Published online Oct 15, 2025. doi: 10.4239/wjd.v16.i10.109568
Revised: June 3, 2025
Accepted: August 15, 2025
Published online: October 15, 2025
Processing time: 153 Days and 20 Hours
Diabetic kidney disease (DKD) stands as the key contributor to chronic kidney disease worldwide. Clinical studies have shown that Kunkui Baoshen decoction (KKBS) effectively reduces proteinuria and enhances renal function in DKD pa
To evaluate the nephroprotective efficacy of KKBS in DKD and explore the un
Liquid chromatography-tandem mass spectrometry was utilized to analyze the chemical constituents of KKBS. Metabonomic and transcriptomic analyses were conducted to identify key targets and pathways associated with the therapeutic effects of KKBS on DKD. The nephroprotective effects of KKBS were assessed both in high glucose-induced human kidney-2 cells and in db/db mice. A variety of assays were performed, including Cell Counting Kit-8, Western blot, quan
The glutathione metabolic pathway emerged as the most prominent metabolic pathway in the metabonomic analysis of KKBS. Transcriptomic and bioinformatic analyses revealed that nuclear receptor coactivator 4 (NCOA4) was instrumental in regulating ferroptosis within renal tubules of mice with DKD. Both in vitro and in vivo experiments showed that KKBS ameliorated renal dysfunction, mitigated renal tissue damage, and repressed the expression of autophagy-dependent fer
KKBS confers nephroprotection in DKD by modulating HERC2/NCOA4-me
Core Tip: Kunkui Baoshen decoction (KKBS), a traditional Chinese herbal formula, exerts renoprotective effects in diabetic kidney disease (DKD) by inhibiting autophagy-dependent ferroptosis. Through integrated transcriptomic and metabolomic analyses, we identified the homologous to E6-AP C-terminus and RCC1-like domain-containing E3 ubiquitin protein ligase/nuclear receptor coactivator 4 axis as a critical therapeutic target. KKBS enhances homologous to E6-AP C-terminus and RCC1-like domain-containing E3 ubiquitin protein ligase-mediated K48-linked ubiquitination of nuclear receptor coactivator 4, promoting its degradation and thereby suppressing ferroptosis, renal inflammation, and fibrosis in high glucose-induced human kidney-2 cells and db/db mice. This research reveals a novel pathway through which KKBS regulates ferroptosis to prevent DKD progression.
- Citation: Song SY, Shan CC, Zhou PP, Xu WL, Tan Y, Zhou XQ, Huang LJ, Yan QH, Yu JY. Nephroprotective mechanism of Kunkui Baoshen decoction in diabetic kidney disease: Targeting the HERC2/NCOA4-mediated autophagy-dependent ferroptosis pathway. World J Diabetes 2025; 16(10): 109568
- URL: https://www.wjgnet.com/1948-9358/full/v16/i10/109568.htm
- DOI: https://dx.doi.org/10.4239/wjd.v16.i10.109568
Diabetic kidney disease (DKD) continues to be the predominant driver of chronic kidney disease, accounting for nearly 50% of chronic kidney disease cases globally[1]. Although therapies such as sodium-glucose cotransporter-2 inhibitors, renin-angio
According to 2023 data from the China National Renal Data System, DKD now accounts for 29.9% of newly initiated hemodialysis cases in China, surpassing primary glomerular diseases (28.7%) for the first time[4]. These statistics highlight an urgent need to enhance DKD prevention and treatment. In this context, it is critical to provide deeper insight into the underlying molecular mechanisms of DKD and to develop innovative therapeutic strategies that can effectively manage its progression.
Traditional Chinese medicine (TCM) is broadly utilized for DKD treatment in China. KKBS, formulated by Song and Yu[5], two renowned TCM practitioners from Jiangsu Province, is a clinically established herbal prescription composed of Astragali Radix, Abelmoschi Corolla, Tripterygium hypoglaucum root, and Corni Fructus Preparata. The formula is derived from Jisheng Shenqi Wan, itself rooted in Jisheng Fang, a prescription widely used during the Southern Song Dynasty. Traditionally, KKBS has been used to warm the kidney, regulate qi, promote diuresis, and alleviate edema. Its clinical indications include kidney deficiency syndromes characterized by edema and soreness in the lower back and knees.
Modern pharmacological research has validated the multifaceted therapeutic potential of KKBS. Abelmoschi Corolla alleviates DKD-related proteinuria and renal injury by reducing tubular damage and interstitial fibrosis, oxidative stress, inflammatory responses, and podocyte apoptosis[6]. Tripterygium hypoglaucum root improves renal function by protecting glomerular endothelial cells, suppressing inflammatory factor expression, enhancing immunosuppression, and reducing proteinuria and hematuria[7]. Astragali Radix has been demonstrated to lower blood glucose levels, reduce proteinuria, and exert nephroprotective effects in DKD animal models[8]. The main active compound in Corni Fructus Preparata, loganin, mitigates inflammation and organelle damage in DKD cell models by inhibiting advanced glycation end product-triggered oxidative stress, suppressing mesangial cell proliferation, and alleviating endoplasmic reticulum stress[9]. Clinical research has additionally demonstrated that KKBS effectively reduces proteinuria and maintains kidney function in patients with DKD[5]. However, the specific molecular targets and mechanisms through which KKBS exerts its therapeutic effects remain uncertain.
Ferroptosis is a controlled type of cell death reliant on iron, marked by the buildup of reactive oxygen species (ROS) and the polyunsaturated fatty acids’ depletion in cellular membranes[10]. This process is mediated by complex molecular pathways, including those related to iron[11], lipid[12], and amino acid metabolism[13], along with signaling pathways associated with coenzyme Q[14], p53[15], inositol trisphosphate 3-kinase[16], and mitochondrial voltage-dependent anion channels[17]. In terms of morphology, ferroptosis is marked by mitochondrial shrinkage or swelling, raised membrane density, and a loss of cristae, distinctive features differentiating it from other cell death types[18]. An increasing volume of evidence implicates ferroptosis in the pathogenesis of DKD[19,20], suggesting that targeting this process may reduce renal fibrosis and slow disease progression.
To examine the anti-fibrotic mechanism of KKBS in DKD, we conducted metabolomic analysis to identify its relevant metabolic pathways. Glutathione (GSH) metabolism emerged as the most significantly altered pathway following KKBS treatment. Given the pivotal role of GSH homeostasis in regulating ferroptosis, these results suggest that KKBS may exert its nephroprotective impact by modulating ferroptosis. Building on this hypothesis, both in vitro and in vivo experiments were performed utilizing high glucose (HG)-induced human kidney-2 (HK-2) cells and db/db mice to investigate the impact of KKBS on ferroptosis in DKD. Our findings may offer a novel therapeutic approach for alleviating renal injury in DKD.
The four Chinese herbal medicine used in the KKBS (Table 1) were sourced from Jiangsu Province Hospital of Chinese Medicine, Nanjing, China.
| No. | Chinese name | Scientific name | Weight |
| 1 | Huangqi | Astmgali Radix | 30 g |
| 2 | Huobahuagen | Tripterygium hypoglaucum | 15 g |
| 3 | Jiuyurou | Fructus Ligustri Lucidi | 10 g |
| 4 | Huangshukuihua | Corni Fructus Preparata | 30 g |
Preparation and liquid chromatography-tandem mass spectrometry analysis of KKBS: Following the original for
Metabonomics analysis: To clarify the metabolic pathways influenced by KKBS in DKD, differential metabolites were analyzed in plasma samples from untreated db/db mice compared to those treated with KKBS. The raw LC-MS data underwent processing with Progenesis QI software (Waters Corporation, Milford, MA, United States), which included baseline correction, peak detection, data alignment, and retention time calibration. This yielded a data matrix consisting of retention time, mass-to-charge ratio, and intensity values. Metabolite identification was conducted by spectral ma
The remaining data were transformed using log10 conversion to stabilize variance and normalize the distribution for downstream analyses. Multivariate statistical analysis was conducted utilizing the ropls package (version 1.6.2), where principal component analysis and orthogonal projections to latent structures-discriminant analysis models were assessed through 7-fold cross-validation. Metabolite significance was assessed based on variable importance in projection scores obtained from the orthogonal projections to latent structures-discriminant analysis model as follows:
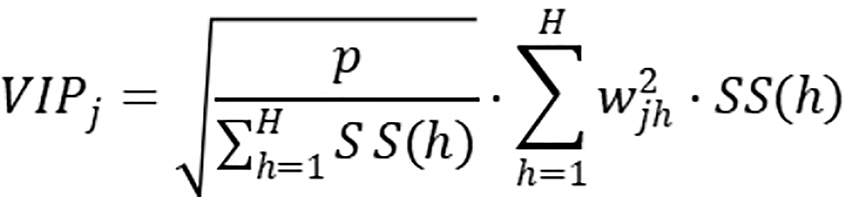
where P represents the total number of metabolites, H denotes the number of principal components included in the model, Wjh refers to the weight of the jth metabolite in the hth principal component, and SS(h) indicates the sum of squares of the hth principal component.
A two-tailed hypothesis test was applied, and the false discovery rate correction was used to adjust for multiple comparisons. Statistical significance was set utilizing Student’s t-test (P < 0.05) in combination with variable importance in projection scores > 1. Pathway enrichment analysis was performed utilizing the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (https://www.kegg.jp/kegg/pathway.html)[26], and pathway relevance was evaluated using Fisher’s exact test, implemented via the scipy.stats module in Python.
Molecular docking: Compound structures were retrieved from PubChem (https://pubchem.ncbi.nlm.nih.gov/), and corresponding protein targets were obtained from the PDB database (https://www.rcsb.org/)[27]. Protein preparation was performed using AutoDock 4.2.6, which included the elimination of water molecules, incorporation of hydrogen atoms, and allocation of partial charges, followed by conversion of the structures to the PDBQT format. Molecular docking simulations were performed with AutoDock Vina version 1.1.2[28], and the docking outcomes were visualized with PyMOL 2.3.0.
RNA isolation: Total RNA was extracted from renal tissues of the db/db group and the KKBS treatment group (n = 3 per group; detailed grouping information is available in section 2.3.9.1) using TRIzol® reagent (Magen, Guangzhou, China). RNA purity and integrity were assessed utilizing a Nanodrop ND-2000 (Thermo Scientific, MA, United States) for A260/280 ratios and the Agilent 4150 Bioanalyzer (RIN evaluation). Only samples meeting quality criteria proceeded to library construction.
Library preparation and sequencing: Paired-end libraries were constructed following standard protocols using the mRNA-seq Library Prep Kit (ABclonal, Wuhan, China). Shortly, 1 μg of total RNA was subjected to poly(A)+ RNA enrichment utilizing oligo(dT) magnetic beads. Moreover, fragmentation was performed with ABclonal’s proprietary cation-containing buffer under elevated temperatures. First-strand complementary DNA was produced utilizing random hexamer primers and an RNase H-active reverse transcriptase, which was then followed by second-strand synthesis employing DNA polymerase I and RNase H. Sequencing adapters were attached to the resulting double-stranded complementary DNA fragments. Indexed libraries were amplified and size-selected using AMPure XP beads. Library quality was validated via electrophoretic analysis on the Agilent 4150 system before sequencing on either the Illumina NovaSeq 6000 or the MGI MGISEQ-T7 platform.
Data analysis: Raw sequencing data were analyzed utilizing the DESeq2 package (http://bioconductor.org/packages/release/bioc/html/DESeq2.html)[29] to identify differentially expressed genes (DEGs). Genes with |log2 fold change| > 1 and P < 0.05 were deemed statistically significant. Moreover, DEGs’ functional enrichment analysis was performed using the KEGG database. Gene Ontology and KEGG pathway enrichment analyses were carried out utilizing the clusterProfiler package in R, with Benjamini-Hochberg adjusted P value < 0.05 supposed statistically significant[30].
For integrative pathway analysis, we utilized the “Joint Pathway Analysis” module in MetaboAnalyst 5.0, which combines transcriptomic and metabolomic data by mapping DEGs and metabolites onto KEGG pathways. Enrichment was determined using a hypergeometric test followed by false discovery rate correction, with pathways meeting an adjusted P value < 0.05 considered significantly enriched.
Culturing conditions: HK-2 cells were obtained from Wuhan Procell Life Science and Technology Co., Ltd. (CL-0109, Wuhan, China) in March 2024. Cells were cultured in low-glucose Dulbecco’s modified Eagle medium/F12 medium enriched with 10% foetal bovine serum (PM150312B, Procell, Wuhan, China) at 37 °C in a humidified atmosphere comprising 5% CO2. The experimental groups and treatment conditions were as follows: Normal glucose group: 5.5 mmol/L glucose; HG group: 50 mmol/L glucose to simulate diabetic conditions; mannitol group: 50 mmol/L mannitol to control for osmotic effects; HG + KKBS group: 50 mmol/L glucose with 14% KKBS; HG + ferrostatin-1 (Fer-1) group: 50 mmol/L glucose with 10 μM Fer-1; HG + Fer-1 + KKBS group: 50 mmol/L glucose with 10 μM Fer-1 and 14% KKBS. When HK-2 cells reached approximately 95% confluence, treatments were applied and maintained for 24 hours.
