Published online Mar 15, 2025. doi: 10.4251/wjgo.v17.i3.97673
Revised: October 30, 2024
Accepted: December 4, 2024
Published online: March 15, 2025
Processing time: 254 Days and 7 Hours
Activation of the epithelial-mesenchymal transition (EMT), a pivotal process in tumor metastasis and evasion, as well as the NLRP3 inflammasome, both promote colorectal cancer (CRC) progression. Recent studies have shown that Trans
To investigate the role of TMEM176B in modulating NLRP3 inflammasome and its implications on EMT and tumor progression in CRC.
CRC in situ mouse and co-cultured cell models were established using CT26 cells, BALB/c mice, and primary cultured mouse natural killer (NK) cells. Short hairpin RNA knocked down TMEM176B and NLRP3 expression in CT26 cells. Fluo
Silencing TMEM176B in CRC mice significantly reduced tumor metastasis, proliferation, and EMT, while activating apoptosis, NLRP3 inflammasome, and NK cell activity. Furthermore, silencing TMEM176B in co-cultured cell models inhibited cell migration and invasion, and promoted apoptosis. The interference of NLRP3 reversed these effects by modulating key proteins such as phosphorylated nuclear factor kappa B subunit 1 p65, matrix metallopeptidase 9, and transforming growth factor-β.
This study highlights the critical role of TMEM176B/NLRP3 in CRC progression and provides a basis for targeting this axis as a novel therapeutic approach to manage CRC progression and metastasis.
Core Tip: Silencing Transmembrane protein 176B (TMEM176B) inhibited tumor metastasis, proliferation, and epithelial-mesenchymal transition in colorectal cancer (CRC) in situ, in mice co-cultured with natural killer (NK) cells, while activating apoptosis, NLRP3 inflammasome, and NK cell function. The interference of NLRP3 reversed these effects by modulating key proteins such as phosphorylated nuclear factor kappa B subunit 1 p65, matrix metallopeptidase 9, and transforming growth factor-β. This study highlights the critical role of TMEM176B/NLRP3 in CRC management, providing a basis for targeting this axis as a novel therapeutic approach to manage CRC progression and metastasis.
- Citation: Qian W, Xu CY, Hong W, Li ZM, Xu DG. Transmembrane protein 176B promotes epithelial-mesenchymal transition in colorectal cancer through inflammasome inhibition. World J Gastrointest Oncol 2025; 17(3): 97673
- URL: https://www.wjgnet.com/1948-5204/full/v17/i3/97673.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i3.97673
Colorectal cancer (CRC) is one of the most prevalent cancers globally, with high incidence and mortality rates, according to the World Health Organization[1,2]. Early diagnosis and surgical resection significantly improve patient survival rates. However, treatment outcomes are often poor in cases of metastasis or invasion. Chemotherapy and radiotherapy play crucial roles in the treatment of CRC; however, toxicity and resistance limit long-term usage[3]. Pathogenesis of CRC involves various biological processes, including genetic, metastasis-initiating cells, epithelial-mesenchymal transition (EMT), and tumor microenvironment (TME)[4], among which EMT is a key factor. EMT is a process in which epithelial cells lose their polarity and transform into more invasive. It is crucial in CRC progression and metastasis[5]. Long et al[6] designed a targeted delivery system for shikonin inhibiting CRC and reported that it inhibits EMT by regulating lactate and transforming growth factor-β (TGF-β), suggesting that inhibiting EMT-related transcription factors may reduce the cancer progression risk and offer a potential therapeutic strategy. Extensive research has demonstrated the pivotal role of TGF-β and tumor necrosis factor-α (TNF-α) in the induction of EMT in CRC. TGF-β is an effective inducer of cancer-related EMT and is known to participate in various cellular processes. EMT not only promotes tumor invasion and metastasis but also interacts with the TME and contributes to immune evasion. Xu et al[7] found that EMT-related gene expression positively correlates with tumor-infiltrating immune cells and immune checkpoints. Thus, understanding and controlling the EMT process is crucial for the treatment of CRC.
Transmembrane protein 176B (TMEM176B), a transmembrane protein, has recently garnered attention in cancer biology for its potential role in modulating immune responses and influencing TME dynamics[8]. Initially identified for its upregulation in a rat model of allograft tolerance, TMEM176B was termed a tolerance-related induced transcript[8]. TMEM176B is involved in various critical functions, particularly in cancer, where it regulates cellular processes critical for cancer progression, including cell proliferation, apoptosis, and EMT[9,10]. TMEM176B plays an important role in bolstering antitumor immunity. It enhances the effectiveness of immune checkpoint blockade therapies by activating the inflammasomes[10]. However, the exact mechanism by which TMEM176B influences these processes in CRC remains unclear.
Recent studies have revealed a notable correlation between two prevalent single nucleotide polymorphisms (SNPs) in the NLRP3 gene, and poorer CRC outcomes in patients monitored for a decade post-surgery[11]. These genes encode NLRP3 receptors, which are integral to the innate immune system and initiate the assembly of the inflammasome, a pro-inflammatory signaling complex. This assembly is triggered by the recognition of cytosolic pathogens or cellular disturbances, leading to receptor oligomerization and recruitment of caspase-1 through the ASC adaptor protein. This inflammasome facilitates caspase-1 activation and subsequent cleavage of the pro-interleukin (IL)-1b, IL-18, and the pore-forming protein gasdermin-D (GSDMD), which drives an inflammatory response characterized by the secretion of IL-1b and IL-18 and, in some cases, triggers pyroptosis through GSDMD[12]. Activating NLRP3 in myeloid cells promotes CRC[13], while inflammatory factors activate NLRP3-dependent pyroptosis in CRC cells[14]. NLRP3 inflammasome plays a paradoxical role in CRC.
Genetic variations in the components of the inflammasome, particularly sensory proteins, can modify the activation threshold or magnitude of the inflammatory response, influencing susceptibility to various conditions, including certain cancers[15]. Previous research has identified SNPs in NLRP1 (rs11651270) and NLRP3 (rs35829419), which are gain-of-function mutations[11] that enhance inflammasome activation and cytokine production, thereby aggravating the inflammatory microenvironment conducive to CRC progression, as corroborated by animal studies[16]. Conversely, continuous IL-18 release through inflammasome in intestinal epithelial cells is crucial for gut homeostasis[17], and Il18 gene loss-of-function mutations increase CRC susceptibility[11]. These findings suggests that excessive inflammasome activity in immune cells, but not in epithelial cells, might adversely affect CRC prognosis.
Moreover, high stromal expression of TMEM176B, a cationic channel protein, is correlated with reduced survival rates in patients with CRC[10]. In murine models, TMEM176B modulates NLRP3 inflammasome activity and IL-1b production in dendritic cells, influencing CD8+ T cell tumor infiltration and activation[10]. A deficiency in Tmem176b in murine models led to heightened inflammasome activity, particularly in tumor-draining lymph nodes rather than in the tumor itself, inhibiting tumor growth through IL-1b and caspase-1-dependent mechanisms[10].
Exploring the mechanisms by which TMEM176B influences EMT and CRC progression is vital for a comprehensive understanding of its role in cancer biology. This study aimed to elucidate the intricate relationship between TMEM176B and NLRP3 inflammasome, shedding light on their combined impact on CRC metastasis. Through detailed molecular and cellular analyses, this study explored how TMEM176B-mediated inhibition of NLRP3 inflammasome activation contributes to EMT in CRC cells.
Thirty eight-week-old male BALB/c mice, each weighing 22 g, were purchased from Shanghai SLAC Laboratory Animal Co., Ltd. A standard mouse pellet diet and unlimited access to drinking water were supplied during the breeding season, along with a 12-hour light/dark cycle. The mice were acclimated for one week. Under, the mice were housed in a controlled environment (24 °C, humidity at 55%, and 12-hour light/dark cycle) at Zhejiang Eyong Pharmaceutical Research and Development Co., Ltd [experimental license number SYXK (Zhe) 2021-0033].
