Published online Mar 15, 2025. doi: 10.4251/wjgo.v17.i3.100739
Revised: December 8, 2024
Accepted: January 3, 2025
Published online: March 15, 2025
Processing time: 169 Days and 19.9 Hours
Helicobacter pylori (H. pylori) infection induces pathological changes via chronic inflammation and virulence factors, thereby increasing the risk of gastric cancer development. Compared with invasive examination methods, H. pylori-related serum indicators are cost-effective and valuable for the early detection of gastric cancer (GC); however, large-scale clinical validation and sufficient understanding of the specific molecular mechanisms involved are lacking. Therefore, a comprehensive review and analysis of recent advances in this field is necessary. In this review, we systematically analyze the relationship between H. pylori and GC and discuss the application of new molecular biomarkers in GC screening. We also summarize the screening potential and application of anti-H. pylori immunoglobulin G and virulence factor-related serum antibodies for identifying GC risk. These indicators provide early warning of infection and enhance screening accuracy. Additionally, we discuss the potential combination of multiple scr
Core Tip: Traditional serum tumor markers exhibit low diagnostic efficacy. Although gastrin and pepsin reflect the current state of the stomach, their clinical application remains impractical. New molecular biomarkers, such as noncoding genomes, have potential utility in diagnosing and screening for gastric cancer (GC); however, further validation is required. Serum anti-Helicobacter pylori antibodies have been used in various combined examination methods for GC screening, and the relationship between specific antibodies to virulence factors and GC risk has been thoroughly researched. The application of multiple serological detection and prediction models has expanded the pool of candidate biomarkers for GC screening.
- Citation: Sun HT. Helicobacter pylori-related serum indicators: Cutting-edge advances to enhance the efficacy of gastric cancer screening. World J Gastrointest Oncol 2025; 17(3): 100739
- URL: https://www.wjgnet.com/1948-5204/full/v17/i3/100739.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i3.100739
Gastric cancer (GC) is the fifth most common neoplasm and fourth leading cause of cancer-related deaths worldwide[1]. Despite the availability of various treatment methods, including endoscopic therapy, surgical intervention, targeted therapy, radiotherapy, and chemotherapy, the prognosis of patients with GC remains poor, with a 5-year survival rate of < 30% for patients with advanced GC[2]. As GC often lacks specific clinical manifestations in its early stages[3], identifying individuals and subgroups at higher-than-average risk is the most effective way to reduce cancer risk. However, current methods for diagnosing and screening GC are typically invasive and carry a considerable economic burden[4]. Thus, there is an urgent need for clinically relevant noninvasive diagnostic markers.
Helicobacter pylori (H. pylori) is a gram-negative bacterium that colonizes the gastric mucosa and causes various digestive system diseases, such as chronic gastritis, peptic ulcers, and gastric adenocarcinoma, via multiple pathways[5]. Its infection rate impacts 50% of the global population, with higher rates in developing countries[6,7]. H. pylori is a class I carcinogen and the most important risk factor for GC occurrence and development. For example, inflammatory tissue lesions caused by its infection affect approximately 75% of patients with GC[8-10]. Therefore, targeted monitoring and eradication of H. pylori could reduce the incidence of GC[11,12].
Population-based screening for H. pylori offers health benefits at a low cost and can effectively reduce the increased risk of GC[13]. At different baseline levels of GC, H. pylori eradication can considerably reduce the incidence of GC, with benefits unaffected by research design, sex, or follow-up period[10]. Short-term treatment can provide long-term protection against GC for high-risk groups[14]. Therefore, the global consensus conference proposed that eradicating H. pylori at the individual level can reduce the risk of GC in asymptomatic individuals. Moreover, they proposed large-scale H. pylori screening and treatment requirements in high-risk populations for GC[15]. In regions with a high incidence of GC, screening and treatment strategies for H. pylori infection in young individuals are the most cost-effective, although still beneficial for those > 50 years old[16,17].
Serological testing is widely used and recognized as one of the most effective H. pylori screening methods. A systematic review demonstrated that screening for H. pylori is cost-effective in different contexts and that serologic testing is the most effective of all screening methods[18]. The current diagnostic efficacy of conventional serum tumor markers is low and incapable of early detection. Although gastrin and pepsin can reflect the condition of the stomach, their sensitivity and specificity require further improvement. Thus, there is an urgent need to explore the combined use of multiple efficient serum markers and develop standardized assays to enhance the accuracy of noninvasive tests for diagnosing early GC and precancerous lesions.
