INTRODUCTION
Type 2 diabetes (T2D) is a metabolic disorder impacting around 25 million individuals in South America, with a global T2D population estimated at 387 million, projected to reach 592 million by 2035[1].
In T2D, endothelial dysfunction manifests in distinct patterns of injury across various organ systems. Notably, the pancreas, vasculature, brain, and heart are particularly susceptible to T2D-induced endothelial damage, leading to a spectrum of secondary complications. This multi-faceted dysfunction can lead to additional health complications including hypertension, diabetic cardiomyopathy, coronary artery disease, and cancer. Research indicates that individuals with T2D have an increased predisposition to specific cancer types[2], including breast cancer (BC)[3].
Numerous pathophysiological processes intricately linked to T2D actively contribute to the onset and progression of BC. These processes encompass hyperglycemia, insulin resistance[4], elevated free fatty acids, heightened reactive oxygen species, inflammation, and vascular fibrosis[5]. Despite the recognition of these interconnected pathways, the precise cellular and molecular mechanisms bridging T2D to BC remain largely elusive, especially those intertwined with the regulatory functions to microRNAs (miRNAs). It is important to point out that miRNAs comprise molecules ranging from 18 to 25 nucleotides, wield significant influence by either inhibiting or degrading specific target messenger RNAs, thereby modulating gene expression at a post-transcriptional level[6].
Given the increased BC risk in T2D patients, this review aims to identify deregulated miRNAs implicated in both diseases, searching for shared miRNA signatures and their influence on common signaling pathways implicated in BC development.
Pathophysiology of T2D
The pathophysiology of T2D has undergone extensive investigation in recent decades, yielding significant insights. This condition is primarily characterized by a sustained elevation in glycemic levels stemming from insulin resistance across diverse tissues such as the liver, muscle, and adipose tissue, alongside dysfunction in the pancreatic β-cells[7-12]. Evidence indicates that the dysfunction of pancreatic β-cells is in part, a consequence of prolonged and excessive exposure to fatty acids and glucose[13,14]. Moreover, the presence of intracellular and extracellular amyloid deposits in pancreatic islets can accelerate β-cell apoptosis, with an estimated 40%-100% of individuals with T2D exhibiting such deposits in these islets[15].
A theoretical proposition suggests that individuals with T2D may experience compromised glucose absorption and utilization attributable to mitochondrial dysfunction[16]. Additionally, glucotoxicity and lipotoxicity are posited to play a role in impairing β-cell mass, potentially culminating in β-cell apoptosis[13,17]. Furthermore, an observable surge in glucagon secretion has been documented with elevated concentrations observed during both fasting and the postprandial period. This heightened secretion contributes to an upswing in hepatic glucose production. Notably, the origin of postprandial glucagon production may extend beyond the pancreas, implicating tissues such as the intestine rather than being exclusive to pancreatic sources[8].
Taking into account that emergence of T2D is intricate and interplays within both early and late-life environments, coupled with the neurohormonal network predominantly orchestrated in the brain to regulate weight[18]. It is crucial to emphasize that this entire sequence of changes exhibits considerable variations from one individual to another, underscoring the inherent heterogeneity among those with T2D. Landmark genome-wide association studies have identified > 500 genetic loci contributing to T2D largely focused on β-cell function and mass. This has provided unprecedented insights into the polygenic architecture of the disease. Remarkably, the prevailing genetic variants linked to T2D exhibit similarities across populations of Asia and Europe. This suggests a broad and genetic predisposition to T2D among diverse ethnicities[15].
Numerous studies consistently indicate that a family history of T2D stands out as one of the most robust risk factors for the onset of this condition. Individuals with a family predisposition of T2D may face up to four times higher likelihood of developing the disease compared to those without such a family history[19,20]. Evidence also suggests a potential correlation between high birth weight in children and an elevated risk of development. This association is believed to be influenced by the impact of maternal hyperglycemia during pregnancy. Specifically, the excessive transfer of glucose across the placenta from mothers with elevated blood sugar levels is not only linked to increased fetal growth but also contributes to a direct, non-genetic escalation in the likelihood of their offspring developing T2D. This heightened risk may be attributable to the additional metabolic strain imposed on the developing endocrine pancreas during fetal development[21].
Morze et al[22] conducted a meta-analysis, incorporating data from 61 prospective observational studies involving approximately 72000 participants. Their objective was to explore potential associations between baseline metabolite levels and the subsequent risk of developing T2D. The results unveiled significant correlations between heightened circulating levels of glycerolipids (diacylglycerols and triacylglycerols), phosphatidylethanolamines, ceramides, and specific amino acids-especially branched-chain amino acids – with an increased risk of T2D.
Recent data also suggest that the pathogenesis of T2D may involve the activation of central counterregulatory neuroendocrine and autonomic responses. Within this framework, dysfunction of agouti-related peptide and pro-opiomelanocortin neurons, leading to a subsequent reduction in melanocortin signaling, could contribute to the development of T2D[13]. However, research in this area remains limited, and further studies are necessary to elucidate these potential mechanisms involved in the pathogenesis of T2D.