Using the provided sequences, short hairpin RNA (shRNA) targeting NLRP3 (sh-NLRP3) (#1: 5’-CCAGGAGAGAACCTCTTATTT-3’; #2: 5’-TTGGGTGAAATGTACTTAAAT-3’; #3: 5’-CCAAGGCCTCTCTGCTCATAA-3’) and sh-TMEM176B (#1: 5’-CTAGCTGGAGTTGGTACTATT-3’, #2: 5’-TCTGGGTGTGAACAGCTTAAT-3’; #3: 5’-CTCAGGCCAGAATCCACTATG-3’) were constructed and compared to their negative controls (sh-NC). All shRNAs were designed and prepared by Genechem Co., Ltd., Shanghai, China. Five groups of mice were established as follows: Model, sh-NC, TMEM176B knockdown (sh-TMEM176B), control shRNA (sh-TMEM176B + sh-NC), and TMEM176B knockdown with NLRP3 knockdown (sh-TMEM176B + sh-NLRP3). Two days before establishing the in situ model, adenoviral vectors or controls carrying the mouse TMEM176B/NLRP3 coding sequence (2.5 × 109 PFU/60 μL) were injected into the tail veins using a 29 G ´ 8 mm syringe[18]. Quantitative reverse transcription PCR (qRT-PCR) was used to assess transfection efficiency.
CT26-LUC-GFP tumor cells (iCell-m092, icell Bioscience, China) were cultured in a medium containing 1000 µg/mL G418 for 24 hours, or until clearly discernible monoclonal colonies appeared. Subsequently, monoclonal colonies were selected and grown in 96-well plates until they attained confluence rates of 70%-90%. The cells were then photographed using bioluminescence imaging equipment to identify those that stably expressed luciferase, and DMEM. Under aseptic conditions, 5 × 105 CT26-LUC-GFP tumor cells were suspended in a 1:1:0.1 combination of phosphate-buffered saline (PBS), Matrigel, and ink, and injected into the submucosal layer of the cecum wall. According to a previous study[19], the cecum was carefully adjusted after injection, and the abdominal cavity was sutured. The ethical endpoints included 20% weight loss, ulceration, or severe necrosis at the surgical site. All animal experiments were performed using random control and double-blind principles, approved by the Animal Experimentation Ethics Committee of the Zhejiang Eyong Pharmaceutical Research and Development Center (approval number: ZJEY-20231130-01) and conducted according to the appropriate guidelines.
After a month, the mice were anesthetized with isoflurane to monitor tumor spread using an in vivo imaging system (AniView 100; BLT, Guangzhou, China). Intestinal tumor tissue samples (tumor-infiltrated tissues, corresponding to the tumor mass) and blood samples (from tail vein) were collected for histological labeling, identification of biomarkers and cell culture.
Formalin-fixed colon tumor tissues were embedded in paraffin. The implanted tissues were sliced to a thickness of 15 μm. Sections were dehydrated using two changes of absolute ethanol, 75% alcohol, and tap water. After staining for 3-5 minutes with Hematoxylin (H3136; Sigma, United States), the sections were rinsed, separated, and rinsed again. After soaking the blue pieces in tap water for 5 minutes each, they were dehydrated using 85% and 95% alcohol. Eosin (Sigma, E4009) was used for staining for 5 minutes. Three changes of 100% ethanol were used for dehydration, and then cleared in xylene before mounting with neutral resin. Each segment was examined under a Nikon Eclipse Ci-L microscope.
Post antigen retrieval, primary antibodies against NLRP3 (DF7438; Affinity, United States), E-cadherin (AF0131; Affinity), and antigen identified by monoclonal antibody Ki 67 (Ki67) (AF0198; Affinity) were used to stain colon tumor tissue sections. The secondary antibody (ab97080, Abcam, United Kingdom) was conjugated to horseradish peroxidase. Examination was done using a Nikon Eclipse Ci-L fluorescence microscope (Tokyo, Japan).
Colon tumor tissue sections were prepared and permeabilized, followed by incubation with the TUNEL reaction mixture from Roche cell death detection kit (11684795910; Switzerland). Mounting medium containing 4’,6-Diamidino-2’-phenylindole/DAPI (ab104139, Abcam) was added, and fluorescence detection was performed. The apoptotic cells (green fluorescence) were examined using a fluorescent microscope (Nikon Eclipse Ci-L; Nikon, Tokyo, Japan), and their proportion in the total cell count is used to assess apoptosis.
Proteins were extracted from CT26 cells and colon tumor tissue samples. Proteins were separated using SDS-PAGE and transferred onto polyvinylidene fluoride membranes (10600023, GE Healthcare Life, United States). To block nonspecific binding, the membranes were incubated with Bovine Serum Albumin (4240GR100; BioFRoxx, Germany). Primary and secondary antibodies (Table 1) were applied to detect target proteins. Chemiluminescence (610020-9Q, Qinxiang) was used for visualization. The GAPDH was used as a reference control.
| Antibody | Company | Cat# | Dilution ratio |
| NLRP3 antibody | Affinity | DF7438 | 1:1000 |
| ASC antibody | Santa Curz Biotechnology | sc-22514-R | 1:1000 |
| Caspase1 antibody | Affinity | AF5418 | 1:1000 |
| IL18 antibody | Affinity | DF6252 | 1:1000 |
| TMEM176B antibody | Bioss | bs-11876R | 1:1000 |
| p-p65 antibody | Affinity | AF2006 | 1:1000 |
| p65 antibody | Affinity | AF5006 | 1:1000 |
| N-cadherin antibody | Affinity | AF4039 | 1:1000 |
| E-cadherin antibody | Proteintech | 20874-1-AP | 1:1000 |
| Vimentin antibody | Abcam | ab20346 | 1:1000 |
| MMP-9 antibody | Proteintech | 10375-2-AP | 1:1000 |
| TGF-β antibody | Affinity | AF1027 | 1:1000 |
| Anti-rabbit IgG, HRP-linked antibody | CST | 7074 | 1:6000 |
| Anti-mouse IgG, HRP-linked antibody | CST | 7076 | 1:6000 |
| GAPDH antibody | Proteintech | 10494-1-AP | 1:10000 |
CT26 cell lysates and tumor tissues were prepared for analysis. Enzyme-linked immunosorbent assay (ELISA) kits (RX202412M for mouse TNF-α and RX203064M for mouse IL-18) were obtained from Ruixin Biotech, China, while the mouse TGF-β ELISA kit (ml057830) was obtained from Shanghai Meilian biotechnology company, China. Standards and samples were added to the plates, followed by the detection antibodies and a substrate for color development. A microplate reader (CMaxPlus; MD, Shanghai, China) was used to determine the absorbance, and standard curves were used to compute the cytokine concentrations.
Peripheral blood mononuclear cells (PBMC) were collected from peripheral blood using a Mouse PBMC Isolation Kit (P5230; Solarbio, China). Briefly, Ficoll-Hypaque density gradient medium and centrifugation are used to separate PBMC layered. PBMCs were harvested and washed with PBS. The cells were counted, and the concentration was adjusted for further experiments.
PBMCs were extracted from peripheral blood, and then were incubated with allophycocyanin Hamster anti-mouse CD3e (553066; BD Biosciences, United States), R-phycoerythrin anti-mouse CD161 (NK1.1) (E-AB-F0987D; Elabscience, China), or fluorescein isothiocyanate anti-mouse CD107ab (562061; BD Biosciences). After incubation, cleaning, and centrifugation, natural killer (NK) cells were separated by flow cytometer (FCM) (NovoCyte; Agilent, CA, United States), sorting for CD3- NK1.1+ cells. Furthermore, NK cells were activated with IL-2 cytokine (2 nM) for 48 hours at 37 °C in a 5% CO2 atmosphere to increase their cytotoxicity and their ability to kill target cells more effectively. The stimulated and unstimulated NK cells were collected for further experiments.
CT26 cells were cultured and maintained in DMEM supplemented with 10% fetal bovine serum and incubated in a 5% CO2 incubator at 37 °C. CT26 cells (at passage 3) were transfected with three different configurations: An NLRP3 shRNA interference vector, a shRNA interference negative control vector, and a combination of NLRP3 interference and an shRNA negative control. Additionally, cells were transfected with shRNA targeting TMEM176B, interfering with NLRP3 expression. The CT26 cells were harvested during the incubation period for additional experiments. Subsequently, NK cells were co-cultured with CT26 (5:1) to explore the effects of TMEM176B or NLRP3 knockdown on CT26 cell inhibition rate, migration and invasion, and cell apoptosis under NK cells’ exposure.