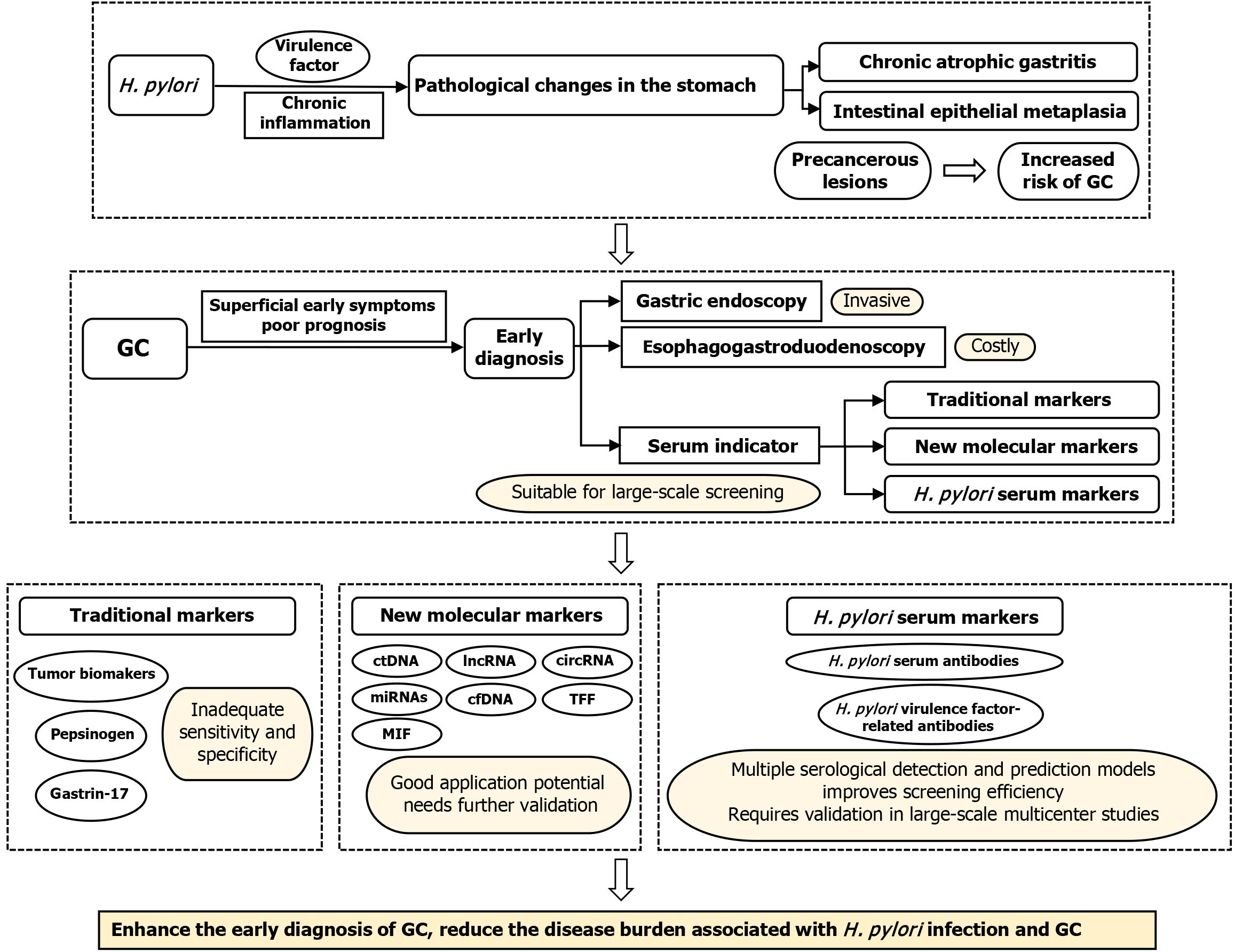
This review summarizes the current serum markers used in GC screening and focuses on the role and recent research progress of H. pylori-associated serum biomarkers in early GC screening and diagnosis (Figure 1). By systematically analyzing recent clinical and basic research, we highlight the molecular mechanisms of H. pylori virulence factors (e.g., vacuolating cytotoxin A [VacA] and cytoxin-associated gene A [CagA]) and their associations with gastric carcinogenesis and progression. Our goal is to explore the potential of these serum markers as noninvasive biomarkers and evaluate their feasibility and clinical value, aiming to identify cost-effective and efficient serum markers that could support the development of more effective strategies for GC screening and early diagnosis.
