TCAGAGGGCCTGTACCTCAT and GGAAGACCCCTCCC
This reaction was performed in triplicate with 10 ng cDNA in SYBR Green master mix and run on an Applied Biosystems QuantStudio 6 system. The mRNA expression levels were normalized by Homo-glyceraldehyde-3-phosphate dehydrogenase.
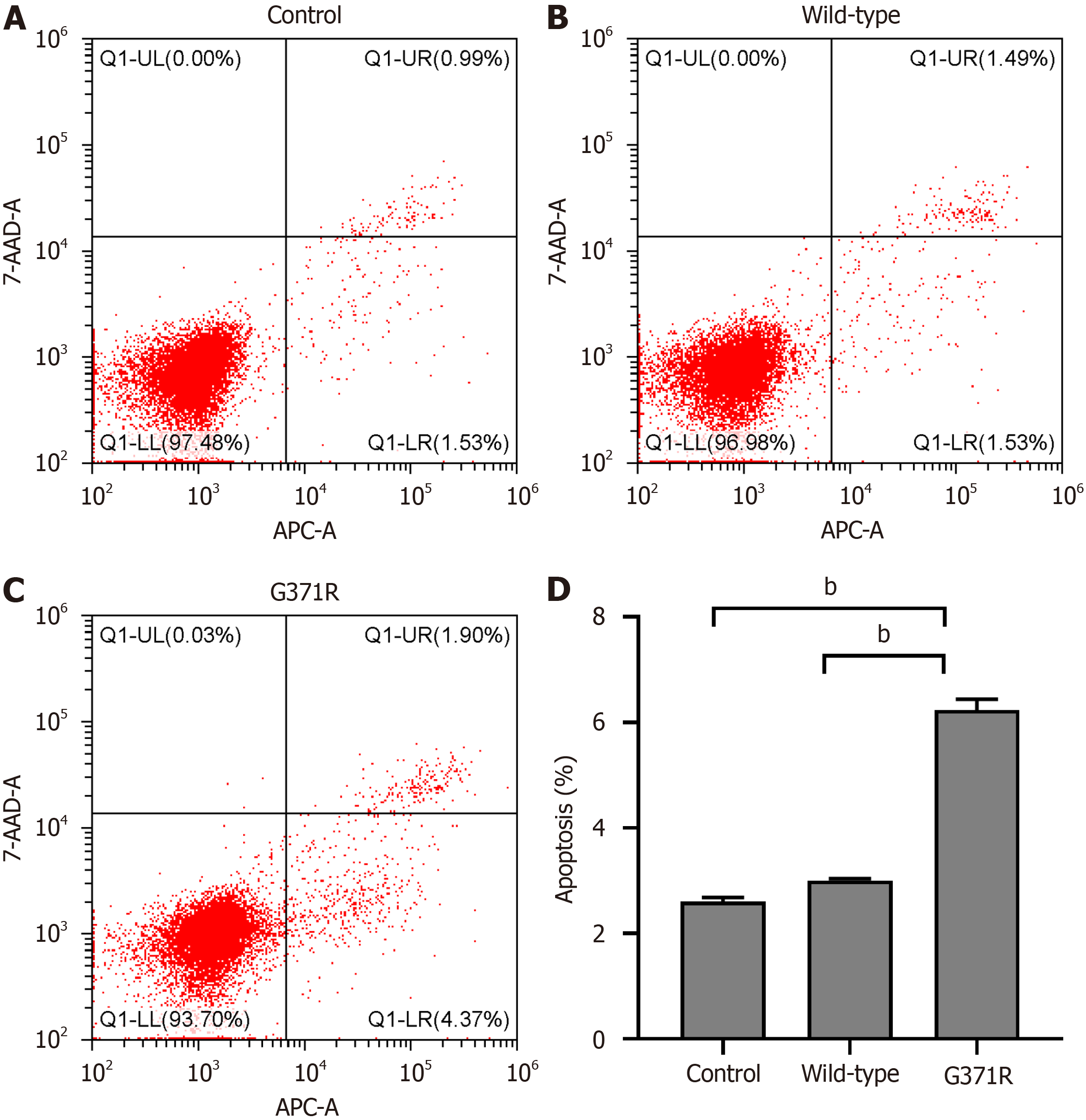
The apoptosis of HEK 293T cells after the transfection of wild-type or G371R was detected by Annexin V-Allophycocyanin (APC)/7-Aminoactinomycin D (7-AAD) apoptosis detection kit (Elab science Biotechnology Co., Ltd, Wuhan, Hubei province, China). Briefly, HEK 293T cells were harvested after 48 hours and pelleted by centrifugation at 1500 rpm for 5 minutes. The resuspended cells (1 × 105 cells/mL) were stained with 5 μL 7-AAD and 1 μL Annexin V-APC at room temperature. The percentage of cell apoptosis was determined by cytoFLEX platform (Beckman coulter, United States).
The one-way analysis of variance multiple comparisons was used for statistical analysis by statistical product and service solutions 19.0 software. A two-tailed P value < 0.05 was considered statistically significant.
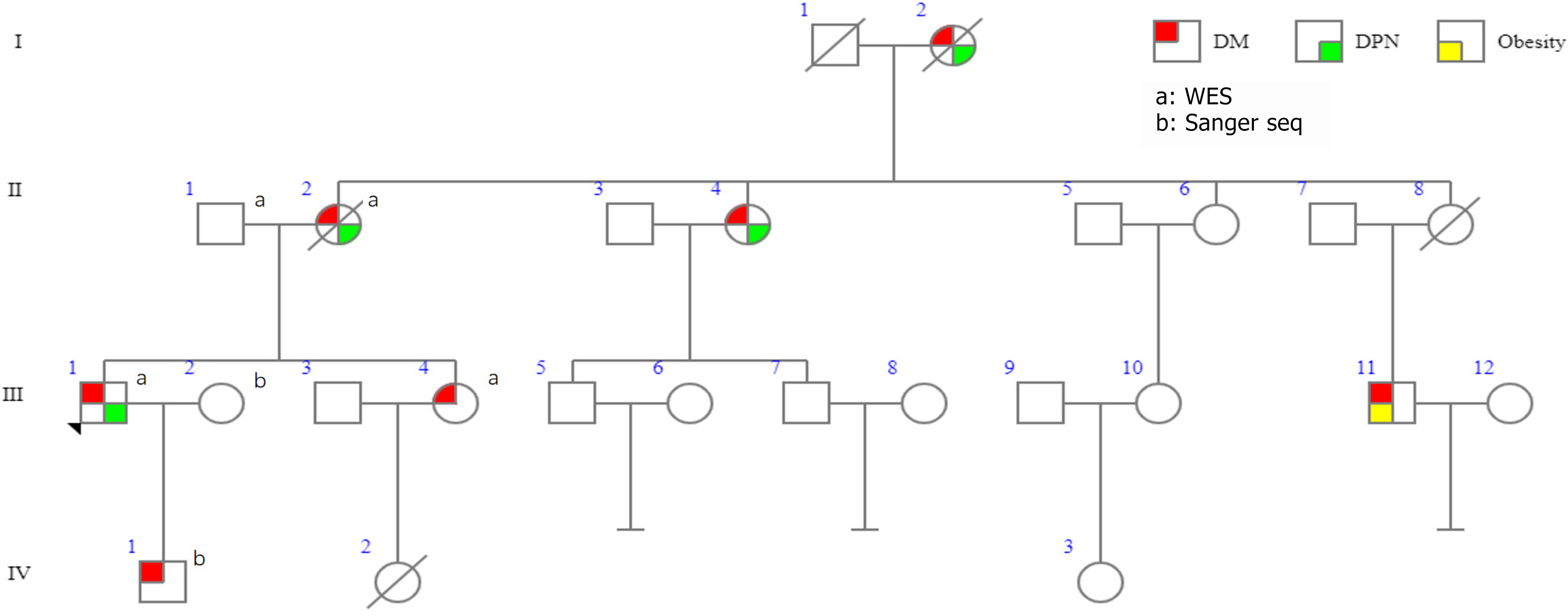
III-1 is the proband of the pedigree, who suffered from T2D at the age of 42 years and was diagnosed with DPN one year later. Surprisingly, the patient’s mother (II-2), maternal aunt (II-4), and grandmother (I-2) were also diagnosed with diabetes and DPN (Figure 1, Table 1 and Supplementary Table 1). Furthermore, the proband’s son (IV-1), sister (III-4), and a younger male cousin (III-11) also have diabetes, whose illness onset ages are 17, 41 and 22 years, respectively. And the III-11 also has obesity (Figure 2).