Using an RNA isolation kit (AG21024; Accurate Biology, China), RNA was extracted from CT26 cell lines. A reverse transcription kit (CW2569M, CW Biology, China) was used to convert RNA to cDNA. PCR reactions were conducted using TMEM176B-specific primers (Table 2). Primer specificity was verified using the Primer-Basic Local Alignment Search Tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome). Expression levels of these genes were determined by qRT-PCR using a SYBR Green kit (11201ES03; Yeasen Biology, China). The expression data for differences in the total RNA amounts were adjusted for GAPDH, a housekeeping gene.
| Gene | Forward primer (5’-3’) | Reverse primer (5’-3’) |
| Mouse Teme176b | AGTCCGCTCACATCAGCATC | ACTTAGTCCCAGGGAAGCCA |
| Mouse NLRP3 | CTACGGCCGTCTACGTCTTC | GGCCAAAGAGGAATCGGACA |
| Mouse GAPDH | GGGTCCCAGCTTAGGTTCAT | CTCGTGGTTCACACCCATCA |
Propidium iodide and Annexin V were used to stain CT26 cells to detect apoptosis, following the guidelines of a staining kit (556547; BD Biosciences, United States). Each test was performed using a NovoCyte FCM. Single cells were gated on FSC-H and SSC-H, and a two-color FCM analysis was performed. The level of cell apoptosis was evaluated based on the proportion of cells in the two quadrants on the right; from top to bottom, the two quadrants represent late and early apoptotic cells.
Transfected CT26 cells were seeded in 96-well plates and allowed to adhere overnight, followed by exposing to NK cells. MTT solution (298-93-1, Sigma) was added in each well following the instructions provided by the manufacturer and incubated for 4 hours. A solubilization solution was used to dissolve formazan crystals produced by metabolically active cells. Cell viability was assessed at 570 nm absorbance using a CMaxPlus microplate reader.
A 6-well plate was prepared with five horizontal lines drawn 0.5 cm apart. After plating the CT26 cells at 5 × 105 cells per well, the cells were allowed to grow until confluence was reached, ensuring that all wells were covered. A scratch was made across the cell monolayer using the tip of a pipetting gun to draw a straight line perpendicular to the premarked lines at the bottom of the wells. To remove any detached cells from the scratched area, the cells were washed thrice with PBS, followed by exposing to NK cells. Serum-free mediums were used to wash cells, and then photographs were taken at a 12-hour time point and used to calculate the rate of wound healing in each cell group by ImageJ software (NIH, MD, United States).
A 24-well Transwell chamber was coated with 50 μL of diluted Matrigel (for invasion) or not (for migration), which was allowed to solidify at 37 °C. The upper chamber was then seeded with 200 μL of CT26 cell suspension at a concentration of 2 × 104/mL. In the lower chamber, 600 μL of fresh culture media enriched with 10% fetal bovine serum were filled. After 24 hours of incubation with NK cells, medium and non-invading cells in the upper chamber were carefully removed. Cells were fixed for 30 minutes with 4% paraformaldehyde. After fixation, the cells were stained with 0.1% crystal violet for visibility, and remaining stain was removed by washing with PBS.
Statistical analysis was conducted using IBM SPSS 20.0 (NY, United States). For normally distributed data with equal variance, one-way ANOVA along with Tukey's post-hoc test were used. Independent t-tests or Dunnett's T3 tests were used for unequal variances. When it came to data that wasn't normally distributed, the Kruskal-Wallis H test was brought into play. Results were shown as mean ± SD, with significance set at a = 0.05. A P value less than 0.05 indicated statistical significance.
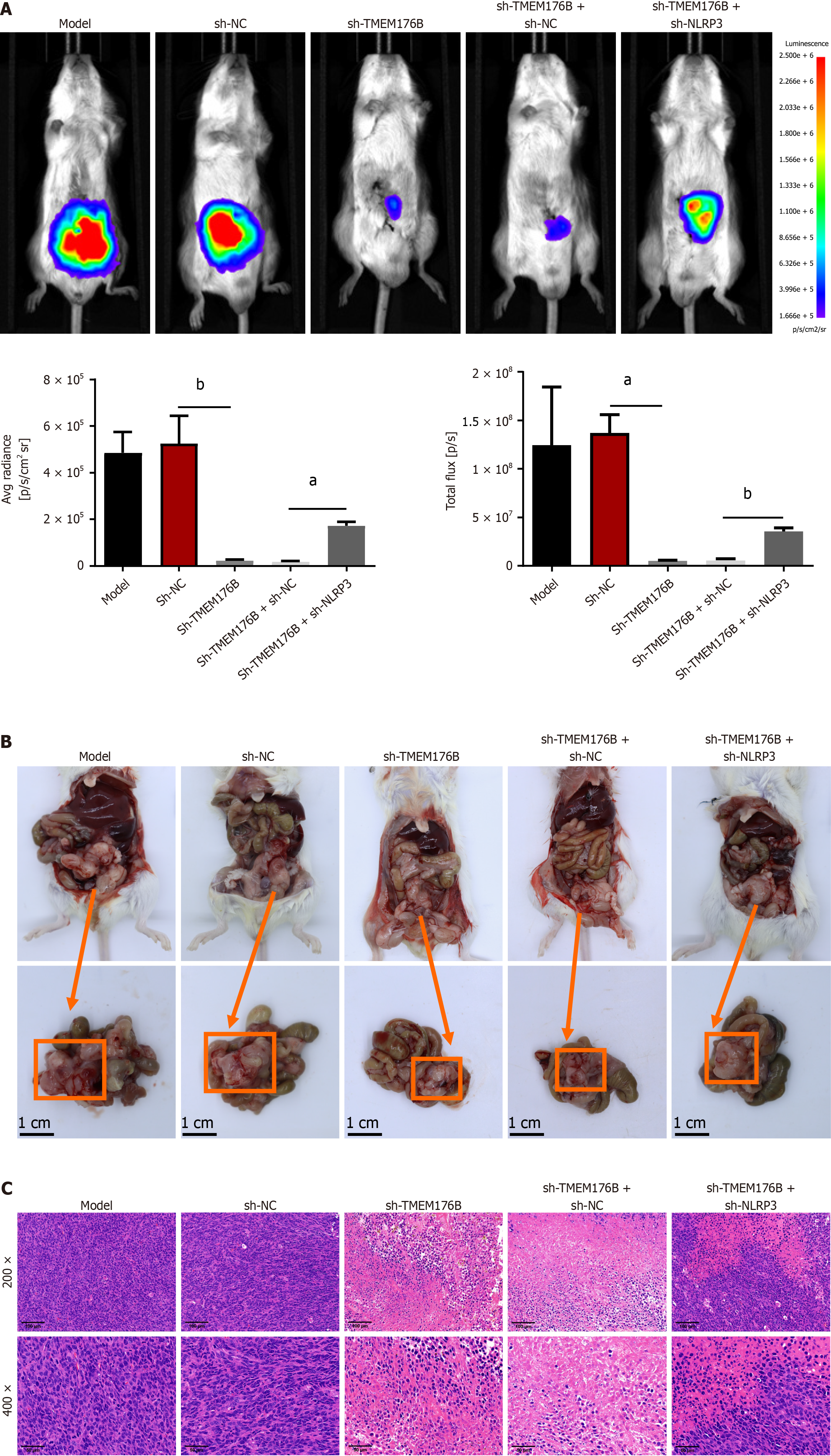
Fluorescence Imaging (Figure 1A) revealed that the mice in the sh-TMEM176B group had reduced tumor metastasis than the sh-NC group due to lower average radiance and total flux (P < 0.05). Conversely, knockdown NLRP3 of tumors increased the average and total fluorescence intensities in the sh-TMEM176B mice compared to its NC mice (sh-TMEM176B + sh-NC) (P < 0.05). As shown in Figure 1B, in the sh-TMEM176B group, smaller tumor tissues were observed in the colon than in sh-NC group. Sh-TMEM176B + sh-NLRP3 group showed a significant increase in tumor volume than the sh-TMEM176B + sh-NC group.
According to Hematoxylin and eosin staining (Figure 1C), the tumor cells were closely organized, and the overall appearance of the tumor tissues in the model mice was intact with no visible damage. Tumor tissues from the sh-NC group did not exhibit any morphological changes compared to those of the model group. In contrast, the sh-TMEM176B mice showed significant necrosis and tumor cell degeneration, along with severe tissue damage. The sh-TMEM176B + sh-NLRP3 animals showed less total tissue damage, including less injury, necrosis, and degeneration, than sh-TMEM176B + sh-NC mice.