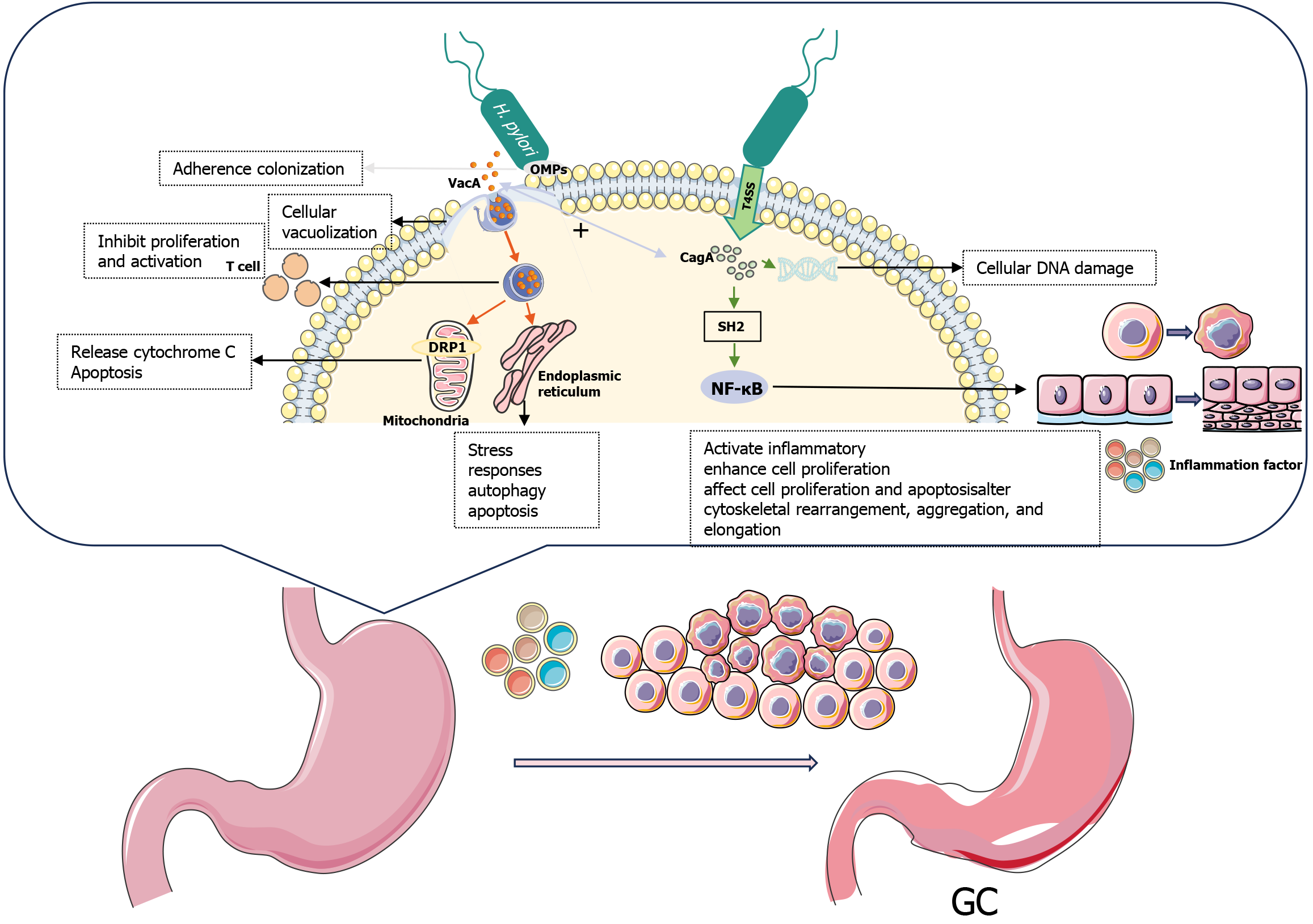
H. pylori infection is a multifactorial process that triggers gastric diseases via chronic inflammation and virulence factors. This bacterium uses flagella to evade the immune system and penetrate the gastric mucosa[19], where it produces urease to protect itself from low pH environments and provide a source of metabolic energy[20,21]. H. pylori colonizes gastric epithelial cells through adhesion factors, such as outer membrane proteins (OMPs)[22]. This process recruits circulating immune cells, such as neutrophils, macrophages, and lymphocytes, to the site of infection and stimulates various signaling pathways within the host to disrupt the gastric mucosal barrier, resulting in its chronic inflammation[23]. During GC development, chronic inflammatory reactions and virulence factors persist and mutually reinforce each other, damaging the DNA of gastric epithelial cells[24,25], activating oncogenes, and inactivating tumor suppressor genes[26], thereby increasing the risk of GC development over time (Figure 2).
Chronic inflammation, mucosal damage, genetic and epigenetic alterations, and altered gene expression are key features of H. pylori infection leading to gastric carcinogenesis[27]. Chronic inflammation induced by H. pylori infection damages cells and causes them to undergo multiple stages of carcinogenesis. Epidemiologically, patients with H. pylori CagA-positive infection exhibit a 4.9-fold increased incidence of GC compared to H. pylori-positive patients without CagA, and a 14.5-fold increased incidence compared to H. pylori-negative patients without CagA[28]. Long-term chronic gastritis triggers an inflammatory response typically associated with the innate immune response, oxidative stress, and the adaptive immune response, leading to chronic atrophic gastritis (CAG) and intestinal epithelial metaplasia (IM) precancerous lesions of significance[29]. The inflammatory response promotes tumor growth, while inflammatory mediators attract immune cells to the lesion, providing favorable conditions for tumor cell growth, invasion, and metastasis, forming a tumor microenvironment (TME). The inflammatory microenvironment promoted by H. pylori leads to increased mutation rates, resulting in DNA damage and other epigenetic changes, triggering a cascade of reactions ranging from chronic gastritis to atrophic gastritis; residual post-eradication DNA methylation may be involved in the development of GC[30]. In the innate immune response, neutrophils produce oxidants to kill pathogens, but the antioxidant enzymes of H. pylori enable the bacteria to survive oxidative stress and induce increased oxidative stress in gastric epithelial cells, enhancing genomic instability[31]. Contrastingly, adaptive immune responses are typically associated with T helper 1 immunity. The presence of serum anti-H. pylori immunoglobulin g (IgG) and IgA suggests that bacterial infection elicits a broad immune response and serves as a useful tool for monitoring patients with gastric atrophy, intestinal metaplasia, and dysplasia[32]. Monitoring H. pylori infection status is a highly accurate, simple, and noninvasive method.