Dysregulated miRNAs in T2D
In recent years, as increase number of circulating miRNAs have exhibited dysregulation in individuals with T2D or those at a pre-T2D stage, shedding light on the potential utility of these small non-coding RNAs as biomarkers for predicting T2D.
In a study by Jiménez-Lucena et al[23] with a median follow-up of 60 months, higher baseline plasma values of miR-30a and miR-150, as well as more attenuated basal values of miR-15a and miR-375, were identified in incident-T2D human subjects compared to non-T2D subjects. miR-150 targets Myb, Elk1, and Etf1, genes pivotal in the regulation of β-cell differentiation and function[24], thereby amplifying insulin resistance in various tissues. Similarly, miR-30a-5p directly suppresses the expression of the Beta2/NeuroD gene, instigating glucotoxicity in β-cells and compromising insulin production and secretion[25]. In contrast, miR-15a inhibits endogenous uncoupling protein-2 expression, thereby improving glucose-induced insulin secretion. Conversely, studies on miR-375 in individuals with T2D have consistently shown upregulation[26-29]. This particular miRNA targets pancreatic genes, including insulin, myotrophin, and phosphoinositide-dependent protein kinase-1, with its overexpression suppressing glucose-induced insulin secretion[30].
In a recent investigation, individuals with T2D and dyslipidemia exhibited the upregulation of plasma miR-218, miR-132, and miR-143, accompanied by the downregulation of miR-21, miR-122, and miR-155[31]. The study further unveiled notable correlations, revealing negative associations between miR-218 and erythrocyte microvesicle levels, miR-132 and erythrocyte and uric acid plasma values, miR-143 and creatinine levels and diastolic blood pressure. Among the downregulated miRNAs, positive correlations were observed between miR-21 and tumor necrosis factor-α plasma levels, as well as between miR-155 and interleucin-1β. Conversely, miR-122 exhibited a negative correlation with creatinine and diastolic blood pressure. It is noteworthy, however, that conflicting findings exist in the literature. While this study reported downregulation of miR-122, other studies have documented its upregulation in the plasma of adults with T2D[32,33] and in children who also displayed elevated levels of miR-29a, a miRNA implicated in glucose homeostasis[34,35]. miR-122, recognized as the most abundant miRNA in the liver, is renowned for its role in regulating lipid homeostasis[36]. Nevertheless, its downregulation has also been associated with the induction of PTP1B, leading to hepatic insulin resistance[37].
In the realm of down-regulated miRNAs associated T2D, Avgeris et al[38] conducted an analysis of 84 circulating miRNAs and identified decreased values of miR-24-3p, miR-214-3p and let-7f-5p in individuals with T2D. Notably, these miRNAs exhibited correlations with serum insulin and hemoglobin A1c (HbA1c) levels. Another study, comparing normal individuals, T2D-susceptible individuals, and those with established T2D, observed significant reductions in miR-126-a miRNA linked to endothelial dysfunction[39]-in both susceptible and T2D subjects[40]. Recently, Ruan et al[41] reported decreased levels of miR-199-3p in the serum of individuals with T2D compared to healthy individuals, accompanied by increases in blood glucose, HbA1c, and Homeostasis Model Assessment of Insulin Resistance. In another study examining plasma from prediabetic, T2D, and healthy individuals, miR-148b-3p was found to be upregulated, while miR-27a-3p was downregulated in both diabetes and prediabetes. Intriguingly, miR-148b-3p appears to contribute to T2D development by augmenting the expression of pro-inflammatory cytokines and influencing DNA methylation, potentially disrupting metabolic homeostasis. Conversely, miR-27a-3p directly targets peroxisome proliferator-activated receptors γ, a master regulator of glucose and fatty acid metabolism in T2D, potentially impacting insulin sensitivity and lipid dysregulation, key hallmarks of the disease. Additionally, miR-27a-3p regulates the expression of several genes involved in various metabolic pathways[42].
Intriguingly, beyond miR-375, Monfared et al[29] revealed altered levels of circulating miR-182, miR-503, and miR-320a in both saliva and serum of individuals with T2D. The former two exhibited upregulation, while the latter showed downregulation. Notably, miR-182 has been demonstrated to regulate Sirt1 and NOX4 in patients with diabetic keratopathy[43], while miR-503 regulates mTOR in gestational diabetes mellitus, influencing pancreatic β-cell function[44]. Concerning miR-320a, Jo et al[45] established a correlation between its downregulation in human islets of T2D subjects and an increase in glucagon expression, attributed to miR-320a binding to the glucagon 3′ UTR.
Numerous other circulating miRNAs exhibit dysregulation in the context of T2D. For instance, miR-130a is downregulated in peripheral blood samples from T2D patients, acting on peroxisome proliferator-activated receptors γ and thereby modulating glucose metabolism[46]. In serum samples from T2D patients, miR-128, miR-130b, and miR-374a are upregulated. These elevated miRNAs facilitate the downregulation of their targets-insulin receptor substrate 1, phosphoinositide-3-kinase regulatory subunit 1, suppressor of cytokine signaling 4-thereby attenuating insulin signaling, promoting insulin resistance, and inducing dysregulation of insulin secretion. This cascade contributes to endoplasmic reticulum stress, culminating in long-term β-cell dysfunction and apoptosis[47].
