©The Author(s) 2021.
World J Diabetes. Dec 15, 2021; 12(12): 2058-2072
Published online Dec 15, 2021. doi: 10.4239/wjd.v12.i12.2058
Published online Dec 15, 2021. doi: 10.4239/wjd.v12.i12.2058
Figure 1 Intersection analysis of Kallmann syndrome and normosmic hypogonadotropic hypogonadism pathogenic genes.
nIHH: Normosmic hypogonadotropic hypogonadism; KS: Kallmann syndrome.
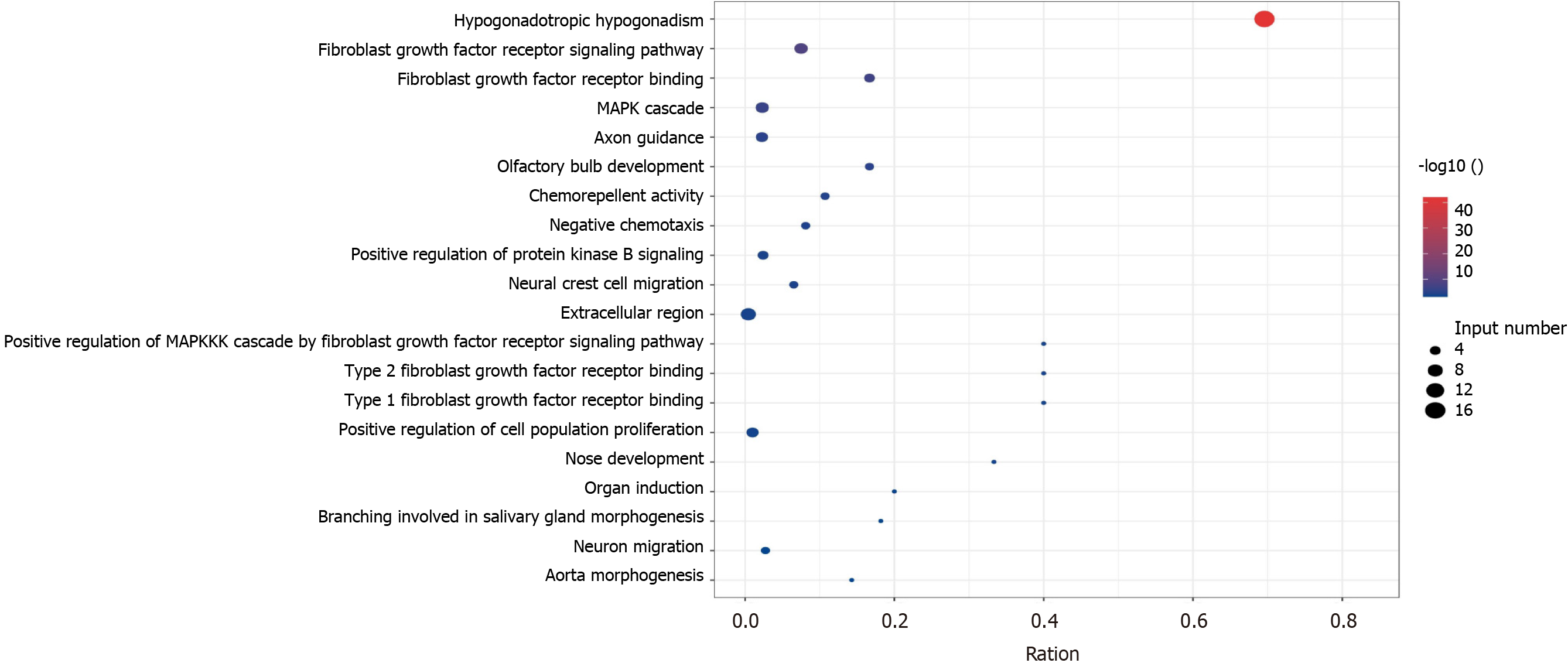
Figure 2 Kallmann syndrome pathogenic gene enrichment pathway and gene ontology, the x-axis represents gene ratio, and the y-axis represents gene ontology term; the size of the dot represents the number of genes, and the color of the dot represents the level of P value.