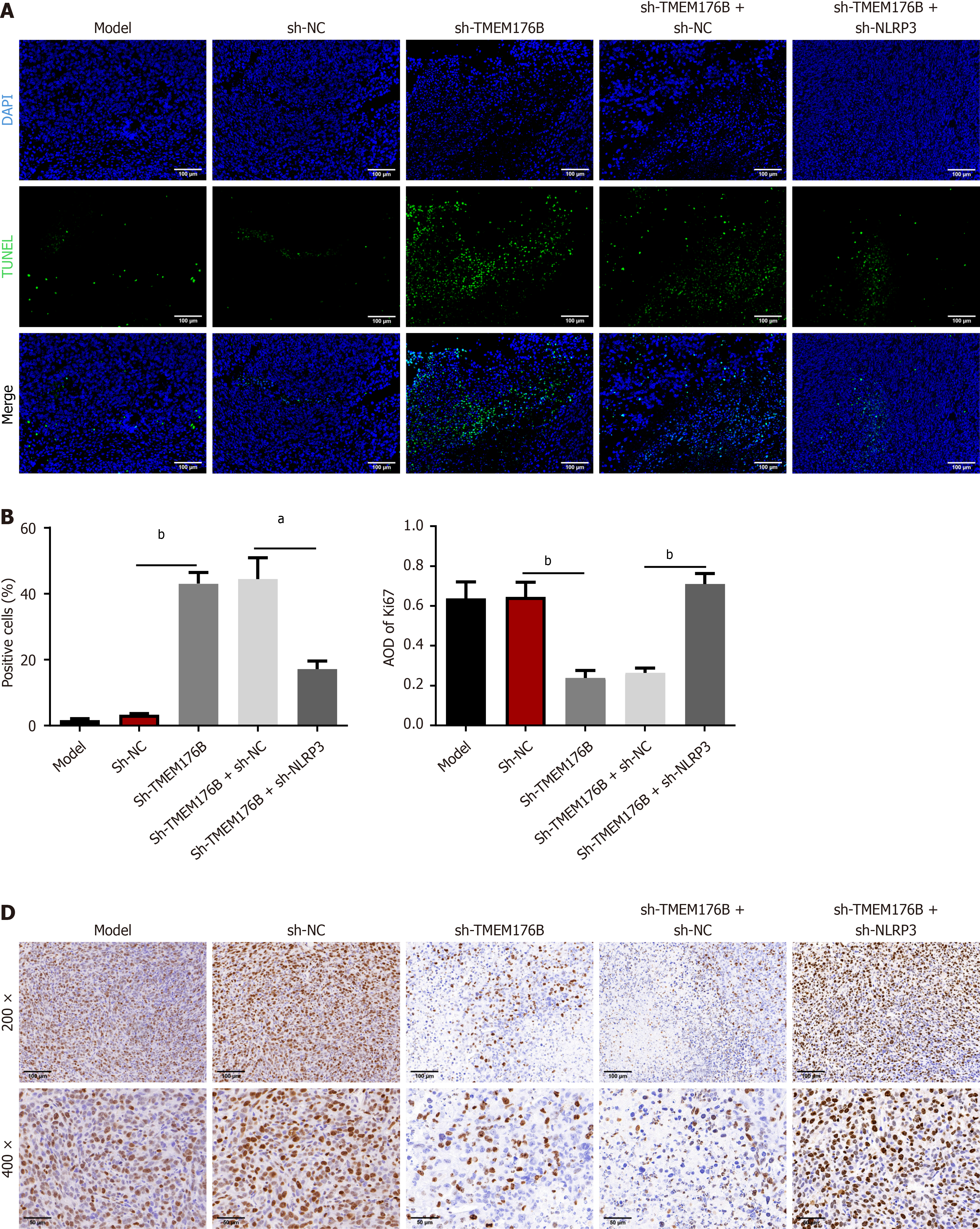
TUNEL and immunohistochemistry (IHC) staining (Figure 2) showed that sh-TMEM176B raised the proportion of TUNEL-positive cells and decreased Ki67 average optical density (AOD) compared to that in the sh-NC group (P < 0.01). These effects were reversed in the sh-TMEM176B + sh-NLRP3 group (P < 0.05).
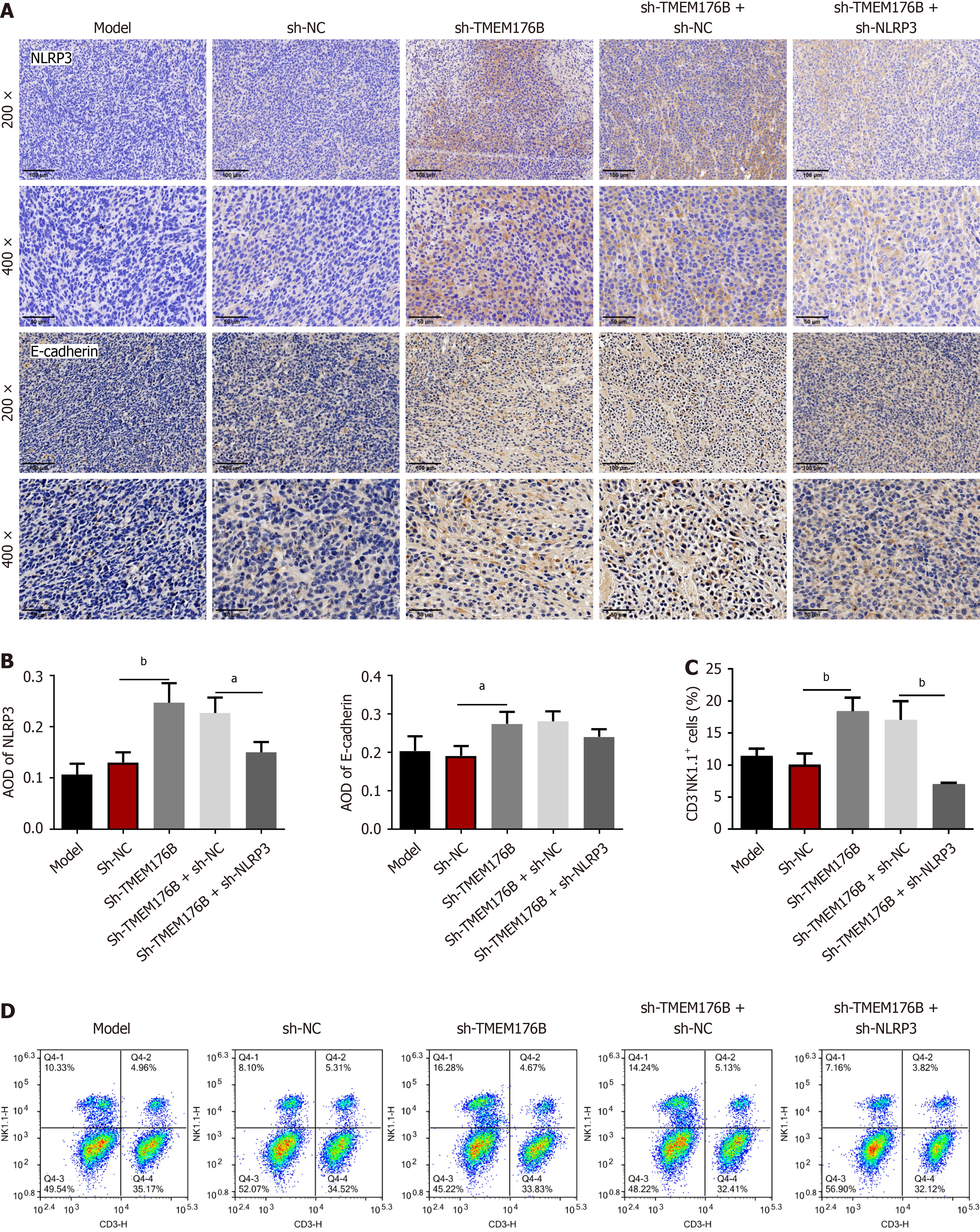
The expression and density of NLRP3 and E-cadherin were quantified using IHC staining (Figure 3A and B). The AOD of NLRP3 and E-cadherin in the tumor tissue was considerably higher in the sh-TMEM176B group than in the sh-NC group (P < 0.05). FCM analysis (Figure 3C and D) showed an increased number of CD3-NK1.1+ cells in the sh-TMEM176B group compared to that in the sh-NC group (P < 0.05). Furthermore, sh-NLRP3 partially antagonized the promotion of sh-TMEM176B on NLRP3 expression and the number of CD3-NK1.1+ cells (P < 0.05).