In the plasma of diabetic patients, miR-572 is overexpressed, while miR-1249 and miR-320b are poorly expressed. The dysregulation of these miRNAs influences the expression of eukaryotic translation initiation factor 2 alpha kinase 3, a gene that governs the differentiation and proliferation of pancreatic β-cells. Eukaryotic translation initiation factor 2 alpha kinase 3 is induced by endoplasmic reticulum stress, contributing to β-cell dysfunction and T2D. The decrease in miR-1249 and miR-320b also promotes an increase in FOXO1, a gene associated with the deregulation of cellular energy metabolism, mitochondrial dysfunction, apoptosis, and potential contributions to cardiac dysfunction[48].
Finally, numerous miRNAs are deregulated, inducing modulation of the expression of some target genes within the insulin signaling pathway. These include miR-21, miR-30d, miR-34a, miR-125b, miR-126, miR-130b, miR-140, miR-142, miR-144, miR-148a, miR-192, miR-195, miR-222, miR-423-5p, miR-503, and miR-532; while some deregulated miRNAs favor the inflammatory process, such as miR-23a, miR-96, miR-144, miR-146a, miR-186, miR-191, miR-409, miR-454, miR-455, miR-665, miR-766, let-7 and let-7i[49].
Pathophysiology of BC
World Cancer Day 2021 brought shocking news from the World Health Organization: The December 2020 IARC statistics revealed BC surpassing lung cancer as the most diagnosed cancer globally. In 2020 alone, BC claimed a staggering 2.3 million diagnoses and 685000 lives worldwide. By year-end, an estimated 7.8 million women were living with a recent diagnosis, solidifying BC position as the most widespread cancer globally[50]. This staggering prevalence underscores the urgency of intensifying research and support for individuals battling this ubiquitous disease.
BC displays significant clinical and molecular diversity. This diversity guides treatment strategies, considering factors like tumor burden and metastatic patterns to optimize effectiveness while minimizing side effects. Early-stage BC, either in situ or invasive offers a high cure probability (70%-95%). In contrast, advanced or metastatic BC isn't curable but is treatable to enhance survival and symptom control. Diagnosis relies on a triad of clinical examination, biopsy analysis, and mammography, with a nearly 100% sensitivity and specificity[51].
Microcalcifications, first identified in 1951[52], are a common mammographic finding and serve as a valuable early indicator of BC. Approximately 30% to 50% of non-palpable tumors detected in screenings are solely identified by the presence of microcalcifications[53,54]. They are also present in 93% of ductal carcinoma in situ cases[55]. Characterization of microcalcifications involves assessing morphology, size, and spatial distribution within the breast tissue[56,57]. Calcifications are also classified based on their chemical composition and association with benign or malignant lesions.
All BC originates in the terminal duct lobular units of the collecting duct, regarded as functional components of the breast. The histological and molecular characteristics of BC have important implications for therapy, leading to the development of several classifications based on molecular and histological characteristics. The most frequent BC histological subtypes include ductal carcinoma, more recently called ‘no special type’, and lobular carcinoma as invasive lesions. However, these early lesions are regularly detected as in situ or in a preinvasive status in microcalcifications or small tumor biopsies.
The Perou and Sorlie subtypes, based on an expression pattern of 50 genes (PAM50), postulate three types of BC: Basal-like (presenting TP53 and BRCA mutations and medullary-like histology with poor differentiation); Claudin low (largely triple-negative, metaplastic and human epidermal growth factor receptor 2 (HER2)-enriched (which presents HER2, GRB7, TOPO2, and/or MYC amplification; PIK3CA mutations; amplification; is non-special type and presents pleomorphic lobular and micropapillary histology[58]. Recent elucidation has changed this conception, regarding these 3 subtypes as phenotypes related to BC[59].