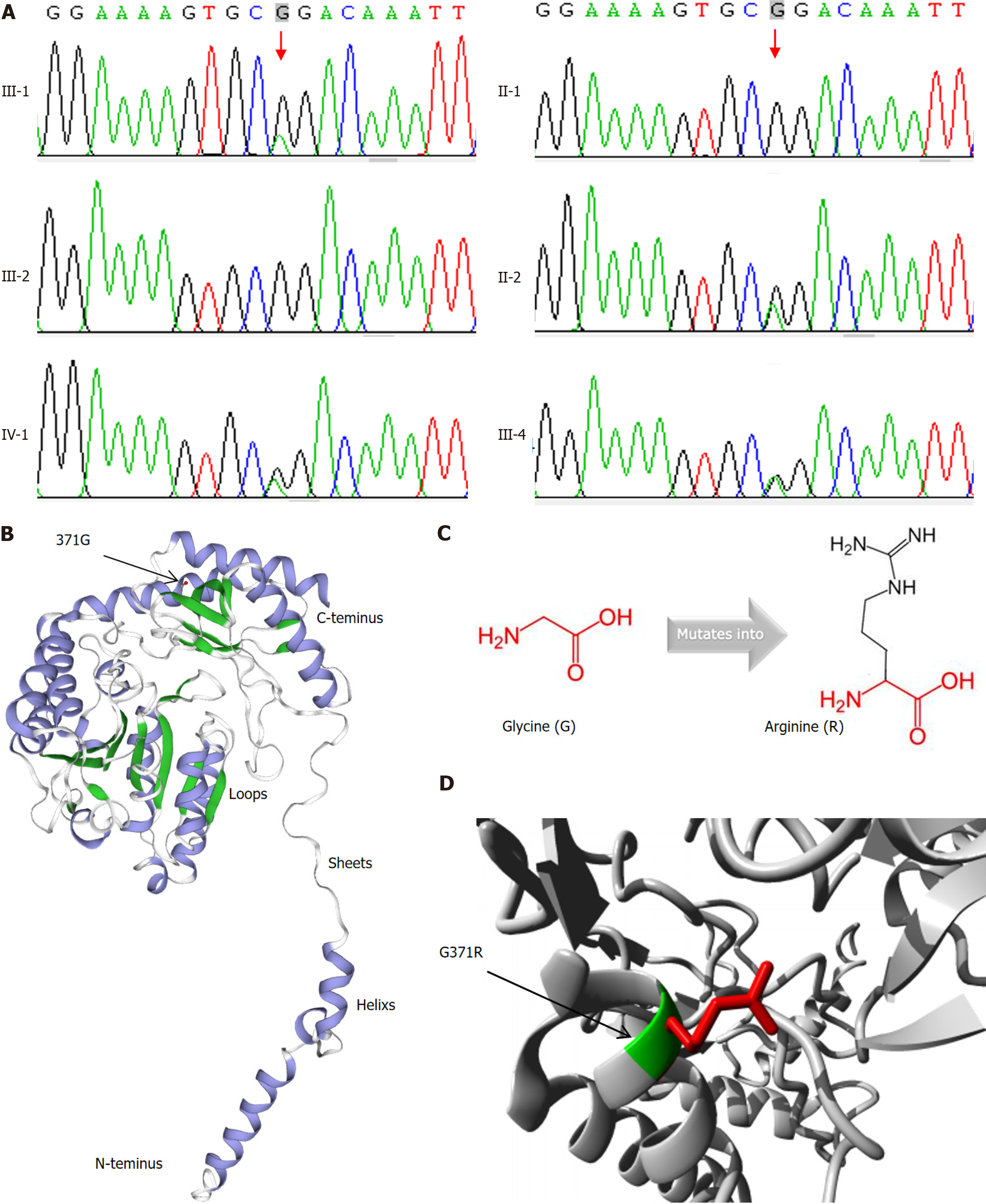
WES analysis did not reveal the mutation of candidate genes of monogenic diabetes, such as GCK, HNF1A, HNF4A, HNF1B, KCNJ11, INS, and ABCC8, etc. However, we found a single nucleotide variation in SPTLC1 (NM-006415: exon12: C.1111G>A: p.G371R) in the proband (Figure 3A). Proband’s mother, sister, and son were confirmed to carry this mutation by Sanger sequencing. However, his wife and father do not carry this mutation. Unfortunately, we don’t have detailed data yet about II-4 or III-11.
The frequencies of this mutation in humans are less than 0.05% in dbSNP, ExAC, genom AD, or 1000 genome databases. Although the interpretation of this mutation is likely benign from dbSNP, pathogenic variant effect on SPTLC1 protein was predicted by functional analysis through hidden Markov models (FATHMM) and mendelian clinically applicable pathogenicity (M-CAP) software. This pathogenicity might be derived from the different amino acid properties (Figures 3B-D): (1) The mutated residue is located in a domain that is important for the interaction with other molecules or the parts of the other protein, but is not essential for the catalytic site; (2) The new residue was bigger and more hydrophilic than wild-type residue; and (3) The mutation introduces a positive charge, instigating repulsion between the mutant residue and neighboring residues (https://www3.cmbi.umcn.nl/hope/report/625d75962412fdabe40cc1ce/). Thus, this mutation was classified into uncertain significance (PM2 + PP1 + PP3) according to American college of medical genetics and genomics (ACMG) criteria.