The presence of specific virulence factors, particularly CagA and VacA, in H. pylori plays an important role in GC development. The global prevalence of CagA positivity in patients with H. pylori infectious gastropathy varies from 43% to 90%[33]. Epidemiological studies in a Moroccan population have shown that patients infected with strains bearing the VacA-d region have a 13.33-fold increased risk of GC. The combination of the active form of the VacA genotype, the duodenal ulcer promoting gene status, and the number of EPIYA-C motifs, may facilitate differentiation among multiple gastric diseases[34]. Patients infected with H. pylori strains expressing both CagA and VacA proteins have nearly a 5-fold increased risk of GC, as determined using serologic tests (enzyme-linked immunosorbent assay [ELISA] and Western blot analysis)[35]. These specific antigens may serve as candidate serum biomarkers for H. pylori-associated GC.
The cytotoxin-associated gene pathogenicity island codes for the production of CagA and the type IV secretion system (T4SS)[36]. The pili formed by T4SS inject the CagA tumor protein, which has cytotoxic effects on host cells, into target cells[37]. Once it enters the host cell, CagA interacts with various host Src homology 2 domains, resulting in tyrosine phosphorylation[38]. Phosphorylated and unphosphorylated CagA interacts with several host proteins to activate inflammatory signaling pathways, such as nuclear factor kappa B[39], and enhances the proliferation of gastric epithelial cells[40]. The CagA protein induces cellular alterations that impair cell motility, cause DNA damage[41], affect cell proliferation and apoptosis, and alter cytoskeletal rearrangement, aggregation, and elongation, in addition to disrupting tumor suppressor mechanisms in gastric epithelial cells[42,43]. The cascade of biochemical and cellular events promotes cancer development. Typically, CagA-positive strains lead to severe clinical outcomes, with odds ratios (ORs) for increased risk of precancerous lesions, such as atrophic gastritis, IM, and heterogeneous hyperplasia of 3.03, 2.98, and 3.16, respectively[44]. Compared to those with gastritis, CagA-positive patients are at an elevated risk of developing GC (OR = 2.53, 95% confidence interval [CI]: 1.15-5.55)[45].
VacA, another key virulence factor of H. pylori, is secreted by the type V autotransporter secretion system and enters host cells via endocytosis[40]. VacA has a wide range of biological activities[46] that help it evade the host immune system, leading to chronic infection. It can form anion-selective membrane channels in the cytoplasmic membrane, causing cellular vacuolization and the formation of a series of large vesicles characterized by the presence of late endosomes and early lysosomes[47]. VacA proteins targeting mitochondria can induce cytochrome C release and apoptosis[48]. Stimulation of dynamin-related protein 1 (DRP1) may play a key role in VacA-induced mitochondrial disruption, as inhibition of DRP1-dependent mitochondrial fission in VacA-treated cells impedes B-cell lymphoma 2-associated X protein activation and mitochondrial outer membrane permeabilization, thus helping to prevent cell death in VacA-treated cells[49]. Toxic proteins targeting the endoplasmic reticulum can trigger stress responses, activate autophagy, and induce apoptosis[46]. Internalized VacA inhibits T-cell proliferation and activation, activates cell signaling, and induces inflammatory responses[50]. These disruptions enable H. pylori to successfully evade the immune system and promote the development of GC. The presence of VacA genotypes strongly predicts the degree of virulence of CagA-positive H. pylori strains; additionally, s1a, s1c, and m1 are reportedly associated with GC, peptic ulcers, and IM[51].
OMPs, such as blood group antigen-binding adhesive (BabA), sialic acid-binding adhesive (SabA), outer inflammatory protein A (OipA), and adhesion-associated lipoproteins, mediate bacterial colonization and enhance CagA translocation via T4SS[52].
GC screening is a secondary prevention measure that can detect early cancer in asymptomatic carriers, early lesions, and precancerous lesions. Several countries have included GC screening in their national cancer screening plans[17]; however, all of them rely primarily on invasive examinations. For example, individuals aged ≥ 40 years in South Korea undergo upper gastrointestinal examinations or endoscopy every 2 years[53]. The screening of GC in Japan primarily relies on radiology and endoscopy. At the local level, H. pylori antibody (Ab) detection and serum pepsinogen (SPG) levels have been used as joint screening approaches but not as primary screening methods[54]. In the European population, gastric test kits for detecting PGI, PGII, gastrin-17 (G-17), and H. pylori IgG group Abs are used as biomarkers for screening and detecting GC[55].
Common invasive screening methods only demonstrate cost-effectiveness in medium-to-high risk populations, and the discomfort and costs associated with the process may hinder the comprehensive promotion of screening[56]. For example, endoscopy depends on the operator’s expertise; however, its diagnostic efficiency for precancerous lesions is limited. In a retrospective study, the sensitivities of endoscopic diagnosis for atrophic gastritis and intestinal metaplasia were 69.5% and 19.4%, respectively[57]. Moreover, esophagogastroduodenoscopy is an expensive diagnostic procedure with reported complications[58]. Therefore, the ideal cancer screening test requires high accuracy and considers the population’s willingness to undergo the examination.
The detection of serum biomarkers could provide a more convenient, cost-effective, and noninvasive screening method suitable for a wider population range. During GC screening, common serum indicators include tumor markers, SPG, G-17, urease, and H. pylori-related Abs[4]. New biomarkers based on novel technologies have the potential for the early and improved detection of GC[59].