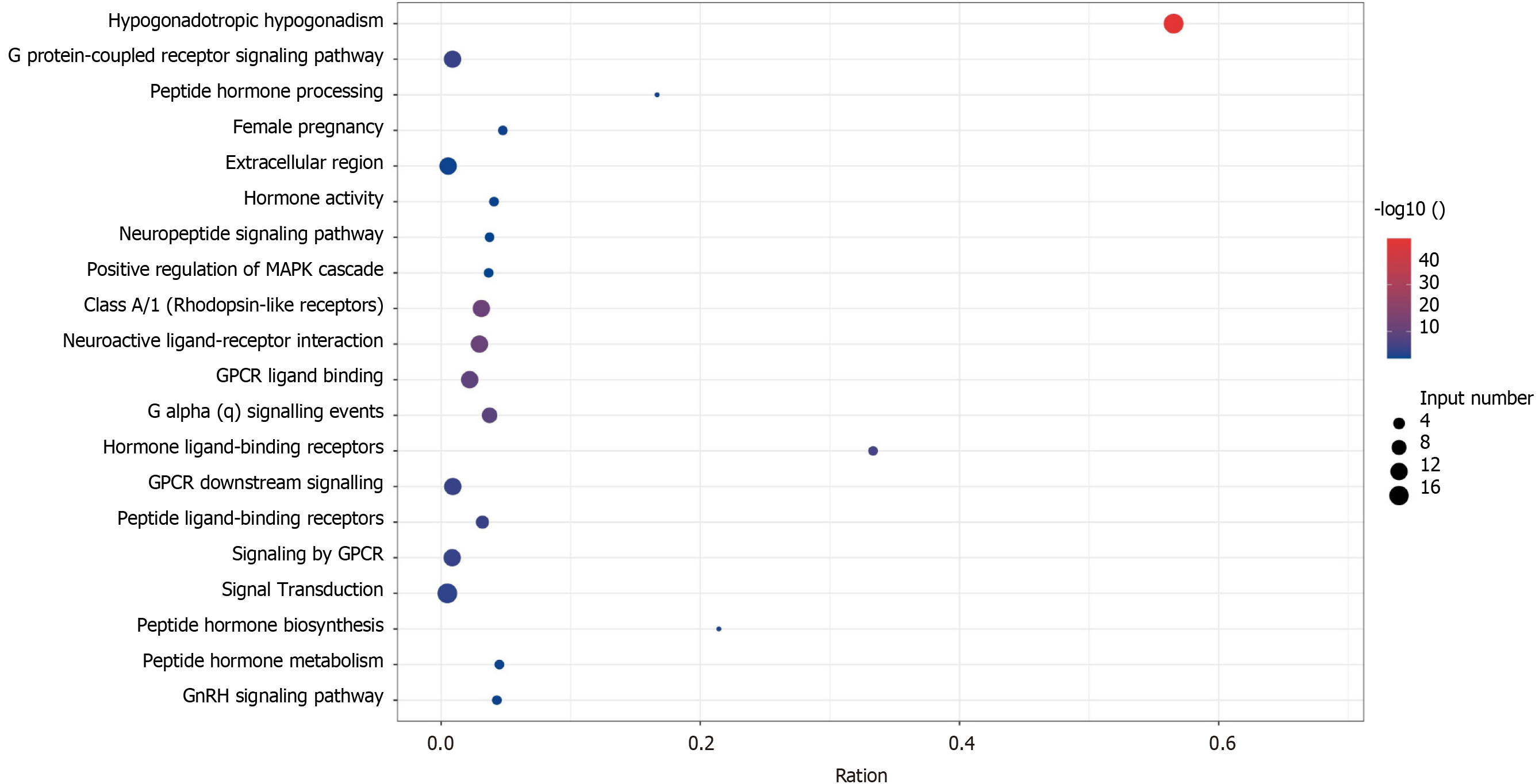
Figure 3 Normosmic hypogonadotropic hypogonadism pathogenic gene enrichment pathway and gene ontology, the x-axis represents gene ratio, and the y-axis represents gene ontology term; the size of the dot represents the number of genes, and the color of the dot represents the level of P value.