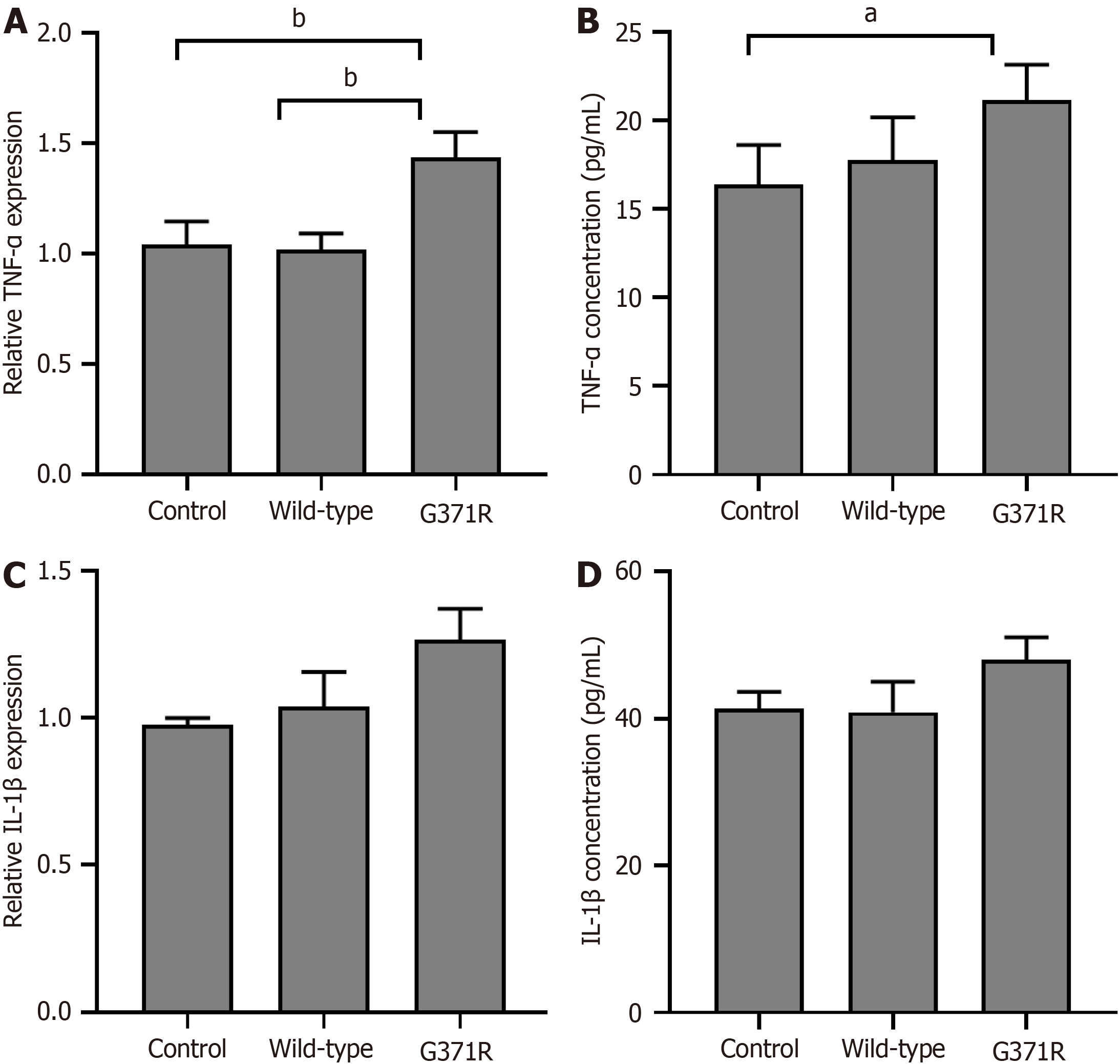
Because the 371 amino acid residue is not essential for catalytic activation of SPTLC1 (https://www.ncbi.nlm.nih.gov/Structure/icn3d/full.html?showanno=1&mmdbid=199148), we tested whether the mutation could give rise to the potential any other functions of SPTLC1. As shown in Figure 4, both mRNA and protein levels of TNF-α were slightly elevated in p.G371R expressing HEK 293T cells (P < 0.05). But the mutation of SPTLC1 did not affect the expression of IL-1β.
Flow cytometric analysis indicated that the percentage of apoptosis in p.G371R expressing cells was nearly three times than that of wild type-expressing cells (Figure 5).
Of some interest, the data of 43125 humans from AMP T2D knowledge also showed a positive correlation between rare variant gene-level associations for SPTLC1 and the risk of T2M (the overall odds ratio = 2.4968, P = 0.0164) (Table 2).
| Mask | Methods | Sample size | Combined allele frequency (%) | Passing variants | Singleton variants | SD | Odds ratio | P value |
| LofTee | Predicted loss of function | 43125 | 0.03942 | 14 | 11 | 0.52284 | 2.1014 | 0.13368 |
| 16/16 | Predicted deleterious by 16 methods | 43125 | 0.04638 | 15 | 11 | 0.47401 | 1.8749 | 0.16848 |
| 11/11 | Predicted deleterious by 11 methods | 43125 | 0.09507 | 29 | 20 | 0.33683 | 2.0718 | 0.025839 |
| 5/5 | Predicted deleterious by 5 methods | 43125 | 0.15072 | 31 | 21 | 0.2571 | 1.3852 | 0.20087 |
| 5/5 + LofTee LC | Predicted deleterious by 5 methods, plus LofTee low confidence | 43125 | 0.16464 | 33 | 21 | 0.24528 | 1.3551 | 0.21151 |
| 5/5 + 1/5, 1% | Predicted deleterious by 5 methods, plus variants with MAF < 1% that are predicted to be deleterious by 1 of 5 methods | 43125 | 0.85565 | 95 | 59 | 0.10614 | 1.0444 | 0.68172 |
| 5/5 + 0/5, 1% | Predicted deleterious by 5 methods, plus variants with MAF < 1% that are not predicted to be deleterious by any of 5 methods | 43125 | 0.92058 | 102 | 63 | 0.10226 | 1.0154 | 0.88112 |
| Total | 43125 | 2.4968 | 0.0164 |
SPTLC1 protein, localized to the endoplasmic reticulum, was found in widely cells and tissue. The median SPTLC1 protein expression in pancreatic islets ranks No. 4 in all 39 human tissues (https://www.proteomicsdb.org/). Im
Early genetic analysis based on familial linkage or Genome-wide association studies have revealed that T2D is a disease of polygenic inheritance[1,5,6,23,24]. However, the role of single gene mutations in T2D with strong family history are still unknown. In this study, we classified a novel likely pathogenic mutation in the SPTLC1 gene from an adult-onset T2D patients with strong family history by WES combinate with functional study.
We initially identified a variant (p.G371R) in the SPTLC1 gene as uncertain significance according to ACMG criteria in a family with dominant adult-onset diabetes (PM2 + PP1 + PP3). Nonetheless, uncertain genetic variants should not be considered benign because they may also confer a risk of related disease. It has been reported that in 36 diabetic patients carrying candidate variants, 27.9% were uncertain significance variants, which is more than likely pathogenic variants (20.9%)[25]. A recent study also reported that cardiomyopathy patients with uncertain variants in actionable cardiac genes had comparable left ventricular internal diameter-diastole and -systole than patients without medically actionable genes[26].