Research resource identifier: Authentication of the HK-2 cell line was conducted via short tandem repeat profiling, yielding a match score exceeding 90% compared to the American Type Culture Collection reference profile. This confirmed the identity of the cell line and ruled out misidentification or cross-contamination. Short tandem repeat profiling was conducted by Wuhan Procell Life Science and Technology Co., Ltd. Additionally, all cell cultures were routinely screened for mycoplasma contamination prior to experimentation utilizing the MycoAlert™ Mycoplasma Detection Kit (Lonza, MD, United States). All tests returned negative results, confirming that the cell lines used in the experiments were free from mycoplasma contamination.
Cell proliferation analysis: Cellular proliferative capability was evaluated utilizing the Cell Counting Kit-8 (CCK-8) colorimetric assay kit (C0037, Beyotime Biotechnology, Shanghai, China). After 24 hours of KKBS treatment, cells were treated with 10% CCK-8 reagent and incubated at 37 °C for 2 to 4 hours. Following incubation, optical density was estimated at 450 nm utilizing a microplate spectrophotometer.
Fe2+/Fe3+, malondialdehyde, ROS, and reduced/oxidized GSH analysis: Intracellular and tissue levels of Fe2+/Fe3+ and malondialdehyde (MDA; A039-2-1 and A003-1-2, respectively; Nanjing Jiancheng Bioengineering Institute, Nanjing, China) and reduced/oxidized GSH (GSH/GSSG) and ROS (S0053 and S0033S, respectively; Beyotime Biotechnology, Shanghai, China) were quantified according to the manufacturers’ protocols.
Scratch assay: Cells were plated in 6-well plates and maintained in culture until approximately 95% confluence. Additionally, a straight-line scratch was made utilizing a 200 μL pipette tip. Furthermore, the wells were rinsed with phosphate buffered saline (C0221A, Beyotime Biotechnology, Shanghai, China) to eliminate detached cells. Complete medium (2 mL per well) was added, and images of the initial wound (0 hour) were captured using an inverted mi
Quantitative reverse transcription-polymerase chain reaction: Total RNA was isolated using the R0017M reagent system (Beyotime Biotechnology, Shanghai, China). Reverse transcription was performed utilizing the R323 kit (Vazyme, Nanjing, China) per the manufacturer’s directions. Primer sequences are listed in Supplementary Table 1.
Western blot and co-immunoprecipitation: Proteins were isolated from both cells and tissues utilizing radio-immunoprecipitation assay lysis buffer (WB3100, New Cell and Molecular Biotech, Suzhou, China). Protein concentrations were measured via the bicinchoninic acid assay (WB6501, same supplier). The samples were resolved by sodium-dodecyl sulfate gel electrophoresis and subsequently transferred onto polyvinylidene fluoride membranes (IPFL00010, Merck Millipore, MA, United States). The membranes were blocked for 30 minutes at room temperature using rapid blocking buffer (P30500), followed by overnight incubation at 4 °C with primary antibodies. After three (10 minutes duration) washes with tris-buffered saline with Tween, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (WB20500) at room temperature for 1 hour. Protein bands were detected using enhanced chemiluminescence substrate (P10300), and signal intensities were quantified employing ImageJ software (National Institutes of Health, MD, United States).
The following primary antibodies were used. Most were sourced from Proteintech, China: Cysteine-aspartic acid protease-3 (Caspase-3) (1:1000, 19677-1-AP), Bcl-2 associated X protein (Bax) (1:10000, 50599-2-Ig), tumor necrosis factor-α (TNF-α) (1:1000, 17590-1-AP), glyceraldehyde-3-phosphate dehydrogenase (1:10000, 10494-1-AP), interleukin-6 (IL-6) (1:1000, 21865-1-AP), epithelial-cadherin (E-cadherin) (1:20000, 20874-1-AP), transforming growth factor-β1 (TGF-β1) (1:10000, 81746-2-RR), α-smooth muscle actin (α-SMA) (1:10000, 14395-1-AP), GSH peroxidase 4 (GPX4) (1:1000, 30388-1-AP), immunoglobulin G (1:1000, 11541-1-AP), and ubiquitin (1:2000, 10201-2-AP). Additional antibodies were obtained from Affinity, Shanghai, China: Solute carrier family 7 member 11 (SLC7A11) (1:1000, DF12509), autophagy related 7 (ATG7) (1:1000, DF6130), sequestosome 1 (SQSTM1) (1:500, AF5384), and microtubule-associated protein 1A/1B-light chain 3 (LC3) (1:1000, AF5402). Moreover, homologous to E6-AP C-terminus and RCC1-like domain-containing E3 ubiquitin protein ligase (HERC2) antibody (1:500, CY-15460R) was sourced from Shanghai Caiyou Industry Co., Ltd., Shanghai, China; ferritin heavy chain 1 (FTH1) antibody (1:500, sc-376594) was acquired from Santa Cruz Biotechnology, CA, United States; and nuclear receptor coactivator 4 (NCOA4) antibodies were procured from Beijing Novus, Beijing, China (1:1000, ab314553, Abcam, MA, United States; 1:1000, H00008031-M04G).
HK-2 cells underwent lysis on ice for 30 minutes employing weak radio-immunoprecipitation assay lysis buffer (P0013D, Beyotime, Shanghai, China). The lysates subsequently were centrifuged for 20 minutes at 4 °C, and the supernatants were collected. These were then incubated for entire night at 4 °C with gentle shaking in the presence of the appropriate primary antibody. Co-immunoprecipitation (Co-IP) was performed according to the Protein A-Agarose kit protocol (P2051, Beyotime, Shanghai, China). Immunoprecipitates were resuspended in 1 × sodium-dodecyl sulfate gel electrophoresis loading buffer (P0015A, Beyotime, Shanghai, China), and protein expression was analyzed by Western blot per standard procedures.
Cell transfection: Plasmids for HERC2 overexpression, NCOA4 knockdown, and their respective negative controls were designed and produced by Sangon Biotech (Shanghai, China). HK-2 cells were transfected per the manufacturer’s protocols (Sangon Biotech, Shanghai, China). Transfection efficiency exceeding 70% was confirmed by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) and immunoblotting, and successfully transfected cells were used for subsequent experiments. After 24 hours of transfection, cells were treated with varying concentrations of KKBS.
Immunofluorescence: Cells were treated with 4% paraformaldehyde (P0099-3 L, Beyotime, Shanghai, China) for fixation and subsequently permeabilized with 0.1% Triton X-100 (ST795, Beyotime, Shanghai, China) for 30 minutes at room temperature. Antigen blocking was performed by incubating cells with fluorescence-blocking solution (P0260, Beyotime, Shanghai, China) for 30 minutes. Moreover, primary antibodies were employed overnight at 4 °C under a humidified environment. After washing, fluorescein isothiocyanate-conjugated (P0196, Beyotime, Shanghai, China) and Cy3-labeled (P0183, Beyotime, Shanghai, China) secondary antibodies were incubated for 1 hour in the dark at 25 °C. Nuclei were counterstained using DAPI (C1005, Beyotime, Shanghai, China) for 10 minutes protected from light, followed by three washes with phosphate buffered saline. Additionally, fluorescent images were captured using a Nikon DS-Fi2 inverted epifluorescence microscope equipped with multiband filter sets.
Animal experimentation: The db/db mouse model, carrying a leptin receptor gene mutation causing obesity and type 2 diabetes mellitus, is widely used to study DKD. These mice develop obesity, hyperglycemia, hyperlipidemia, and glucosuria around 4 weeks, with DKD pathology emerging by about 8 weeks[31], closely mirroring human disease progression. Male db/db and wild-type db/m control mice (8 weeks old) were obtained from Jiangsu Ailingfei Biotechnology Co., Ltd, Jiangsu, China. All animals were housed under specific pathogen-free conditions at 22 ± 2 °C, 35% ± 5% humidity, and 12-hour light/dark cycles, with free access to standard feed and autoclaved water. For this research, all procedures were approved by the Institutional Animal Care Committee of Affiliated Hospital of Nanjing University of Chinese Medicine (No. 2023DW-039-02) and conducted per international animal welfare guidelines.
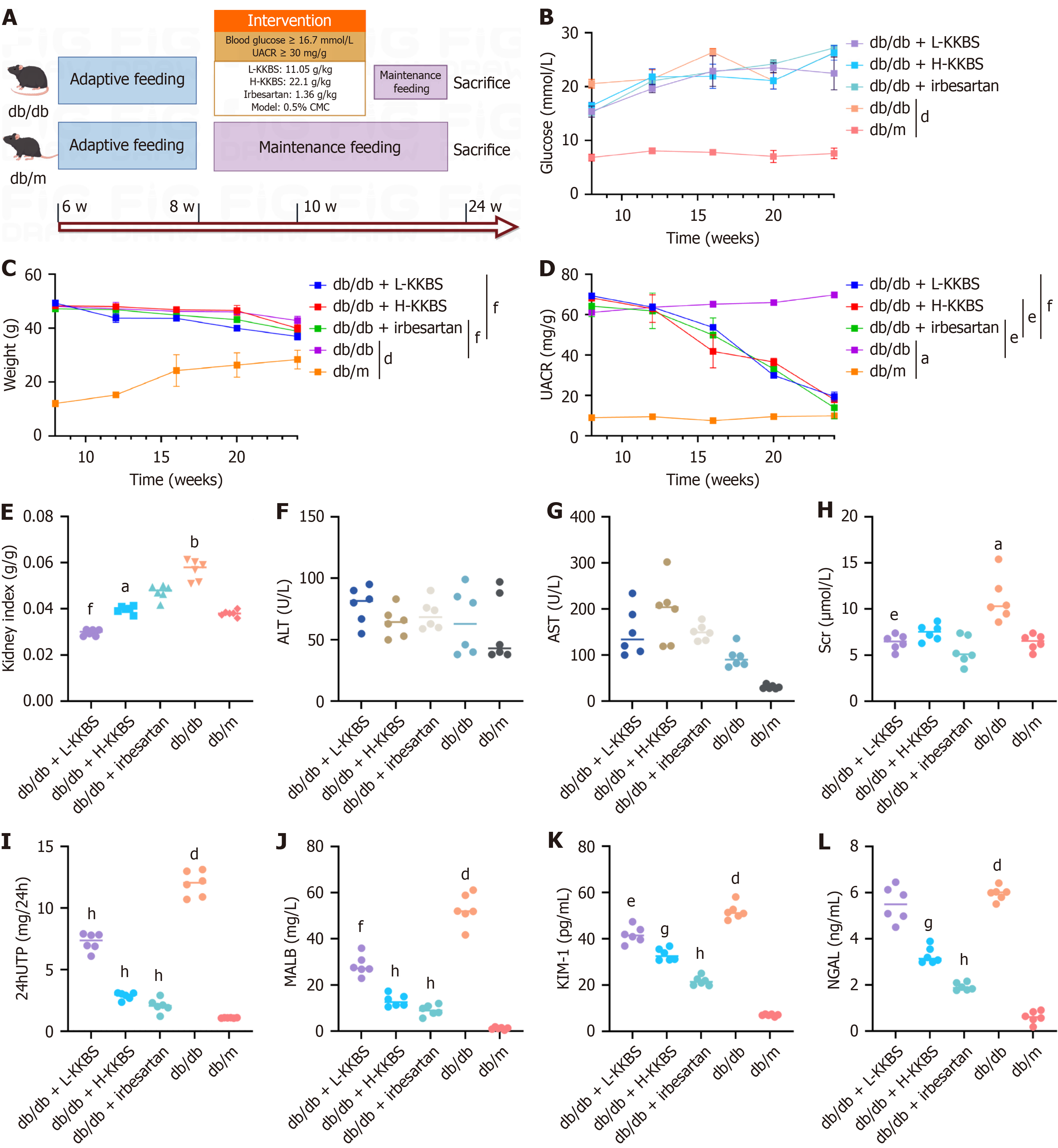
Animal grouping and model establishment: The db/m mice served as non-diabetic controls. The db/db mice were divided into four experimental groups (n = 8 per group): (1) Vehicle-treated model group (0.5% carboxymethylcellulose); (2) Low-dose KKBS (L-KKBS) intervention (11.05 g/kg body weight, oral gavage); (3) High-dose KKBS (H-KKBS) intervention (22.1 g/kg, oral gavage); and (4) Positive control group (irbesartan, 1.36 g/kg, daily gavage). After a 7-day acclimation period, fasting blood glucose levels were determined employing precision glucometry. Simultaneously, 24-hour urine samples were harvested in metabolic cages. DKD was confirmed by meeting both diagnostic criteria: Fasting blood glucose ≥ 16.7 mmol/L and urinary albumin-to-creatinine ratio (UACR) ≥ 30 mg/g, consistent with established guidelines[32].
Histopathological examination of renal tissue: Kidney tissues were paraffin-embedded, sectioned into 4 μm slices, and deparaffinized with xylene. After rehydration, sections were subjected to various histological stains: (1) Hematoxylin and eosin (HE) for cellular structure; (2) Periodic acid-Schiff (PAS) for glycogen detection; (3) Oil Red O for lipid droplet visualization; (4) Terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate-nick end labelling assay to assess apoptosis; and (5) Masson’s trichrome for collagen deposition. All staining procedures followed manufacturer instructions. Sections were scrutinized under the bright-field microscope (Nikon Eclipse Ci-L, Tokyo, Japan), and representative images were captured using NIS-Elements imaging software. Additionally, 1 mm3 kidney samples from each group were collected for ultrastructural analysis of mitochondrial morphology via transmission electron microscopy (TEM).
