Published online Jul 15, 2025. doi: 10.4251/wjgo.v17.i7.105981
Revised: March 26, 2025
Accepted: May 21, 2025
Published online: July 15, 2025
Processing time: 152 Days and 10.5 Hours
UDP-glucose 6-dehydrogenase (UGDH) is a key enzyme in glucuronic acid metabolism and acts as a key mediator in several cancer developmental signaling pathways.
To offer a more systematic and comprehensive elucidation of the involvement of UGDH in the onset and progression of various malignancies.
The role of UGDH in cancer was investigated via public databases. The data were analyzed via various R packages and websites, including TISIDB, cBioPortal, STRING, Cytoscape, GSCALite, and CancerSEA. A rat hepatocellular carcinoma (HCC) model was established via the intraperitoneal injection of diethylnitrosamine. Hematoxylin-eosin staining, Masson staining, Ki67 and UGDH immunohistochemical staining, and ARG1 immunofluorescence staining of liver tissues were performed. Real-time quantitative PCR and Western blotting were used to detect UGDH expression. The UGDH gene was knocked down in Huh7 cells, and CCK8 and nude mouse tumor xenograft assays were performed.
High UGDH expression is associated with poor clinical outcomes in HCC, lung adenocarcinoma, lung squamous cell carcinoma, and sarcoma patients and is differentially expressed across molecular and immune subtypes. UGDH is primarily involved in the pentose and glucuronate interconversion pathway. Its expression is positively correlated with T helper, Tcm, and Th2 cells in most cancers. Moreover, experimental results demonstrated that UGDH expression is elevated in HCC tissues and that its downregulation inhibits HCC cell proliferation.
Our study revealed that UGDH could be a valuable prognostic biomarker and potential therapeutic target in many cancers, especially liver and lung cancer. UGDH could promote HCC cell proliferation, potentially by modulating the pentose and glucuronate interconversion pathways.
Core Tip: Our study identified UDP-glucose 6-dehydrogenase (UGDH) as a potential prognostic biomarker and therapeutic target for many cancers, especially liver and lung cancer. UGDH promotes hepatocellular carcinoma cell proliferation, possibly by modulating the pentose and glucuronate interconversion pathway.
- Citation: Cao X, Zheng SH, Shen JM, Li SZ, Hou L, Zhang JX, Li XK, Du HB, Zhang LP, Ye YA, Zao XB. Pan-cancer analysis of UDP-glucose 6-dehydrogenase in human tumors and its function in hepatocellular carcinoma. World J Gastrointest Oncol 2025; 17(7): 105981
- URL: https://www.wjgnet.com/1948-5204/full/v17/i7/105981.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i7.105981
Cancer is one of the most prevalent causes of mortality worldwide[1], and tumor drug resistance has become more prevalent due to the ongoing advancement of chemotherapy drugs[2]. Cancer is characterized by metabolic repro
UDP-glucose 6-dehydrogenase (UGDH) is an oxidoreductase that catalyzes the NAD+-dependent four-electron oxidation of UDP-glucose to UDPGA[6]. The active site of UGDH contains a highly conserved cysteine residue, which plays a key role in covalent catalysis[7]. During sugar nucleotide metabolism, UGDH is a rate-limiting enzyme that participates in the biosynthesis of glycosaminoglycans, including hyaluronan, chondroitin sulfate, and heparan sulfate[8]. Importantly, these glycosaminoglycans synthesized under UGDH-mediated UGDPA regulation are prevalent in the extracellular matrix and are likely involved in signal transduction, cell migration, cancer growth, and cancer metastasis[9]. Thus, UGDH is considered a molecular indicator of tumor progression in multiple cancer types, a potential prognostic marker in oncology and therapeutic target in cancer biology[10].
In recent years, researchers have devoted significant attention to the role of UGDH in malignant tumors. UGDH accelerates SNAI1 mRNA decay and impairs lung cancer metastasis[11]; it supports autophagy-deficient pancreatic ductal adenocarcinoma growth by increasing hyaluronic acid biosynthesis[12]; UGDH promotes tumor-initiating cells and a fibroinflammatory tumor microenvironment in ovarian cancer, and targeting UGDH inhibits ovarian cancer growth and metastasis[13,14]; UGDH limits prostate androgen availability without impacting hyaluronan levels and is a novel field-specific candidate biomarker of prostate cancer[15,16]; UGDH knockout inhibits glioblastoma growth and migration[17,18]; UGDH knockout impairs breast cancer cell migration and metastatic ability. UGDH is also a prognostic marker in breast cancer patients[19,20]. Additionally, UGDH is closely associated with tumor drug resistance[21], epithelial-mesenchymal transition (EMT)[22], and cellular localization[23]. Our previous study revealed that UGDH is a potential therapeutic target for hepatocellular carcinoma (HCC)[24]. Moreover, other studies have shown that UGDH upregulation is correlated with increased metastatic potential, decreased patient survival, and drug resistance in HCC[25,26]. The above studies indicate that UGDH has broad and diverse regulatory effects on cancer and is a promising target for diagnosing and treating tumors. However, there are no systematic studies on the expression, prognosis, and function of UGDH across cancers, so it is necessary to perform relevant studies.
Tumors with anatomical structures belonging to the same system and tumors from distinct organs but of the same histological type exhibit significant molecular similarities[27]. Therefore, examining the phenotypic characteristics of pan-cancer molecules will help elucidate their intrinsic regulatory mechanisms and their similarities to those of malignancies. In this study, we characterized UGDH expression via pan-cancer analysis across public databases. The expression, diagnostic value, clinical prognosis, and functional enrichment of UGDH were comprehensively analyzed via bioinformatics. Next, the relationships between UGDH and tumor-infiltrating lymphocytes, major histocompatibility complex molecules, immunostimulators, immunoinhibitors, chemokines, and chemokine receptors were investigated. To further assess the expression and impact of UGDH in HCC, we implemented clinical sample testing and in vivo and in vitro assays.
To obtain comprehensive information regarding UGDH RNA and protein expression in human tissues and cell lines, we conducted a systematic query of the human protein atlas (HPA) database (https://www.proteinatlas.org/). Furthermore, we employed TCGA (https://cancergenome.nih.gov) and the GTEx project (https://gtexportal.org/) to obtain comprehensive data on UGDH mRNA expression in tumor samples, corresponding paracancerous tissues, and normal controls. Samples with gene expression values of zero were excluded. Paired sample analyses were exclusively implemented. The Toil process was used to convert and normalize the fragments per kilobase per million format of the RNA sequencing data to transcripts per million reads, which were then log2-transformed for subsequent analysis. R software (version 4.2.1), "stats" (version 4.2.1), and "car" (version 3.1-0) were used to conduct statistical analyses. The "ggplot2" package (version 3.3.6) was used to visualize UGDH gene expression across the cancer categories. Cutoff values were established via the median expression procedure. The Wilcoxon rank-sum test was used to evaluate differences in expression between groups.
The diagnostic value of UGDH was assessed via receiver operating characteristic (ROC) curves in the context of 33 cancer categories. UGDH mRNA expression data in malignant and corresponding normal tissues from the TCGA and GTEx databases were used to construct ROC curves. The curves were depicted via the "ggplot2" package (version 3.3.6), and the "pROC" package (version 1.18.0) in R was used to perform ROC analysis. The diagnostic metrics that were calculated were the area under the curve (AUC), cutoff values, sensitivity, specificity, positive predictive value, and negative predictive value. Superior diagnostic accuracy is suggested by an AUC value close to 1.
The "survival" package (version 3.3.1) in R (version 4.2.1) was used to conduct Kaplan-Meier (K-M) survival analysis. This analysis compared the rates of overall survival (OS), disease-specific survival (DSS), and progression-free interval (PFI) between the high and low UGDH gene expression groups across 33 cancer types. Furthermore, correlations between UGDH and common clinical parameters in tumor types impacted by UGDH were analyzed with respect to OS. The statistical evaluation of survival differences was conducted via Cox regression analysis. The "survminer" (version 3.3.1) and "ggplot2" packages (version 3.3.6) were used to generate and visualize forest plots that depicted HRs, 95%CI, and P values.
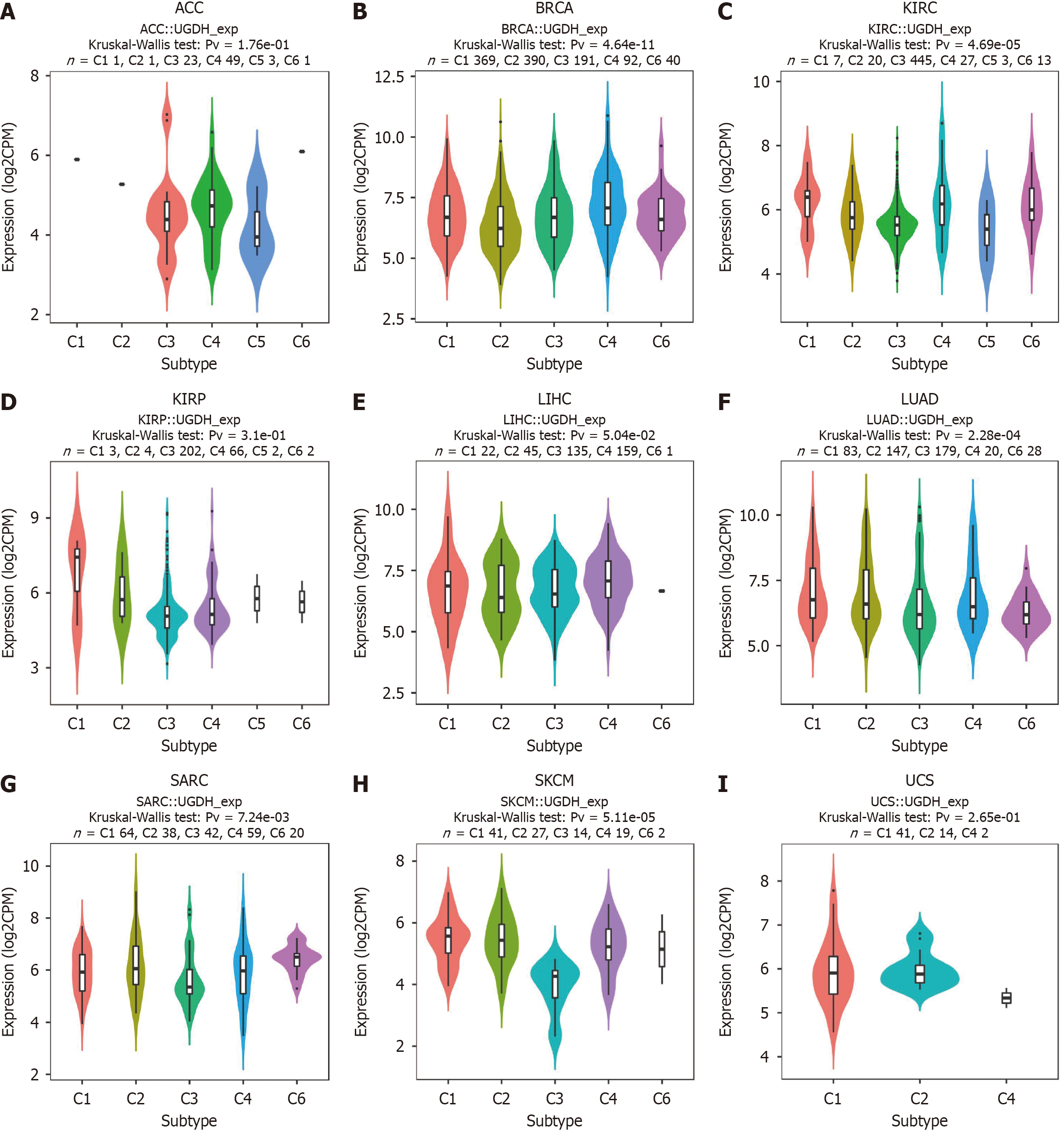
The "subtype" module of the TISIDB database (http://cis.hku.hk/TISIDB/) was employed to examine the relationships between UGDH expression and molecular or immune subtypes in cancers. This database incorporates a variety of datasets to evaluate the interactions between the immune system and cancer. UGDH mRNA expression was examined in various immune subtypes, including C1 (wound healing), C2 (IFN-g dominant), C3 (inflammatory), C4 (lymphocyte-depleted), C5 (immunologically silent), and C6 (TGF-b dominant). The molecular forms of various tumors were distinct.