Furthermore, this study supports that p.G371R mutation is harmful to the function of SPTLC1 by functional study in HEK 293T cells (PS3). The pathogenic effect of p.G371R was predicted by FATHMM and M-CAP software (Figure 3). Additionally, some studies found the crucial role of SPTLC1, especially p.Y164F mutation, on the survival of bone marrow cells and adipocytes[27,28]. Consistently, this study also found that p.G371R of SPTLC1 induced the expression of TNF-α and the percent of apoptosis in HEK 293T cells (Figure 5). Thus, SPTLC1 p.G371R may be a likely pathogenic mutation.
Moreover, there are some other evidences to support the potential pathogenic role of SPTLC1 on adult-onset diabetes with strong family. Firstly, AMP T2D knowledge shows the rare variant of the STPLC1 gene infer the high risk of T2D by seven different masks approaches (Table 2), which were developed to reveal the gene-level correlation of rare variants[6]. Based on this strategy, Flannick et al[6] found that the most vital T2D gene-level signals explain nearly 25% of the heritability of the strongest common single-variant signals.
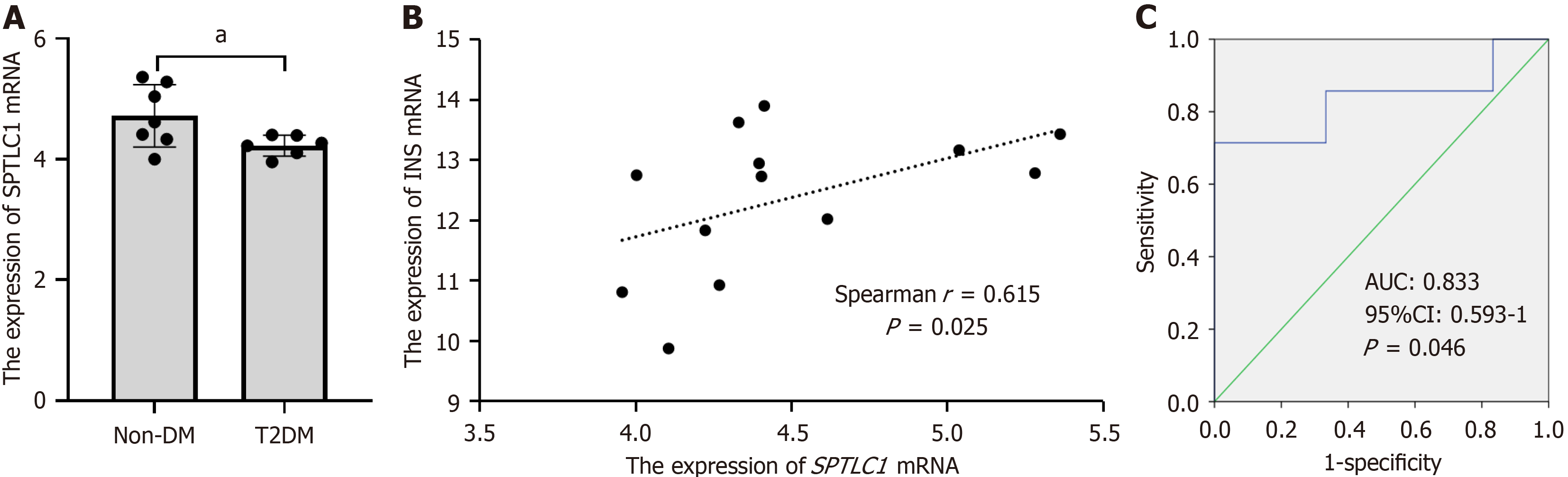
Secondly, this study reported the pro-apoptosis and pro-inflammatory effect of SPTLC1 p.G371R (Figure 4 and Figure 5), which is consistent with the previous studies[27,28]. It has been confirmed that apoptosis of islets β-cells induced by inflammatory cytokines was the key molecular mechanism of diabetes mellitus[29]. Actually, the study found that lower expression of SPTLC1 mRNA in pancreatic islets from diabetic patients was negatively associated with insulin levels (Figure 6). Interestingly, mice with adipocyte-specific deletion of SPTLC1 exhibited insulin resistance and impaired glucose tolerance[15]. Cellular effects of SPTLC1 deletion appear to involve activation of endoplasmic reticulum stress[27], which leads to the progressive β-cell failure[29].
Thirdly, deoxysphingolipids, formed by the pathogenic mutation of STPLC1, participated in the development of T2D and DPN[18-20]. It has been confirmed that the plasma concentration of deoxysphingolipids was significantly elevated in patients with metabolic syndrome, T2D, and DPN[18,19,30,31]. Importantly, deoxysphingolipids were independent risk factors for T2D even after adjusting for glycated hemoglobin, age, and BMI[18,19]. Lowering deoxysphingolipids by L-serine supplementation drastically improved the nerve conduction velocity in streptozotocin induced diabetic rats[17]. Some authors suggested that deoxysphingolipids caused the disassembly of actin stress fibers in Vero cells, increased the intracellular accumulation of filamentous actin in Ins-1 cells, and promoted dose-dependent apoptosis and impaired glucose-stimulated insulin secretion in insulin-producing cells[17,20]. Thus, deoxysphingolipids activated metabolic stress and the apoptosis of the cells may be a possible bridge between SPTLC1 and T2D and DPN.
Despite all our efforts to find the association between SPTLC1 and diabetes, some limitations which may reduce the credibility of the study still exist in this study. Firstly, p.G371R of the SPTLC1 gene is not pathogenic or likely pathogenic according to the criteria from ACMG, but, in vitro experiments showed the pro-apoptosis effect of this mutation. Importantly, several studies demonstrated that uncertain genetic variants also confer a high risk of related disease[25,26]. Secondly, some members of this family did not perform the genetic testing due to a lack of cooperation from these people. Thirdly, because the mutation of 371 amino acid residue is not essential for the catalytic activity site of SPTLC1, the profiles of sphingolipids and deoxysphingolipids were not analyzed in this study. Lastly, this study did not verify the pathological effects of the mutation of SPTLC1 on T2D by animal models. Nevertheless, current literatures suggest that both SPTLC1 deletion and higher deoxysphingolipids (formed by the pathogenic mutation of SPTLC1) induced the cells apoptosis and impaired insulin secretion.
Taken together, the study classified SPTLC1 p.G371R mutation as the likely pathogenic mutation from an adult-onset T2D patients with strong family history by family study, bioinformation analysis and functional study. The study also found the inflammatory cytokines, especially TNF-α, and the increased apoptosis was the potential downstream pathway of p.G371R mutation of the SPTLC1 gene. However, this theory needs more data to support. If so, this will provide a novel perspective to understand the pathology of T2D and associated chronic complications.