The World Health Organization classification for BC recognizes several subtypes of invasive BC, presenting diverse and specific molecular alterations[50,60]. The 10 Histological Types bring assessment around the morphology or appearance of the BC, such as the formation of structures similar or not to normal breast tissue, as well as the appearance of cells and mitotic activity: Lobular pleomorphic or classical, micropapillary, mucinous, tall cell carcinoma with reverse polarity, metaplastic, adenoid cystic, Adenocarcinoma with lymphoid-rich stroma, secretory, and apocrine. Lobular carcinomas mainly harbor CDH1 mutations leading to the loss of E-Cadherin expression in the immunohistochemistry assay. This type presents PIK3CA, PTEN, AKT1, ERBB2, and ERBB3 mutations as gains in copy number in the estrogen receptor (ER) 1 gene. Secretory carcinomas regularly present a very frequent and specific translocation fusing both genes NTRK3–ETV6. Adenoid cystic carcinoma is characterized by the frequent fusion of the MYB–NFIB gene (60%), lacks TP53 and PIK3CA mutations; and shift in MYB or MYBL, including MYB–NFIB fusion. Micropapillary histological type presents frequent PIK3CA, MAP3K1, and GATA3 mutations, presents potency for lymphovascular invasion and difficulty in accurate imaging estimation, and as a component of mixed Non-Specific Type carcinoma. The mucinous type presents an increase in circulating MUC1 breast product CA 15-3[51,61]. Tall cell carcinoma with reverse polarity is often Triple-negative; with focal and low ER expression in 40%; with very frequent IDH2 and PIK3CA mutations. Metaplastic has frequent TP53 and PIK3CA, WNT pathway activation, and Claudin-low: Reduced genomic instability, mutation load and proliferation levels in addition to high levels of inflammatory infiltrate and stromal cells[57]. Adenocarcinoma with lymphoid-rich stroma has frequent TP53 mutations and BRCA inactivated. The secretory type presents NTRK3–ETV6 fusion, and clinicopathologic characteristics and survival outcomes remain to be elaborated[62].
In summary, achieving a complete understanding of the histological and phenotypic characteristics of BC, combined with the profile of genetic mutations and the epigenetic regulation underlying the disease, is a long-term and extremely complex task. However, it appears to be the path toward personalized treatment with tailored approaches for each molecular and histological type. Studies with large-scale or scanning scientific approaches involving genomic, transcriptomic, proteomic, and metabolomic profiles, in association with mechanistic studies, can categorize and characterize BC in pathophysiological terms, pointing to therapies, discoveries of new therapies, and future large-scale therapeutic approaches.
The precise pathophysiological mechanisms initiating BC are not entirely clear. However, it is recognized that genetic and epigenetic regulations of cells are altered in early disease, functioning as a cascade of events at the cell level. Mutations, in interaction with epigenetic changes, lead to the emergence of cancer cells displaying the known Hallmarks of Cancer: Evading growth suppressors, sustaining proliferative signaling, resisting cell death, enabling replicative immortality, genome instability, tumor-promoting inflammation, inducing/accessing vasculature, activating invasion and metastasis, reprogramming cellular metabolism, and avoiding immune destruction[63]. Hanahan et al[63] proposed four emerging hallmarks: Non-mutational epigenetic reprogramming, unlocking phenotypic plasticity, polymorphic microbiomes, and cellular senescence. These hallmarks provide a heuristic tool for categorizing essential and common aspects in the vast diversity of BC and other cancer phenotypes and genotypes into underlying biological attributes. Understanding the main regulatory pathways involved in the Hallmarks of cancer, their commonalities with T2D, and the dynamics of their regulation presents intriguing therapeutic potential in both diseases.
Mapping the involvement of miRNAs in cancer hallmarks and cancer pathophysiology, as well as their response to different types of drugs, is part of the study of non-mutational epigenetics-a growing Hallmark. A comprehensive approach involves observing the interaction with other epigenetic mechanisms to comprehend the diversity of BC in molecular, histological, inflammatory, and metabolic fields, aiming for therapies with greater clinical precision.
Regarding genetic mutations, altered genes exhibiting high levels of amplification and gain-of-function mutations in BC include ER 1, FOXA, MYC (MYC Proto-Oncogene, BHLH Transcription Factor), Retinoic acid receptor alpha, Tumor Protein 53, Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Alpha, Phosphatase and tensin homolog, Cyclin D1, ERBB2 (13%), FGFR1 (11%), GATA transcription factor 3, Lysine Demethylase 5A, and Topoisomerase IIα. On the other hand, genes that have shown homozygous deletions and loss-of-function mutations include Lysine Methyltransferase 2C, and AT-Rich Interaction Domain 1A. These genes are part of crucial pathways for growth, proliferation, angiogenesis, survival, and metabolism[64].
Chromatin state and accessibility play crucial roles in the regulation of gene expression and cellular identity, and modifications in accessibility are implicated in oncogenesis, progression, and metastasis. Epigenetic changes are integral to breast carcinogenesis and progression, involving both global hypomethylation (activating genes, upregulating oncogenes, and causing chromosomal instability) and locus-specific hypermethylation (repressing genes and leading to genetic instability by the silencing of DNA repair genes). Methylation in histone tail modifications induces changes in chromatin structure, silencing gene expression, and remodeling nucleosomes.
A comprehensive study utilizing single-nucleus and single-cell approaches constructed a pan-cancer epigenetic and transcriptome atlas, identifying epigenetic drivers associated with cancer changes. This study revealed universal epigenetic drivers, such as ABCC1 and VEGFA, as well as cancer-specific drivers like regulatory regions of FGF19, ASAP2, EN1, and the PBX3 motif. TP53, TNF signaling and hypoxia were associated to oncogenesis, while pathways involving estrogen response, epithelial to mesenchymal transition, and apical junction were associated with transition to metastasis. This research demonstrated a strong relationship between improving accessibility and suggested cooperation between epigenetic and genetic drivers in cancer, providing evidence of clinically relevant epigenetic mechanisms in cancer initiation and transitions[65].