Genetic alteration data on UGDH were collected by accessing the cBioPortal (https://www.cbioportal.org/). The search encompassed all studies from the TCGA Pan-Cancer Atlas. The "cancer types summary and mutations" and "mRNA vs study" modules were employed to investigate the frequency of somatic mutations and genomic information related to UGDH mutations in cancers. The "mutations" module was employed to identify mutation sites.
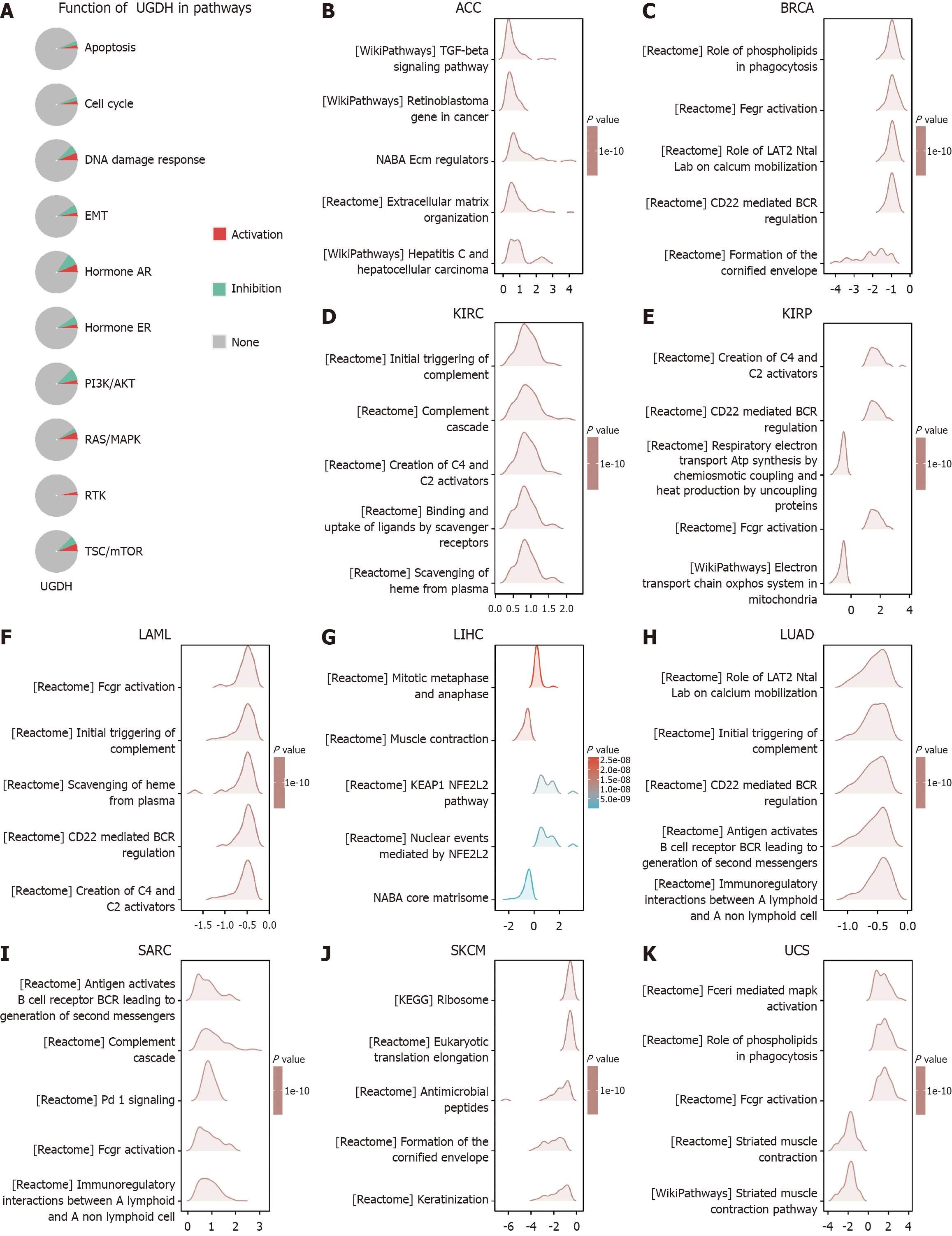
The STRING database (https://string-db.org/) was used to investigate the protein interactions of UGDH. A protein-protein interaction (PPI) network was constructed from the collected data, with a confidence score threshold of greater than 0.7 indicating significance. The data were subsequently imported into Cytoscape (3.8.0) for further analysis and visualization. The cytoHubba plug-ins identified key modules, and the top 10 nodes, as rated by the maximal clique centrality of cytoHubba, were presented as hub genes. The relationship between UGDH and hub gene expression was investigated. A P value < 0.05 was used to establish a significance threshold for the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses of UGDH PPI-related genes in the DAVID database (https://david.ncifcrf.gov/). The results obtained via the "ggplot2" package (version 3.3.6) are presented as a bubble chart. In addition, the GSCALite database (https://guolab.wchscu.cn/GSCA/) was utilized to identify the changes in critical cancer-related pathways in the 33 cancer categories that involve UGDH. These pathways included TSC/mTOR, RTK, RAS/MAPK, PI3K/AKT, hormone ER, hormone AR, EMT, DDR, the cell cycle, and apoptosis.
Variations in biological pathways between the high- and low-UGDH expression groups were elucidated via the "clusterProfiler" package (version 4.4.4) in the Gene Set Enrichment Analysis (GSEA) package. Significantly altered pathways were defined as those with an adjusted P value < 0.05 and a false discovery rate < 0.25. For each analysis, the number of permutations was set to 1000. The "ggplot2" package (version 3.3.6) was used to visualize the five enriched pathways as mountain maps.
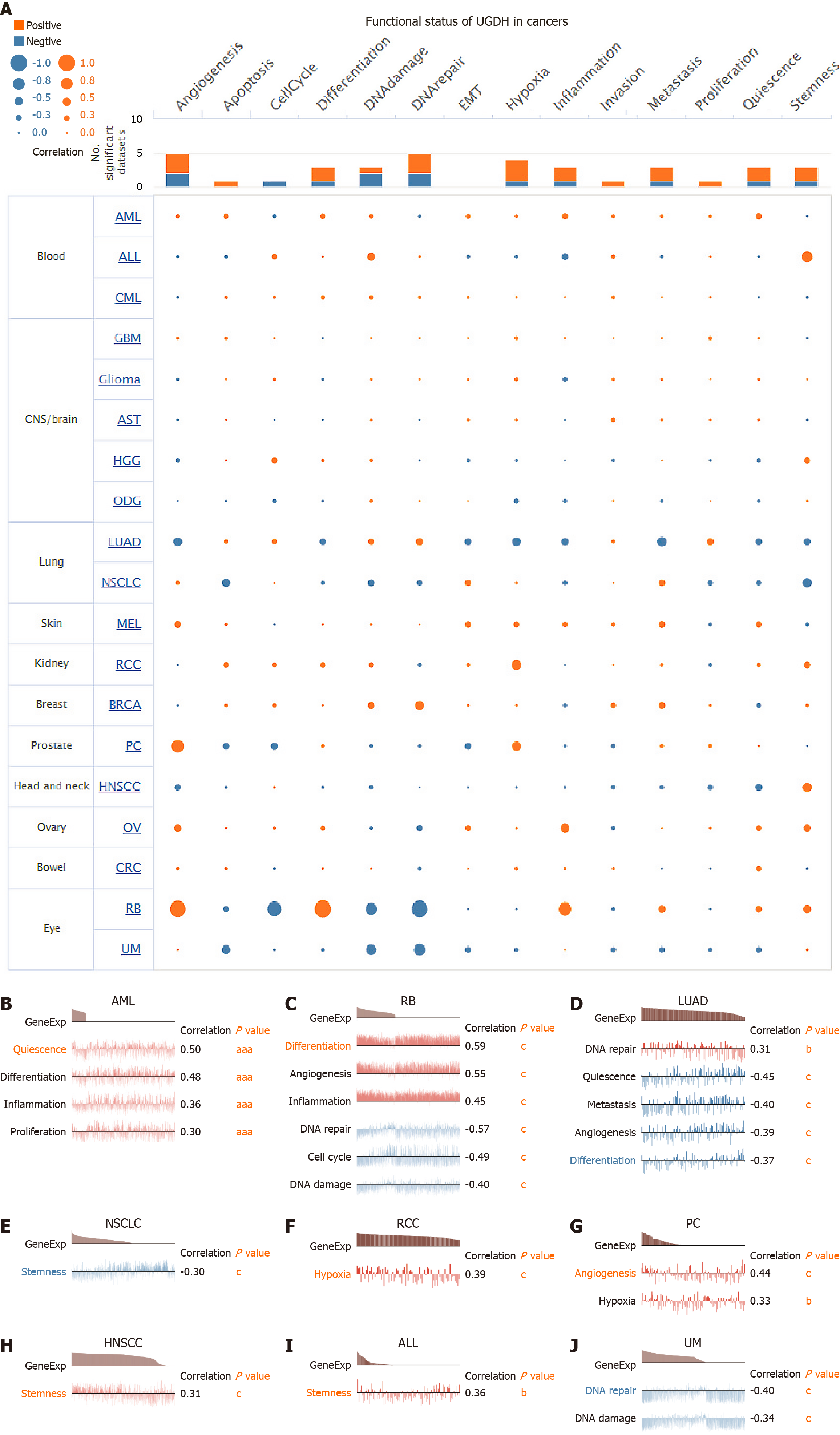
The CancerSEA database (http://biocc.hrbmu.edu.cn/CancerSEA) was used to investigate the functional status of UGDH in a variety of malignancies. The average correlation between UGDH and functional states was examined in 18 malignancies, including cell cycle, differentiation, invasion, metastasis, proliferation, EMT, angiogenesis, apoptosis, DNA damage, DNA repair, hypoxia, inflammation, quiescence, and stemness. The thresholds for significance were established as a correlation strength > 0.3 absolute value and a P value < 0.05.