| 1. | International Diabetes Federation. IDF Diabetes Atlas, 10th edn. Brussels, Belgium: International Diabetes Federation, 2021. [cited November 7, 2024]. Available from: https://www.diabetesatlas.org. |
| 2. | American Diabetes Association Professional Practice Committee. 2. Diagnosis and Classification of Diabetes: Standards of Care in Diabetes-2024. Diabetes Care. 2024;47:S20-S42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 316] [Cited by in RCA: 1014] [Article Influence: 507.0] [Reference Citation Analysis (2)] |
| 3. | Boehm BO, Kratzer W, Bansal V. Whole-genome sequencing of multiple related individuals with type 2 diabetes reveals an atypical likely pathogenic mutation in the PAX6 gene. Eur J Hum Genet. 2023;31:89-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 4. | Yang Y, Zhou TC, Liu YY, Li X, Wang WX, Irwin DM, Zhang YP. Identification of HNF4A Mutation p.T130I and HNF1A Mutations p.I27L and p.S487N in a Han Chinese Family with Early-Onset Maternally Inherited Type 2 Diabetes. J Diabetes Res. 2016;2016:3582616. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 5. | Fuchsberger C, Flannick J, Teslovich TM, Mahajan A, Agarwala V, Gaulton KJ, Ma C, Fontanillas P, Moutsianas L, McCarthy DJ, Rivas MA, Perry JRB, Sim X, Blackwell TW, Robertson NR, Rayner NW, Cingolani P, Locke AE, Tajes JF, Highland HM, Dupuis J, Chines PS, Lindgren CM, Hartl C, Jackson AU, Chen H, Huyghe JR, van de Bunt M, Pearson RD, Kumar A, Müller-Nurasyid M, Grarup N, Stringham HM, Gamazon ER, Lee J, Chen Y, Scott RA, Below JE, Chen P, Huang J, Go MJ, Stitzel ML, Pasko D, Parker SCJ, Varga TV, Green T, Beer NL, Day-Williams AG, Ferreira T, Fingerlin T, Horikoshi M, Hu C, Huh I, Ikram MK, Kim BJ, Kim Y, Kim YJ, Kwon MS, Lee J, Lee S, Lin KH, Maxwell TJ, Nagai Y, Wang X, Welch RP, Yoon J, Zhang W, Barzilai N, Voight BF, Han BG, Jenkinson CP, Kuulasmaa T, Kuusisto J, Manning A, Ng MCY, Palmer ND, Balkau B, Stančáková A, Abboud HE, Boeing H, Giedraitis V, Prabhakaran D, Gottesman O, Scott J, Carey J, Kwan P, Grant G, Smith JD, Neale BM, Purcell S, Butterworth AS, Howson JMM, Lee HM, Lu Y, Kwak SH, Zhao W, Danesh J, Lam VKL, Park KS, Saleheen D, So WY, Tam CHT, Afzal U, Aguilar D, Arya R, Aung T, Chan E, Navarro C, Cheng CY, Palli D, Correa A, Curran JE, Rybin D, Farook VS, Fowler SP, Freedman BI, Griswold M, Hale DE, Hicks PJ, Khor CC, Kumar S, Lehne B, Thuillier D, Lim WY, Liu J, van der Schouw YT, Loh M, Musani SK, Puppala S, Scott WR, Yengo L, Tan ST, Taylor HA Jr, Thameem F, Wilson G Sr, Wong TY, Njølstad PR, Levy JC, Mangino M, Bonnycastle LL, Schwarzmayr T, Fadista J, Surdulescu GL, Herder C, Groves CJ, Wieland T, Bork-Jensen J, Brandslund I, Christensen C, Koistinen HA, Doney ASF, Kinnunen L, Esko T, Farmer AJ, Hakaste L, Hodgkiss D, Kravic J, Lyssenko V, Hollensted M, Jørgensen ME, Jørgensen T, Ladenvall C, Justesen JM, Käräjämäki A, Kriebel J, Rathmann W, Lannfelt L, Lauritzen T, Narisu N, Linneberg A, Melander O, Milani L, Neville M, Orho-Melander M, Qi L, Qi Q, Roden M, Rolandsson O, Swift A, Rosengren AH, Stirrups K, Wood AR, Mihailov E, Blancher C, Carneiro MO, Maguire J, Poplin R, Shakir K, Fennell T, DePristo M, de Angelis MH, Deloukas P, Gjesing AP, Jun G, Nilsson P, Murphy J, Onofrio R, Thorand B, Hansen T, Meisinger C, Hu FB, Isomaa B, Karpe F, Liang L, Peters A, Huth C, O'Rahilly SP, Palmer CNA, Pedersen O, Rauramaa R, Tuomilehto J, Salomaa V, Watanabe RM, Syvänen AC, Bergman RN, Bharadwaj D, Bottinger EP, Cho YS, Chandak GR, Chan JCN, Chia KS, Daly MJ, Ebrahim SB, Langenberg C, Elliott P, Jablonski KA, Lehman DM, Jia W, Ma RCW, Pollin TI, Sandhu M, Tandon N, Froguel P, Barroso I, Teo YY, Zeggini E, Loos RJF, Small KS, Ried JS, DeFronzo RA, Grallert H, Glaser B, Metspalu A, Wareham NJ, Walker M, Banks E, Gieger C, Ingelsson E, Im HK, Illig T, Franks PW, Buck G, Trakalo J, Buck D, Prokopenko I, Mägi R, Lind L, Farjoun Y, Owen KR, Gloyn AL, Strauch K, Tuomi T, Kooner JS, Lee JY, Park T, Donnelly P, Morris AD, Hattersley AT, Bowden DW, Collins FS, Atzmon G, Chambers JC, Spector TD, Laakso M, Strom TM, Bell GI, Blangero J, Duggirala R, Tai ES, McVean G, Hanis CL, Wilson JG, Seielstad M, Frayling TM, Meigs JB, Cox NJ, Sladek R, Lander ES, Gabriel S, Burtt NP, Mohlke KL, Meitinger T, Groop L, Abecasis G, Florez JC, Scott LJ, Morris AP, Kang HM, Boehnke M, Altshuler D, McCarthy MI. The genetic architecture of type 2 diabetes. Nature. 2016;536:41-47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 809] [Cited by in RCA: 795] [Article Influence: 79.5] [Reference Citation Analysis (0)] |
| 6. | Flannick J, Mercader JM, Fuchsberger C, Udler MS, Mahajan A, Wessel J, Teslovich TM, Caulkins L, Koesterer R, Barajas-Olmos F, Blackwell TW, Boerwinkle E, Brody JA, Centeno-Cruz F, Chen L, Chen S, Contreras-Cubas C, Córdova E, Correa A, Cortes M, DeFronzo RA, Dolan L, Drews KL, Elliott A, Floyd JS, Gabriel S, Garay-Sevilla ME, García-Ortiz H, Gross M, Han S, Heard-Costa NL, Jackson AU, Jørgensen ME, Kang HM, Kelsey M, Kim BJ, Koistinen HA, Kuusisto J, Leader JB, Linneberg A, Liu CT, Liu J, Lyssenko V, Manning AK, Marcketta A, Malacara-Hernandez JM, Martínez-Hernández A, Matsuo K, Mayer-Davis E, Mendoza-Caamal E, Mohlke KL, Morrison AC, Ndungu A, Ng MCY, O'Dushlaine C, Payne AJ, Pihoker C; Broad Genomics Platform, Post WS, Preuss M, Psaty BM, Vasan RS, Rayner NW, Reiner AP, Revilla-Monsalve C, Robertson NR, Santoro N, Schurmann C, So WY, Soberón X, Stringham HM, Strom TM, Tam CHT, Thameem F, Tomlinson B, Torres JM, Tracy RP, van Dam RM, Vujkovic M, Wang S, Welch RP, Witte DR, Wong TY, Atzmon G, Barzilai N, Blangero J, Bonnycastle LL, Bowden DW, Chambers JC, Chan E, Cheng CY, Cho YS, Collins