Immunohistochemical staining: Paraffin-embedded tissue sections, processed as for histological staining, underwent antigen retrieval in citrate buffer (pH = 6.0). Endogenous peroxidase activity was inhibited using 3% H2O2, then nonspecific binding sites were blocked with 5% bovine serum albumin at 37 °C for 1 hour. Tissue sections were immersed overnight at 4 °C in a solution of primary antibodies diluted in the suitable buffer. After three washes with tris-buffered saline with Tween, horseradish peroxidase-conjugated secondary antibodies were employed for 30 minutes at room temperature. Chromogenic detection was conducted utilizing diaminobenzidine substrate (DAB0031, Maixin Bio, Fuzhou, China), with reaction time optimized under microscopic observation. Nuclei were counterstained with Mayer’s hematoxylin (G1080, Servicebio, Wuhan, China) for 30 seconds before dehydration through graded ethanol and clearing in xylene. Whole-slide images were acquired with an Olympus VS200 slide scanner using a 40 × objective (0.75 NA) under standardized settings.
Results are shown as the mean ± SD. Statistical analyses were conducted using GraphPad Prism 9.0 (GraphPad Software, San Diego, CA, United States). For comparisons of multiple groups, one-way ANOVA was utilized, followed by Tukey’s post hoc test. Before analysis, data were checked for normality utilizing the Shapiro-Wilk test and for equal variances by the Levene’s test. When these assumptions were not met, non-parametric methods, Kruskal-Wallis test with Dunn’s multiple comparisons, were used. A P value < 0.05 was deemed statistically significant. Missing data were not subjected to imputation. Samples with missing values due to experimental errors were excluded on a case-by-case basis; final sample sizes (n) are reported in figure legends and results. All experiments were independently conducted a minimum of three times to guarantee reproducibility.
To evaluate the renal protective efficacy of KKBS, we used db/db mice as a model of DKD. After 12 weeks of KKBS treatment (Figure 1A), db/db mice had higher body weight, blood glucose levels, and UACR compared to db/m mice (P < 0.05). Both irbesartan and KKBS significantly reduced body weight and UACR in db/db mice, whereas blood glucose levels showed a minimal change (Figure 1B-D). Regarding renal function, the db/db mice displayed a significantly elevated kidney index compared to db/m controls (P < 0.01). KKBS treatment notably decreased the kidney index (P < 0.05), while irbesartan did not cause a statistically significant change (Figure 1E). Similar levels of hepatic transaminases (alanine aminotransferase and aspartate aminotransferase) across all groups indicated the in vivo safety of KKBS treatment (Figure 1F and G). L-KKBS significantly reduced serum creatinine levels (P < 0.05), whereas H-KKBS had no significant effect (Figure 1H). However, both H-KKBS and L-KKBS markedly decreased 24-hour urine total protein and urinary malondialdehyde levels (P < 0.01; Figure 1I and J), suggesting improved renal function in treated db/db mice.
Neutrophil gelatinase-associated lipocalin and kidney injury molecule-1 are established biomarkers of tubular damage[33]. Both markers were markedly increased in the db/db group relative to db/m controls (P < 0.0001). Treatment with KKBS significantly reduced their expression, with a pronounced decrease in kidney injury molecule-1 observed in the L-KKBS group (P < 0.05), while neutrophil gelatinase-associated lipocalin reduction was less marked (Figure 1K and L). These findings support the conclusion that KKBS ameliorates renal function in db/db mice.
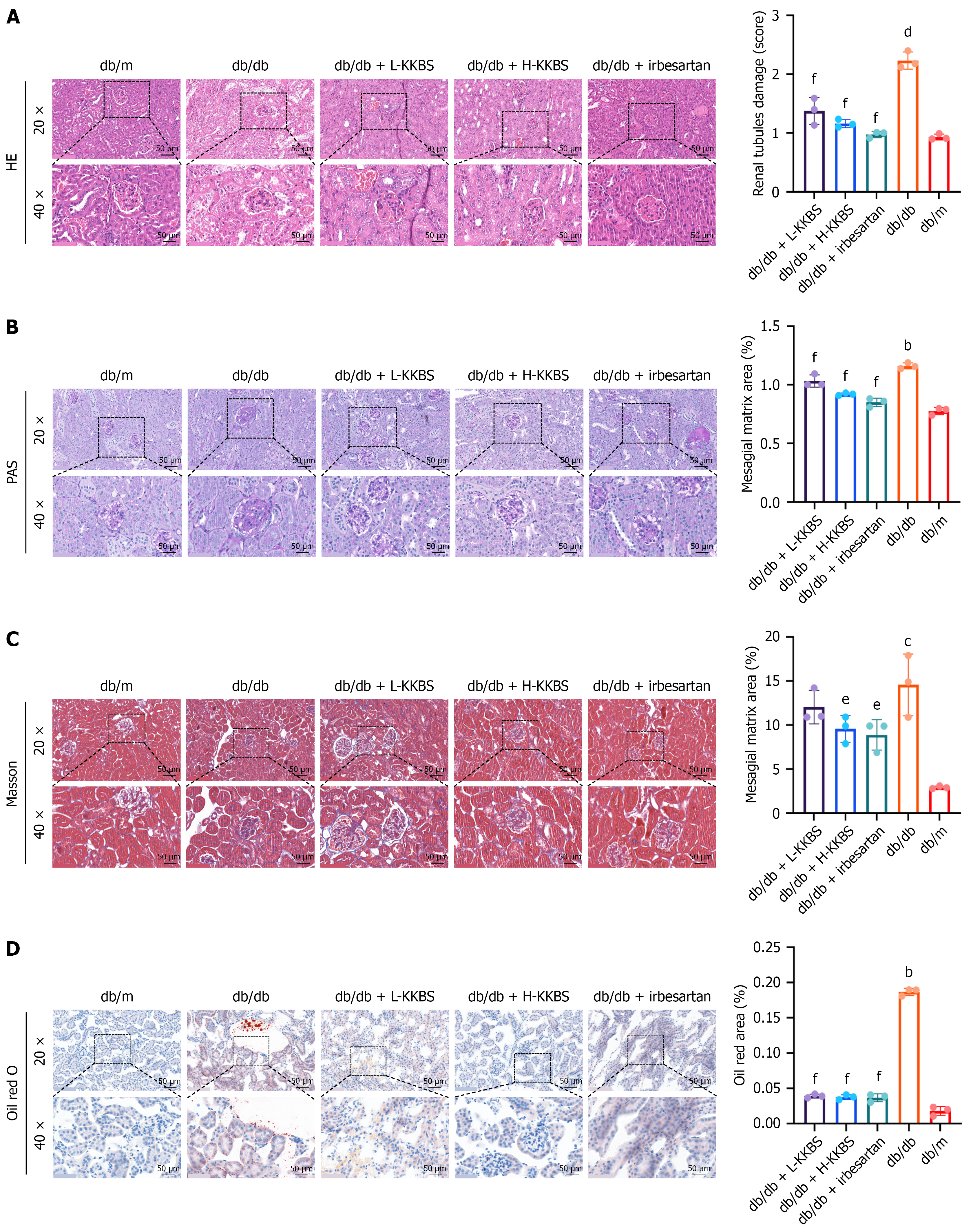
Further, histopathological examinations were performed to assess KKBS’s protective effects on renal tissue. HE staining demonstrated notable pathological alterations in db/db mice compared to db/m controls, including mesangial matrix thickening and expansion, tubular edema with vacuolization, and prominent interstitial inflammatory cell infiltration. In contrast, the KKBS and irbesartan-treated groups showed no evident mesangial proliferation, only mild tubular edema, and reduced inflammatory infiltration (P < 0.01; Figure 2A). PAS staining corroborated these observations: Db/db mice displayed enlarged mesangial cells, matrix proliferation, disrupted tubular architecture, and interstitial inflammation (P < 0.01), all of which were significantly alleviated by KKBS treatment as evidenced by a decreased mesangial matrix area (P < 0.01; Figure 2B). Masson’s trichrome staining indicated enhanced collagen accumulation in the tubular basement membrane and interstitium of db/db mice compared to controls (P < 0.001), while the H-KKBS and irbesartan groups showed significantly reduced collagen accumulation (P < 0.05) (Figure 2C). In addition, Oil Red O staining demonstrated a higher number of lipid droplets in db/db mice relative to db/m mice (P < 0.01). Both KKBS and ir
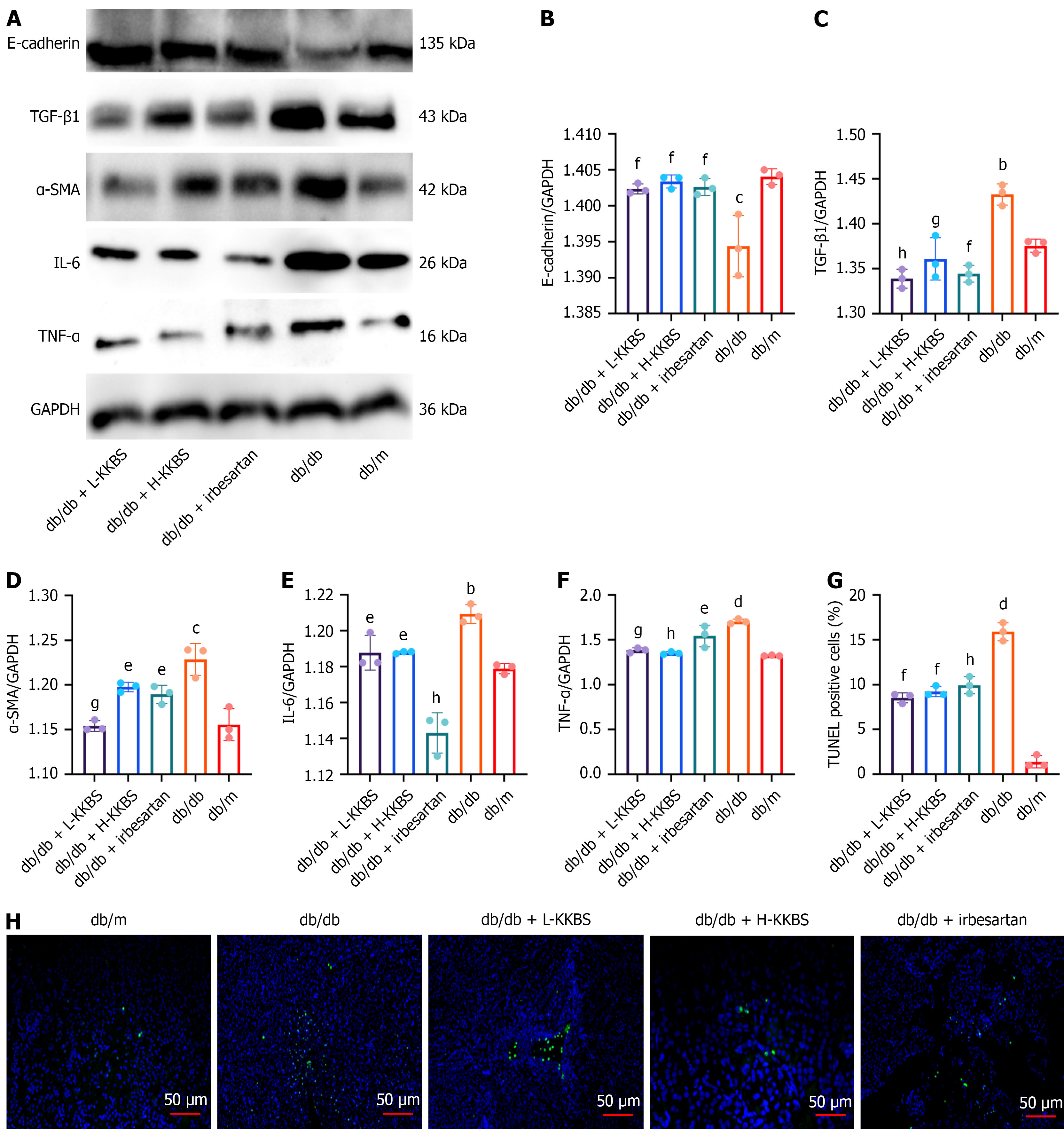
To further validate the anti-fibrotic effects of KKBS in DKD, inflammatory and fibrotic markers (TGF-β1, IL-6, TNF-α, E-cadherin, and α-SMA) were assessed both in vivo and in vitro. In vivo, the db/db group had significantly elevated expression of TGF-β1 (P < 0.01), IL-6 (P < 0.01), TNF-α (P < 0.0001), and α-SMA (P < 0.001), accompanied by decreased E-cadherin levels (P < 0.001) compared to the db/m group. KKBS treatment markedly reduced the expression of these fibrotic and inflammatory markers (P < 0.01). Terminal deoxynucleotidyl transferase-mediated deoxyuridine tri