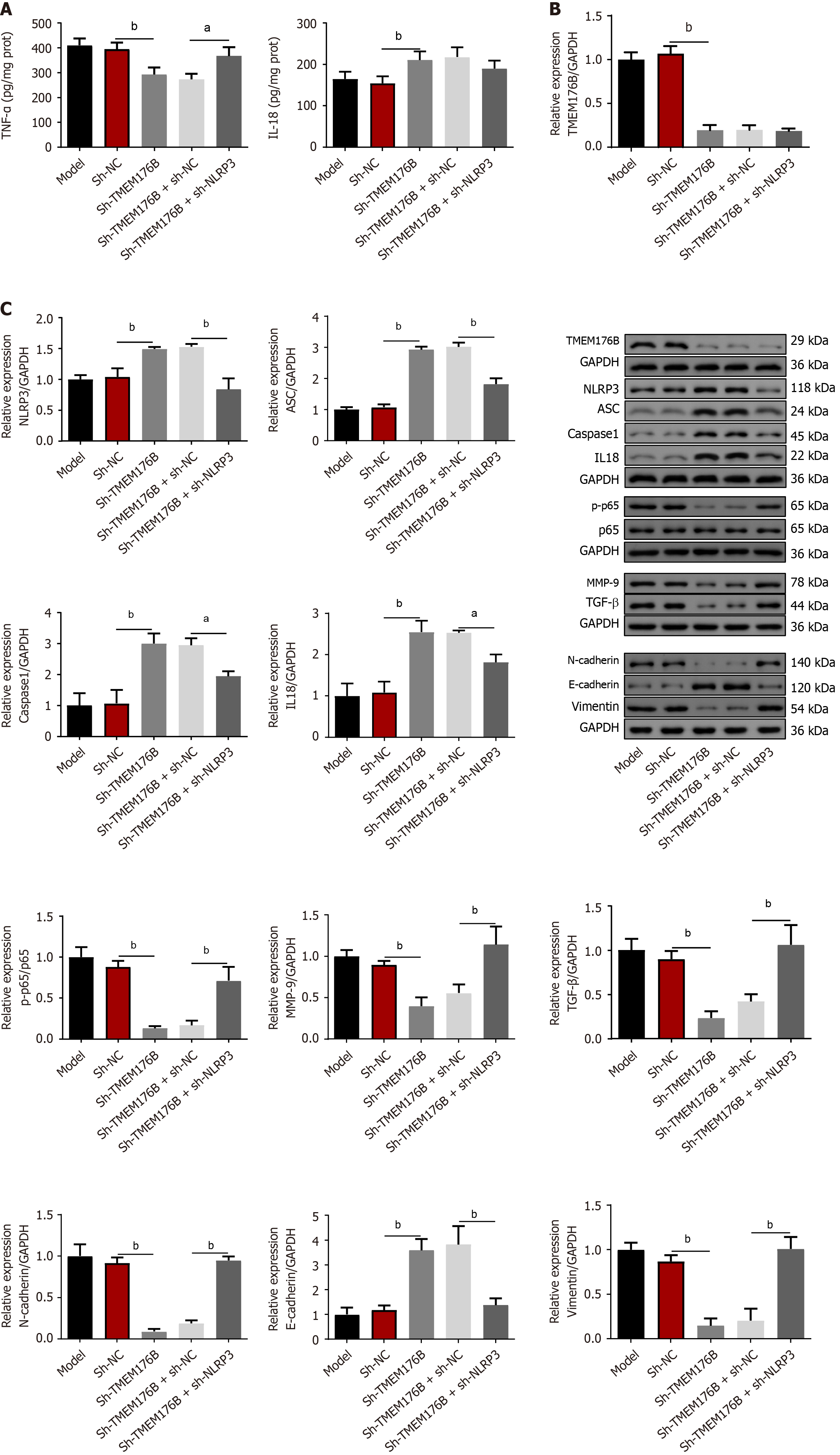
As illustrated in Figure 4A, ELISA revealed that the sh-TMEM176B group had a lower level of TNF-α, while having a higher IL-18 level (P < 0.01). However, NLRP3 knockdown increased TNF-α levels in the sh-TMEM176B mice (P < 0.05). Figure 4B and C demonstrate that in the sh-TMEM176B group the expression of TMEM176B, p-p65, p65, MMP-9, TGF-β, N-cadherin, and Vimentin proteins were significantly reduced, whereas NLRP3, ASC, caspase-1, IL-18 and E-cadherin were enhanced (P < 0.05). Conversely, NLRP3 knockdown decreased the expression of NLRP3, ASC, caspase-1, IL-18, and E-cadherin in the sh-TMEM176B mice (P < 0.05), whereas TMEM176B, p-p65, p65, MMP-9, TGF-β, N-cadherin, and Vimentin proteins increased (P < 0.01).
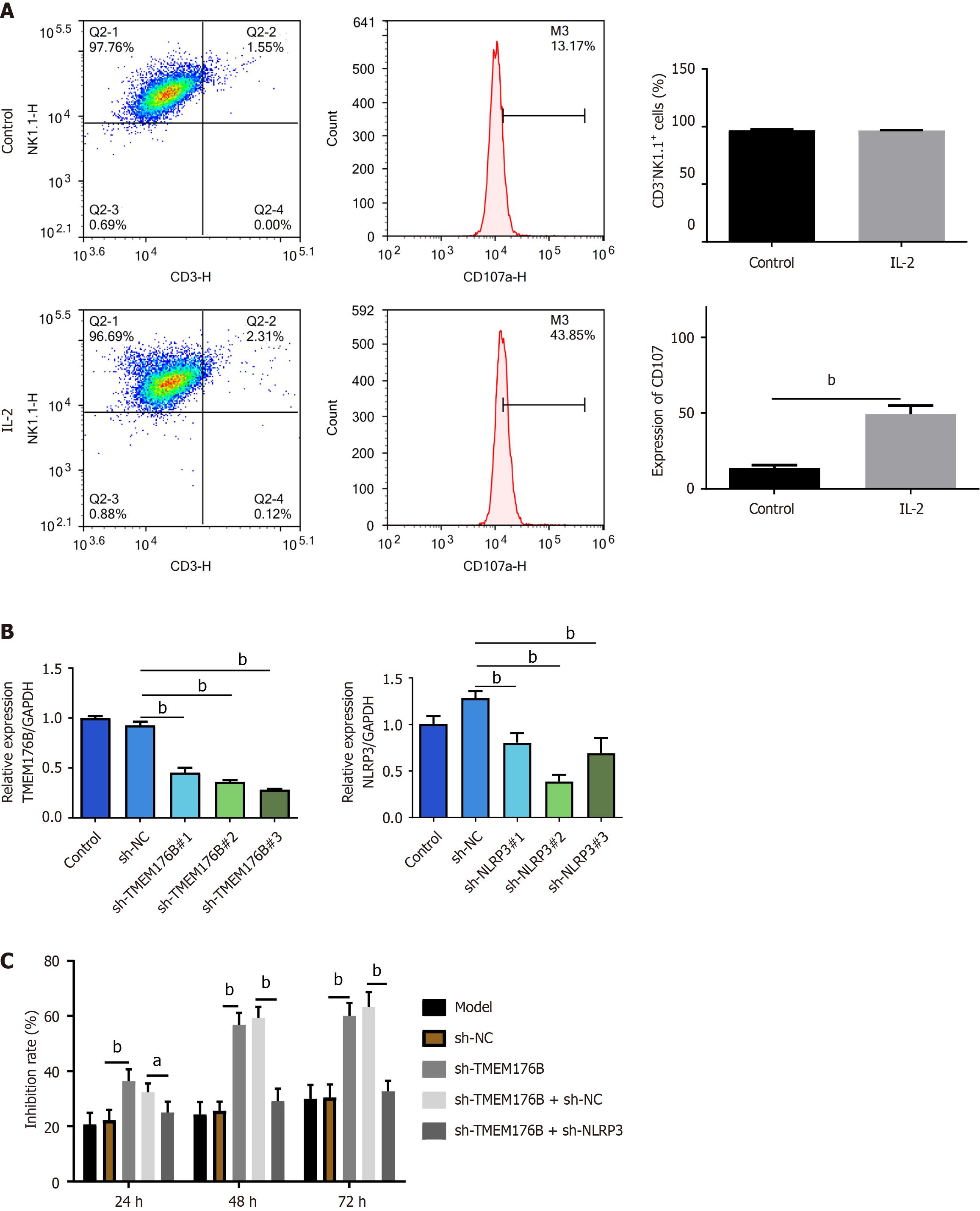
IL-2 was used to activate NK cells. FCM analysis (Figure 5A) highlights the increased CD107 expression in primary mouse NK cells (CD3-NK1.1+ cells) exposed to IL-2 compared to controls (P < 0.01). Figure 5B shows the results of qRT-PCR analysis to test the transfection efficiency: Sh-TMEM176B#1/#2/#3 decreased TMEM176B mRNA in CT26 cells and sh-NLRP3#1/#2/#3 also inhibited NLRP3 mRNA (P < 0.01). The efficiency of sh-TMEM176B#3 and sh-NLRP3#2 in transfection is the highest, exceeding 50%, so they are being used for subsequent research (named sh-TMEM176B and sh-NLRP3). The MTT assay (Figure 5C) showed the inhibition rate of IL-2-exposed NK cells on CT26 cells with TMEM176B and NLRP3 knockdown. Results of Figure 5C demonstrated that the inhibition rate of NK cells on sh-TMEM176B treated CT26 cells at 24, 48, and 72 hours was increased compared to sh-NC treated CT26 cells (P < 0.01). Conversely, at 24 hours, 48 hours, and 72 hours, the inhibition rate in the sh-TMEM176B + sh-NLRP3 group was considerably lower than that in the sh-TMEM176B + sh-NC group (P < 0.05).