The "GSVA" package (version 1.44.5) with the ssGSEA algorithm was used to evaluate the correlation between UGDH expression and tumor-infiltrating lymphocytes, MHC molecules, immunostimulators, immunoinhibitors, chemokines, and chemokine receptors in 33 malignancies. Spearman's correlation was employed to determine statistical significance, with P < 0.05 indicating a significant correlation. These correlations were represented as heatmaps via the "ggplot2" package (version 3.3.6).
Six-week-old male Wistar rats and 4-week-old male BALB/C nude mice were acquired from Beijing Vital River Laboratory Animal Technology Co., Ltd. (license: SCXK (Beijing) 2016-0006). All animals were raised in the Barrier Environmental Animal Laboratory of Dongzhimen Hospital of Beijing University of Chinese Medicine (license: SYXK (Beijing) 2015-0001) and were maintained by the National Standards for Laboratory Animals of China (GB14925-2010). The animals were maintained in a distinct SPF laboratory, where they were subjected to a breeding environment that included a temperature of 25 ± 1°C, a humidity of 50% ± 10%, a 12-hour day and night cycle, and adaptable feeding for 5 days. The ARRIVE 2.0 guidelines were followed during the investigation. The Ethics Committee of Laboratory Animals of Dongzhimen Hospital of Beijing University of Chinese Medicine approved this investigation (No. 21-10-01 and 21-46-01).
The HCC rat model was established by administering diethylnitrosamine (50 mg/kg/week, Psaitong, N60001, CN) intraperitoneally for 16 consecutive weeks. After week 16, the liver condition of experimental animals was evaluated. Following a 12-hour fast, the rats were anesthetized via intraperitoneal injection of 0.2 mL/100 g 3% pentobarbital sodium solution. The largest lobe of liver tissue was removed and stored in tissue fixation fluid as described below, while the remaining liver tissue was stored at -80°C.
Tissues were collected and fixed in 4% formalin (Servicebio, G1101, CN) and embedded in paraffin. The tissue sections were subjected to hematoxylin and eosin staining (Servicebio, G1003, CN), Masson staining (Servicebio, G1006, CN), or periodic acid-Schiff staining (Servicebio, G1008, CN). The stained sections were observed and imaged under a microscope, and quantification of the positively stained area was performed with ImageJ software.
For immunohistochemistry, the tissue sections were deparaffinized and rehydrated before antigen retrieval, endogenous peroxidase activity was removed, and the sections were blocked with normal goat serum. To detect proliferating cells, a Ki67 rabbit polyclonal antibody (GB111499, Servicebio, CN) was added dropwise, and the samples were incubated in a wet chamber overnight at 4°C. To detect UGDH, the paraffin-embedded sections were incubated overnight at 4°C in primary anti-UGDH (rabbit pAb, Proteintech, 13151-1-AP, United States). The next day, the reaction mixture and HRP-labeled anti-rabbit secondary antibody (GB23303, Servicebio, CN) were added and incubated at 37°C for 30 minutes. Diaminobenzidine (G1212, Servicebio, CN) was used for color reaction. Images of the stained sections were acquired on a microscope, and quantification of the positively stained area was performed with ImageJ software.
For immunofluorescence, tissue sections were deparaffinized and rehydrated before antigen retrieval, endogenous peroxidase activity was removed, and the sections were blocked with normal goat serum. To detect ARG1, paraffin-embedded sections were incubated overnight in primary anti-ARG1 (rabbit pAb, Servicebio, GB11285, CN) at 4°C. The next day, the reaction mixture and Alexa Fluor 488-labeled anti-rabbit secondary antibody (GB25303, Servicebio, CN) were added dropwise, and the mixture was incubated at 37 °C for 30 minutes. DAPI dye solution (G1012, Servicebio, CN) was added, and the samples were incubated at room temperature for 10 minutes. The sections were placed under a scanner for image collection. Images of stained sections were acquired on a microscope, and quantification of the positively stained area was performed with ImageJ software.
Huh7 and other hepatoma cell lines were acquired from the ATCC (Manassas, VA, United States) and maintained in DMEM medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin, 100 μg/mL streptomycin, and 5%
A total of 2 × 105 cells were inoculated in 400 μL of antibiotic-free medium at a confluency of 60%-80% for transfection 24 hours prior. Lipofectamine 2000 was used to transfect the siRNAs at a concentration of 50 nM. To dilute the siRNA, 50 μL of Opti-MEM was added, and the mixture was delicately pipetted 3-5 times. In a separate tube, the transfection reagent (1.0 μL) was diluted into 50 μL Opti-MEM, delicately mixed by pipetting 3-5 times and allowed to sit at room temperature for 5 minutes. Lipofectamine 2000 was combined with siRNA, and the mixture was gently pipetted 3-5 times. The mixture was then incubated at room temperature for 20 minutes. The transfection complex was uniformly distributed across the cell plate at a concentration of 100 μL/well. Supplementary Table 1 lists the siRNAs used in this investigation.
A total of 5 × 103 cells were inoculated in 96-well plates in six replicates after UGDH was knocked down according to the instruction manual. The cells were incubated for 24, 48, or 72 hours. To evaluate cell proliferation, absorbance at 450 nm was measured via a TECAN Infinite M200 Multimode microplate reader (Tecan, Mechelen, Belgium) with a CCK-8 assay kit (Solarbio, CN).
Nude mouse tumor xenografts were performed as described previously[28]. Briefly, a total of 5 × 106 Huh7 cells were injected into the underarm epidermis of 4-week-old male BALB/c nude mice[28]. Transplanted tumors developed 2 weeks after injection. After 2 weeks of consistent feeding, the rodents were sacrificed, and the tumors were harvested.
The reverse transcription of total RNA to cDNA was conducted via a quantitative-PCR RT Master Mix kit (TOYOBO, JAN). Quantitative real-time-PCR (qRT-PCR) was conducted using SYBR green real-time PCR Master Mix (TOYOBO, Japan) and a real-time PCR detection system (Agilent Technologies, United States). GAPDH and Actb served as internal control genes. The experiments were conducted in triplicate and repeated three times. Supplementary Table 2 lists the primers used in this investigation.
Protein lysates were separated via 10% SDS-PAGE (Epizyme Biomedical Technology, PG112, CN) and subsequently electrophoretically transferred onto polyvinylidene fluoride membranes (Epizyme Biomedical Technology, WJ001, CN). Anti-UGDH (Proteintech, 67360-1-Ig, 1:1000, United States) and anti-GAPDH (MBL, M171-3, 1:5000, JPN) primary antibodies were used. The secondary antibody used was HRP-conjugated AffiniPure goat anti-mouse IgG (H + L) (Proteintech, SA00001-1, 1:8000, United States). The integrated density of the protein bands was analyzed via ImageJ software.
For all the bar graphs, data were presented as mean ± SD. GraphPad Prism software (GraphPad Software, Inc., La Jolla, CA, United States) was used for survival curves, charts, and statistical analysis. For comparisons of quantitative data between groups, a student’s t-test was used. Otherwise, independent sample rank sum test was used; survival was calculated using K-M’s method and compared using log-rank test; Spearman rank correlation test was used to calculate the correlation between two nonparametric values.
UGDH mRNA and protein were expressed in a variety of organs and tissues, with the liver, gallbladder, and gastrointestinal tract exhibiting the highest expression levels (Supplementary Figure 1A). The highest levels of UGDH mRNA were found in the liver, colon, rectum, urinary bladder, adipose tissue, stomach, duodenum, small intestine, gallbladder, and placenta, as demonstrated by the consensus dataset, which included 375 normal tissues in the HPA database and 13084 samples in GTEx (Supplementary Figure 1B). The HPA database, which contains 144 individuals and 44 samples of various normal tissue categories, was used to obtain the UGDH protein data. UGDH protein was predominantly expressed in the stomach, esophagus, bronchus, colon, liver, prostate, endometrium, and appendix (Supplementary Figure 1C). The highest UGDH mRNA expression was observed in liver cancer, gallbladder cancer, kidney cancer, noncancerous, and lung cancer tissues (Supplementary Figure 1D). Supplementary Table 3 displays the abbreviations and cancer categories.