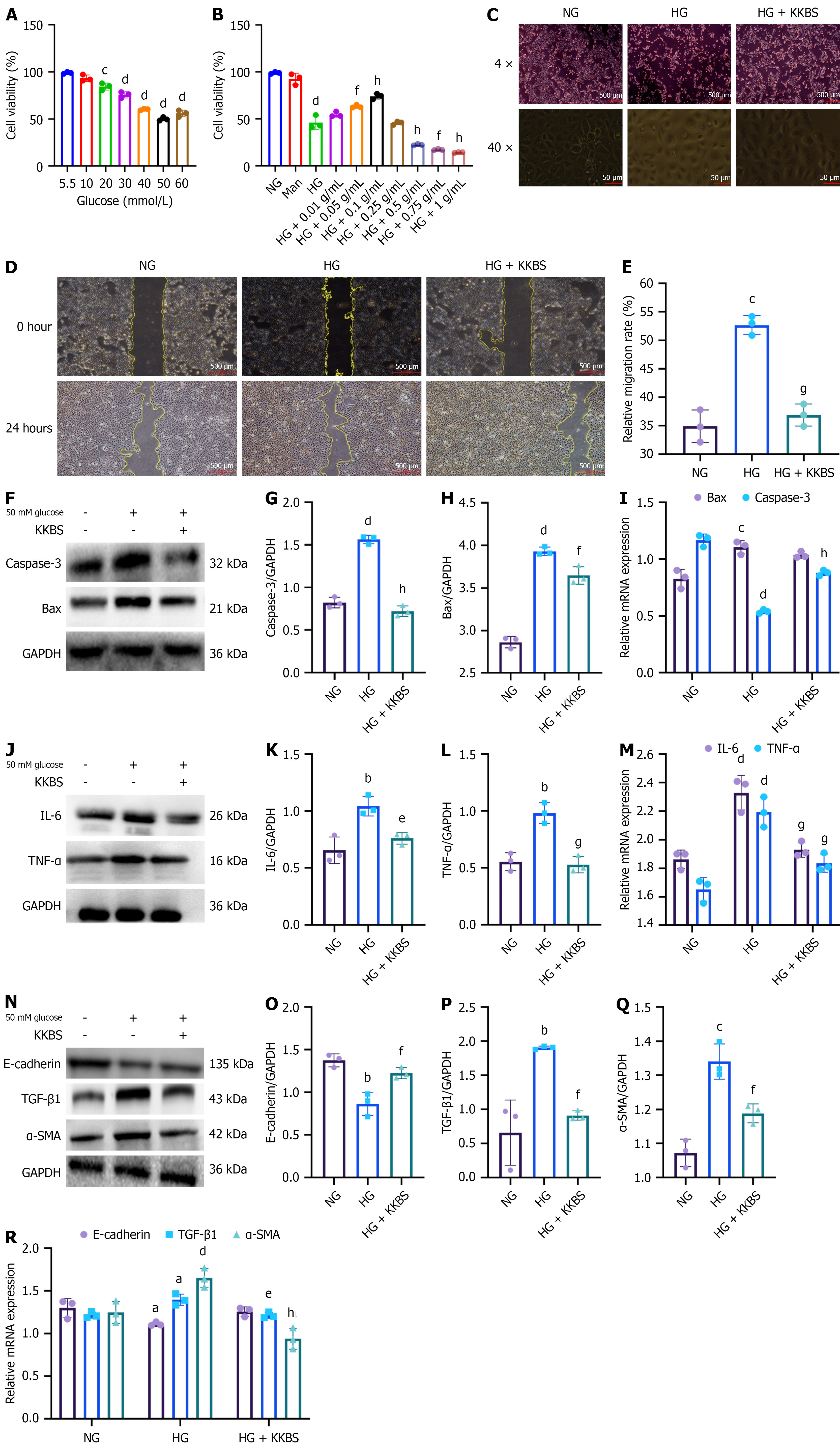
In vitro, the renal protective effects of KKBS were evaluated in HK-2 cells using a CCK-8 assay under varying glucose conditions. Cell viability was significantly reduced at 50 mmol/L glucose (P < 0.0001; Figure 4A), which was therefore selected for subsequent experiments. A 50 mmol/L mannitol group was included to control for potential osmotic effects. KKBS treatment significantly improved cell viability under HG conditions, with the greatest effect observed at 0.1 g/mL (P < 0.0001) compared to the HG group (Figure 4B). Morphological analysis showed that cells in the normal glucose (5.5 mmol/L) group maintained the typical cobblestone-like morphology, whereas cells exposed to HG (50 mmol/L glucose) exhibited irregular, spindle-shaped morphology with increased intercellular spacing. KKBS treatment (50 mmol/L glucose + 14% KKBS) largely restored the cobblestone-like morphology and reduced cell spacing (Figure 4C). Scratch assays demonstrated that HG induced HK-2 cell migration (P < 0.001), which was significantly inhibited by KKBS treatment (P < 0.001; Figure 4D and E).
Western blot and qRT-PCR analyses revealed significant upregulation of the apoptotic markers Caspase-3 and Bax under HG conditions (P < 0.0001). KKBS treatment significantly decreased Caspase-3 and Bax protein levels (P < 0.01; Figure 4F-I), although Bax mRNA expression was not significantly altered. These results suggest that KKBS reduces HG-induced apoptosis by downregulating key apoptotic proteins. Furthermore, pro-inflammatory cytokines (TNF-α and IL-6) were raised in the HG group (P < 0.01) but significantly decreased following KKBS treatment (P < 0.05; Figure 4J-L), with qRT-PCR data confirming similar trends (P < 0.001; Figure 4M). Fibrosis-related markers were also affected by HG, with decreased E-cadherin (P < 0.01) and increased α-SMA and TGF-β1 expression (P < 0.01). KKBS treatment reversed these changes, restoring marker levels toward normal (P < 0.01; Figure 4N-Q), supported by corresponding qRT-PCR results (Figure 4R). Collectively, these results demonstrate that KKBS exerts renoprotective effects in HG-induced HK-2 cells by inhibiting apoptosis, inflammation, migration, and fibrosis-associated marker expression.
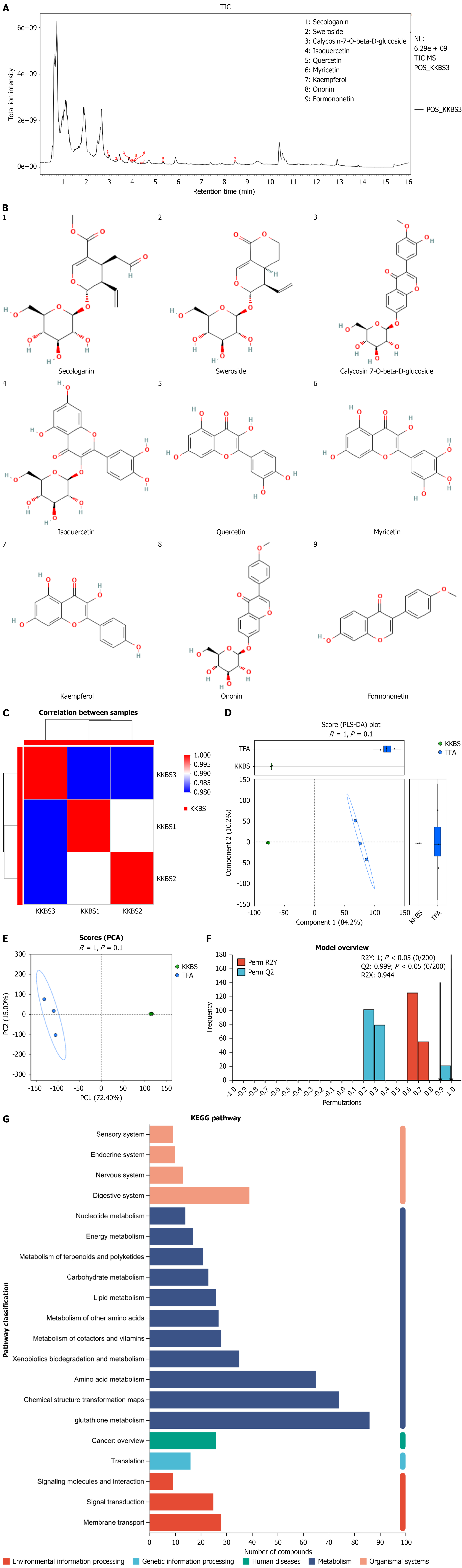
To investigate the mechanism of KKBS against DKD, metabolomics analysis was performed. LC-MS/MS was employed in full scan mode to generate a compound fingerprint of KKBS (Supplementary Figure 1A and B). Following systematic data matching and manual validation, nine compounds were identified: Secologanin, sweroside, calycosin 7-O-β-D-glucoside, isoquercetin, quercetin, myricetin, kaempferol, ononin, and formononetin (Figure 5A and B). These com
| Peak name | Retention time (minutes) | Mass-to-charge ratio (experiment) | Precursor type | Compound name | Pubchem number |
| 1 | 2.990916667 | 389.1450240 | M+H | Secologanin | 161276 |
| 2 | 3.543550000 | 359.1343285 | M+H | Sweroside | 161036 |
| 3 | 4.055166667 | 447.1292902 | M+H | Calycosin 7-O-beta-D-glucoside | 5318267 |
| 4 | 4.055166667 | 465.1038809 | M+H | Isoquercetin | 5280804 |
| 5 | 4.055166667 | 303.0504691 | M+H | Quercetin | 5280343 |
| 6 | 4.076500000 | 301.0350797 | M+H-H2O | Myricetin | 5281672 |
| 7 | 4.319766667 | 287.0556821 | M+H | Kaempferol | 5280863 |
| 8 | 5.334866667 | 431.1343269 | M+H | Ononin | 442813 |
| 9 | 8.458666667 | 269.0813292 | M+H | Formononetin | 5280378 |
Metabolomics analysis revealed that the sample correlation heatmap (Figure 5C) showed a strong resemblance in both metabolic composition and abundance among the three KKBS plasma samples from db/db mice (KKBS1, KKBS2, and KKBS3). Principal component analysis further demonstrated clear separation between the KKBS and control groups, with principal component 1, which accounts for 72.40% of the variance and principal component 2, which accounts for 15.0% (Figure 5D). Increased intergroup distances corresponded to greater metabolic differences. Additionally, partial least squares discriminant analysis highlighted a significant classification distinction between the KKBS and control groups (Figure 5E). The partial least squares discriminant analysis permutation test used R2Y and Q2 values to evaluate the model’s explanatory and predictive power, respectively, where higher cumulative R2Y and Q2 values indicate better model stability and reliability. Notably, an R2Y value of 0.05 suggested that the explanatory power of the random model exceeded that of the original model during permutation testing (Figure 5F). Pathway analysis revealed that KKBS mainly affected GSH metabolism, lipid metabolism, and amino acid metabolism and the production of secondary metabolites (Figure 5G). Importantly, GSH metabolism is known to be a key pathway altered during ferroptosis[34].
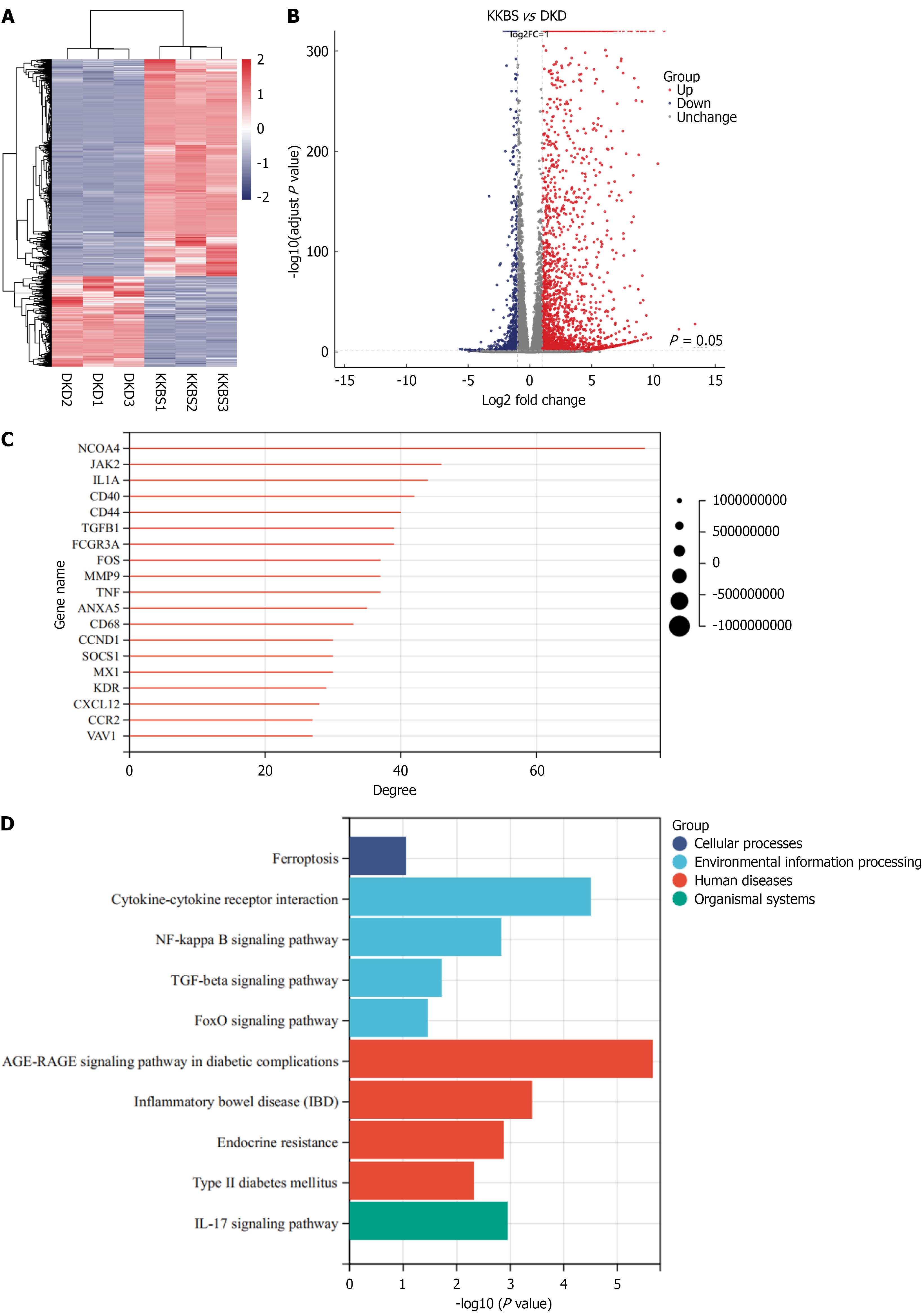
Transcriptomic analysis provided deeper insight into the mechanisms by which KKBS affects DKD. The results of transcriptomic sequencing of renal tissue from KKBS-treated and db/db mice are summarized in Figure 6A. The volcano plot comparing the two groups identified 2354 upregulated and 984 downregulated genes (Figure 6B). Protein-protein interaction (PPI) network and KEGG pathway analyses revealed key DEGs including NCOA4, Janus kinase 2, CD44, CD40, and TNF, with NCOA4 emerging as the most significant hub in the PPI network (Figure 6C). These DEGs were significantly enriched in pathways such as ferroptosis, nuclear factor-κB signaling, and forkhead box O signaling (Figure 6D).