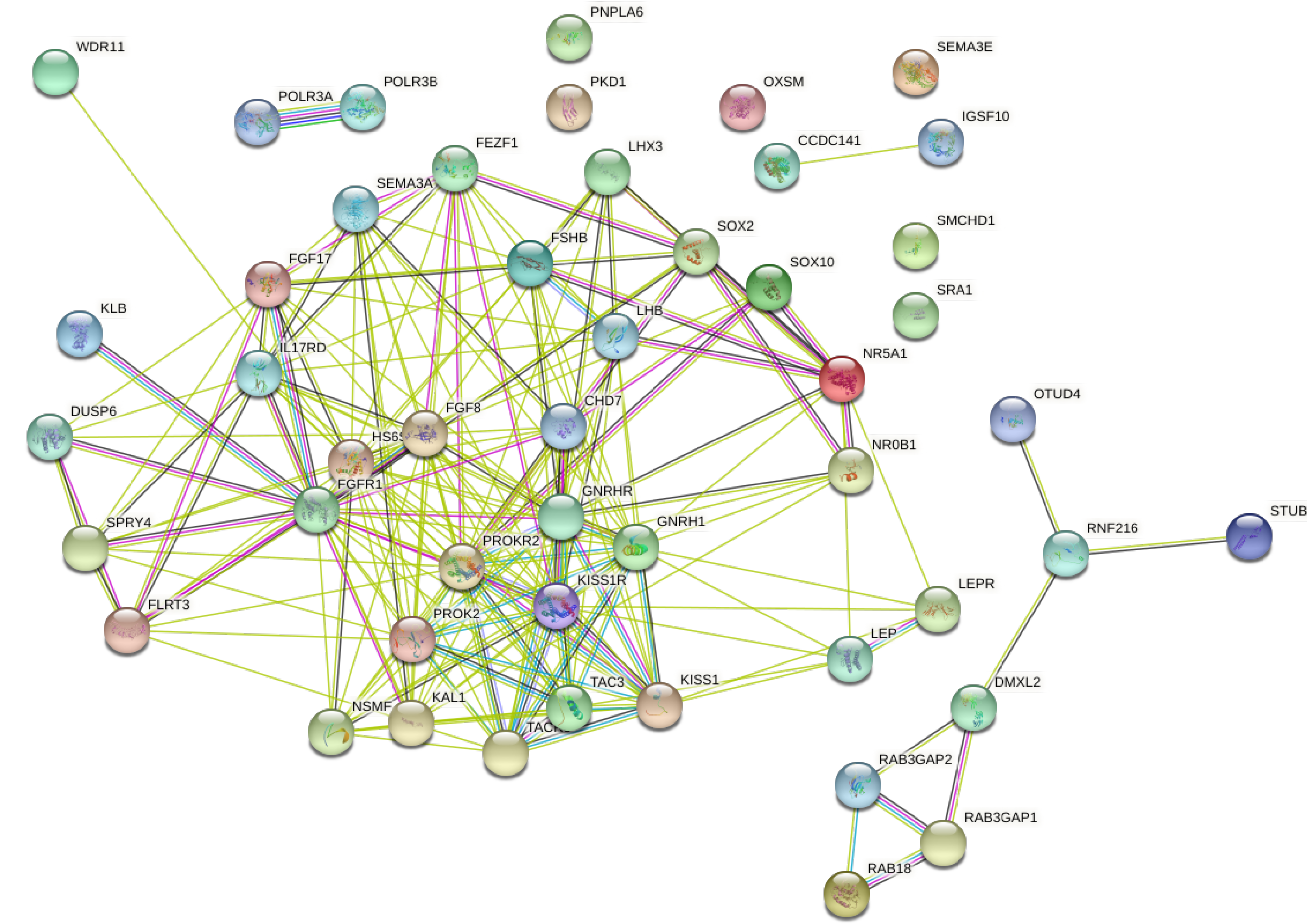
Figure 4 Normosmic hypogonadotropic hypogonadism interacts with Kallmann syndrome pathogenic gene proteins.
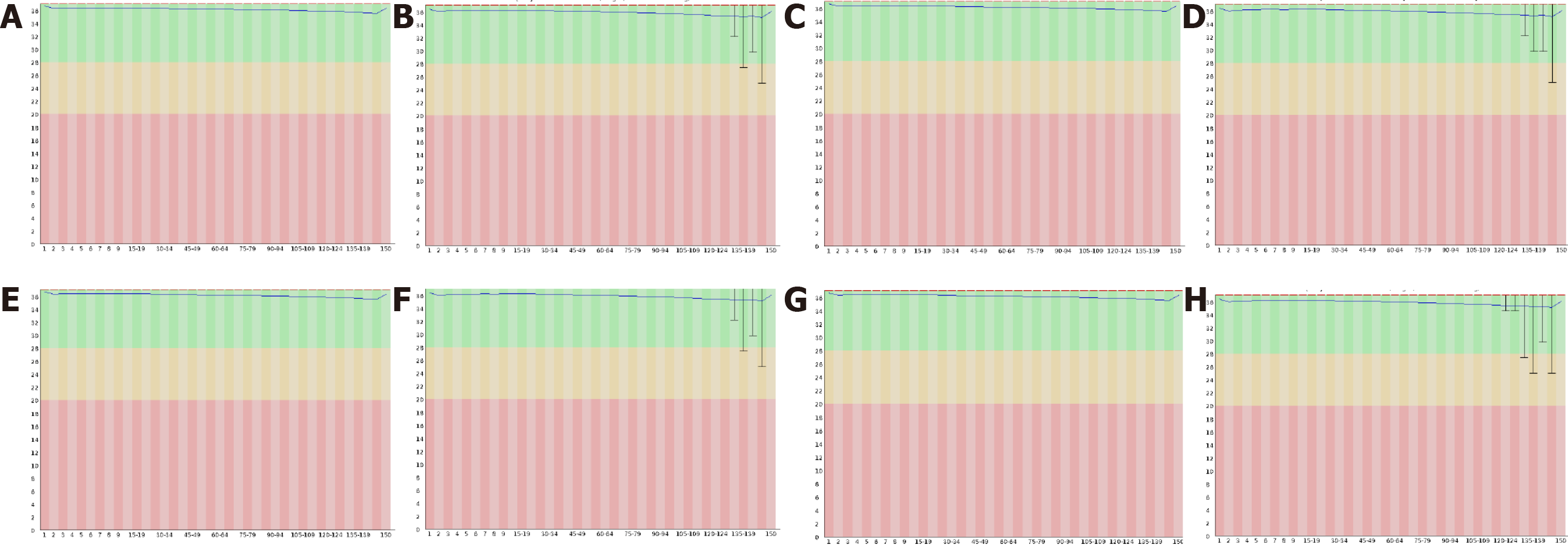
Figure 5 Original results of exome sequencing.
A and B: The sequence quality distribution map of reads 1 and 2 of patient 1; C and