FS, de Vries PS, Duggirala R, Glaser B, Gonzalez C, Gonzalez ME, Groop L, Kooner JS, Kwak SH, Laakso M, Lehman DM, Nilsson P, Spector TD, Tai ES, Tuomi T, Tuomilehto J, Wilson JG, Aguilar-Salinas CA, Bottinger E, Burke B, Carey DJ, Chan JCN, Dupuis J, Frossard P, Heckbert SR, Hwang MY, Kim YJ, Kirchner HL, Lee JY, Lee J, Loos RJF, Ma RCW, Morris AD, O'Donnell CJ, Palmer CNA, Pankow J, Park KS, Rasheed A, Saleheen D, Sim X, Small KS, Teo YY, Haiman C, Hanis CL, Henderson BE, Orozco L, Tusié-Luna T, Dewey FE, Baras A, Gieger C, Meitinger T, Strauch K, Lange L, Grarup N, Hansen T, Pedersen O, Zeitler P, Dabelea D, Abecasis G, Bell GI, Cox NJ, Seielstad M, Sladek R, Meigs JB, Rich SS, Rotter JI; DiscovEHR Collaboration; CHARGE; LuCamp; ProDiGY; GoT2D; ESP; SIGMA-T2D; T2D-GENES; AMP-T2D-GENES, Altshuler D, Burtt NP, Scott LJ, Morris AP, Florez JC, McCarthy MI, Boehnke M. Exome sequencing of 20,791 cases of type 2 diabetes and 24,440 controls. Nature. 2019;570:71-76. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 274] [Cited by in RCA: 229] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 7. | Sirdah MM, Reading NS. Genetic predisposition in type 2 diabetes: A promising approach toward a personalized management of diabetes. Clin Genet. 2020;98:525-547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 8. | Dassie F, Favaretto F, Bettini S, Parolin M, Valenti M, Reschke F, Danne T, Vettor R, Milan G, Maffei P. Alström syndrome: an ultra-rare monogenic disorder as a model for insulin resistance, type 2 diabetes mellitus and obesity. Endocrine. 2021;71:618-625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 9. | Sun Y, Qiang W, Wu R, Yin T, Yuan J, Yuan J, Gu Y. A glycogen storage disease type 1a patient with type 2 diabetes. BMC Med Genomics. 2022;15:205. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 10. | Boso F, Armirotti A, Taioli F, Ferrarini M, Nobbio L, Cavallaro T, Fabrizi GM. Deoxysphingolipids as candidate biomarkers for a novel SPTLC1 mutation associated with HSAN-I. Neurol Genet. 2019;5:e365. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 11. | Gantner ML, Eade K, Wallace M, Handzlik MK, Fallon R, Trombley J, Bonelli R, Giles S, Harkins-Perry S, Heeren TFC, Sauer L, Ideguchi Y, Baldini M, Scheppke L, Dorrell MI, Kitano M, Hart BJ, Cai C, Nagasaki T, Badur MG, Okada M, Woods SM, Egan C, Gillies M, Guymer R, Eichler F, Bahlo M, Fruttiger M, Allikmets R, Bernstein PS, Metallo CM, Friedlander M. Serine and Lipid Metabolism in Macular Disease and Peripheral Neuropathy. N Engl J Med. 2019;381:1422-1433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 178] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 12. | Johnson JO, Chia R, Miller DE, Li R, Kumaran R, Abramzon Y, Alahmady N, Renton AE, Topp SD, Gibbs JR, Cookson MR, Sabir MS, Dalgard CL, Troakes C, Jones AR, Shatunov A, Iacoangeli A, Al Khleifat A, Ticozzi N, Silani V, Gellera C, Blair IP, Dobson-Stone C, Kwok JB, Bonkowski ES, Palvadeau R, Tienari PJ, Morrison KE, Shaw PJ, Al-Chalabi A, Brown RH Jr, Calvo A, Mora G, Al-Saif H, Gotkine M, Leigh F, Chang IJ, Perlman SJ, Glass I, Scott AI, Shaw CE, Basak AN, Landers JE, Chiò A, Crawford TO, Smith BN, Traynor BJ; FALS Sequencing Consortium; American Genome Center; International ALS Genomics Consortium; and ITALSGEN Consortium, Smith BN, Ticozzi N, Fallini C, Gkazi AS, Topp SD, Scotter EL, Kenna KP, Keagle P, Tiloca C, Vance C, Troakes C, Colombrita C, King A, Pensato V, Castellotti B, Baas F, Ten Asbroek ALMA, McKenna-Yasek D, McLaughlin RL, Polak M, Asress S, Esteban-Pérez J, Stevic Z, D'Alfonso S, Mazzini L, Comi GP, Del Bo R, Ceroni M, Gagliardi S, Querin G, Bertolin C, van Rheenen W, Rademakers R, van Blitterswijk M, Lauria G, Duga S, Corti S, Cereda C, Corrado L, Sorarù G, Williams KL, Nicholson GA, Blair IP, Leblond-Manry C, Rouleau GA, Hardiman O, Morrison KE, Veldink JH, van den Berg LH, Al-Chalabi A, Pall H, Shaw PJ, Turner MR, Talbot K, Taroni F, García-Redondo A, Wu Z, Glass JD, Gellera C, Ratti A, Brown RH Jr, Silani V, Shaw CE, Landers JE, Dalgard CL, Adeleye A, Soltis AR, Alba C, Viollet C, Bacikova D, Hupalo DN, Sukumar G, Pollard HB, Wilkerson MD, Martinez EM, Abramzon Y, Ahmed S, Arepalli S, Baloh RH, Bowser R, Brady CB, Brice A, Broach J, Campbell RH, Camu W, Chia R, Cooper-Knock J, Ding J, Drepper C, Drory VE, Dunckley TL, Eicher JD, England BK, Faghri F, Feldman E, Floeter MK, Fratta P, Geiger JT, Gerhard G, Gibbs JR, Gibson SB, Glass JD, Hardy J, Harms MB, Heiman-Patterson TD, Hernandez DG, Jansson L, Kirby J, Kowall NW, Laaksovirta H, Landeck N, Landi F, Le Ber I, Lumbroso S, MacGowan DJL, Maragakis NJ, Mora G, Mouzat K, Murphy NA, Myllykangas L, Nalls MA, Orrell RW, Ostrow LW, Pamphlett R, Pickering-Brown S, Pioro EP, Pletnikova O, Pliner HA, Pulst SM, Ravits JM, Renton AE, Rivera A, Robberecht W, Rogaeva E, Rollinson S, Rothstein JD, Scholz SW, Sendtner M, Shaw PJ, Sidle KC, Simmons Z, Singleton AB, Smith N, Stone DJ, Tienari PJ, Troncoso JC, Valori M, Van Damme P, Van Deerlin VM, Van Den Bosch L, Zinman L, Landers JE, Chiò A, Traynor BJ, Angelocola SM, Ausiello FP, Barberis M, Bartolomei I, Battistini S, Bersano E, Bisogni G, Borghero G, Brunetti M, Cabona C, Calvo A, Canale F, Canosa A, Cantisani TA, Capasso M, Caponnetto C, Cardinali P, Carrera P, Casale F, Chiò A, Colletti T, Conforti FL, Conte A, Conti E, Corbo M, Cuccu S, Dalla Bella E, D'Errico E, DeMarco G, Dubbioso R, Ferrarese C, Ferraro PM, Filippi M, Fini N, Floris G, Fuda G, Gallone S, Gianferrari G, Giannini F, Grassano M, Greco L, Iazzolino B, Introna A, La Bella V, Lattante S, Lauria G, Liguori R, Logroscino G, Logullo FO, Lunetta C, Mandich P, Mandrioli J, Manera U, Manganelli F, Marangi G, Marinou K, Marrosu MG, Martinelli I, Messina S, Moglia C, Mora G, Mosca L, Murru MR, Origone P, Passaniti C, Petrelli C, Petrucci A, Pozzi S, Pugliatti M, Quattrini A, Ricci C, Riolo G, Riva N, Russo M, Sabatelli M, Salamone P, Salivetto M, Salvi F, Santarelli M, Sbaiz L, Sideri R, Simone I, Simonini C, Spataro R, Tanel R, Tedeschi G, Ticca A, Torriello A, Tranquilli S, Tremolizzo L, Trojsi F, Vasta R, Vacchiano V, Vita G, Volanti P, Zollino M, Zucchi E. Association of Variants in the SPTLC1 Gene With Juvenile Amyotrophic Lateral Sclerosis. JAMA Neurol. 