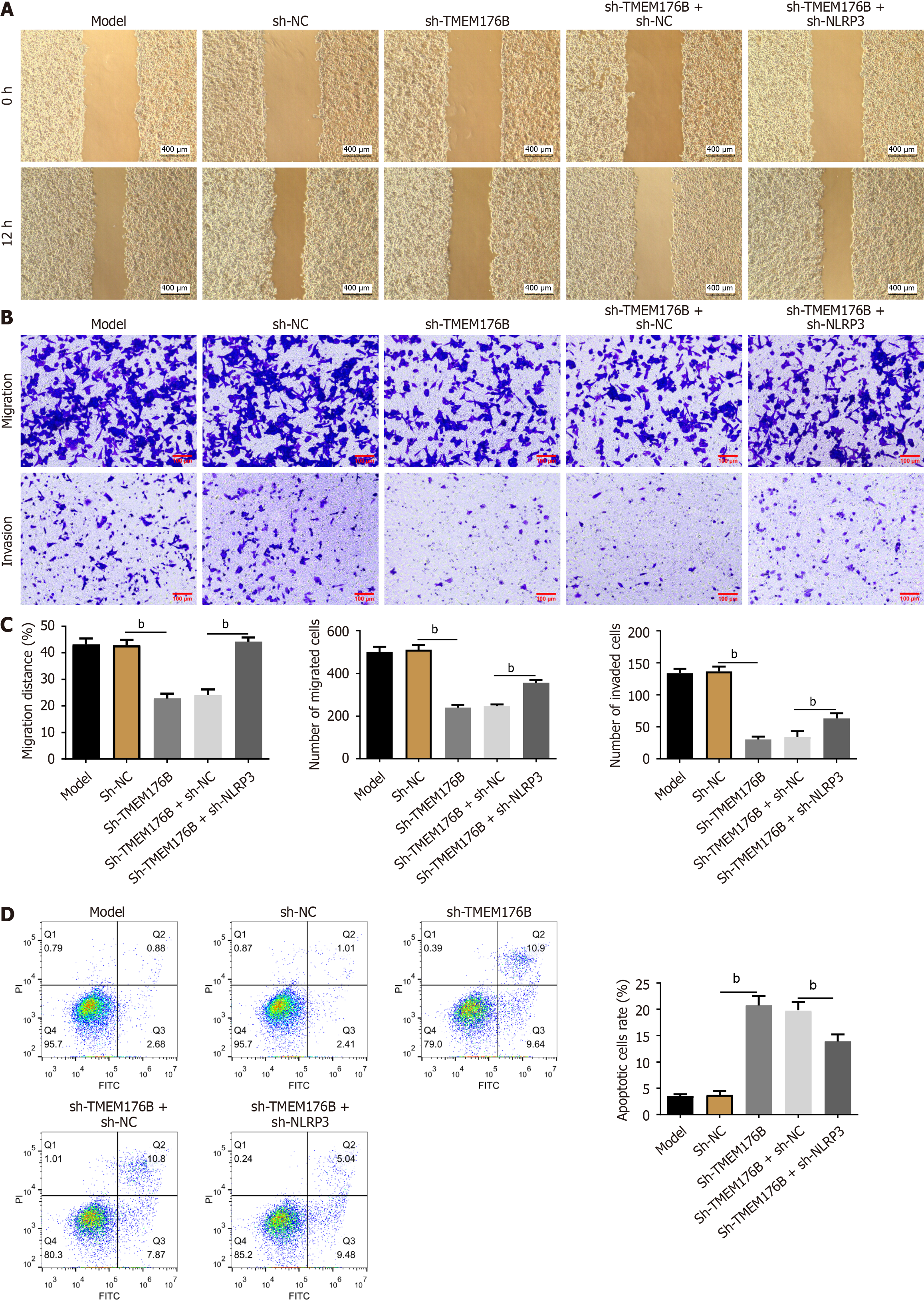
CT26 cells were co-cultured with IL-2 exposed NK cells for 12 hours and the cell migration and invasion abilities were evaluated using wound healing and Transwell assays (Figure 6A-C). The 12-hour migration rate and the number of migrated and invaded sh-TMEM176B cells were considerably lower than sh-NC cells (P < 0.01). In contrast, NLRP3 knockdown increased the number of migrated and invaded sh-TMEM176B cells (P < 0.01).
FCM was used to examine CT26 cell apoptotic rate (Figure 6D). TMEM176B knockdown increased the CT26 cell apoptosis rate (P < 0.01), while NLRP3 knockdown partially antagonized it (P < 0.01).
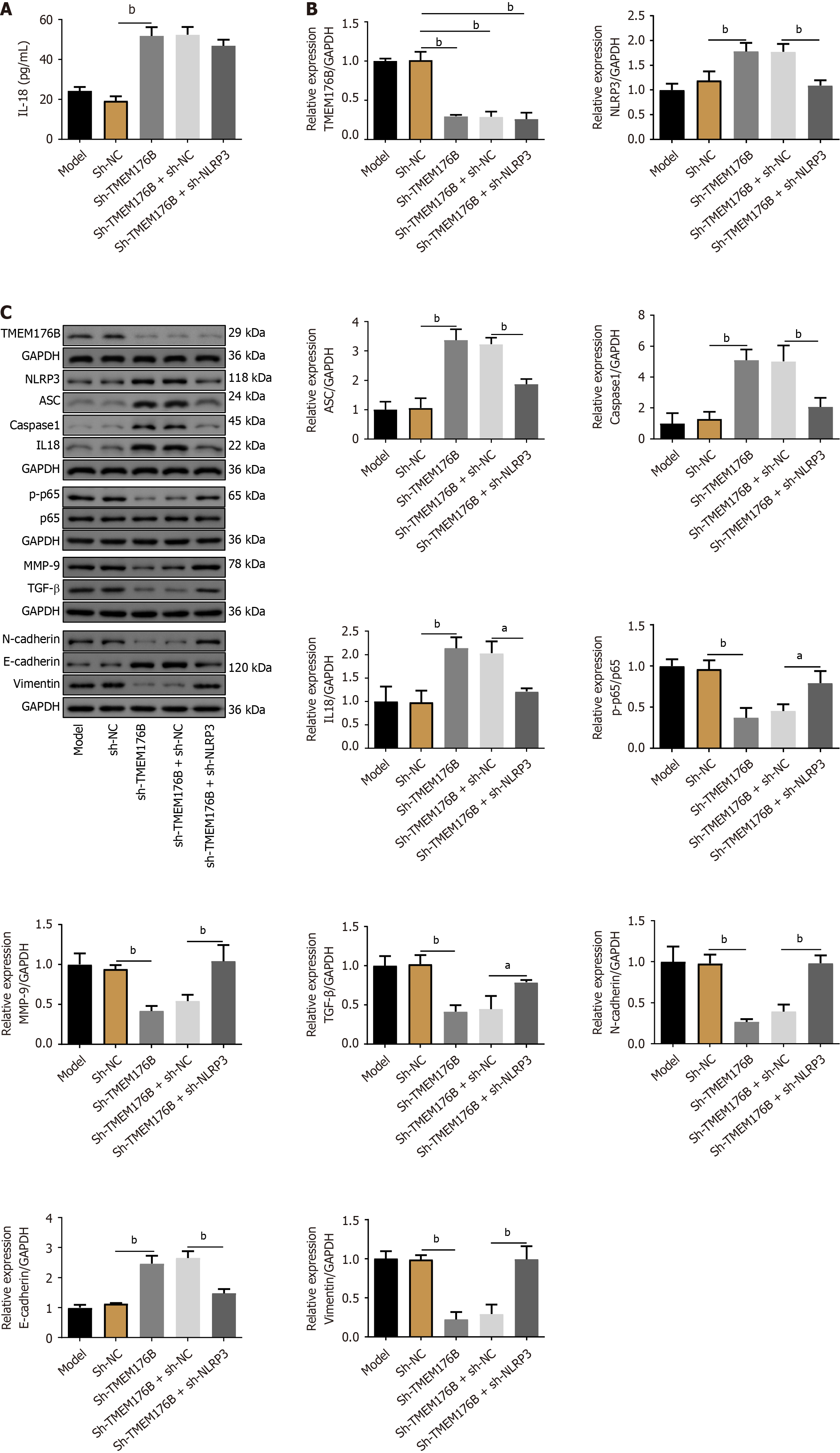
Sh-TMEM176B increased IL-18 concentration in CT26 cells (P < 0.01) as determined by ELISA (Figure 7A). Figure 7B and C showed that sh-TMEM176B inhibited the expression of TMEM176B, p-p65, p65, MMP-9, TGF-β, N-cadherin, and Vimentin proteins, whereas NLRP3, ASC, caspase-1, IL-18, and E-cadherin were enhanced (P < 0.05). In contrast, the NLRP3 knockdown partially antagonized the effects of sh-TMEM176B on the above protein expression (P < 0.05).