Traditional serum tumor markers include carbohydrate antigen (CA) 72-4, alpha fetoprotein (AFP), CA12-5, carcinoembryonic antigen (CEA), and CA19-9[60]. These are the most commonly used biomarkers in clinical practice[61]; however, due to low sensitivity and specificity, their use in diagnostics is limited[62]. When used alone, the sensitivity and specificity of CA72-4 to diagnose GC are 49% and 96%, respectively, which are superior to other tumor biomarkers, such as CEA (sensitivity: 41% and specificity: 93%), CA19-9 (sensitivity: 44% and specificity: 92%), and CA242 (sensitivity: 38% and specificity: 97%)[63]. However, CA72-4 is highly expressed in normal tissues, resulting in false positives in patients with atrophic gastritis[64]. Moreover, measuring AFP levels during the initial diagnosis of GC may provide information about prognosis, but elevated AFP is not as effective as CEA in predicting stage IV disease and 1-year mortality[65]. In certain studies, the combined detection of various tumor markers may improve their diagnostic value[66,67]; however, more accurate serum biomarkers are urgently required.
As a precursor of gastric protease, SPG is one of the most extensively investigated biomarkers for predicting precancerous lesions of GC[68]. Human PG is classified into two groups based on biochemistry and immunohistochemistry. PGI is secreted by primary and mucosal cervical cells in the gastric fundus gland, whereas PGII is produced by gastric fundus cells, pyloric cells, and the Bruner gland[69]. Serum PGI and PGII levels increase with the progression of gastritis. As gastritis reduces gastric fundus mucosa, serum PGI levels gradually decrease while serum PGII levels remain stable. Therefore, serum PGI levels and the PGI/PGII ratio decrease with gastritis progression[55] and may serve as predictive indicators for gastritis and gastric atrophy when PGI ≤ 70 ng/mL and the PGI/II ratio ≤ 3[70]. These indicators, which Japan has included in GC screening[71], play an important role in identifying precancerous lesions, such as CAG, IM, and dysplasia[68], with acceptable sensitivity and specificity[72]. However, SPG identification has a better predictive effect for gastric atrophy than for GC, as its sensitivity for GC is significantly less[73]. Eradicating H. pylori can significantly alter SPG levels; thus, the predictive value of SPG as a risk marker for GC following treatment decreases[74].
G-17 is a polypeptide hormone produced by G cells in the gastric antrum, which regulates gastric acid secretion and mucosal growth. The secretion of G-17 is regulated by a feedback system, reflecting the progression and severity of gastric mucosal diseases[75]. During chronic H. pylori infection, G-17 levels increase[76], and hypergastrinemia occurs before atrophy and cancer development, rendering it a good serological marker for autoimmune gastritis[77]. When multifocal atrophy occurs in the stomach, loss of gastric antral glands may lead to a reduced number of G cells and lower G-17 secretion levels. One study reported that G-17 levels demonstrated a sensitivity, specificity, and overall accuracy of 36.8%, 86.5%, and 82.6%, respectively, in diagnosing antral atrophy[78]. In another meta-analysis, the diagnostic sensitivity and specificity of G-17 for predicting atrophic gastritis were 48% and 79%, respectively[79]. In clinical practice, the low sensitivity of the G-17 test limits its utility as the sole biomarker for diagnosing gastric atrophy.
Overall, combining G-17 detection with SPG ideally reflects the functional status of the stomach and its atrophy; however, the performance of this test in practical applications has not met expectations.
With the rapid development of next-generation sequencing technology, the emergence of noncoding molecules, such as circulating tumor DNA (ctDNA), microRNA (miRNA), long noncoding RNA (lncRNA), and circular RNA (circRNA), has enhanced the understanding of GC molecular pathogenesis and holds great potential for early diagnosis of GC. Noncoding genes are used to assess prognosis, monitor tumor burden, predict treatment resistance, quantify small residual lesions, and enable real-time cancer management[80].
ctDNA is a fragment of genetic material released into the bloodstream after shedding from necrotic or apoptotic tumor cells, carrying the same genetic information as the originating tumor cells. It has emerged as a candidate biomarker for screening cancer patients[81] and is used to monitor cancer recurrence[82,83] and identify somatic mutations in cancer patients[84]. During GC treatment, ctDNA can be used to monitor the clinical response of immunotherapy[85]. For example, a meta-analysis revealed that the sensitivity and specificity of ctDNA testing for diagnosing GC were 62% and 95%, respectively[86]. However, in patients with early GC, the ctDNA serum load is lower and mutation sequences are fewer, which leads to false negative results[87]. Additionally, DNA fragments produced by nontumor hematopoietic stem cells may obscure ctDNA detection, resulting in false positive results[88]. Currently, significant differences in sensitivity and specificity have been reported among studies, and corresponding ctDNA detection methods lack reliability and stability, thereby limiting their potential use in clinical testing.