While the transcriptomic data substantially contributed to identifying potential targets and pathways modulated by KKBS, several limitations should be noted. The small sample size used for RNA sequencing may reduce the robustness and reproducibility of differential expression results, potentially hindering detection of subtle gene expression changes and increasing the risk of false positives and negatives. Additionally, confounding factors such as biological variability, batch effects, or minor differences in sample handling may have influenced gene expression profiles. Although standard procedures were employed to minimize these sources of variability, they remain inherent limitations of transcriptomic studies. Future research should include larger sample cohorts, technical replicates, and validation of key findings by quantitative PCR or protein-level assays to reinforce these conclusions.
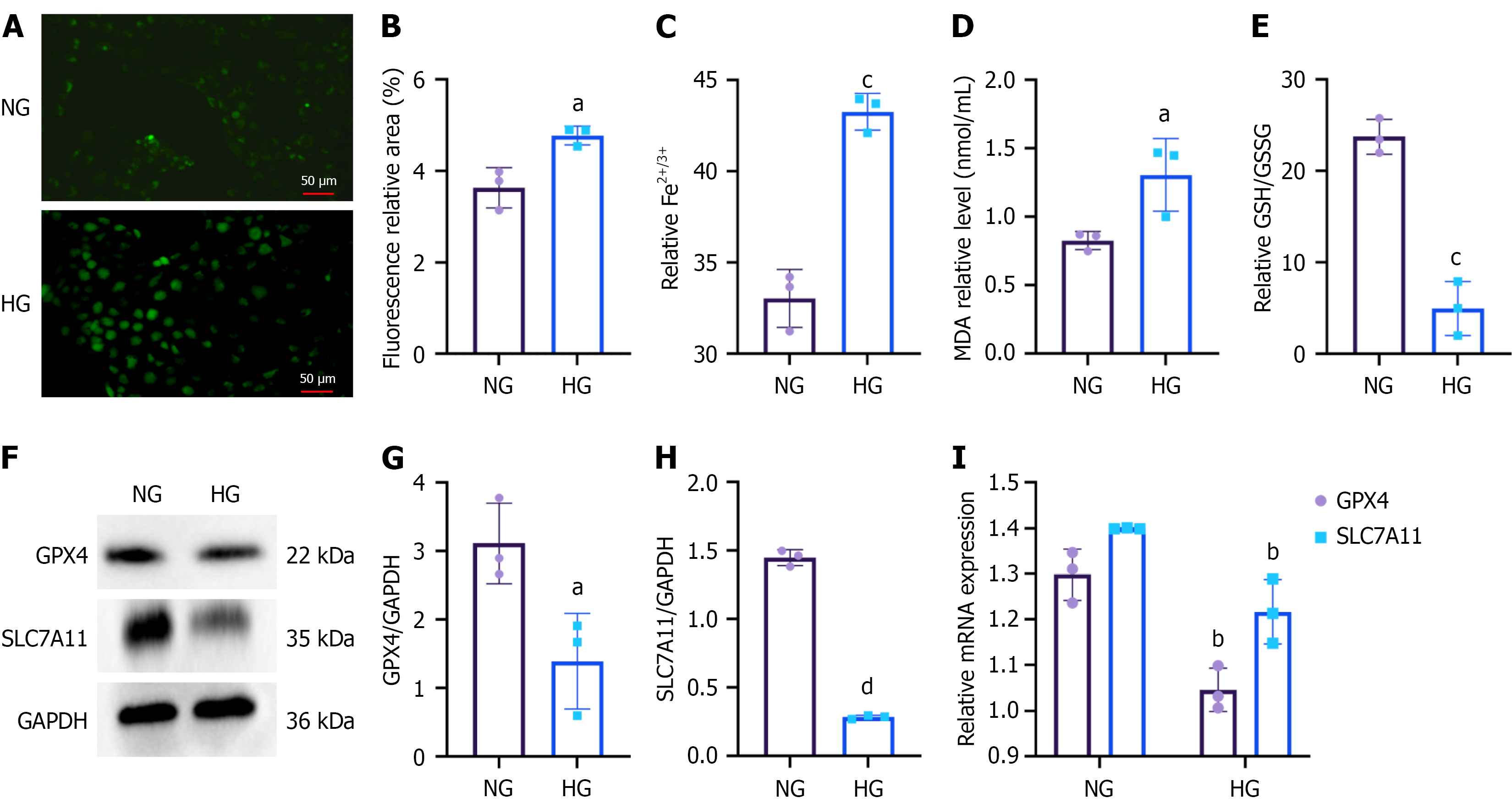
Comprehensive metabolomics analysis identified the GSH metabolic pathway as the most significant metabolic pathway associated with KKBS, which is also a key metabolic feature of ferroptosis. Integrating these results with transcriptomic data, we hypothesized that KKBS exerts its protective effects against DKD by inhibiting ferroptosis. Our experiments confirmed that, under HG conditions, HK-2 cells exhibited a significant rise in ROS (P < 0.05), iron ions (Fe2+/Fe3+; P < 0.001), and MDA (P < 0.05), accompanied by a marked decrease in the GSH/GSSG ratio (P < 0.001; Figure 7A-E). Both mRNA and protein expression levels of the ferroptosis markers GPX4 and SLC7A11 were significantly downregulated (P < 0.05), with GPX4 and SLC7A11 transcripts showing a particularly pronounced reduction (P < 0.01; Figure 7F-I). Collectively, these results suggest that ferroptosis is very substantial in fibrosis development in HG-induced HK-2 cells.
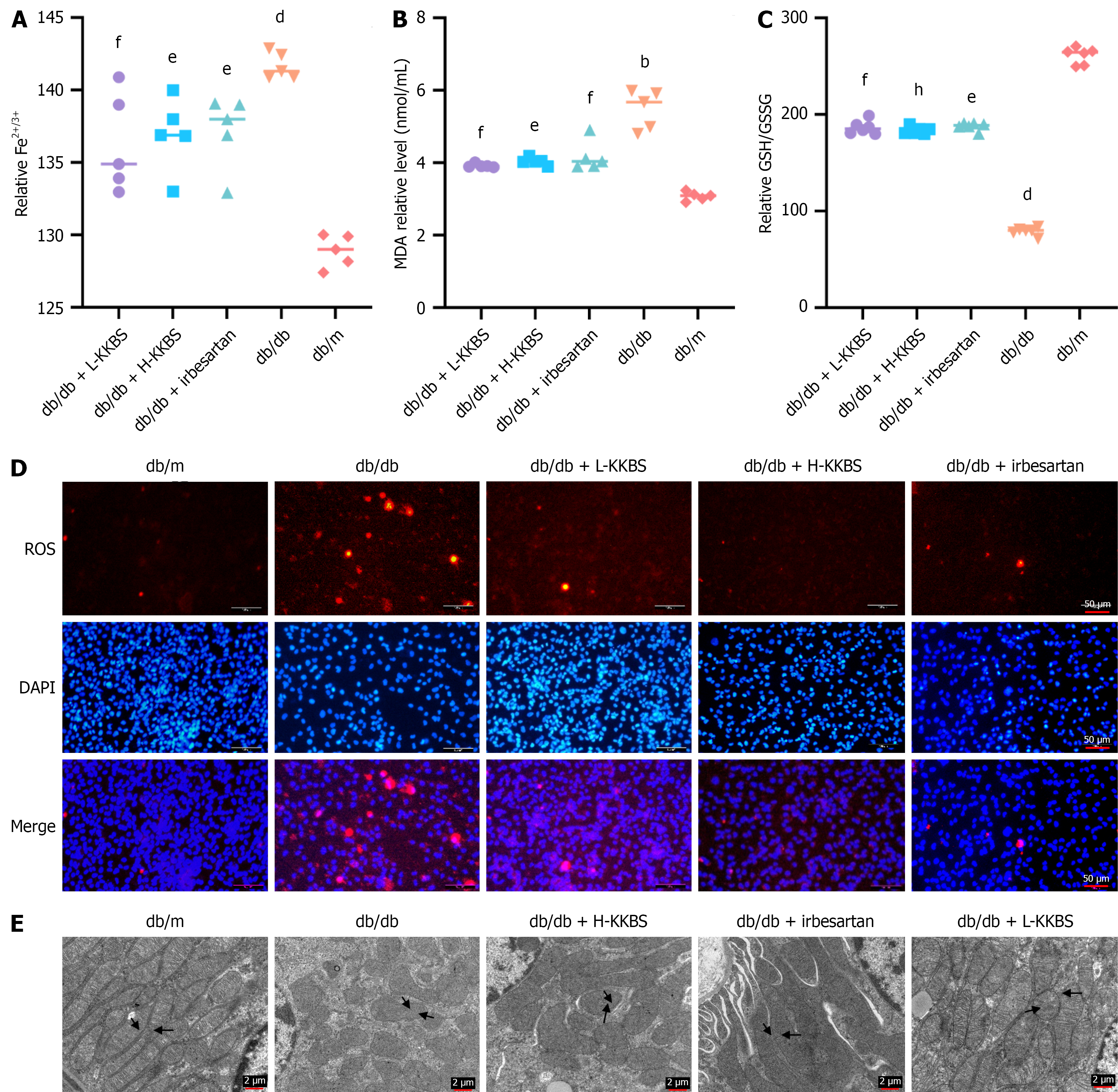
To investigate the effect of KKBS on the ferroptosis pathway, we measured ferroptosis biomarkers in renal tissues. Iron ions (Fe2+/Fe3+) were significantly elevated in the db/db group compared to the db/m group (P < 0.0001), accompanied by elevated levels of MDA (P < 0.01) and ROS, along with a marked reduction in the GSH/GSSG ratio (P < 0.0001). Treatment with KKBS and irbesartan significantly reduced Fe2+/Fe3+ (P < 0.05), MDA (P < 0.05), and ROS levels, while increasing the GSH/GSSG ratio (P < 0.05; Figure 8A-D). TEM analysis further demonstrated that KKBS treatment significantly ameliorated ferroptosis-associated mitochondrial morphological changes, including organelle shrinkage, cristae loss, fragmentation, and increased membrane density (Figure 8E)[35].