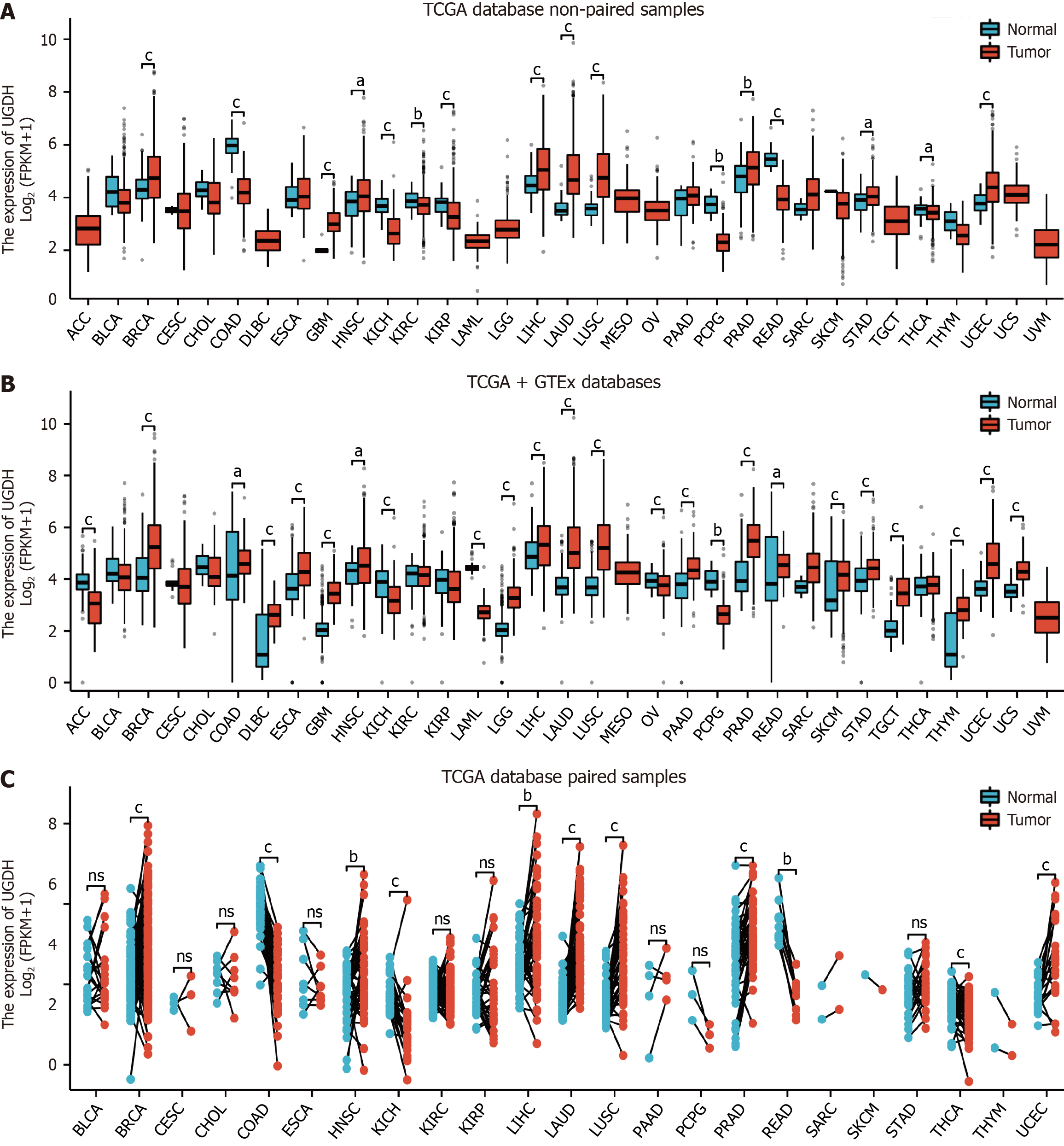
In the TCGA database, compared with nontumor tissues, statistically significant differences in UGDH mRNA expression were observed in BRCA, COAD, GBM, HNSC, KICH, KIRC, KIRP, LIHC, LUAD, LUSC, PCPG, PRAD, READ, STAD, THCA, and UCEC (Figure 1A). In the TCGA and GTEx databases, we observed statistically significant differences in UGDH mRNA expression in ACC, BRCA, COAD, DLBC, ESCA, GBM, HNSC, KICH, LAML, LGG, LIHC, LUAD, LUSC, OV, PAAD, PCPG, PRAD, READ, SKCM, STAD, TGCT, THYM, UCEC, and UCS (Figure 1B). In the TCGA database, we identified statistically significant differences in UGDH mRNA expression between carcinomas and paired paracarcinomas in BRCA, COAD, HNSC, KICH, LIHC, LUAD, LUSC, PRAD, READ, THCA, and UCEC (Figure 1C).
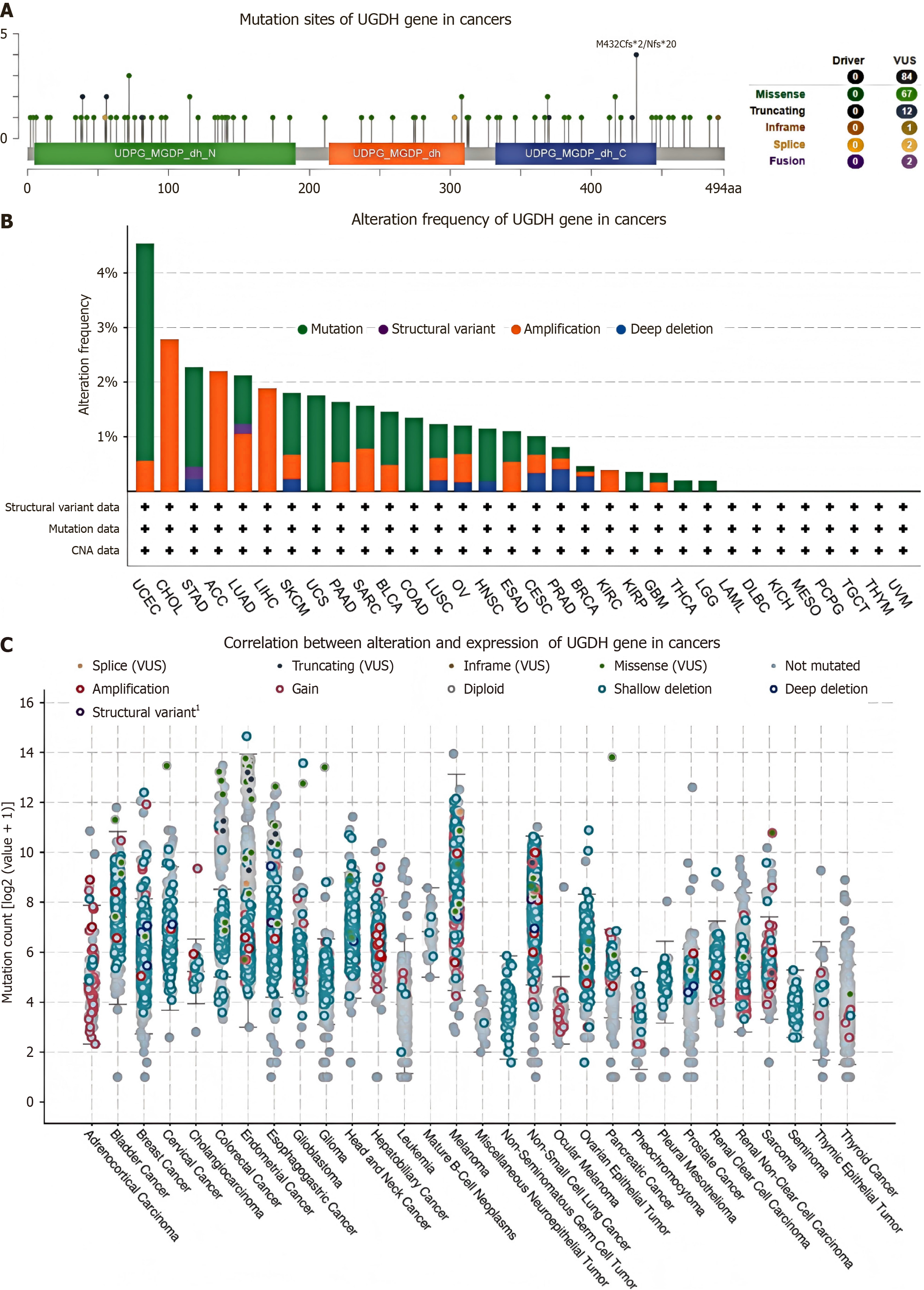
The cBioPortal database was used to analyze mutations in UGDH that are expressed during malignancy. We identified 84 mutation sites between amino acids 0 and 494, which included 67 missense mutations, 12 truncating mutations, 1 in-frame mutation, 2 splices, and 2 fusions. The most prevalent mutation sites were M432Cfs and Nfs (Figure 2A). The top 10 malignancies carrying the most UGDH mutations were UCEC, CHOL, STAD, ACC, LUAD, LIHC, SKCM, UCS, PAAD, and SARC (Figure 2B). Almost all 32 cancer categories exhibited shallow deletions in UGDH mRNA expression. Adrenocortical cancer frequently exhibited gains, whereas hepatobiliary cancer more frequently exhibited amplifications (Figure 2C).