miRNAs are endogenous small noncoding RNA. Abnormal miRNA expression can regulate pathological and biological processes of GC, mimicking the functions of tumor suppressor genes and oncogenes[89]. They can be used to evaluate therapeutic intervention efficacy as well as the diagnosis and prognosis of patients with GC[90]. Increased expression of miR-200 family members is associated with the occurrence of early GC[91]. Moreover, miR-652, miR-629, and miR-627 have a sensitivity and specificity of 86.7% and 85.5%, respectively, for diagnosing GC[92]. A prospective validation study in Singapore, based on miRNAs, showed an overall sensitivity and specificity of 87.0% and 68.4%, respectively, for GC diagnosis[93]. Compared with those in healthy control groups, preoperative circulating miR-196a and miR-196b levels were elevated in patients with GC, and their expression levels decreased following surgical tumor resection[94]. miRNAs have potential as biomarkers for diagnosing GC, but the interactions between miRNAs-whether synergistic or antagonistic-have not yet been fully elucidated.
lncRNAs are a type of RNA that lack protein-coding ability and play a role in regulating various cellular processes. They affect tumor development through complex signaling pathways and interact with miRNAs. Several lncRNAs have been identified as abnormally expressed in GC and participate in proliferation, invasion, metastasis, and drug resistance by regulating different target genes[95]. For example, the lncRNA C1RL-AS1 promotes GC proliferation and metastasis via the AKT/β-catenin pathway[96]. Moreover, the m6A modification-mediated lncRNA TP53 target 1 inhibits GC progression by regulating the stability of cancerous inhibitor of protein phosphatase 2A[97]. The hypoxia-inducible cystathionine beta synthase (CBS) mRNA stabilizing lncRNA modulates ferroptosis via the m6A-YTH N6-methyladenosine RNA-binding protein F2-dependent module of CBS in GC[98]. The expression of lnc-SLC2A12-10:1 in the extracellular vesicles of patients with GC is significantly upregulated and correlates with tumor size, tumor-node-metastasis staging, lymph node metastasis, and differentiation degree. Lnc-SLC2A12-10:1 can be used for dynamic monitoring during GC treatment[99]. However, due to the numerous and complex mechanisms of lncRNAs, our understanding of lncRNAs in GC remains limited, restricting their use in clinical practice.
circRNAs have high abundance and stability, evolutionary species conservation, and tissue specificity[100]. They can regulate gene expression at the transcriptional, post-transcriptional, and translational levels, playing an important role in cancer growth, metastasis, recurrence, and treatment resistance[101]. Extensive clinical testing of tissue and serum samples from patients with GC and healthy controls, along with analyses of clinical pathological factors, prognosis, and survival rates, has led to the discovery of circRNA biomarkers for the early diagnosis and prediction of recurrence and metastasis in GC. For example, circCEACAM5 and circCOL1A1 interact with miRNAs associated with GC[102]. The area under the curve (AUC), sensitivity, and specificity of plasma hsa_circ_0000520 in GC are 0.897%, 82.4%, and 84.4%, respectively, which are higher than those in normal tissue (0.613%, 53.6%, and 85.7%, respectively)[103]. The sensitivity and specificity of has_circ_0001017 and has_circ_0061276 in tissues and plasma are > 95%[104], demonstrating excellent application prospects. In addition, extracellular vesicles could expand the research and application of circRNAs, and further exploration is required to determine whether they can be developed as new GC biomarkers and therapeutic targets.
cfDNA is a heterogeneous mixture, primarily comprising nuclear and mitochondrial DNA that is present in exosomes or other extracellular vesicles. cfDNA carries genetic and epigenetic information regarding tumor-specific changes, which are associated with the origin, invasiveness, and metastatic potential of tumors[105]. Moreover, cfDNA is abundant in patients with advanced or metastatic tumors and is a promising component in tumor liquid biopsy[106]; however, further research is needed to explore its properties, subtypes, release mechanisms, and clearance rates in patients with cancer[107].
The trefoil factor family (TFF) is a peptide family of small-molecule soluble proteins, primarily expressed in the normal gastrointestinal tract[108]. TFF1 is a gastric tumor suppressor, TFF2 is a candidate gene for GC suppression, and TFF3 is a single tripeptide expressed throughout the intestine that can promote the development of GC[109]. The concentrations of serum TFF1, TFF2, and TFF3 are significantly increased in patients with GC[110]. In patients with early-stage GC, serum TFF3 can serve as an independent biomarker for detecting tumor staging and distant metastasis[111,112]. Currently, the specific mechanism of action of TFFs is unknown.
The macrophage migration inhibitory factor is overexpressed in almost all types of cancer and contributes to tumor progression[113]. Tumor-educated platelets absorb tumor-related biomolecules and vesicles when interacting with tumor cells in the TME. They can be used alone or in combination with other diagnostic biomolecules, such as circulating tumor cells, cancer-associated fibroblasts, or ctDNAs, for the early diagnosis and screening of patients with cancer[114]. Extracellular vesicle proteins may carry tumor-specific biomarkers into circulation, which can be used for diagnostic and prognostic applications. These specific biomarkers reflect the cellular origin of extracellular vesicles and contribute to cancer detection, rendering them powerful candidates for revealing the disease status of patients with cancer[115].