2021;78:1236-1248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 65] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 13. | Mohassel P, Donkervoort S, Lone MA, Nalls M, Gable K, Gupta SD, Foley AR, Hu Y, Saute JAM, Moreira AL, Kok F, Introna A, Logroscino G, Grunseich C, Nickolls AR, Pourshafie N, Neuhaus SB, Saade D, Gangfuß A, Kölbel H, Piccus Z, Le Pichon CE, Fiorillo C, Ly CV, Töpf A, Brady L, Specht S, Zidell A, Pedro H, Mittelmann E, Thomas FP, Chao KR, Konersman CG, Cho MT, Brandt T, Straub V, Connolly AM, Schara U, Roos A, Tarnopolsky M, Höke A, Brown RH, Lee CH, Hornemann T, Dunn TM, Bönnemann CG. Childhood amyotrophic lateral sclerosis caused by excess sphingolipid synthesis. Nat Med. 2021;27:1197-1204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 121] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 14. | Xie Y, Lin Z, Liu L, Li X, Huang S, Zhao H, Wang B, Zeng S, Cao W, Li L, Zhu X, Huang S, Yang H, Wang M, Hu Z, Wang J, Guo J, Shen L, Jiang H, Zuchner S, Tang B, Zhang R. Genotype and phenotype distribution of 435 patients with Charcot-Marie-Tooth disease from central south China. Eur J Neurol. 2021;28:3774-3783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 15. | Alexaki A, Clarke BA, Gavrilova O, Ma Y, Zhu H, Ma X, Xu L, Tuymetova G, Larman BC, Allende ML, Dunn TM, Proia RL. De Novo Sphingolipid Biosynthesis Is Required for Adipocyte Survival and Metabolic Homeostasis. J Biol Chem. 2017;292:3929-3939. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 45] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 16. | Fridman V, Zarini S, Sillau S, Harrison K, Bergman BC, Feldman EL, Reusch JEB, Callaghan BC. Altered plasma serine and 1-deoxydihydroceramide profiles are associated with diabetic neuropathy in type 2 diabetes and obesity. J Diabetes Complications. 2021;35:107852. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 17. | Othman A, Bianchi R, Alecu I, Wei Y, Porretta-Serapiglia C, Lombardi R, Chiorazzi A, Meregalli C, Oggioni N, Cavaletti G, Lauria G, von Eckardstein A, Hornemann T. Lowering plasma 1-deoxysphingolipids improves neuropathy in diabetic rats. Diabetes. 2015;64:1035-1045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 74] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 18. | Othman A, Rütti MF, Ernst D, Saely CH, Rein P, Drexel H, Porretta-Serapiglia C, Lauria G, Bianchi R, von Eckardstein A, Hornemann T. Plasma deoxysphingolipids: a novel class of biomarkers for the metabolic syndrome? Diabetologia. 2012;55:421-431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 117] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 19. | Othman A, Saely CH, Muendlein A, Vonbank A, Drexel H, von Eckardstein A, Hornemann T. Plasma 1-deoxysphingolipids are predictive biomarkers for type 2 diabetes mellitus. BMJ Open Diabetes Res Care. 2015;3:e000073. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 64] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 20. | Zuellig RA, Hornemann T, Othman A, Hehl AB, Bode H, Güntert T, Ogunshola OO, Saponara E, Grabliauskaite K, Jang JH, Ungethuem U, Wei Y, von Eckardstein A, Graf R, Sonda S. Deoxysphingolipids, novel biomarkers for type 2 diabetes, are cytotoxic for insulin-producing cells. Diabetes. 2014;63:1326-1339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 102] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 21. | Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405-424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19696] [Cited by in RCA: 24406] [Article Influence: 2218.7] [Reference Citation Analysis (0)] |
| 22. | Dominguez V, Raimondi C, Somanath S, Bugliani M, Loder MK, Edling CE, Divecha N, da Silva-Xavier G, Marselli L, Persaud SJ, Turner MD, Rutter GA, Marchetti P, Falasca M, Maffucci T. Class II phosphoinositide 3-kinase regulates exocytosis of insulin granules in pancreatic beta cells. J Biol Chem. 2011;286:4216-4225. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 126] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 23. | American Diabetes Association Professional Practice Committee. 2. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes-2022. Diabetes Care. 2022;45:S17-S38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 689] [Cited by in RCA: 1596] [Article Influence: 399.0] [Reference Citation Analysis (1)] |