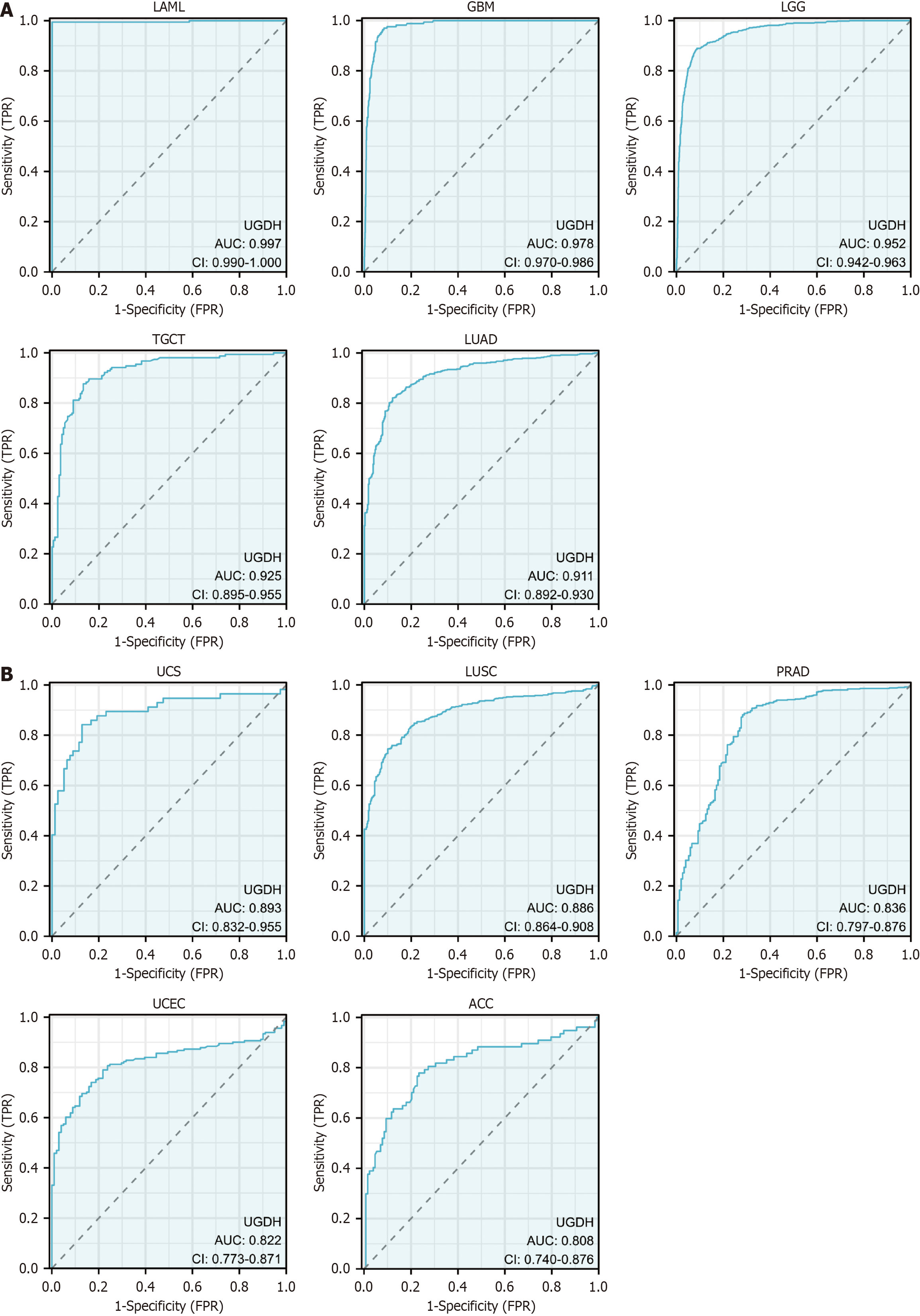
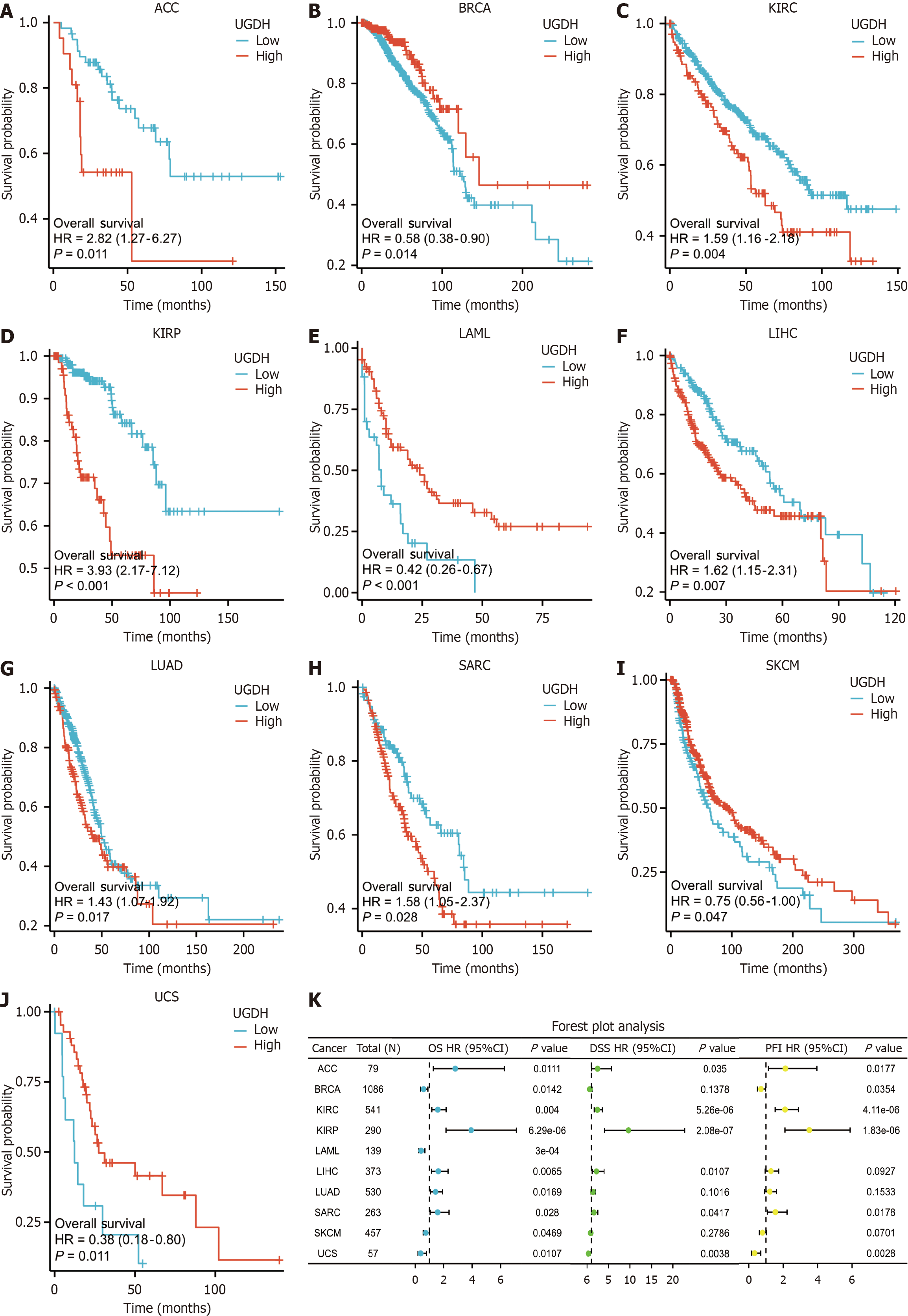
UGDH demonstrated substantial diagnostic potential for a wide range of malignancies, whose AUC was greater than 0.9 for five cancers (Figure 3A), 0.8 < AUC < 0.9 for five cancers (Figure 3B), and 0.7 < AUC < 0.8 for seven cancers (Supplementary Figure 2). Furthermore, we performed K-M analysis on 33 malignancies to assess the prognostic value of UGDH expression. UGDH expression was correlated with OS in 10 cancer types, whereas the effects of UGDH on 23 other cancer types were not apparent. The survival rate was lower for ACC, KIRC, KIRP, LICH, LUAD, and SARC when UGDH was highly expressed and higher for BRCA, LAML, SKCM, and UCS when UGDH was barely expressed (Figure 4A-J). The forest plot displayed DSS and PFI data for ten malignancies in which UGDH expression was significantly correlated with OS, as determined by Cox regression analyses of the cancers (Figure 4K). Lack of information regarding LAML, DSS, and PFI. BRCA, LUAD, and SKCM were not significantly different in terms of DSS, and LUAD and SKCM were not significantly different in terms of PFIs. Supplementary Figure 2 illustrates the correlation between UGDH and prevalent clinical parameters in patients with malignancies.
In ten malignancies, we discovered that UGDH expression had an impact on OS, regardless of whether it was high or low, as indicated by our previous findings. Consequently, we analyzed UGDH expression in the immune and molecular subtypes of these ten tumors. The results revealed that UGDH is significantly differentially expressed in ACC, BRCA, KIRC, KIRP, LIHC, LUAD, SARC, SKCM, and UCS (Figure 5).
To further evaluate the correlation between UGDH expression and immune infiltration and regulation, we produced heatmaps of UGDH expression and markers of immune cells or factors. We found that the infiltration levels of T helper cells, Tcm cells, and Th2 cells across 33 cancer types positively correlated with UGDH expression, whereas the infiltration levels of dendritic cells (DCs) and cytotoxic cells were negatively correlated (Figure 6A). UGDH was positively correlated with the majority of MHC molecules in KICH, PCPG, and UVM but negatively correlated with the majority of MHC molecules in LUAD, LUSC, and TGCT (Figure 6B). Interestingly, there was a positive correlation in ACC, KICH, and UVM in terms of immune activation and suppression, while there was a negative correlation in LUAD and LUSC (Figure 6B). UGDH expression was positively correlated with the most immunostimulators in nearly all malignancies (Figure 6C) and with the expression of the immunosuppressive factors TGFBR1 and KDR in nearly all malignancies (Figure 6D). Most cytokines in KICH, KIRC, PCPG, and UVM were positively correlated with UGDH, whereas negative correlations were observed in BRCA, LUAD, and LUSC (Figure 6E). UGDH exhibited positive correlations with most cytokine receptors in BLCA, DLBC, KICH, KIRC, PCPG, PRAD, and UVM, as well as negative correlations with LUAD and LUSC (Figure 6F).