Considering that cancer arises from cumulative genetic mutations, along with epigenetic alterations and environmental factors, epigenomic events, including miRNAs related to cancer, can be valuable in oncological practice as therapeutic tools. They have the potential to modify gene expression without causing permanent changes in the genomic sequence.
Dysregulated miRNAs in BC
Significant efforts have been directed towards developing and validating innovative approaches for BC patients. BC is characterized by the aberrant expression of multiple genes and miRNAs[66,67]. The exploration of miRNAs in BC aims to reveal novel biomarkers and therapeutic targets, providing valuable insights into high-scale epigenetic gene regulation. Recently, numerous studies have delved into the role of miRNAs in BC gene regulation[67-69]. However, the effectiveness of these emerging biomarkers relies heavily on their validation through clinical and analytical assessments. This validation is essential to pave the way for personalized treatment, early diagnosis, and prognostic biomarkers, aligning with established approaches used in clinical practice[66].
Garrido-Palacios et al[67] highlighted the crucial impact of early diagnosis on significant improving long-term survival rates for BC[70]. The predominant diagnostic techniques for BC screening involve imaging methods, with mammography considered the gold standard and ultrasounds[71] is regarded an effective complementary technique to identify false-negative.
However, mammography faces severe limitations outside the age range of 40 to 59 years, particularly in patients under 40 years of age, leading to potential underdiagnosis[72]. Additionally, the incidence of triple-negative tumors, associated with a worse prognosis, is higher in the 40 to 59 age group. Notably, cancer such as pregnancy-associated BC, with a mortality rate of about 50%-60%, are prevalent in this age group[73]. Early diagnosis before metastasis significantly improves the overall cancer survival rate[74]. Biomarker analysis emerges as a valuable strategy to overcome the limitations of imaging techniques and facilitate early disease diagnosis. Commonly used biomarkers for prognosis and guidance regarding systemic treatment in BC include HER2, the KI-67 protein, ER, and progesterone receptor (PR).
MiRNAs have emerged as promising biomarkers for BC and have garnered increase attention in recent years[67]. MiRNAs constitute a class of molecules extensively studied for their crucial role in post-transcriptional regulation of protein expression. These small, short-stranded RNAs, approximately 17 to 22 nucleotides in length, do not encode proteins but act by binding to mRNA, thereby repressing protein translation. MiRNAs are ubiquitously found in various organisms, including animals and plants, and play a pivotal role in regulating numerous physiological and pathological processes; evidence suggests that at least one-third of all biological pathways are under the influence of miRNAs[75].
MiRNAs implicated in promoting neoplasms are termed "onco-miRNAs". Circulating miRNAs, which regulate gene expression and mediate intercellular communication, have emerged as ideal candidates to be considered as non-invasive biomarkers for cancer diagnosis, prognosis, and therapeutic analysis[67]. Various justifications support this approach: MiRNAs identified in serum and plasma exhibit high stability, making blood collection non-invasive and reproducible[67,76], and dysregulation of miRNA expression has been associated with cancer.
Numerous studies have highlighted the involvement of miRNAs in various types of cancer. The literature indicates that miRNAs exhibit altered expression in several cancer types, and ongoing studies aim to elucidate their role in cancer physiopathology control[77].
Evidence strongly indicates that miRNAs play a pivotal role in processes commonly referred to as cancer hallmarks[78], and the cancer phenotype can be modulated through miRNA expression[79]. Recognizing this, the scientific community is actively engaged in comprehending the intricate mechanisms involving miRNAs and cancer to develop specific gene therapies. The objectives of these therapies include promoting better responses to conventional drug treatments and precisely suppressing oncogenic processes, thereby acting as a treatment to ameliorate disease prognosis. This paradigm is particularly relevant for BC.
MiRNA-200c emerges as a significant marker for a favorable prognosis in BC treatment. This specific miRNAs is readily detectable in circulation, and its absence serves as an early indicator that an individual may be developing metastases[80]. MiRNA-200c plays a critical role in suppressing proteins that promote the epithelial-mesenchymal transition. Notably, it competes with the binding site of an important long non-coding RNA, ATB. The interaction between miRNA-200c and ATB leads to increased expression of proteins that facilitate the promotion of metastasis[79].
In a study by Chen et al[68] miRNAs were identified as differentially expressed in the blood serum of MMTV-PyMT BC mice. Among these, miRNA-486 was selected for further investigation due to its significant decrease in circulation, accompanied by a decrease in skeletal and cardiac muscle tissue. MiRNA-486 is known to regulate the expression of proteins controlling protein degradation, such as PTEN and FOXO1a. The study suggests a connection between the expression of circulating miRNA-486 and its decrease in muscles, resulting in decreased protein synthesis. Furthermore, the study examined the expression of miRNA-486 in the blood serum of humans with BC and also found a decrease in its expression, implying a potential role in muscle atrophy in individuals with BC mediated by miRNAs[68].