D: The sequence quality distribution map of reads 1 and 2 of patient 2; E and F is the sequence quality distribution map of reads 1 and 2 of patient 3; G and H is the sequence quality distribution map of reads 1 and 2 of patient 4.
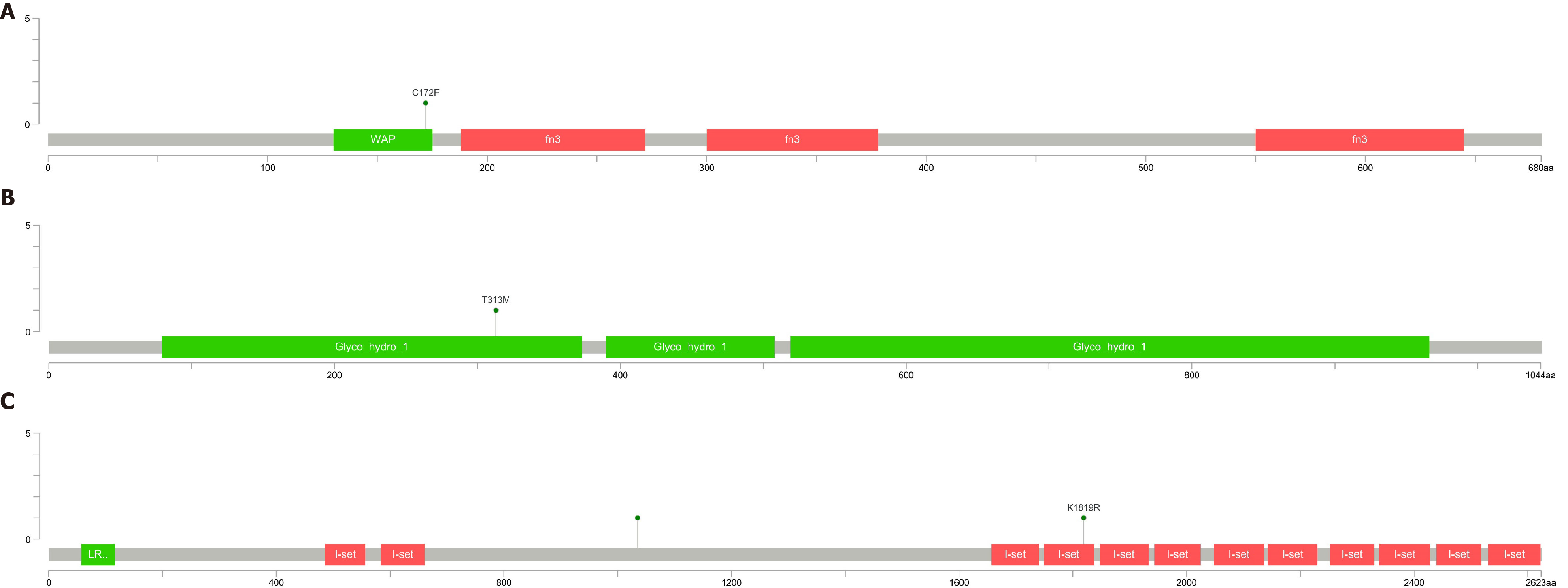
Figure 6 The location of four mutation sites.