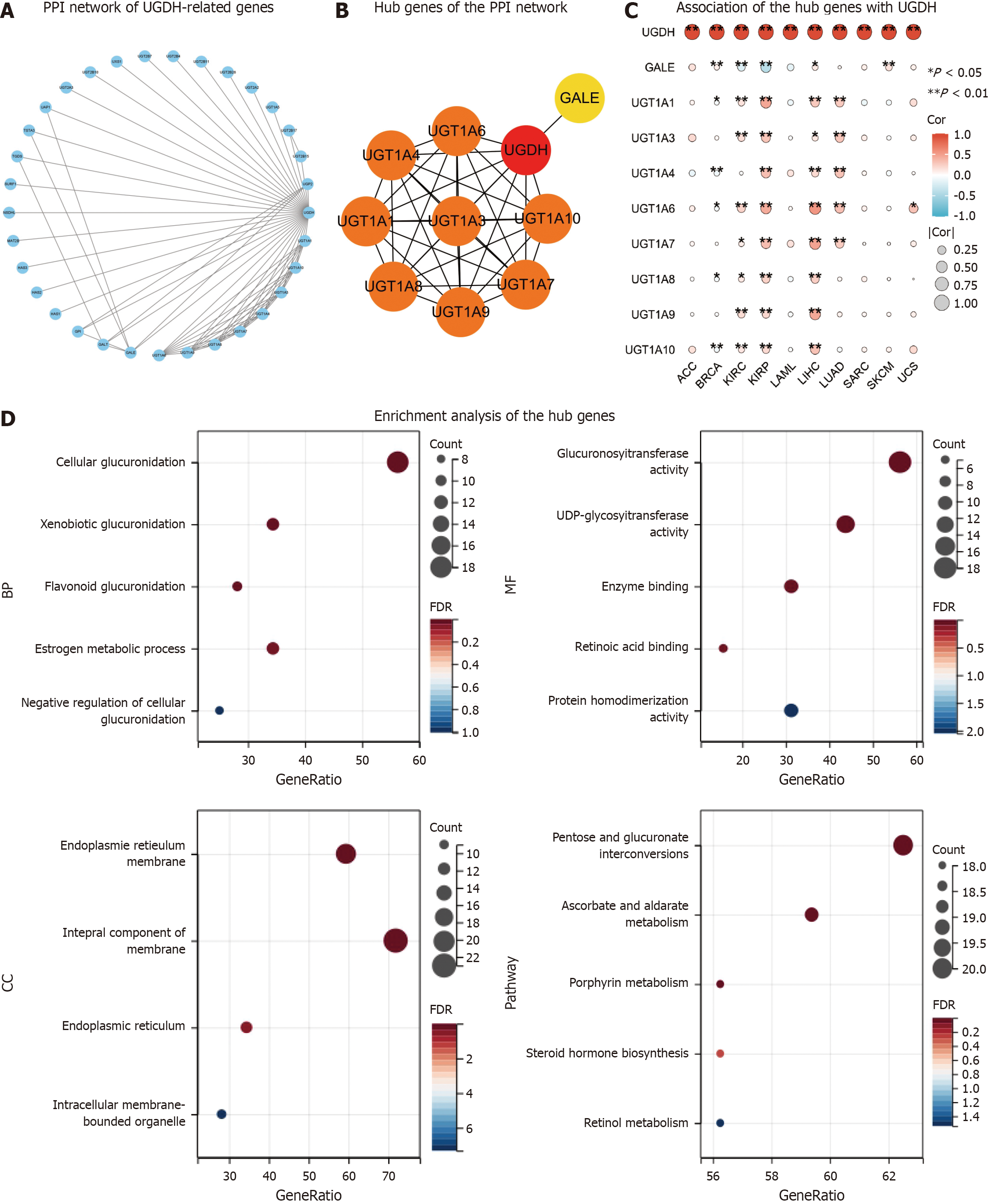
We constructed a PPI network by obtaining a total of 32 genes that were closely related to UGDH from String (Figure 7A). UGDH, UGT1A1, UGT1A3, UGT1A4, UGT1A6, UGT1A7, UGT1A8, UGT1A9, UGT1A10, and GALE were the top 10 pivotal genes (Figure 7B). The top 10 hub genes were associated primarily with BRCA, KIRC, KIRP, LIHC, and LUAD, and OS prognosis was influenced by UGDH expression (P < 0.05) (Figure 7C). The functions of these genes were analyzed across three categories, namely, molecular function (MF), biological process (BP), and cellular component (CC), through GO and KEGG enrichment. Cellular glucuronidation, xenobiotic glucuronidation, and flavonoid glucuronidation were the most frequently used GO terms for BP. The MF was characterized by enzyme binding, UDP-glycosyltransferase activity, and glucuronosyltransferase activity. Furthermore, the endoplasmic reticulum membrane, and the endoplasmic reticulum are referred to as CCs. The primary KEGG pathways were ascorbate and alternate metabolism, pentose and glucuronate interconversion, and porphyrin metabolism (Figure 7D). In addition, UGDH was activated and repressed in the DNA damage response, EMT, hormone AR, hormone ER, P13K/AKT, RAS/MAPK, and TSC/mTOR pathways (Figure 8A). The GSEA results for the ten cancer types revealed that UGDH is primarily associated with pathways related to cell cycle regulation, the immune response, metabolism, and cell signaling (Figure 8B-K). These common pathways indicate that UGDH expression plays a coordinated role in specific cellular processes across various cancer types.
We next analyzed the correlation between UGDH and the functional states of cancer cells at the single-cell level. The results suggested that the functional status of various malignancies was significantly influenced by UGDH expression (Figure 9A). Additional investigations of the correlation between UGDH and specific malignancies revealed that LUAD is positively correlated with DNA repair and negatively correlated with quiescence, metastasis, angiogenesis, and differentiation. Non-small cell lung cancer is negatively correlated with stemness, whereas acute lymphoblastic leukemia and head and neck squamous cell carcinoma are positively correlated with stemness. Quiescence, differentiation, inflammation, and proliferation are positively correlated with acute myelocytic leukemia. Renal cell carcinoma and prostatic cancer are positively correlated with hypoxia, whereas RB is negatively correlated with DNA repair, the cell cycle, and DNA damage and positively correlated with differentiation, inflammation, and angiogenesis. DNA repair and DNA damage were negatively correlated with UM (Figure 9B-J).
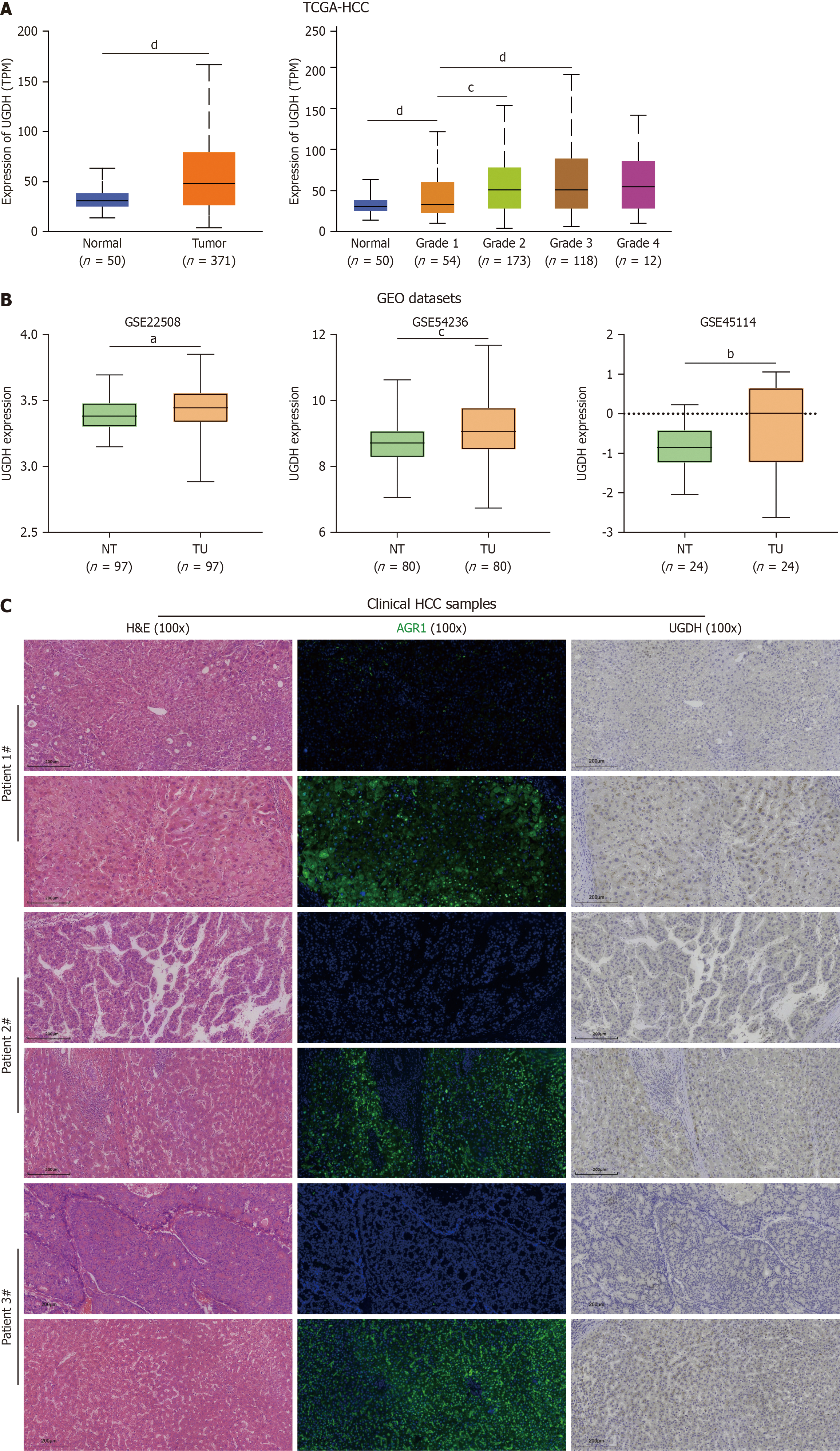
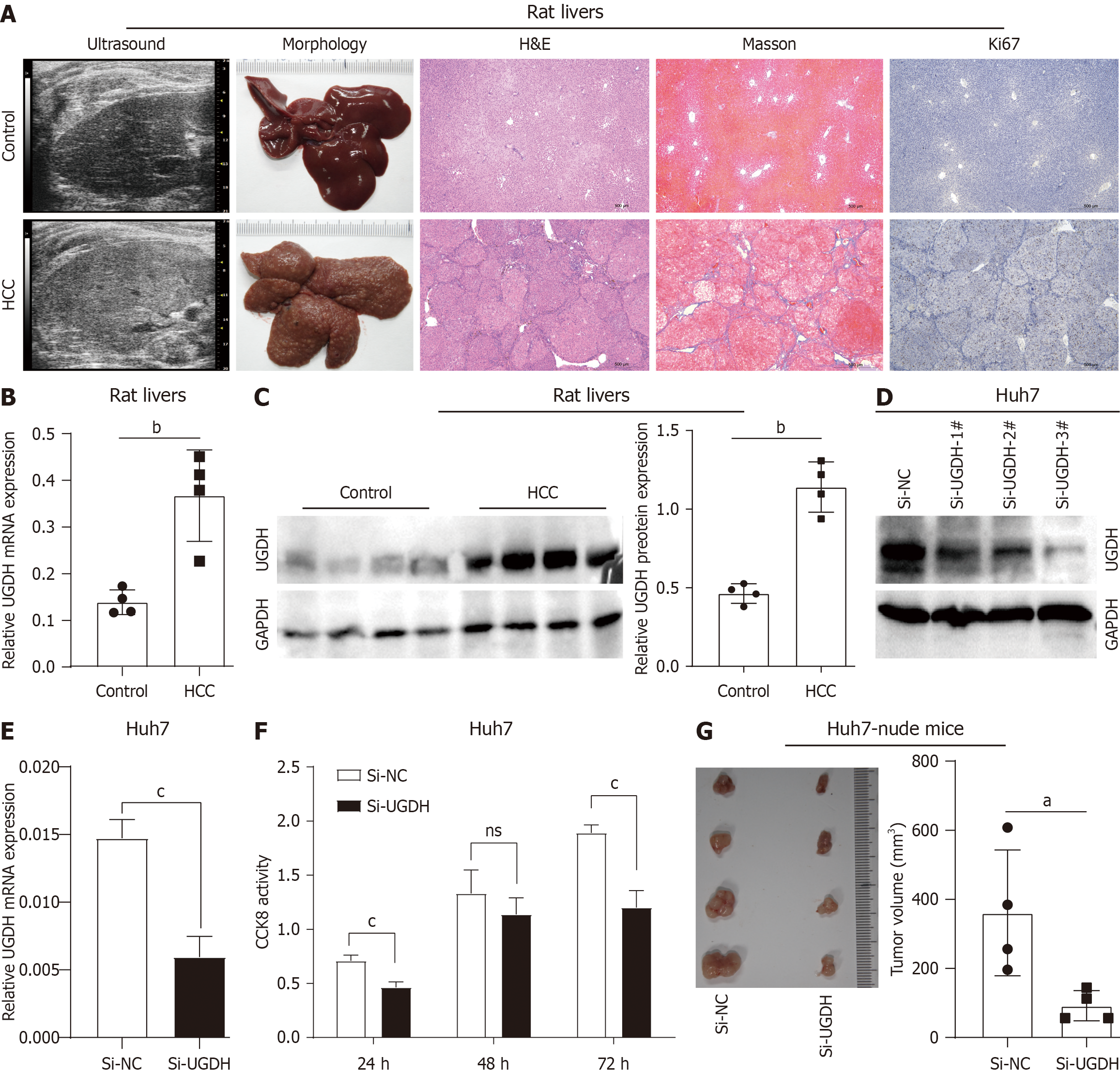
We further analyzed UGDH expression in HCC. In the TCGA database, UGDH mRNA expression was significantly upregulated in HCC tissues compared to nontumor tissues and increased with increasing tumor G stage (Figure 10A). In the GEO datasets, UGDH mRNA expression was significantly upregulated in paired HCC tissues compared with paired non-HCC tissues (Figure 10B). The detection of clinical HCC samples suggested that UGDH protein expression is upregulated in HCC tissues compared to nontumor tissues (Figure 10C). In the HCC rat model (Figure 11A), qRT-PCR and Western blot results indicated that UGDH expression was significantly upregulated at the mRNA and protein levels compared to the control group, (Figure 11B and C).