This study presents significant findings regarding the role of sh-TMEM176B in inhibiting tumor metastasis and promoting apoptosis in mice with colon cancer, highlighting the involvement of the NLRP3 signaling pathway. Evidence supports that targeting TMEM176B enhances immunity and improves immune checkpoint blocker efficacy by inhibiting inflammasome activation[10,20,21]. Cambui et al[20] claimed that the Ala134Thr variant (rs2072443) in TMEM176B serves as a protector against CRC, linked to lower TMEM176B gene expression and heightened NLRP3 inflammasome in monocytes and dendritic cells. Additionally, previous studies have shown that TMEM176B promotes invasion of cancer cells via the phosphoinositide 3-kinase pathway[8]. In our study, fluorescence imaging and morphological analyses demonstrated that sh-TMEM176B treatment reduced tumor metastasis and promoted the necrosis and degeneration of tumor cells. Moreover, inhibition of the NLRP3 pathway, in conjunction with sh-TMEM176B treatment (sh-TMEM176B + sh-NLRP3 group), resulted in increased tumor volume and reduced apoptosis, suggesting that NLRP3 plays a crucial role in mediating the effects of sh-TMEM176B in NK cells.
Decreased levels of TMEM176B appear to be positively correlated with improved overall survival and heightened inflammasome activity[20,22,23]. While extensive research has been conducted on tumorigenesis, fewer studies have explored prognosis of CRC, particularly in animal models and, to a lesser extent, in human subjects[24]. To date, only a handful of independent association studies have examined the role of inflammasomes in determining CRC outcomes. Notably, two studies have identified NLRP3 mutation as an influential player in poor CRC prognosis[11,25]. Another study has found a correlation between increased NLRP3 expression and reduced overall survival in patients[26], further highlighting the potential impact of inflammasome activity in cancer progression and patient outcomes. In this study, the IHC and molecular findings further supported the notion that sh-TMEM176B modulates the TME and cell signaling pathways. Specifically, the decreased expression of Ki67 and increased apoptosis in the sh-TMEM176B group indicated potent anti-proliferative and pro-apoptotic effects[27]. However, modulation of the NLRP3 pathway altered these effects, indicating a complex interaction between TMEM176B and NLRP3 in tumor biology. Consistently, Jing et al[28] reported that CD146 activated NLRP3 inflammasome-mediated activation of macrophages in the TME via inhibiting TMEM176B and treatment with a TMEM176B inhibitor boosts antitumor effects. This study also supports the idea that interfering with TMEM176B in CRC may be beneficial for the activation of immune cells and antitumor effects, making it a potential target for adjuvant therapy in CRC.
Furthermore, we explored the effect of sh-TMEM176B on EMT, a key process in cancer metastasis, by assessing the expression of E-cadherin and NLRP3. These results suggested that sh-TMEM176B inhibited EMT, further contributing to its anti-metastatic properties. This was corroborated by the malignant phenotype analysis of CT26 cells, where sh-TMEM176B impeded cell migration and invasion and enhanced apoptosis. The changes in cytokine levels, such as IL-18, TNF-α, and TGF-β, along with the expression of various proteins like MMP-9, N-cadherin, and Vimentin, provide additional insight into the molecular mechanisms through which sh-TMEM176B exerts its effects[10]. These alterations in cytokine and protein expression levels emphasize the anti-inflammatory and antitumor properties of sh-TMEM176B. Additionally, EMT is positively correlated with the expression of tumor immune cell biomarkers and immune che
Following the successful anticancer effects of immunosuppressants in solid tumors, interest and expectations for immunotherapy have increased[32]. As silencing TMEM176B in CRC can enhance the inhibitory effects of NK cells on CRC malignant phenotypes, such as EMT and invasion, it can be inferred that TMEM176B has the potential to serve as an adjuvant therapy for immunotherapy. However, because laboratory animal and cell models cannot fully mirror human CRC, the application of experimental results in clinical practice requires substantial amounts of clinical and laboratory research. Additionally, although this study provides substantial insights into the role of TMEM176B in regulating EMT and apoptosis via the NLRP3 inflammasome, a complete understanding requires further exploration.
In summary, this study elucidates the significant role of sh-TMEM176B in suppressing tumor growth and metastasis in colon cancer, primarily through the modulation of apoptosis and EMT process, with the NLRP3 signaling pathway playing a pivotal role. These findings not only enhance our understanding of the molecular mechanisms underlying colon cancer progression but also suggest potential therapeutic targets for treatment. The interaction between sh-TMEM176B and the NLRP3 pathway offers a promising avenue for further research, particularly for the development of targeted therapies for colon cancer.