MiR-145 and BC have also attracted attention. Jiang et al[69] demonstrated that miR-145 functions as a tumor-attenuating miRNA and its expression was reduced in BC tissues. The restoration of miR-145 can attenuate the proliferation of BC cells through the negative regulation of the HBXIP gene. Zou et al[81] showed that the expression of miR-145 in BC tissues was significantly reduced than that in normal breast tissues. In vitro experiments identified that miR-145 attenuated angiogenesis and tumor growth by reducing N-RAS and VEGF, thus playing a function in tumor suppression. However, there was no difference in the expression of miR-145 in normal breast tissues when compared to BC tissues at different clinical stages[81]. These findings suggest that the relationship between miRNAs and BC is complex and may vary in different contexts.
Given the conflicting data in the literature, Yoruker et al[82] conducted a systematic meta-analysis of miR-145 expression in BC. The study concluded that the expression of miR-145 in ER-positive BC patients was reduced compared to patients with negative ER. However, there was no significant difference in the expression of miR-145 between PR-positive and PR-negative BC patients. Furthermore, compared with HER-2 negative patients, miR-145 expression was decreased in HER-2-positive patients.
In current studies focusing on the interconnection between miRNAs and tumors, the abnormal expression of miR-155 in BC patients has received great attention. As an oncogene, the high expression of miR-155 has been considered a risk factor for BC. The suppressor of cytokine signaling-1 (SOCS1) is a consistent target gene of miR-155 in BC cells in the human evolutionary process. The expression of miR-155 and SOCS1 is negatively correlated[83]. Kong et al[84] showed that miR-155 acts in the epithelial-to-mesenchymal transition process and infiltration in NMuMG cells, demonstrating that miR-155 could be used as a diagnostic biomarker for BC metastasis.
The meta-analysis by Zeng et al[85] reported multiple clusters of miRNA networks involving antiestrogen resistance in BC; miR-155 was one of the main dysregulated miRNAs, showing that miR-155 can be used as a diagnostic marker for endocrine therapy in BC. Furthermore, the expression level of miR-155 was correlated with ER, PR, HER-2, tumor size, lymph node metastasis, and p53 status. In the analysis of different BC subtypes with ER and PR status, miR-155 had lower expression in ER+ or PR+ BC, but it was highly expressed in HER-2+ positive or lymph node metastasis BC compared to negative lymph node metastasis or HER-2-BC[86]. Sun et al[86] demonstrated that serum miR-155 levels were significantly increased in BC patients compared to healthy individuals. Moreover, serum from BC patients was collected after surgery and after four cycles of chemotherapy to assess the clinical treatment effects on candidate miRNA serum miR-155 levels decreased. Results showed that 79% of patients exhibited a response or stable disease after therapy, decreasing miR-155 serum levels.
MiRNA-10b is another miRNA utilized as a biomarker. Studies involving a combination of mouse and human cells have demonstrated that miR-10b has high expression in metastatic BC cells and positively regulates cell migration and invasion[87]. The elevated expression of miR-10b in non-metastatic breast tumors initiates robust invasion and metastasis, and the level of miR-10b expression in primary breast carcinomas correlates with clinical progression[88]. In recent years, many studies have assessed the prognostic value of miR-10b expression in BC, yielding conflicting results. Some studies concluded that miR-10b expression did not significantly influence survival[89], while others reported that miR-10b expression was predictive of decreased survival outcomes for BC[90].
The meta-analysis conducted by Wang et al[91] aimed to evaluate the prognostic value of miR-10b in BC. Analyzing overall survival and/or disease-free survival of 770 patients across 7 studies, the meta-analysis revealed that the overexpression of miR-10b is associated with lower disease-free survival. The expression of miR-10b is significantly linked to a worse prognosis, particularly in terms of lower disease-free survival in BC patients.
Other diverse miRNAs are deregulated in cellular and pathophysiological processes in BC. For example, miR-1207; miR-492; miR-135b induces cell proliferation and cell cycle progression, whereas miR-15a, miR-16, miR-22, miR-26a, miR-30b, miR-30c, miR-143, miR-206, miR-365, miR-424, miR-455, miR-483, miR-543, miR-708 act as anti-proliferative and cell cycle regulators. Other miRNAs such as miR-200c; miR-141, miR-331; miR-200b; miR-122; miR-374a induce metastasis and invasion; on the other hand, miR-497, miR-124a, miR-26b, miR-195, miR-148a, miR-340, miR-34a, miR-138, miR-494, miR-33b, miR-421, miR-193a, miR-211, miR-335, miR-133a, miR-124, miR-204 have anti-metastasis and anti-invasion effects. Finally, miR-21 and miR-203 have an oncogenic and pro survivor effect, while miR-148a, miR-34a, miR-204, miR-101 induce apoptotic and tumor suppressor response[92].