In in vitro experiments, Western blot and qRT-PCR analyses demonstrated that UGDH was successfully knocked down via siRNA (Figure 11D and E). The proliferative activity of Huh7 cells was substantially inhibited by UGDH knockdown at 24 and 72 hours, as demonstrated by CCK-8 assay (Figure 11F). Photographs of xenograft tumors from Huh7-nude mice demonstrated that siRNA-mediated silencing of UGDH resulted in a significant reduction in tumor size (Figure 11G). Collectively, these findings indicate that UGDH is a critical factor in HCC progression and that targeting it may be a viable therapeutic approach.
In this study, we first analyzed UGDH expression across different types of cancers. We discovered that UGDH mutation sites are primarily missense, truncating, in-frame, splice, and fusion mutations in cancers. UGDH has been genetically altered in 24 cancer types, with UCEC, CHOL, STAD, ACC, and LUAD having alteration frequencies greater than 2%. Mutation is the primary form of UGDH gene alteration in most cancers, followed by amplification.
To further investigate the biological functions of UGDH, we established a PPI network and identified hub genes. We investigated the relationships between the hub genes and cancer types and demonstrated that UGDH expression influences the prognosis of cancer patients. Among the hub genes, GALE has been associated with the differentiation grade of gastric cancer[29], can promote glioma cell proliferation and migration[30], and serve as a potential marker for papillary thyroid carcinoma[31,32]. The other hub genes are members of the UDP glucuronosyltransferase family, which encodes UDP-glucuronosyltransferase, an enzyme in the glucuronidation pathway that converts small lipophilic molecules, including steroids, bilirubin, hormones, and pharmaceuticals, into water-soluble, excretable metabolites[33,34].
UGDH expression manifests varying functional states at the single-cell level in a variety of cancers. In general, UGDH expression is positively correlated with cell apoptosis, invasion, and proliferation but negatively correlated with the cell cycle. GSEA functional enrichment analysis was implemented for cancer types in which UGDH expression had an impact on overall prognosis. The results of the analysis were replicated more than twice and were deemed to be common enrichment pathways. Most of the enriched pathways were immunologically related. The adaptive immune system is influenced by immunoregulatory interactions between lymphoid and nonlymphoid cell pathways[35]. UGDH expression varies among a variety of molecular or immune subtypes of malignancies, with differences in immune subtypes present in 30 cancer types and differences in molecular subtypes present in 17 cancers. The cancer types that exhibited disparities in UGDH expression and immune subtypes were consistent with the cancer types whose expression affected survival prognosis, with the exception of LAML. These findings suggest that UGDH may influence cancer prognosis by affecting immune subtypes.
In addition, we concentrated on the role of UGDH expression on immunomodulatory factors and major histocompatibility complex molecules, as well as the targeting of immune lymphocytes. Nevertheless, there are specific subtypes of cancer in which genes are aberrantly expressed, and the results may not be representative of the overall patient population. Consequently, the survival of certain cancers is not influenced by variations in gene expression, whereas the prognosis of distinct cancers may be influenced by the varied expression of genes in different molecular or immune subtypes[36]. In the majority of malignancies, UGDH expression was positively correlated with T helper cells, Tcm cells, and Th2 cells but was negatively correlated with pDCs and cytotoxic cells. Upon activation, primary T cells differentiate into memory T cells and effector T cells. Cytotoxic T cells and helper T (Th) cells are examples of effector T cells[37]. DCs activate T cells, which ultimately differentiate into a variety of subtypes of T cells, such as Th2 cells. Th2 cells elicit an immunosuppressive protumorigenic response. Poor clinical outcomes in human malignancies are frequently linked to Th2 cell infiltration in the tumor microenvironment[38]. Research has demonstrated that Tcm cells provide superior antitumor immunity in CAR-T-cell therapy compared with effector memory T cells and effector T cells[39]. UGDH was correlated with the type of malignancy and other immune characteristics. Therefore, the function of UGDH in cancer immunity is multifaceted, and its positive correlation with Th2 cells and negative correlation with cytotoxic cells may be linked to the negative prognosis of certain malignancies.
We conducted GO and KEGG analyses of UGDH and its associated genes. Cellular glucuronidation was the most significant BP enrichment result, followed by xenobiotic glucuronidation. According to the MF analysis results, glucuronosyltransferase activity ranked first, followed by UDP-glycosyltransferase activity. The enriched CC terms were related primarily to the endoplasmic reticulum membrane and its associated membrane, which is a component of the membrane and an intracellular membrane-bound organelle. In contrast, the most significant pathway in KEGG analysis was pentose and glucuronate interconversion, indicating a high degree of correlation between the results of the GO enrichment analysis. UDP-glucuronosyltransferases are located in the endoplasmic reticulum, and the active transport of the metabolite UDPGA, which plays important role in drug resistance and cancer progression[40]. Consequently, the significance of UGDH in HCC should be seriously considered. Through the detection of clinical and rat HCC samples, we confirmed UGDH upregulation in HCC. The in vitro experiments further revealed the important role of UGDH in regulating HCC cell proliferation. However, the detailed regulatory mechanism of UGDH in HCC needs further study.
Our investigation highlights the role of UGDH across cancers from a variety of angles, such as its immunomodulation, mutation, and biological functions. Potential diagnostic markers for lung adenocarcinoma, testicular cancer, and gliomas include UGDH. It has the potential to serve as a prognostic indicator for sarcomas, lung malignancies, and HCC. HCC cell proliferation is inhibited by decreased UGDH expression.