Liquid biopsy, a technique that uses a series of tumor-related cellular, subcellular, and molecular analytes, has become a powerful new tool for the early diagnosis of various malignant tumors[116]. Overall, these novel biomarkers have significant potential to diagnose and screen for GC. However, preliminary studies involved small sample sizes, and require further explorations, larger sample sizes, and global population studies to validate current findings. Multi-analyte studies have the potential to reduce statistical bias in patient and biomarker diversity. Currently, no unified standard is available for the extraction, transportation, storage, and testing methods of samples. Selection of key targeted novel biomarkers from numerous candidate sequences remains a significant challenge.
H. pylori is an important risk factor in the occurrence and development of GC, resulting in a high overlap between patients with GC and those with H. pylori infection[117]. In high-risk areas around the world, H. pylori serum marker testing has been proposed as a GC screening tool, either alone or in combination with other markers, with unique advantages in large-scale epidemiological investigations[118]. This section elaborates on the indicative role of various serum indicators related to H. pylori in GC and their current application in screening, with the aim of identifying candidate biomarkers for GC screening.
The method of measuring serum anti-H. pylori Ab (HpAb) titers is simple, economical, and easy to obtain. It is widely used in population screening and evaluating treatment efficacy. Serological testing accuracy is not affected by ulcer bleeding, gastric atrophy, or the use of proton pump inhibitors or antibiotics, often resulting in false-negative tests[119]. Chronic inflammation caused by H. pylori infection primarily results in the production of IgG and IgA in the body, with IgM being rare. When the titer of anti-H. pylori IgG is ≥ 20 U/mL, the body is currently or has previously been infected with H. pylori. The sensitivity, specificity, and accuracy of detecting H. pylori infection using this critical value are 97.8%, 58.0%, and 78.7%, respectively[120]. Positive serum Abs against H. pylori are strongly associated with GC[121], and the risk of GC lesions increases increasing anti-H. pylori IgG titers[122], which can be used as an early screening indicator for GC. Notably, Tatemichi et al[123], conducted a prospective study and reported that patients with atrophic gastritis and serum HpAb titers, as low as 10 U/mL, had a higher risk of developing GC than those with high HpAb titers. As serum Abs can persist following H. pylori eradication, serological testing cannot distinguish between active and past infections. IgG Abs alone are not effective as a serological indicator for diagnosing GC; however, using multiple specific antigens can improve the diagnostic efficiency of Ab testing[124].
Serum anti-H. pylori IgG has been applied in various combined methods for GC screening. For example, Japan launched a new large-scale screening method for GC, called the ABC method. This method divides the population into four groups by jointly measuring serum anti-H. pylori IgG Abs and SPG levels: Groups A (H. pylori [-], PG [-]), B (H. pylori [+], PG [-]), C (H. pylori [+], PG [+]), and D (H. pylori [-], PG [+]). The risk of GC is highest in Group D, and gradually decreases from Groups C to A. The plan for endoscopic examination and eradication therapy is determined based on the risk level of each group with the goal of preventing GC[125]. Using serum HpAb titers as quantitative, rather than qualitative or categorical, parameters can enhance the ability of the ABC method for future GC risk screening. Moreover, its effectiveness was confirmed in a long-term prospective cohort study targeting the Japanese population[126].
A new method, called GastroPanel, is used to diagnose atrophic gastritis and detects gastric specific biomarkers, including HpAbs and SPG and G-17 levels[127]. In two meta-analyses, the sensitivity of GastroPanel in diagnosing advanced GC was 70.2% and 74.7%, with specificities as high as 93.9% and 95.6%[128,129], demonstrating its reliability for diagnosing atrophic gastritis and its suitability for screening individuals or populations at a high risk of GC. In addition to gastric indicators, the combined detection of serum anti-HpAbs and cytokines has good application potential in the screening of GC[130]. Notably, these serological tests can detect the gastric environment that is prone to GC, thereby individuals at a higher risk of cancer can be identified; however, specificity for detecting GC is negligible.
H. pylori is genetically variability, producing various virulence factors that differ with respect to their degrees of pathogenicity[131]. The GC-related virulence factors of H. pylori may be potential candidates for biomarkers and could aid in serological screening to detect and predict the outcome of H. pylori infection[132]. Several virulent antigens of H. pylori have been suggested for serological screening in GC.
According to reports, the staging of GC is correlated with the Ab titers of CagA. Thus, CagA is the most representative virulence factor. The serum positivity rate of CagA in early GC is higher than that in late GC, indicating that anti-CagA Abs can be used for early GC detection[133]. In a case-control study, serum positivity for CagA and VacA was an individual predictor of noncardia GC risk[134]. In populations with a high risk of GC and prevalence of H. pylori in northeastern Iran, only CagA and VacA Abs are associated with GC risk[135]. CagA-positive H. pylori infection is extremely common in East Asian populations; however, not everyone will develop GC. This may be due to different gene sequences and CagA expression levels. For example, strains with low levels of CagA expression or those lacking the “AATAAGATA” motif typically have a better prognosis[136]. Therefore, in the Asia Pacific region, CagA is not an useful predictive biomarker for GC risk[137], and evaluating other toxic H. pylori proteins may better identify individuals at a high risk of GC[138]. However, certain scholars hold the opposite view. Reports from Japan, South Korea, and China have demonstrated that positive CagA serum is associated with an increased risk of GC[139-141]. The different results may be due to different detection methods or changes in the antigen used to detect anti-CagA Abs, which can serve as a biomarker for GC, even in East Asian countries[142].