Overlapping miRNAs in T2D and BC
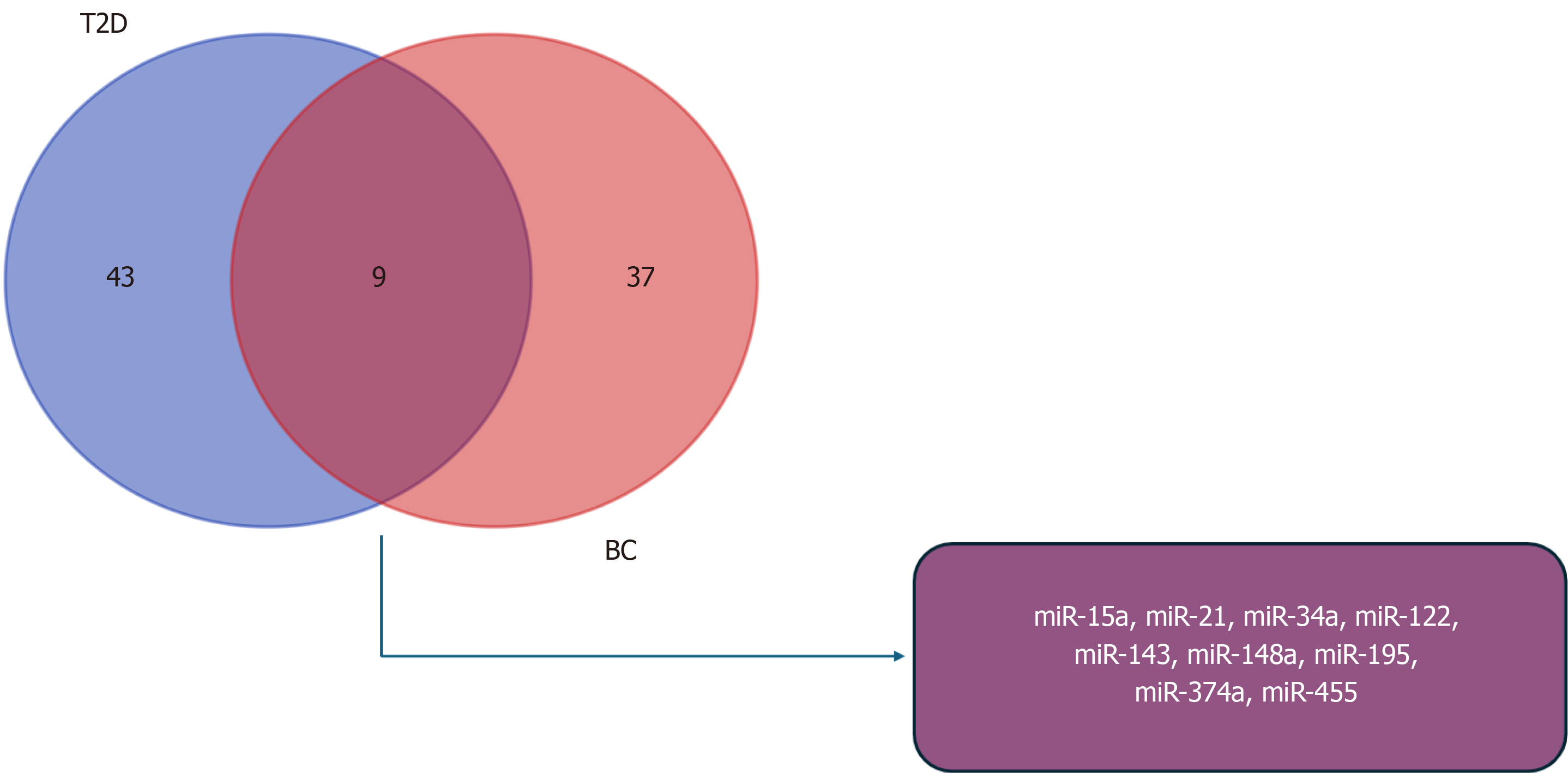
We performed a comprehensive miRNAs expression analysis in both T2D and BC, leading to the creation of a Venn diagram. The diagram revealed 52 miRNAs deregulated in T2D and 46 miRNAs deregulated in BC. Interestingly, 9 miRNAs were identified as overlapping in both conditions: miR-15a, miR-21, miR-34a, miR-122, miR-143, miR-148a, miR-195, miR-374a, and miR-455 (Figure 1). This observation underscores demonstrates the potential pivotal role of these 9 miRNAs in regulating signaling pathways and biological processes associated with the development or progression of T2D and BC. Additionally, it suggests the intriguing possibility that these miRNAs may contribute to pathophysiological processes in T2D that could lead to the development of BC.
Figure 1 Overlap of dysregulated miRNAs in type 2 diabetes and breast cancer.
Venn diagram shows dysregulated miRNAs in type 2 diabetes (blue) and in breast cancer (pink), including the overlapping miRNAs (purple). T2D: Type 2 diabetes; BC: Breast cancer.