| 1. | Zhou M, Wang H, Zeng X, Yin P, Zhu J, Chen W, Li X, Wang L, Wang L, Liu Y, Liu J, Zhang M, Qi J, Yu S, Afshin A, Gakidou E, Glenn S, Krish VS, Miller-Petrie MK, Mountjoy-Venning WC, Mullany EC, Redford SB, Liu H, Naghavi M, Hay SI, Wang L, Murray CJL, Liang X. Mortality, morbidity, and risk factors in China and its provinces, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2019;394:1145-1158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2594] [Cited by in RCA: 2604] [Article Influence: 372.0] [Reference Citation Analysis (2)] |
| 2. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51728] [Cited by in RCA: 48731] [Article Influence: 3248.7] [Reference Citation Analysis (12)] |
| 3. | Lane AN, Higashi RM, Fan TW. Metabolic reprogramming in tumors: Contributions of the tumor microenvironment. Genes Dis. 2020;7:185-198. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 71] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 4. | Park JH, Pyun WY, Park HW. Cancer Metabolism: Phenotype, Signaling and Therapeutic Targets. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 335] [Article Influence: 55.8] [Reference Citation Analysis (0)] |
| 5. | Doshi MB, Lee N, Tseyang T, Ponomarova O, Goel HL, Spears M, Li R, Zhu LJ, Ashwood C, Simin K, Jang C, Mercurio AM, Walhout AJM, Spinelli JB, Kim D. Disruption of sugar nucleotide clearance is a therapeutic vulnerability of cancer cells. Nature. 2023;623:625-632. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 34] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 6. | Zimmer BM, Barycki JJ, Simpson MA. Integration of Sugar Metabolism and Proteoglycan Synthesis by UDP-glucose Dehydrogenase. J Histochem Cytochem. 2021;69:13-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 7. | Egger S, Chaikuad A, Kavanagh KL, Oppermann U, Nidetzky B. UDP-glucose dehydrogenase: structure and function of a potential drug target. Biochem Soc Trans. 2010;38:1378-1385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 65] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 8. | Chen J, Yang S. Catalytic mechanism of UDP-glucose dehydrogenase. Biochem Soc Trans. 2019;47:945-955. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 9. | Morla S. Glycosaminoglycans and Glycosaminoglycan Mimetics in Cancer and Inflammation. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 162] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 10. | Price MJ, Nguyen AD, Byemerwa JK, Flowers J, Baëta CD, Goodwin CR. UDP-glucose dehydrogenase (UGDH) in clinical oncology and cancer biology. Oncotarget. 2023;14:843-857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 11. | Wang X, Liu R, Zhu W, Chu H, Yu H, Wei P, Wu X, Zhu H, Gao H, Liang J, Li G, Yang W. UDP-glucose accelerates SNAI1 mRNA decay and impairs lung cancer metastasis. Nature. 2019;571:127-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 158] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 12. | Fan M, Huo S, Guo Y, Wang R, Hao W, Zhang Z, Wang L, Zhao Y. UDP-glucose dehydrogenase supports autophagy-deficient PDAC growth via increasing hyaluronic acid biosynthesis. Cell Rep. 2024;43:113808. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 13. | Harrington BS, Kamdar R, Ning F, Korrapati S, Caminear MW, Hernandez LF, Butcher D, Edmondson EF, Traficante N, Hendley J; Australian Ovarian Cancer Study, Gough M, Rogers R, Lourie R, Shetty J, Tran B, Elloumi F, Abdelmaksoud A, Nag ML, Mazan-Mamczarz K, House CD, Hooper JD, Annunziata CM. UGDH promotes tumor-initiating cells and a fibroinflammatory tumor microenvironment in ovarian cancer. J Exp Clin Cancer Res. 2023;42:270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 14. | Lin LH, Chou HC, Chang SJ, Liao EC, Tsai YT, Wei YS, Chen HY, Lin MW, Wang YS, Chien YA, Yu XR, Chan HL. Targeting UDP-glucose dehydrogenase inhibits ovarian cancer growth and metastasis. J Cell Mol Med. 2020;24:11883-11902. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 15. | Huang D, Casale GP, Tian J, Lele SM, Pisarev VM, Simpson MA, Hemstreet GP 3rd. Udp-glucose dehydrogenase as a novel field-specific candidate biomarker of prostate cancer. Int J Cancer. 2010;126:315-327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 34] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 16. | Wei Q, Galbenus R, Raza A, Cerny RL, Simpson MA. Androgen-stimulated UDP-glucose dehydrogenase expression limits prostate androgen availability without impacting hyaluronan levels. Cancer Res. 2009;69:2332-2339. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 38] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 17. | Oyinlade O, Wei S, Lal B, Laterra J, Zhu H, Goodwin CR, Wang S, Ma D, Wan J, Xia S. Targeting UDP-α-D-glucose 6-dehydrogenase inhibits glioblastoma growth and migration. Oncogene. 2018;37:2615-2629. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 18. | Zhan D, Yalcin F, Ma D, Fu Y, Wei S, Lal B, Li Y, Dzaye O, Laterra J, Ying M, Lopez-Bertoni H, Xia S. Targeting UDP-α-d-glucose 6-dehydrogenase alters the CNS tumor immune microenvironment and inhibits glioblastoma growth. Genes Dis. 2022;9:717-730. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 19. | Teoh ST, Ogrodzinski MP, Lunt SY. UDP-glucose 6-dehydrogenase knockout impairs migration and decreases in vivo metastatic ability of breast cancer cells. Cancer Lett. 2020;492:21-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 20. | Vitale DL, Caon I, Parnigoni A, Sevic I, Spinelli FM, Icardi A, Passi A, Vigetti D, Alaniz L. Initial Identification of UDP-Glucose Dehydrogenase as a Prognostic Marker in Breast Cancer Patients, Which Facilitates Epirubicin Resistance and Regulates Hyaluronan Synthesis in MDA-MB-231 Cells. Biomolecules. 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 21. | Zhang X, Liu F, Fang Q, Sun C, Fan J. E3 ligase HERC5-catalyzed UGDH isgylation promotes SNAI1-mediated tumor metastasis and cisplatin resistance in oral squamous cell carcinoma. Biol Direct. 2025;20:27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 22. | Wang Q, Karvelsson ST, Johannsson F, Vilhjalmsson AI, Hagen L, de Miranda Fonseca D, Sharma A, Slupphaug G, Rolfsson O. UDP-glucose dehydrogenase expression is upregulated following EMT and differentially affects intracellular glycerophosphocholine and acetylaspartate levels in breast mesenchymal cell lines. Mol Oncol. 2022;16:1816-1840. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 23. | Hagiuda D, Nagashio R, Ichinoe M, Tsuchiya B, Igawa S, Naoki K, Satoh Y, Murakumo Y, Saegusa M, Sato Y. Clinicopathological and prognostic significance of nuclear UGDH localization in lung adenocarcinoma. Biomed Res. 2019;40:17-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 24. | Zao X, Cao X, Liang Y, Zhang J, Chen H, Zhang N, Liu R, Jin Q, Chen Y, Li X, Du H, Chen G, Hou L, Ye Y. The Chinese herbal KangXianYiAi formula inhibits hepatocellular carcinoma by reducing glutathione and inducing ferroptosis. Pharmacol Res Mod Chinese Med. 2023;8:100276. [RCA] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 2] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 25. | Gao Q, Cheng B, Chen C, Lei C, Lin X, Nie D, Li J, Huang L, Li X, Wang K, Huang A, Tang N. Dysregulated glucuronic acid metabolism exacerbates hepatocellular carcinoma progression and metastasis through the TGFβ signalling pathway. Clin Transl Med. 2022;12:e995. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 26. | Guo B, Xu X, Shao M, Yang X, He G, Qi K, Gu J, Wang L. UDP-glucose 6-dehydrogenase lessens sorafenib sensitivity via modulating unfolded protein response. Biochem Biophys Res Commun. 2022;613:207-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 27. | Shen S, Li H, Liu J, Sun L, Yuan Y. The panoramic picture of pepsinogen gene family with pan-cancer. Cancer Med. 2020;9:9064-9080. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 28. | Zao X, Cheng J, Shen C, Guan G, Feng X, Zou J, Zhang J, Liu T, Zheng H, Zhang T, Wang J, Liu J, Li D, Lu F, You F, Chen X. NFATc3 inhibits hepatocarcinogenesis and HBV replication via positively regulating RIG-I-mediated interferon transcription. Oncoimmunology. 2021;10:1869388. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 29. | de Souza MFD, da Silva Filho AF, de Barros Albuquerque AP, Quirino MWL, de Souza Albuquerque MS, Cordeiro MF, Martins MR, da Rocha Pitta I, Lucena-Araujo AR, da Rocha Pitta MG, de Melo Rêgo MJB. Overexpression of UDP-Glucose 4-Epimerase Is Associated with Differentiation Grade of Gastric Cancer. Dis Markers. 2019;2019:6325326. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 30. | Sun X, Xue H, Xiong Y, Yu R, Gao X, Qian M, Wang S, Wang H, Xu J, Chen Z, Deng L, Li G. GALE Promotes the Proliferation and Migration of Glioblastoma Cells and Is Regulated by miR-let-7i-5p. Cancer Manag Res. 2019;11:10539-10554. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 31. | Ye G, Zhang X, Li M, Lin Z, Xu Y, Dong H, Zhou J, Zhang J, Wang S, Zhu Y, Yu X, Qian X. Integrated analysis of circulating and tissue proteomes reveals that fibronectin 1 is a potential biomarker in papillary thyroid cancer. BMC Cancer. 2023;23:412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 32. | Fujarewicz K, Jarzab M, Eszlinger M, Krohn K, Paschke R, Oczko-Wojciechowska M, Wiench M, Kukulska A, Jarzab B, Swierniak A. A multi-gene approach to differentiate papillary thyroid carcinoma from benign lesions: gene selection using support vector machines with bootstrapping. Endocr Relat Cancer. 2007;14:809-826. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 53] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 33. | Meech R, Mackenzie PI. Structure and function of uridine diphosphate glucuronosyltransferases. Clin Exp Pharmacol Physiol. 1997;24:907-915. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 168] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 34. | Mano ECC, Scott AL, Honorio KM. UDP-glucuronosyltransferases: Structure, Function and Drug Design Studies. Curr Med Chem. 2018;25:3247-3255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 35. | Nedvetzki S, Sowinski S, Eagle RA, Harris J, Vély F, Pende D, Trowsdale J, Vivier E, Gordon S, Davis DM. Reciprocal regulation of human natural killer cells and macrophages associated with distinct immune synapses. Blood. 2007;109:3776-3785. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 201] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 36. | Liang X, Li L, Fan Y. Diagnostic, Prognostic, and Immunological Roles of HELLS in Pan-Cancer: A Bioinformatics Analysis. Front Immunol. 2022;13:870726. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 37. | Basu A, Ramamoorthi G, Albert G, Gallen C, Beyer A, Snyder C, Koski G, Disis ML, Czerniecki BJ, Kodumudi K. Differentiation and Regulation of T(H) Cells: A Balancing Act for Cancer Immunotherapy. Front Immunol. 2021;12:669474. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 242] [Article Influence: 48.4] [Reference Citation Analysis (0)] |
| 38. | Renaude E, Kroemer M, Borg C, Peixoto P, Hervouet E, Loyon R, Adotévi O. Epigenetic Reprogramming of CD4(+) Helper T Cells as a Strategy to Improve Anticancer Immunotherapy. Front Immunol. 2021;12:669992. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 39. | Klebanoff CA, Gattinoni L, Torabi-Parizi P, Kerstann K, Cardones AR, Finkelstein SE, Palmer DC, Antony PA, Hwang ST, Rosenberg SA, Waldmann TA, Restifo NP. Central memory self/tumor-reactive CD8+ T cells confer superior antitumor immunity compared with effector memory T cells. Proc Natl Acad Sci U S A. 2005;102:9571-9576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 676] [Cited by in RCA: 787] [Article Influence: 37.5] [Reference Citation Analysis (0)] |
| 40. | Allain EP, Rouleau M, Lévesque E, Guillemette C. Emerging roles for UDP-glucuronosyltransferases in drug resistance and cancer progression. Br J Cancer. 2020;122:1277-1287. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 114] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/