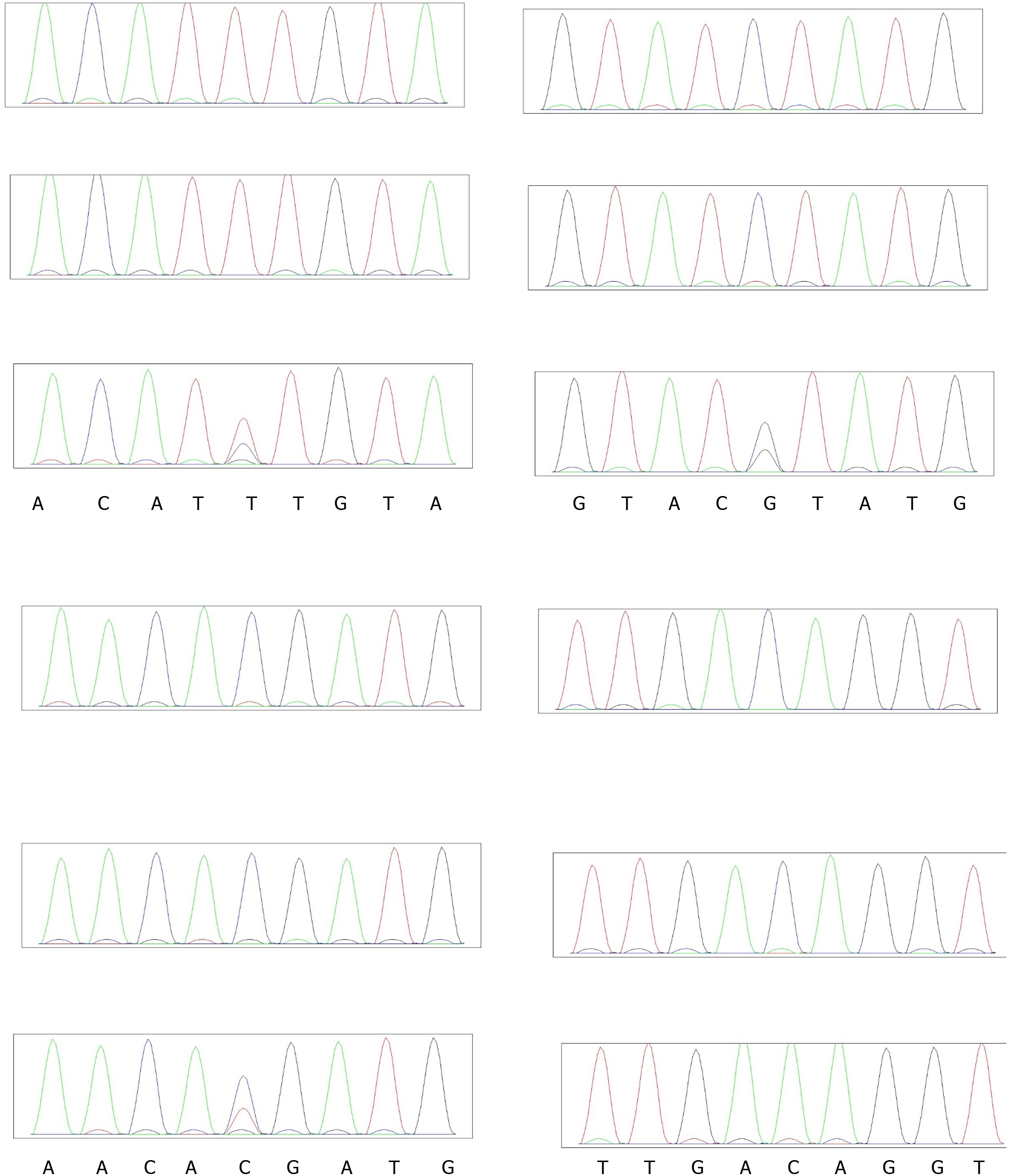
Figure 7 Sanger sequencing of 4 Kallmann syndrome family results.
- Citation: Sun SS, Wang RX. Molecular diagnosis of Kallmann syndrome with diabetes by whole exome sequencing and bioinformatic approaches. World J Diabetes 2021; 12(12): 2058-2072
- URL: https://www.wjgnet.com/1948-9358/full/v12/i12/2058.htm
- DOI: https://dx.doi.org/10.4239/wjd.v12.i12.2058