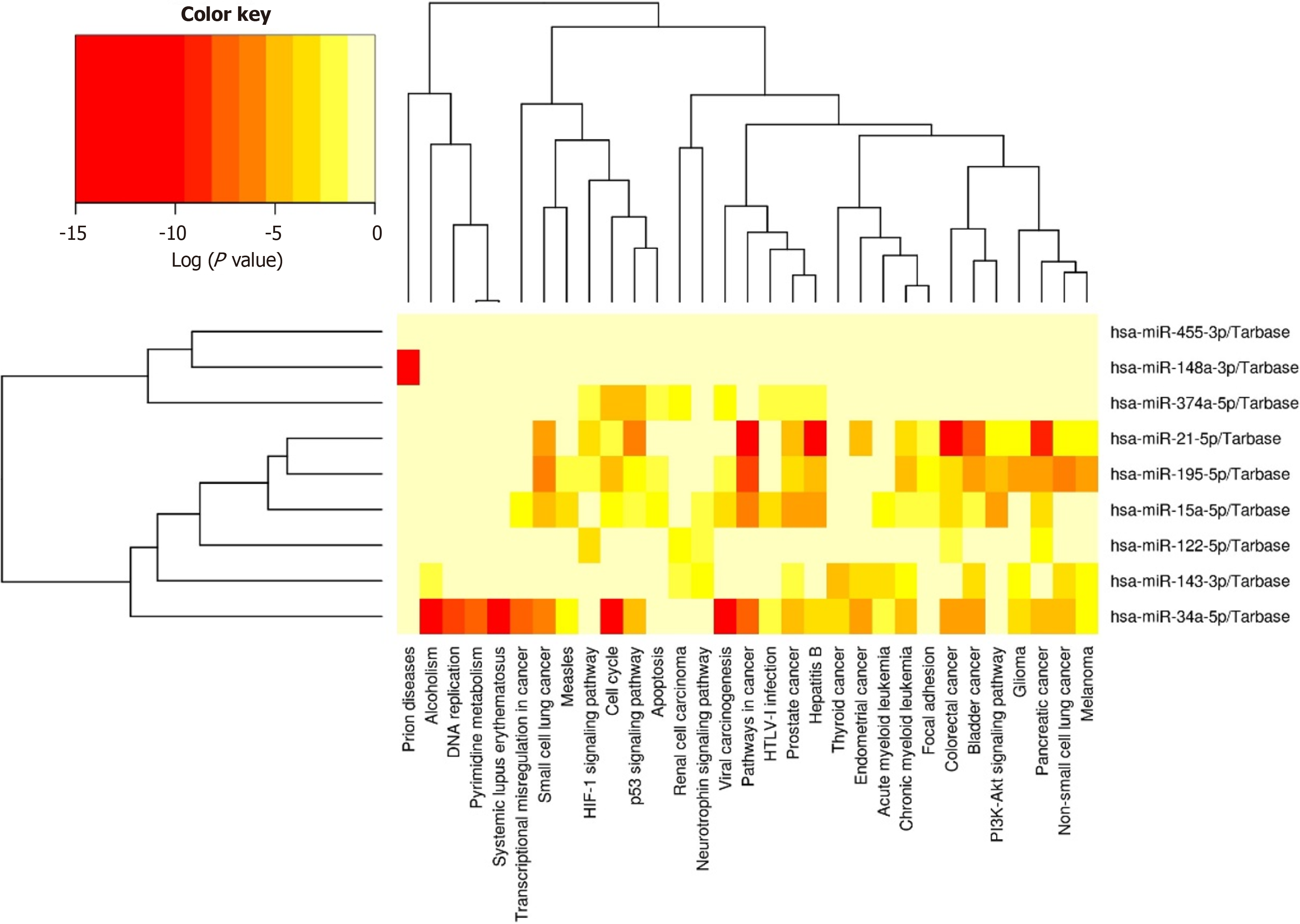
Finally, we conducted an exploratory analysis to elucidate the potential associations of these 9 miRNAs in the two diseases. Utilizing DIANA Tools miRPath V.3 integrated with TarBase and KEGG analysis (Figure 2), we aimed to identify both predicted and validated biological processes linked to these miRNAs.
Figure 2 Bioinformatics analysis of selected miRNAs regulating biological processes.
Nine miRNAs regulating various biological processes associated with type 2 diabetes and breast cancer using TarBase with KEGG analysis.
In the present heatmap, we can observe that 8 miRNAs play a regulatory role in biological processes associated with T2D and BC, with the exception of miR-455-3p, which does not exhibit regulation in these processes. Initially, miRNA has drawn considerable attention, showcasing its involvement in regulating DNA replication and the cell cycle-indicating that its deregulation support cell proliferation. Furthermore, this miRNA is implicated in inducing transcriptional deregulation in cancer and modulating signaling pathways in cancer. Both miR-195 and miR-374a also demonstrate regulatory roles in the cell cycle, potentially promoting the proliferation of tumor cells. miR-21 exhibits activity in cancer-related signaling pathways, demonstrating strong regulation not only in BC but also in other cancer types such as pancreatic, colorectal, and bladder cancer.
Despite uncovering the associations of these 9 miRNAs with T2D and BC, it is crucial to acknowledge that future studies are imperative to fully comprehend the nuanced and likely bidirectional roles of these miRNAs in the complex interplay between T2D and BC.