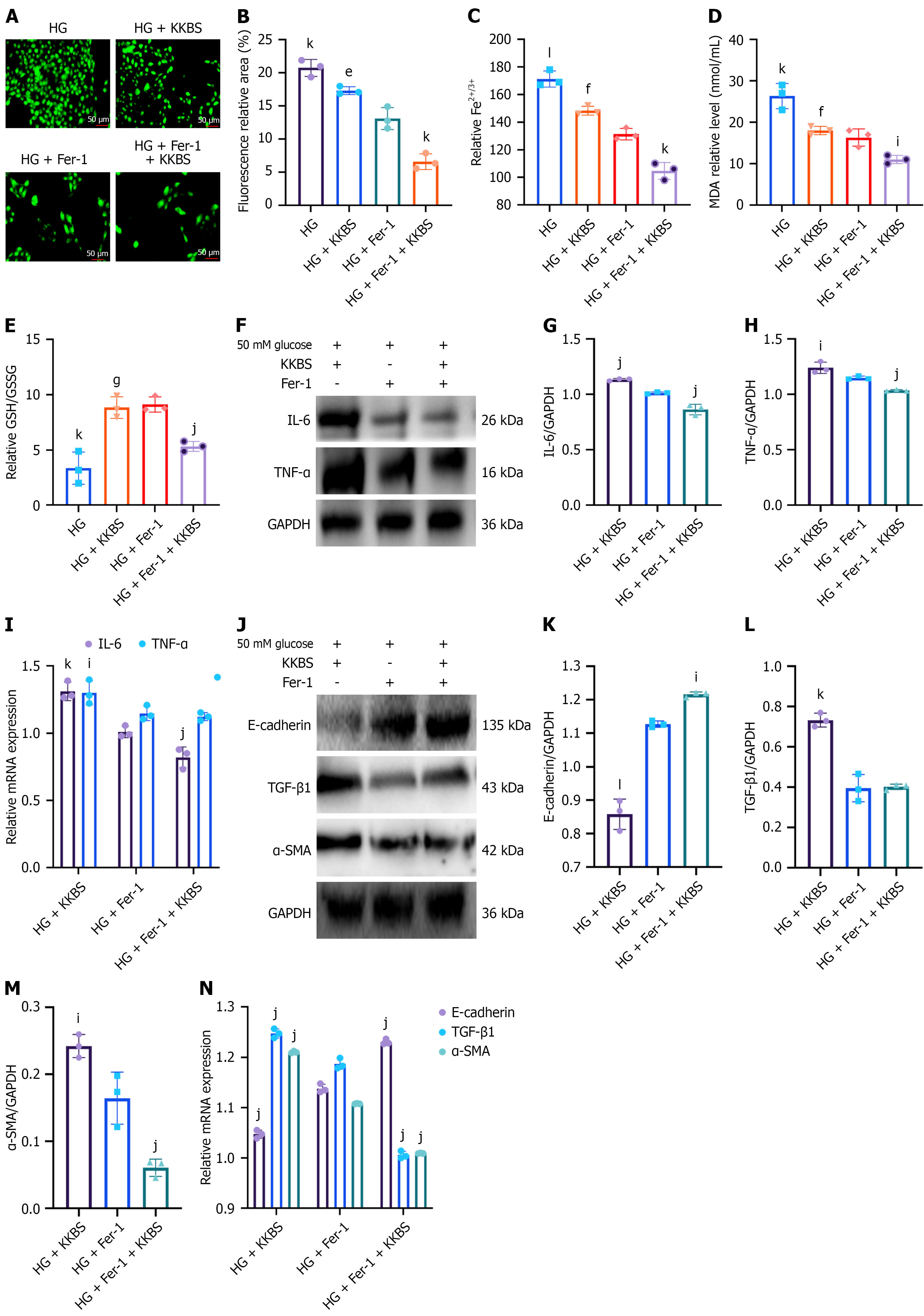
In vitro experiments showed that combining KKBS with the ferroptosis inhibitor Fer-1 (10 μmol/L), used as a positive control, resulted in the HG + Fer-1 + KKBS group exhibiting significantly reduced ROS (P < 0.05), iron ions (Fe2+/Fe3+; P < 0.001), and MDA levels (P < 0.001) compared to the HG + Fer-1 group (Figure 9A-D). Additionally, the GSH/GSSG ratio was increased significantly (P < 0.001; Figure 9E). The expression of IL-6 (protein and mRNA) and TNF-α (protein) was also significantly decreased in the HG + Fer-1 + KKBS group (P < 0.01), although TNF-α mRNA levels remained un
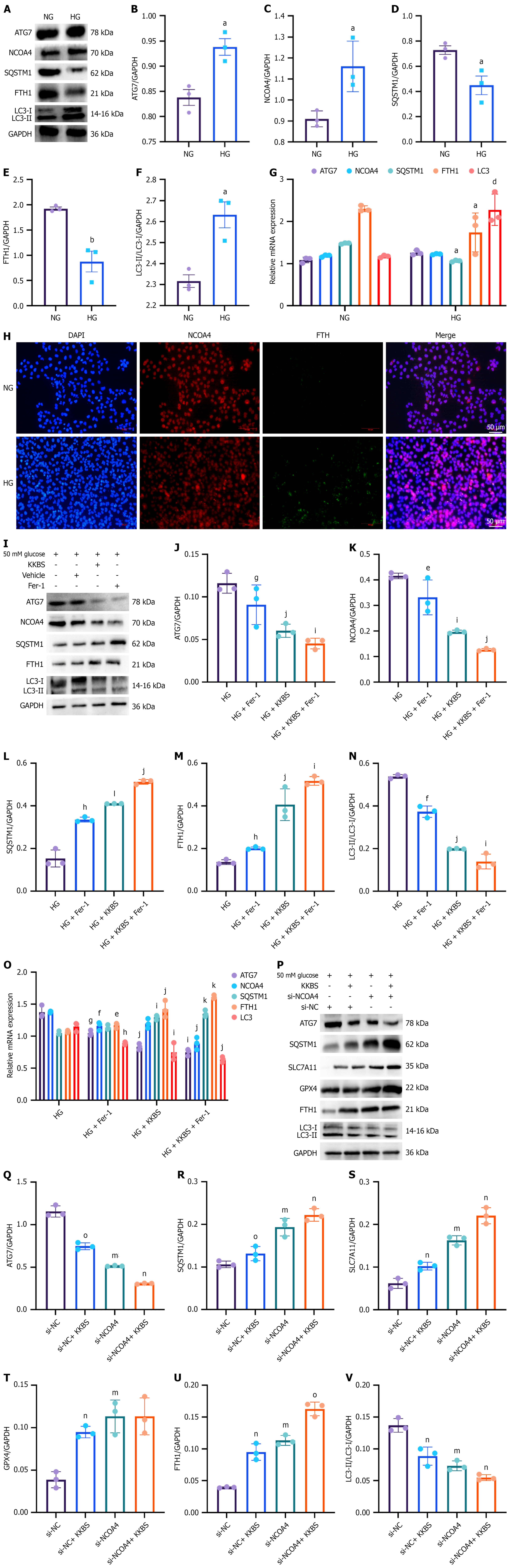
To elucidate how KKBS influences ferroptosis in HK-2 cells, we focused on NCOA4, identified by transcriptomic analysis as a key gene within the PPI network closely associated with the ferroptosis pathway. NCOA4, a selective autophagic cargo receptor, transports the NCOA4-ferritin complex to lysosomes, promoting ferritin degradation and releasing free iron, thereby inducing ferroptosis[36]. We assessed autophagic markers in HG-induced HK-2 cells and observed in
To determine whether KKBS inhibits HG-induced ferroptosis by modulating ferritinophagy, we used Fer-1 as a positive control. Western blot results demonstrated that HG + KKBS treatment significantly downregulated ATG7, LC3, and NCOA4 Levels while upregulating SQSTM1/p62 and FTH1 (P < 0.05). The HG + KKBS + Fer-1 group showed even greater suppression of ATG7, LC3, and NCOA4, along with higher SQSTM1/p62 and FTH1 Levels compared to the HG + Fer-1 group (P < 0.05; Figure 10I-O). Stable transfection with NCOA4 small interfering RNA effectively knocked down NCOA4 expression in HK-2 cells, with small interfering RNA3 showing the highest efficiency (Supplementary Figure 2A-C). Compared to the si-NCOA4 (NCOA4-targeting small interfering RNA) group, the si-NCOA4 + KKBS group exhibited further reductions in ATG7 and LC3 protein expression (P < 0.05), increased FTH1 and SLC7A11 expression (P < 0.05), and no significant changes in GPX4 Levels (Figure 10P-V). These findings suggest that KKBS modulates fer
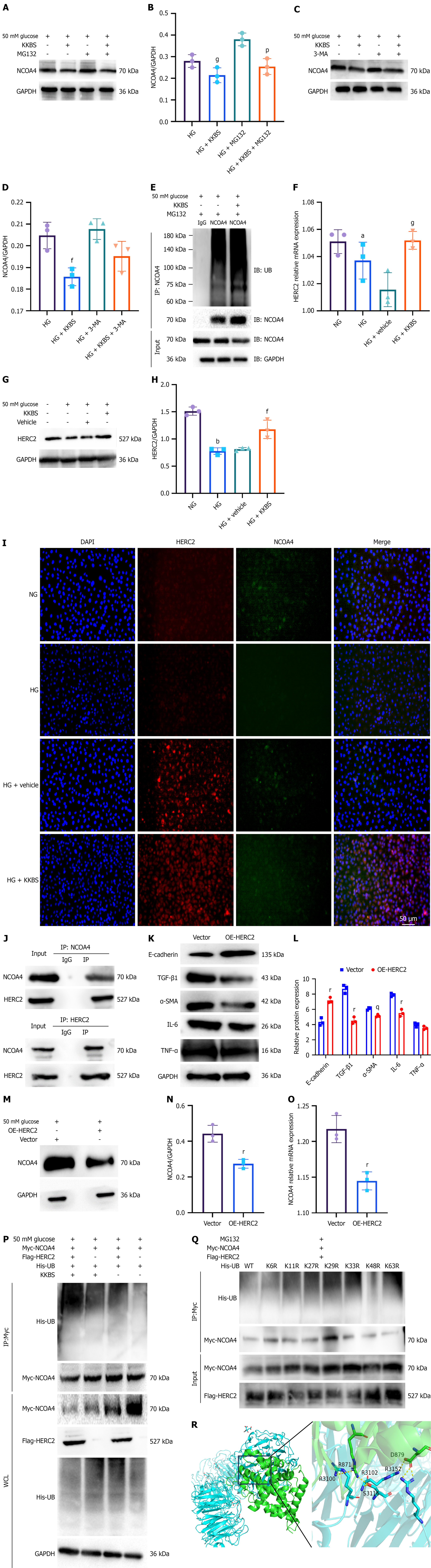
As shown in Figure 10O, KKBS did not affect the mRNA levels of NCOA4, indicating that its suppression primarily occurs at the protein level without altering transcription. In eukaryotic cells, protein degradation predominantly proceeds via two pathways: Ubiquitin-proteasomal degradation and autophagic-lysosomal clearance[37]. To investigate the mechanism further, HK-2 cells under HG conditions were co-incubated with MG132 (a proteasome inhibitor) and 3-MA (an autophagy inhibitor)[38]. The KKBS-induced downregulation of NCOA4 was reversed by MG132 (P < 0.01), but not by 3-MA (Figure 11A-D). Additionally, Co-IP assays demonstrated that KKBS markedly enhanced NCOA4 ubiquitination in HG-induced HK-2 cells, especially in the presence of MG132 (Figure 11E). These results specify that KKBS primarily reduces NCOA4 protein levels through ubiquitin-dependent proteasomal degradation.
Next, we explored the direct targets of KKBS in modulating autophagy-dependent ferroptosis in HG-induced HK-2 cells. Previous studies have shown that HERC2, an E3 ubiquitin ligase, ubiquitinates and degrades NCOA4, thereby inhibiting autophagy-dependent ferroptosis[39]. We hypothesized that KKBS might promote NCOA4 ubiquitination and degradation by upregulating HERC2 under HG conditions. Indeed, in vitro analyses confirmed that KKBS significantly increased HERC2 expression at both the protein (P < 0.01) and mRNA levels (P < 0.001) (Figure 11F-H). Co-IP and IF co-localization assays further validated the interaction between HERC2 and NCOA4 (Figure 11I and J).
To elucidate the functional role of HERC2 in KKBS’s ferroptosis-inhibitory effects, we established an HERC2-overexpressing cell model (P < 0.01; Supplementary Figure 3A-C). Molecular docking of KKBS’s core bioactive compounds, identified via LC-MS/MS, with HERC2 revealed strong binding affinities (Supplementary Figure 4A and B). Overexpression of HERC2 protected HK-2 cells from HG-induced damage (P < 0.05; Figure 11K and L) and led to a significant decline in NCOA4 expression (P < 0.01; Figure 11M-O). Moreover, co-transfection of HK-2 cells with myc-NCOA4, flag-HERC2 (F3), and histidine-ubiquitin plasmids, followed by KKBS treatment, significantly enhanced the interaction between NCOA4 and ubiquitin under HG conditions (Figure 11P).