| 1. | Zhu J, Jiang C, Hui H, Sun Y, Tao M, Liu Y, Qian X. Overexpressed lncRNA LINC00893 Suppresses Progression of Colon Cancer by Binding with miR-146b-3p to Upregulate PRSS8. J Oncol. 2022;2022:8002318. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 2. | Gao Z, Jiang J, Hou L, Zhang B. Dysregulation of MiR-144-5p/RNF187 Axis Contributes To the Progression of Colorectal Cancer. J Transl Int Med. 2022;10:65-75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 34] [Reference Citation Analysis (0)] |
| 3. | Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18694] [Cited by in RCA: 21455] [Article Influence: 1950.5] [Reference Citation Analysis (6)] |
| 4. | Shin AE, Giancotti FG, Rustgi AK. Metastatic colorectal cancer: mechanisms and emerging therapeutics. Trends Pharmacol Sci. 2023;44:222-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 412] [Reference Citation Analysis (0)] |
| 5. | Buhrmann C, Yazdi M, Popper B, Kunnumakkara AB, Aggarwal BB, Shakibaei M. Induction of the Epithelial-to-Mesenchymal Transition of Human Colorectal Cancer by Human TNF-β (Lymphotoxin) and its Reversal by Resveratrol. Nutrients. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 57] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 6. | Long L, Xiong W, Lin F, Hou J, Chen G, Peng T, He Y, Wang R, Xu Q, Huang Y. Regulating lactate-related immunometabolism and EMT reversal for colorectal cancer liver metastases using shikonin targeted delivery. J Exp Clin Cancer Res. 2023;42:117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 48] [Reference Citation Analysis (0)] |
| 7. | Xu J, Yang X, Pan J, Fan H, Mei J, Hua D. Biochanin A Suppresses Tumor Progression and PD-L1 Expression via Inhibiting ZEB1 Expression in Colorectal Cancer. J Oncol. 2022;2022:3224373. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 8. | Li J, Fang Z, Dal E, Zhang H, Yu K, Ma M, Wang M, Sun R, Lu M, Wang H, Li Y. Transmembrane protein 176B regulates amino acid metabolism through the PI3K-Akt-mTOR signaling pathway and promotes gastric cancer progression. Cancer Cell Int. 2024;24:95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 9. | Jiang L, Yang Y, Liu F, Ma M, Gao J, Sun L, Chen Y, Shen Z, Wu D. A Potential Diagnostic and Prognostic Biomarker TMEM176B and Its Relationship With Immune Infiltration in Skin Cutaneous Melanoma. Front Cell Dev Biol. 2022;10:859958. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 10. | Segovia M, Russo S, Jeldres M, Mahmoud YD, Perez V, Duhalde M, Charnet P, Rousset M, Victoria S, Veigas F, Louvet C, Vanhove B, Floto RA, Anegon I, Cuturi MC, Girotti MR, Rabinovich GA, Hill M. Targeting TMEM176B Enhances Antitumor Immunity and Augments the Efficacy of Immune Checkpoint Blockers by Unleashing Inflammasome Activation. Cancer Cell. 2019;35:767-781.e6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 119] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 11. | Cambui RAG, do Espírito Santo GF, Fernandes FP, Leal VNC, Galera BB, Fávaro EGP, Rizzo LA, Elias RM, Pontillo A. Double-edged sword of inflammasome genetics in colorectal cancer prognosis. Clin Immunol. 2020;213:108373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 12. | Zheng D, Liwinski T, Elinav E. Inflammasome activation and regulation: toward a better understanding of complex mechanisms. Cell Discov. 2020;6:36. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 526] [Cited by in RCA: 734] [Article Influence: 122.3] [Reference Citation Analysis (0)] |
| 13. | Wang X, Jia Y, Wen L, Mu W, Wu X, Liu T, Liu X, Fang J, Luan Y, Chen P, Gao J, Nguyen KA, Cui J, Zeng G, Lan P, Chen Q, Cheng B, Wang Z. Porphyromonas gingivalis Promotes Colorectal Carcinoma by Activating the Hematopoietic NLRP3 Inflammasome. Cancer Res. 2021;81:2745-2759. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 142] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 14. | Feng WQ, Zhang YC, Xu ZQ, Yu SY, Huo JT, Tuersun A, Zheng MH, Zhao JK, Zong YP, Lu AG. IL-17A-mediated mitochondrial dysfunction induces pyroptosis in colorectal cancer cells and promotes CD8 + T-cell tumour infiltration. J Transl Med. 2023;21:335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 62] [Reference Citation Analysis (1)] |
| 15. | Fernandes FP, Leal VNC, Souza de Lima D, Reis EC, Pontillo A. Inflammasome genetics and complex diseases: a comprehensive review. Eur J Hum Genet. 2020;28:1307-1321. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 16. | Kolb R, Liu GH, Janowski AM, Sutterwala FS, Zhang W. Inflammasomes in cancer: a double-edged sword. Protein Cell. 2014;5:12-20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 162] [Cited by in RCA: 210] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 17. | Rathinam VAK, Chan FK. Inflammasome, Inflammation, and Tissue Homeostasis. Trends Mol Med. 2018;24:304-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 147] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 18. | Zhao R, Zhang Q, Liu W, Lin Y, He Y, Chang D, Li S, Xu W, Lin Y, Zheng Y, Zhou X, Huang M. Pien Tze Huang attenuated acetaminophen-induced liver injury by autophagy mediated-NLRP3 inflammasome inhibition. J Ethnopharmacol. 2023;311:116285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 19. | Hou Y, Jin J, Duan H, Liu C, Chen L, Huang W, Gao Z, Jin M. Targeted therapeutic effects of oral inulin-modified double-layered nanoparticles containing chemotherapeutics on orthotopic colon cancer. Biomaterials. 2022;283:121440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 74] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 20. | Cambui RAG, Fernandes FP, Leal VNC, Reis EC, de Lima DS, do Espírito Santo GF, Elias RM, Pontillo A. The Ala134Thr variant in TMEM176B exerts a beneficial role in colorectal cancer prognosis by increasing NLRP3 inflammasome activation. J Cancer Res Clin Oncol. 2023;149:3729-3738. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 21. | Segovia M, Russo S, Girotti MR, Rabinovich GA, Hill M. Role of inflammasome activation in tumor immunity triggered by immune checkpoint blockers. Clin Exp Immunol. 2020;200:155-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 22. | Kang C, Rostoker R, Ben-Shumel S, Rashed R, Duty JA, Demircioglu D, Antoniou IM, Isakov L, Shen-Orr Z, Bravo-Cordero JJ, Kase N, Cuajungco MP, Moran TM, LeRoith D, Gallagher EJ. TMEM176B Regulates AKT/mTOR Signaling and Tumor Growth in Triple-Negative Breast Cancer. Cells. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 23. | Liang R, Qi X, Cai Q, Niu L, Huang X, Zhang D, Ling J, Wu Y, Chen Y, Yang P, Liu J, Zhang J, Yu P. The role of NLRP3 inflammasome in aging and age-related diseases. Immun Ageing. 2024;21:14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 38] [Reference Citation Analysis (0)] |
| 24. | Hill M, Russo S, Olivera D, Malcuori M, Galliussi G, Segovia M. The intracellular cation channel TMEM176B as a dual immunoregulator. Front Cell Dev Biol. 2022;10:1038429. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 25. | Ungerbäck J, Belenki D, Jawad ul-Hassan A, Fredrikson M, Fransén K, Elander N, Verma D, Söderkvist P. Genetic variation and alterations of genes involved in NFκB/TNFAIP3- and NLRP3-inflammasome signaling affect susceptibility and outcome of colorectal cancer. Carcinogenesis. 2012;33:2126-2134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 121] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 26. | Wang B, Li H, Wang X, Zhu X. The association of aberrant expression of NLRP3 and p-S6K1 in colorectal cancer. Pathol Res Pract. 2020;216:152737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 27. | Lowe M, Lage J, Paatela E, Munson D, Hostager R, Yuan C, Katoku-Kikyo N, Ruiz-Estevez M, Asakura Y, Staats J, Qahar M, Lohman M, Asakura A, Kikyo N. Cry2 Is Critical for Circadian Regulation of Myogenic Differentiation by Bclaf1-Mediated mRNA Stabilization of Cyclin D1 and Tmem176b. Cell Rep. 2018;22:2118-2132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 28. | Jing L, An Y, Cai T, Xiang J, Li B, Guo J, Ma X, Wei L, Tian Y, Cheng X, Chen X, Liu Z, Feng J, Yang F, Yan X, Duan H. A subpopulation of CD146(+) macrophages enhances antitumor immunity by activating the NLRP3 inflammasome. Cell Mol Immunol. 2023;20:908-923. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 34] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 29. | Figueiredo J, Ferreira RM, Xu H, Gonçalves M, Barros-Carvalho A, Cravo J, Maia AF, Carneiro P, Figueiredo C, Smith ML, Stamenović D, Morais-de-Sá E, Seruca R. Integrin β1 orchestrates the abnormal cell-matrix attachment and invasive behaviour of E-cadherin dysfunctional cells. Gastric Cancer. 2022;25:124-137. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 30. | Yuan Z, Li Y, Zhang S, Wang X, Dou H, Yu X, Zhang Z, Yang S, Xiao M. Extracellular matrix remodeling in tumor progression and immune escape: from mechanisms to treatments. Mol Cancer. 2023;22:48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 542] [Reference Citation Analysis (0)] |
| 31. | Jang HR, Shin SB, Kim CH, Won JY, Xu R, Kim DE, Yim H. PLK1/vimentin signaling facilitates immune escape by recruiting Smad2/3 to PD-L1 promoter in metastatic lung adenocarcinoma. Cell Death Differ. 2021;28:2745-2764. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 66] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 32. | Fan A, Wang B, Wang X, Nie Y, Fan D, Zhao X, Lu Y. Immunotherapy in colorectal cancer: current achievements and future perspective. Int J Biol Sci. 2021;17:3837-3849. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 158] [Cited by in RCA: 293] [Article Influence: 58.6] [Reference Citation Analysis (1)] |