| 24. | DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium; Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium; South Asian Type 2 Diabetes (SAT2D) Consortium; Mexican American Type 2 Diabetes (MAT2D) Consortium; Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in muylti-Ethnic Samples (T2D-GENES) Consortium, Mahajan A, Go MJ, Zhang W, Below JE, Gaulton KJ, Ferreira T, Horikoshi M, Johnson AD, Ng MC, Prokopenko I, Saleheen D, Wang X, Zeggini E, Abecasis GR, Adair LS, Almgren P, Atalay M, Aung T, Baldassarre D, Balkau B, Bao Y, Barnett AH, Barroso I, Basit A, Been LF, Beilby J, Bell GI, Benediktsson R, Bergman RN, Boehm BO, Boerwinkle E, Bonnycastle LL, Burtt N, Cai Q, Campbell H, Carey J, Cauchi S, Caulfield M, Chan JC, Chang LC, Chang TJ, Chang YC, Charpentier G, Chen CH, Chen H, Chen YT, Chia KS, Chidambaram M, Chines PS, Cho NH, Cho YM, Chuang LM, Collins FS, Cornelis MC, Couper DJ, Crenshaw AT, van Dam RM, Danesh J, Das D, de Faire U, Dedoussis G, Deloukas P, Dimas AS, Dina C, Doney AS, Donnelly PJ, Dorkhan M, van Duijn C, Dupuis J, Edkins S, Elliott P, Emilsson V, Erbel R, Eriksson JG, Escobedo J, Esko T, Eury E, Florez JC, Fontanillas P, Forouhi NG, Forsen T, Fox C, Fraser RM, Frayling TM, Froguel P, Frossard P, Gao Y, Gertow K, Gieger C, Gigante B, Grallert H, Grant GB, Grrop LC, Groves CJ, Grundberg E, Guiducci C, Hamsten A, Han BG, Hara K, Hassanali N, Hattersley AT, Hayward C, Hedman AK, Herder C, Hofman A, Holmen OL, Hovingh K, Hreidarsson AB, Hu C, Hu FB, Hui J, Humphries SE, Hunt SE, Hunter DJ, Hveem K, Hydrie ZI, Ikegami H, Illig T, Ingelsson E, Islam M, Isomaa B, Jackson AU, Jafar T, James A, Jia W, Jöckel KH, Jonsson A, Jowett JB, Kadowaki T, Kang HM, Kanoni S, Kao WH, Kathiresan S, Kato N, Katulanda P, Keinanen-Kiukaanniemi KM, Kelly AM, Khan H, Khaw KT, Khor CC, Kim HL, Kim S, Kim YJ, Kinnunen L, Klopp N, Kong A, Korpi-Hyövälti E, Kowlessur S, Kraft P, Kravic J, Kristensen MM, Krithika S, Kumar A, Kumate J, Kuusisto J, Kwak SH, Laakso M, Lagou V, Lakka TA, Langenberg C, Langford C, Lawrence R, Leander K, Lee JM, Lee NR, Li M, Li X, Li Y, Liang J, Liju S, Lim WY, Lind L, Lindgren CM, Lindholm E, Liu CT, Liu JJ, Lobbens S, Long J, Loos RJ, Lu W, Luan J, Lyssenko V, Ma RC, Maeda S, Mägi R, Männisto S, Matthews DR, Meigs JB, Melander O, Metspalu A, Meyer J, Mirza G, Mihailov E, Moebus S, Mohan V, Mohlke KL, Morris AD, Mühleisen TW, Müller-Nurasyid M, Musk B, Nakamura J, Nakashima E, Navarro P, Ng PK, Nica AC, Nilsson PM, Njølstad I, Nöthen MM, Ohnaka K, Ong TH, Owen KR, Palmer CN, Pankow JS, Park KS, Parkin M, Pechlivanis S, Pedersen NL, Peltonen L, Perry JR, Peters A, Pinidiyapathirage JM, Platou CG, Potter S, Price JF, Qi L, Radha V, Rallidis L, Rasheed A, Rathman W, Rauramaa R, Raychaudhuri S, Rayner NW, Rees SD, Rehnberg E, Ripatti S, Robertson N, Roden M, Rossin EJ, Rudan I, Rybin D, Saaristo TE, Salomaa V, Saltevo J, Samuel M, Sanghera DK, Saramies J, Scott J, Scott LJ, Scott RA, Segrè AV, Sehmi J, Sennblad B, Shah N, Shah S, Shera AS, Shu XO, Shuldiner AR, Sigurđsson G, Sijbrands E, Silveira A, Sim X, Sivapalaratnam S, Small KS, So WY, Stančáková A, Stefansson K, Steinbach G, Steinthorsdottir V, Stirrups K, Strawbridge RJ, Stringham HM, Sun Q, Suo C, Syvänen AC, Takayanagi R, Takeuchi F, Tay WT, Teslovich TM, Thorand B, Thorleifsson G, Thorsteinsdottir U, Tikkanen E, Trakalo J, Tremoli E, Trip MD, Tsai FJ, Tuomi T, Tuomilehto J, Uitterlinden AG, Valladares-Salgado A, Vedantam S, Veglia F, Voight BF, Wang C, Wareham NJ, Wennauer R, Wickremasinghe AR, Wilsgaard T, Wilson JF, Wiltshire S, Winckler W, Wong TY, Wood AR, Wu JY, Wu Y, Yamamoto K, Yamauchi T, Yang M, Yengo L, Yokota M, Young R, Zabaneh D, Zhang F, Zhang R, Zheng W, Zimmet PZ, Altshuler D, Bowden DW, Cho YS, Cox NJ, Cruz M, Hanis CL, Kooner J, Lee JY, Seielstad M, Teo YY, Boehnke M, Parra EJ, Chambers JC, Tai ES, McCarthy MI, Morris AP. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46:234-244. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 899] [Cited by in RCA: 823] [Article Influence: 68.6] [Reference Citation Analysis (0)] |
| 25. | Ding Y, Li N, Lou D, Zhang Q, Chang G, Li J, Li X, Li Q, Huang X, Wang J, Jiang F, Wang X. Clinical and genetic analysis in a Chinese cohort of children and adolescents with diabetes/persistent hyperglycemia. J Diabetes Investig. 2021;12:48-62. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 26. | Pottinger TD, Puckelwartz MJ, Pesce LL, Robinson A, Kearns S, Pacheco JA, Rasmussen-Torvik LJ, Smith ME, Chisholm R, McNally EM. Pathogenic and Uncertain Genetic Variants Have Clinical Cardiac Correlates in Diverse Biobank Participants. J Am Heart Assoc. 2020;9:e013808. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 27. | Parthibane V, Acharya D, Srideshikan SM, Lin J, Myerscough DG, Abimannan T, Vijaykrishna N, Blankenberg D, Bondada L, Klarmann KD, Fox SD, Andresson T, Tessarollo L, Acharya U, Keller JR, Acharya JK. Sptlc1 is essential for myeloid differentiation and hematopoietic homeostasis. Blood Adv. 2019;3:3635-3649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 28. | Taouji S, Higa A, Delom F, Palcy S, Mahon FX, Pasquet JM, Bossé R, Ségui B, Chevet E. Phosphorylation of serine palmitoyltransferase long chain-1 (SPTLC1) on tyrosine 164 inhibits its activity and promotes cell survival. J Biol Chem. 2013;288:17190-17201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 29. | Ikegami H, Babaya N, Noso S. β-Cell failure in diabetes: Common susceptibility and mechanisms shared between type 1 and type 2 diabetes. J Diabetes Investig. 2021;12:1526-1539. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 30. | Dohrn MF, Othman A, Hirshman SK, Bode H, Alecu I, Fähndrich E, Karges W, Weis J, Schulz JB, Hornemann T, Claeys KG. Elevation of plasma 1-deoxy-sphingolipids in type 2 diabetes mellitus: a susceptibility to neuropathy? Eur J Neurol. 2015;22:806-814, e55. [PubMed] [DOI] [Full Text] |
| 31. | Hammad SM, Baker NL, El Abiad JM, Spassieva SD, Pierce JS, Rembiesa B, Bielawski J, Lopes-Virella MF, Klein RL; DCCT/EDIC Group of Investigators. Increased Plasma Levels of Select Deoxy-ceramide and Ceramide Species are Associated with Increased Odds of Diabetic Neuropathy in Type 1 Diabetes: A Pilot Study. Neuromolecular Med. 2017;19:46-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 35] [Article Influence: 3.5] [Reference Citation Analysis (0)] |