Ubiquitin can form diverse linkages on substrates, leading to distinct cellular outcomes. To investigate the types of ubiquitin linkages involved in NCOA4 ubiquitination, we used ubiquitin mutants with lysine (K) to arginine (R) substitutions. The results showed that the ubiquitin variants K6R, K11R, K27R, K29R, K33R, and K63R increased HERC2/NCOA4 ubiquitination, whereas the K48R mutant significantly inhibited NCOA4 ubiquitination (Figure 11Q). Addi
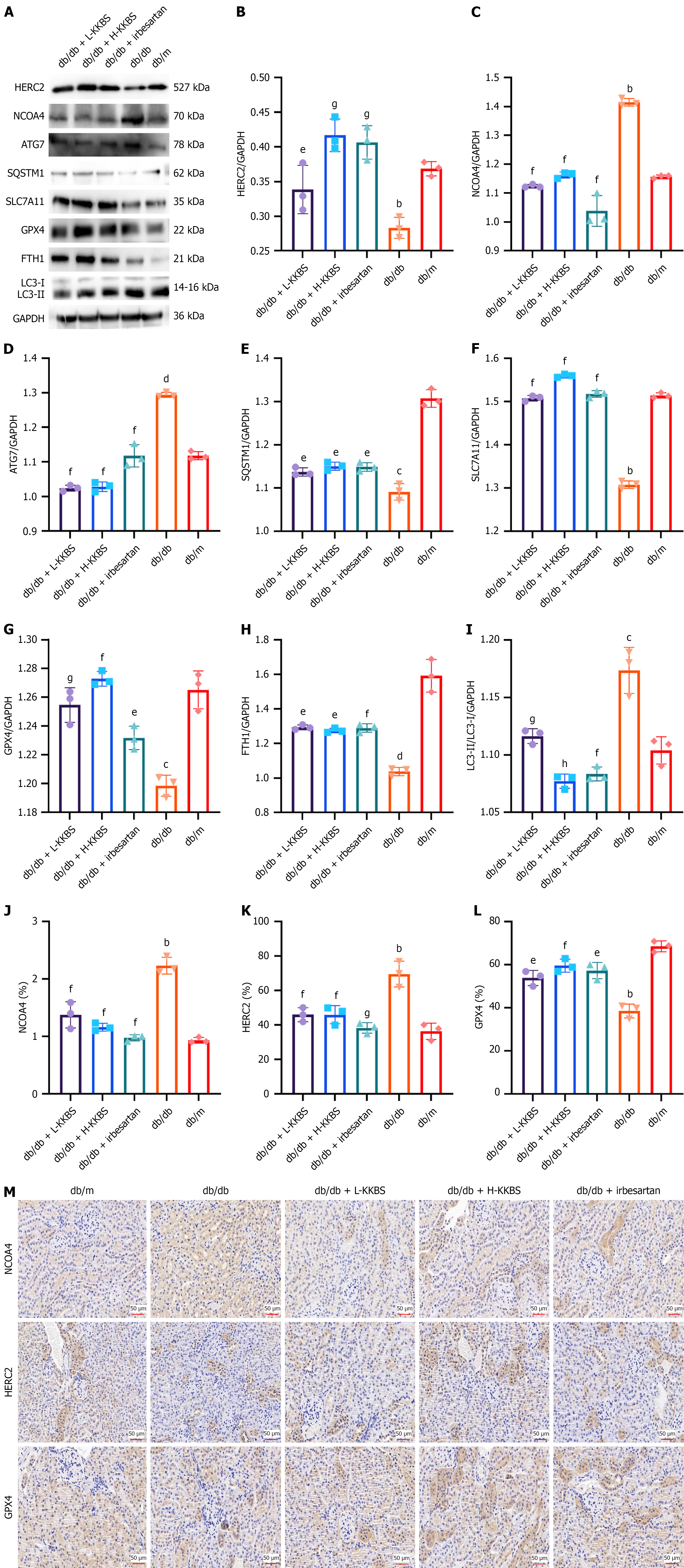
Building on the in vitro findings, we further examined the in vivo effects of KKBS on the HERC2/NCOA4-mediated autophagy-dependent ferroptosis pathway. Renal tissue expression of HERC2, NCOA4, and key ferroptosis-related autophagy markers (ATG7, SQSTM1, FTH1, LC3, SLC7A11, and GPX4) was assessed. Compared to the db/m group, the db/db group exhibited significantly elevated levels of ATG7 (P < 0.0001) and LC3 (P < 0.001), alongside reduced expression of HERC2 (P < 0.01), SQSTM1 (P < 0.001), SLC7A11 (P < 0.01), GPX4 (P < 0.001), and FTH1 (P < 0.0001). Treatment with KKBS significantly reversed these alterations (Figure 12A-L). Immunohistochemical staining corroborated these results (Figure 12M). Collectively, these findings indicate that KKBS alleviates renal injury by modulating the HERC2/NCOA4-mediated autophagy-dependent ferroptosis pathway in db/db mice.
DKD has emerged as a major global health priority due to its high prevalence, multiple comorbidities, significant mortality burden, and substantial economic impact. This urgent situation demands the development of improved treatment strategies and more precise, personalized diagnostic and therapeutic approaches to enhance the prevention and management of DKD[40]. In this study, LC-MS/MS analysis of KKBS identified several key quality control compounds, including secologanin, sweroside, calycosin 7-O-beta-D-glucoside, isoquercetin, quercetin, myricetin, kaempferol, ononin, and formononetin.
Metabonomic analysis revealed the GSH metabolic pathway as the most significant metabolic pathway associated with KKBS, which also represents a central metabolic feature in ferroptosis. Ferroptosis and ubiquitin-mediated proteolysis pathways are potential signaling mechanisms through which KKBS exerts therapeutic effects against DKD. Trans
Integrating metabolomic and transcriptomic findings, the renal protective mechanisms of KKBS were investigated using HG-induced HK-2 cells and db/db mice. The results demonstrated that KKBS provides renal protection by suppressing cellular activity, migration, inflammation (IL-6 and TNF-α), the expression of fibrosis-related factors (E-cadherin, TGF-β1, and α-SMA), and apoptosis in HG-induced HK-2 cells. Elevated lipid peroxide levels and reduced GPX4 expression are two key indicators of ferroptosis[44,45], which is critically involved in the advancement of DKD-associated renal fibrosis in HG-induced HK-2 cells. KKBS mitigated HG-induced renal fibrosis by inhibiting ferroptosis-related biomarkers, including Fe2+/Fe3+, MDA, ROS, GSH/GSSG ratio, GPX4, and SLC7A11. We further examined the expression of autophagy markers (ATG7, SQSTM1, FTH1, and LC3) in HG-induced HK-2 cells. The data suggested that ferroptosis in these cells occurs via ferritinophagy, as indicated by the co-localization of NCOA4 and ferritin. Fer
Importantly, KKBS primarily reduces NCOA4 protein levels through ubiquitin-dependent degradation. Protein ubiquitination is essential for maintaining proteostasis by regulating substrate stability[48]. This process involves a cascade of enzymes: E3 Ligases, E1 activating enzymes, and E2 conjugating enzymes, which confer substrate specificity[49,50]. Previous studies have shown that HERC2-mediated ubiquitination targets NCOA4 for proteasomal degradation, thereby inhibiting autophagy-dependent ferroptosis[51-53]. Co-IP and IF co-localization assays confirmed the interaction between HERC2 and NCOA4. To explore HERC2’s role in KKBS’s ferroptosis-inhibitory effects, we established an HERC2-overexpressing cell model. Overexpression of HERC2 protected HK-2 cells from HG-induced cytotoxicity and correspondingly reduced NCOA4 expression.
Additionally, we transfected HK-2 cells with myc-NCOA4, flag-HERC2 (F3), and His-ubiquitin constructs containing K and R mutations. The results indicated that HERC2 induces K48-linked ubiquitination, leading to NCOA4 degradation. Molecular docking simulations identified three hydrogen bonds (D879-R3152, R871-R3102, and R3100-S3116) at the HERC2-NCOA4 interface, enhancing the stability of their interaction. Next, db/db mice were employed as a DKD model to evaluate KKBS’s therapeutic efficacy in vivo. KKBS demonstrated a favorable safety profile and alleviated renal function impairment in DKD mice. Histological assessments using HE, PAS, and Masson staining revealed that KKBS effectively protected against renal injury in db/db mice. Furthermore, Oil Red O staining showed markedly reduced lipid deposits in KKBS-treated mice compared to db/db controls, suggesting KKBS’s capacity to attenuate lipid accumulation. Given that lipid peroxidation drives ferroptosis progression[54,55], these findings support the hypothesis that KKBS exerts anti-ferroptotic effects partially through lipid metabolism modulation. Finally, analyses of ferroptosis-related biomarkers via Western blot, TEM, and immunohistochemical demonstrated that KKBS ameliorates DKD-induced renal injury by modulating the HERC2/NCOA4-mediated autophagy-dependent ferroptosis pathway in vivo.
Taken together, transcriptomic and metabolomic analyses identify ferroptosis as the central mechanism underlying KKBS’s anti-renal fibrosis effects, with NCOA4 as a pivotal target. KKBS showed significant therapeutic efficacy against DKD in both cellular and animal models. Specifically, KKBS modulates the HERC2/NCOA4-mediated autophagy-dependent ferroptosis pathway, suppresses inflammatory and fibrotic factor secretion, and reduces cellular apoptosis, thereby mitigating renal damage. Furthermore, targeting the interaction between HERC2 and NCOA4 may offer a promising therapeutic strategy for DKD.
The findings align with recent studies implicating ferroptosis in the progression of DKD. For example, Tang et al[18] reported that enhanced ferritinophagy promotes iron overload and lipid peroxidation in DKD models, identifying NCOA4 as a critical driver of ferroptosis. Similarly, Zhou et al[56] highlighted the protective roles of GPX4 and SLC7A11 against oxidative stress-induced renal damage in DKD, emphasizing the therapeutic potential of targeting ferroptosis pathways. However, unlike these studies, our research uniquely identifies HERC2 as a novel upstream regulator of NCOA4 stability via ubiquitin-dependent degradation, a mechanism not previously described in DKD. Moreover, whereas prior pharmacological studies have primarily focused on ferroptosis inhibitors such as Fer-1, our findings provide novel evidence supporting the use of the traditional Chinese herb KKBS to modulate ferroptosis indirectly through regulation of the autophagy machinery. This broadens the spectrum of ferroptosis-targeting therapies and hi
Our research provides novel evidence that KKBS exerts anti-fibrotic effects in DKD by targeting the HERC2/NCOA4-mediated autophagy-dependent ferroptosis pathway. These findings offer new mechanistic insights into DKD pa
We acknowledge all animals involved in this study who gave their lives for the advancement of human health.
| 1. | Mohandes S, Doke T, Hu H, Mukhi D, Dhillon P, Susztak K. Molecular pathways that drive diabetic kidney disease. J Clin Invest. 2023;133:e165654. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 233] [Reference Citation Analysis (0)] |
| 2. | Xue X, Jin XY, Ye XL, Li KY, Li JX, Liu XH, Bai J, Liu Q, Zhang BR, Zou XR, Yuan J, Lu CL, Zhao FF, Liu JP, Wang XQ. Ophiocordyceps sinensis preparations combined with the renin-angiotensin system inhibitor for diabetic kidney disease treatment: an umbrella review of systematic reviews and network meta-analysis. Front Pharmacol. 2024;15:1360633. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 3. | Wang Y, Quan F, Cao Q, Lin Y, Yue C, Bi R, Cui X, Yang H, Yang Y, Birnbaumer L, Li X, Gao X. Quercetin alleviates acute kidney injury by inhibiting ferroptosis. J Adv Res. 2021;28:231-243. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 282] [Cited by in RCA: 463] [Article Influence: 92.6] [Reference Citation Analysis (0)] |
| 4. | Xie D, Esmaiel H, Sun H, Qi J, Qasem ZAH. Feature Extraction of Ship-Radiated Noise Based on Enhanced Variational Mode Decomposition, Normalized Correlation Coefficient and Permutation Entropy. Entropy (Basel). 2020;22:468. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 5. | Song S, Yu J. Integrating Network Pharmacology, Bioinformatics, and Mendelian Randomization Analysis to Identify Hub Targets and Mechanisms of Kunkui Baoshen Decoction in Treating Diabetic Kidney Disease. Curr Pharm Des. 2024;30:3367-3393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 6. | Wei C, Wang C, Li R, Bai Y, Wang X, Fang Q, Chen X, Li P. The pharmacological mechanism of Abelmoschus manihot in the treatment of chronic kidney disease. Heliyon. 2023;9:e22017. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 7. | Tan Y, Li R, Zhou P, Li N, Xu W, Zhou X, Yan Q, Yu J. Huobahuagen tablet improves renal function in diabetic kidney disease: a real-world retrospective cohort study. Front Endocrinol (Lausanne). 2023;14:1166880. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 8. | Han H, Cao A, Wang L, Guo H, Zang Y, Li Z, Zhang X, Peng W. Huangqi Decoction Ameliorates Streptozotocin-Induced Rat Diabetic Nephropathy through Antioxidant and Regulation of the TGF-β/MAPK/PPAR-γ Signaling. Cell Physiol Biochem. 2017;42:1934-1944. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 9. | Xu H, Shen J, Liu H, Shi Y, Li L, Wei M. Morroniside and loganin extracted from Cornus officinalis have protective effects on rat mesangial cell proliferation exposed to advanced glycation end products by preventing oxidative stress. Can J Physiol Pharmacol. 2006;84:1267-1273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 72] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 10. | Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, Morrison B 3rd, Stockwell BR. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060-1072. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4711] [Cited by in RCA: 13250] [Article Influence: 946.4] [Reference Citation Analysis (2)] |
| 11. | Fang X, Ardehali H, Min J, Wang F. The molecular and metabolic landscape of iron and ferroptosis in cardiovascular disease. Nat Rev Cardiol. 2023;20:7-23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 727] [Article Influence: 242.3] [Reference Citation Analysis (0)] |
| 12. | Liang D, Minikes AM, Jiang X. Ferroptosis at the intersection of lipid metabolism and cellular signaling. Mol Cell. 2022;82:2215-2227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 906] [Article Influence: 226.5] [Reference Citation Analysis (0)] |
| 13. | Wu X, Li Y, Zhang S, Zhou X. Ferroptosis as a novel therapeutic target for cardiovascular disease. Theranostics. 2021;11:3052-3059. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 465] [Article Influence: 93.0] [Reference Citation Analysis (0)] |
| 14. | Doll S, Freitas FP, Shah R, Aldrovandi M, da Silva MC, Ingold I, Goya Grocin A, Xavier da Silva TN, Panzilius E, Scheel CH, Mourão A, Buday K, Sato M, Wanninger J, Vignane T, Mohana V, Rehberg M, Flatley A, Schepers A, Kurz A, White D, Sauer M, Sattler M, Tate EW, Schmitz W, Schulze A, O'Donnell V, Proneth B, Popowicz GM, Pratt DA, Angeli JPF, Conrad M. FSP1 is a glutathione-independent ferroptosis suppressor. Nature. 2019;575:693-698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1492] [Cited by in RCA: 2411] [Article Influence: 344.4] [Reference Citation Analysis (0)] |
| 15. | Ji H, Wang W, Li X, Han X, Zhang X, Wang J, Liu C, Huang L, Gao W. p53: A double-edged sword in tumor ferroptosis. Pharmacol Res. 2022;177:106013. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 16. | Kraft VAN, Bezjian CT, Pfeiffer S, Ringelstetter L, Müller C, Zandkarimi F, Merl-Pham J, Bao X, Anastasov N, Kössl J, Brandner S, Daniels JD, Schmitt-Kopplin P, Hauck SM, Stockwell BR, Hadian K, Schick JA. GTP Cyclohydrolase 1/Tetrahydrobiopterin Counteract Ferroptosis through Lipid Remodeling. ACS Cent Sci. 2020;6:41-53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 317] [Cited by in RCA: 972] [Article Influence: 162.0] [Reference Citation Analysis (0)] |
| 17. | Jiang X, Stockwell BR, Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol. 2021;22:266-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2184] [Cited by in RCA: 5233] [Article Influence: 1046.6] [Reference Citation Analysis (0)] |
| 18. | Tang D, Chen X, Kang R, Kroemer G. Ferroptosis: molecular mechanisms and health implications. Cell Res. 2021;31:107-125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1891] [Cited by in RCA: 2943] [Article Influence: 588.6] [Reference Citation Analysis (1)] |
| 19. | Feng Q, Yang Y, Qiao Y, Zheng Y, Yu X, Liu F, Wang H, Zheng B, Pan S, Ren K, Liu D, Liu Z. Quercetin Ameliorates Diabetic Kidney Injury by Inhibiting Ferroptosis via Activating Nrf2/HO-1 Signaling Pathway. Am J Chin Med. 2023;51:997-1018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 104] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 20. | Wang L, Wu J, Wang Y. The Prevalence and Treatment of Diabetes in China From 2013 to 2018-Reply. JAMA. 2022;327:1706-1707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 21. | Alam MA, Al-Jenoobi FI, Al-Mohizea AM. Everted gut sac model as a tool in pharmaceutical research: limitations and applications. J Pharm Pharmacol. 2012;64:326-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 142] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 22. | Bian Y, Gao C, Kuster B. On the potential of micro-flow LC-MS/MS in proteomics. Expert Rev Proteomics. 2022;19:153-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 44] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 23. | Wishart DS, Guo A, Oler E, Wang F, Anjum A, Peters H, Dizon R, Sayeeda Z, Tian S, Lee BL, Berjanskii M, Mah R, Yamamoto M, Jovel J, Torres-Calzada C, Hiebert-Giesbrecht M, Lui VW, Varshavi D, Varshavi D, Allen D, Arndt D, Khetarpal N, Sivakumaran A, Harford K, Sanford S, Yee K, Cao X, Budinski Z, Liigand J, Zhang L, Zheng J, Mandal R, Karu N, Dambrova M, Schiöth HB, Greiner R, Gautam V. HMDB 5.0: the Human Metabolome Database for 2022. Nucleic Acids Res. 2022;50:D622-D631. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 760] [Cited by in RCA: 1454] [Article Influence: 363.5] [Reference Citation Analysis (0)] |
| 24. | Baker ES, Hoang C, Uritboonthai W, Heyman HM, Pratt B, MacCoss M, MacLean B, Plumb R, Aisporna A, Siuzdak G. METLIN-CCS: an ion mobility spectrometry collision cross section database. Nat Methods. 2023;20:1836-1837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 36] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 25. | Ren Y, Yu G, Shi C, Liu L, Guo Q, Han C, Zhang D, Zhang L, Liu B, Gao H, Zeng J, Zhou Y, Qiu Y, Wei J, Luo Y, Zhu F, Li X, Wu Q, Li B, Fu W, Tong Y, Meng J, Fang Y, Dong J, Feng Y, Xie S, Yang Q, Yang H, Wang Y, Zhang J, Gu H, Xuan H, Zou G, Luo C, Huang L, Yang B, Dong Y, Zhao J, Han J, Zhang X, Huang H. Majorbio Cloud: A one-stop, comprehensive bioinformatic platform for multiomics analyses. Imeta. 2022;1:e12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 350] [Cited by in RCA: 481] [Article Influence: 120.3] [Reference Citation Analysis (0)] |
| 26. | Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18868] [Cited by in RCA: 26642] [Article Influence: 1024.7] [Reference Citation Analysis (0)] |
| 27. | Karuppasamy MP, Venkateswaran S, Subbiah P. PDB-2-PBv3.0: An updated protein block database. J Bioinform Comput Biol. 2020;18:2050009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 28. | Forli S, Huey R, Pique ME, Sanner MF, Goodsell DS, Olson AJ. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc. 2016;11:905-919. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 785] [Cited by in RCA: 1622] [Article Influence: 162.2] [Reference Citation Analysis (0)] |
| 29. | Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34752] [Cited by in RCA: 63557] [Article Influence: 5777.9] [Reference Citation Analysis (1)] |
| 30. | Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480-D484. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3661] [Cited by in RCA: 4649] [Article Influence: 244.7] [Reference Citation Analysis (0)] |
| 31. | Tesch GH, Lim AK. Recent insights into diabetic renal injury from the db/db mouse model of type 2 diabetic nephropathy. Am J Physiol Renal Physiol. 2011;300:F301-F310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 114] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 32. | Ni B, Xiao Y, Wei R, Liu W, Zhu L, Liu Y, Ruan Z, Li J, Wang S, Zhao J, Huang W. Qufeng tongluo decoction decreased proteinuria in diabetic mice by protecting podocytes via promoting autophagy. J Tradit Complement Med. 2024;14:312-320. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 33. | Cui Y, Yu L, Cong W, Jiang S, Qiu X, Wei C, Zheng G, Mao J, Liu R, Patzak A, Persson PB, Chen J, Zhao L, Lai EY. Irisin preserves mitochondrial integrity and function in tubular epithelial cells after ischemia-reperfusion-induced acute kidney injury. Acta Physiol (Oxf). 2024;240:e14211. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 10] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 34. | Zheng J, Conrad M. Ferroptosis: when metabolism meets cell death. Physiol Rev. 2025;105:651-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 97] [Article Influence: 97.0] [Reference Citation Analysis (0)] |
| 35. | Wei S, Qiu T, Yao X, Wang N, Jiang L, Jia X, Tao Y, Wang Z, Pei P, Zhang J, Zhu Y, Yang G, Liu X, Liu S, Sun X. Arsenic induces pancreatic dysfunction and ferroptosis via mitochondrial ROS-autophagy-lysosomal pathway. J Hazard Mater. 2020;384:121390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 317] [Article Influence: 52.8] [Reference Citation Analysis (0)] |
| 36. | Fang Y, Chen X, Tan Q, Zhou H, Xu J, Gu Q. Inhibiting Ferroptosis through Disrupting the NCOA4-FTH1 Interaction: A New Mechanism of Action. ACS Cent Sci. 2021;7:980-989. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 380] [Article Influence: 76.0] [Reference Citation Analysis (0)] |
| 37. | Raffeiner M, Zhu S, González-Fuente M, Üstün S. Interplay between autophagy and proteasome during protein turnover. Trends Plant Sci. 2023;28:698-714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 61] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 38. | Chen Y, Shen J, Yuan M, Li H, Li Y, Zheng S, Han B, Zhang C, Liu S, Sun Q, Wu J. Dehydrocostus lactone suppresses gastric cancer progression by targeting ACLY to inhibit fatty acid synthesis and autophagic flux. J Adv Res. 2025;67:331-348. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 24] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 39. | Anandhan A, Dodson M, Shakya A, Chen J, Liu P, Wei Y, Tan H, Wang Q, Jiang Z, Yang K, Garcia JG, Chambers SK, Chapman E, Ooi A, Yang-Hartwich Y, Stockwell BR, Zhang DD. NRF2 controls iron homeostasis and ferroptosis through HERC2 and VAMP8. Sci Adv. 2023;9:eade9585. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 158] [Cited by in RCA: 346] [Article Influence: 115.3] [Reference Citation Analysis (0)] |
| 40. | Wang H, Yu X, Liu D, Qiao Y, Huo J, Pan S, Zhou L, Wang R, Feng Q, Liu Z. VDR Activation Attenuates Renal Tubular Epithelial Cell Ferroptosis by Regulating Nrf2/HO-1 Signaling Pathway in Diabetic Nephropathy. Adv Sci (Weinh). 2024;11:e2305563. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 65] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 41. | Ma J, Chen S, Liu J, Liao Y, Li L, Wang CC, Song S, Feng R, Hu H, Quan S. Cryptochrome 1 regulates ovarian granulosa cell senescence through NCOA4-mediated ferritinophagy. Free Radic Biol Med. 2024;217:1-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 42. | Qin Y, Qiao Y, Wang D, Tang C, Yan G. Ferritinophagy and ferroptosis in cardiovascular disease: Mechanisms and potential applications. Biomed Pharmacother. 2021;141:111872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 100] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 43. | Han Z, Luo Y, Chen H, Zhang G, You L, Zhang M, Lin Y, Yuan L, Zhou S. A Deep Insight into Ferroptosis in Renal Disease: Facts and Perspectives. Kidney Dis (Basel). 2024;10:224-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 19] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 44. | Dai E, Han L, Liu J, Xie Y, Kroemer G, Klionsky DJ, Zeh HJ, Kang R, Wang J, Tang D. Autophagy-dependent ferroptosis drives tumor-associated macrophage polarization via release and uptake of oncogenic KRAS protein. Autophagy. 2020;16:2069-2083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 351] [Cited by in RCA: 482] [Article Influence: 80.3] [Reference Citation Analysis (0)] |
| 45. | Tang M, Chen Z, Wu D, Chen L. Ferritinophagy/ferroptosis: Iron-related newcomers in human diseases. J Cell Physiol. 2018;233:9179-9190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 232] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 46. | Mancias JD, Wang X, Gygi SP, Harper JW, Kimmelman AC. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509:105-109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 728] [Cited by in RCA: 1603] [Article Influence: 133.6] [Reference Citation Analysis (0)] |
| 47. | Santana-Codina N, Gikandi A, Mancias JD. The Role of NCOA4-Mediated Ferritinophagy in Ferroptosis. Adv Exp Med Biol. 2021;1301:41-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 199] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 48. | Ji YX, Zhang P, Zhang XJ, Zhao YC, Deng KQ, Jiang X, Wang PX, Huang Z, Li H. The ubiquitin E3 ligase TRAF6 exacerbates pathological cardiac hypertrophy via TAK1-dependent signalling. Nat Commun. 2016;7:11267. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 155] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 49. | Rong Y, Fan J, Ji C, Wang Z, Ge X, Wang J, Ye W, Yin G, Cai W, Liu W. USP11 regulates autophagy-dependent ferroptosis after spinal cord ischemia-reperfusion injury by deubiquitinating Beclin 1. Cell Death Differ. 2022;29:1164-1175. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 159] [Cited by in RCA: 162] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 50. | Tyagi A, Haq S, Ramakrishna S. Redox regulation of DUBs and its therapeutic implications in cancer. Redox Biol. 2021;48:102194. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 51. | Ma F, Shao X, Zhang Y, Li J, Li Q, Sun H, Wang T, Liu H, Zhao F, Chen L, Chen J, Zhou S, Ji Q, Yu P. An arterial spin labeling-based radiomics signature and machine learning for the prediction and detection of various stages of kidney damage due to diabetes. Front Endocrinol (Lausanne). 2024;15:1333881. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 52. | Mancias JD, Pontano Vaites L, Nissim S, Biancur DE, Kim AJ, Wang X, Liu Y, Goessling W, Kimmelman AC, Harper JW. Ferritinophagy via NCOA4 is required for erythropoiesis and is regulated by iron dependent HERC2-mediated proteolysis. Elife. 2015;4:e10308. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 197] [Cited by in RCA: 433] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 53. | Zhang SJ, Zhang YF, Bai XH, Zhou MQ, Zhang ZY, Zhang SX, Cao ZJ, Wang L, Ding SW, Zheng HJ, Liu YN, Yu GY, Liu WJ. Integrated Network Pharmacology Analysis and Experimental Validation to Elucidate the Mechanism of Acteoside in Treating Diabetic Kidney Disease. Drug Des Devel Ther. 2024;18:1439-1457. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 54. | Bell HN, Stockwell BR, Zou W. Ironing out the role of ferroptosis in immunity. Immunity. 2024;57:941-956. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 119] [Article Influence: 59.5] [Reference Citation Analysis (0)] |
| 55. | Ursini F, Maiorino M. Lipid peroxidation and ferroptosis: The role of GSH and GPx4. Free Radic Biol Med. 2020;152:175-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 382] [Cited by in RCA: 1359] [Article Influence: 226.5] [Reference Citation Analysis (0)] |
| 56. | Zhou H, Zhou YL, Mao JA, Tang LF, Xu J, Wang ZX, He Y, Li M. NCOA4-mediated ferritinophagy is involved in ionizing radiation-induced ferroptosis of intestinal epithelial cells. Redox Biol. 2022;55:102413. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 151] [Article Influence: 37.8] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/