Serum VacA Abs are significantly correlated with an increased risk of GC development[143], indicating that serum VacA Abs may be a potential biomarker[144]. Research has shown that H. pylori seropositive individuals with high Ab reactivity to CagA and VacA are at increased risk of developing noncardia GC[134,145]. In the Moroccan population, VacA genotyping revealed that the VacA i1m1 and VacA i1m2 genotypes are strongly associated with GC risk, whereas VacAs1/cagA+ is negatively correlated with GC[146].
In addition to the classic CagA and VacA Ab tests, the relationship between specific Abs against other H. pylori-related virulence factors and GC risk has also been extensively investigated. The serum positivity rate of H. pylori N ethylmaleimide sensitive factor attachment protein alpha (NapA)-specific Abs in patients with GC is higher than that in patients with chronic gastritis and healthy controls[147]. NapA indicates a significant downregulation effect on the occurrence and development of GC, while low Ab titers of H. pylori NapA can predict susceptibility to GC. However, a low AUC indicates that NapA cannot serve as an independent screening and diagnostic criterion for GC[148]. The high expression levels of adhesins in H. pylori associated with GC suggest a higher host response. The combination of OipA, BabA, and SabA can effectively distinguish patients with GC from patients with duodenal ulcer and healthy individuals, providing good discriminatory ability for GC screening[149]. In Shandong, China, individuals who are seropositive for both are seven times more likely to develop precancerous lesions than those who are seronegative for OMP and HP0305[150].
The H. pylori multiplex serological assay is a noninvasive test with high throughput, based on the combined quantitative detection of Ab reactivity against different antigens via multiplex immunoblotting and ELISA[151]. The use of H. pylori biomarkers as well as other predictors allows for risk stratification, leading to the development of targeted prevention strategies. Identifying new virulence factors for H. pylori may help pinpoint high-risk populations among a large group of infected individuals, guiding clinical intervention or surveillance efforts[152].
Multiplex serological assays have been developed to detect the levels of Abs against 15 H. pylori immunogenic proteins[138], including UreA, catalase, NapA, CagA, HP0231, VacA, and HpaA, based on known immunogenicity in two-dimensional immunoblots and surface exposures[153]. These are specifically recognized in H. pylori Cagδ and CagM, and GroEl, Cad, HyuA, OMP, HcpC, HP0305, and CagM, which are serologically associated with GC and gastric ulcers[154]. Germany was the first country to use this technique to compare the association of seropositivity for these 15 proteins with GC risk, finding significant associations between Abs against eight individual H. pylori proteins with an OR of 2.34 and identifying the chaperone protein GroEL as a new independent risk marker for distal GC[155]. The method has since been applied in several regions, including the United States, Latin America, East Asia, Europe, and the Middle East, to assess the correlation of H. pylori serostatus and/or its antigenic profile with GC. CagA, HP0305, HyuA, OMP, and VacA were associated with noncardia GC risk, with ORs of 3.22, 1.72, 1.42, 1.83, and 2.05, respectively[118]. The test efficacy of the combined serum levels of EFNA1 and MMP13 for the diagnosis of GC is significantly superior than that of the single biomarkers used[156]. In China, five protein biomarkers, cadherin related family member 2, intercellular adhesion molecule 4, protein tyrosine phosphatase receptor type M, cell division cycle 27, and fms related receptor tyrosine kinase 1, were found to show good specificity in differentiating individuals with cardia GC and precancerous lesions from healthy controls via multiplexed serologic testing[157]. These studies corroborated that some antigen combinations can serve as candidate markers of infection and disease[151], emphasizing the diagnostic potential of serological markers and proteomic analyses in the detection of GC.
The application of various mathematical models has improved the efficiency for candidate biomarker screening. Seropositivity for six H. pylori antigens (CagA, VacA, GroEL, UreA, HcpC, and gGT) was determined via recomLine analysis on a subset of samples from Linqu County; of these, CagA and GroEL are thought to predict the progression of gastric lesions[158]. A systematic entropy-based approach to identify cancer-associated residues in CagA-intervening regions could further help understand the relationship between CagA sequences and their virulence to GC, providing a useful tool for predicting the correlation between novel CagA strains and diseases[159]. By combining weighted gene co-expression network analysis with machine learning, researchers identified 206 proteins associated with CGC progression and determined that three candidate proteins (glutathione S-transferase P1, cysteine and glycine rich protein 1, and lymphocyte antigen 6 family member G6F) could collectively differentiate individuals with cardia GC, GC, and those in the precancer stage from healthy individuals, with accuracies above 0.9[160]. The approach of combining artificial intelligence with mathematical modeling could pave the way for the development of reliable noninvasive diagnostic tests that may be useful in the context of large-scale population screening.
Compared to single-indicator tests, multiple serological tests have improved the efficiency of risk screening, considerably expanded the candidate biomarkers for GC screening, and helped in identifying individuals at the highest risk of GC development, thereby facilitating targeted H. pylori eradication programs[10]. These studies have some limitations such as limited sample size, geographic variation, and possible bias; thus, more large-scale, multicenter studies are needed to confirm the validity and reliability of these candidate biomarkers. Additionally, the lack of uniform assays and standardized processes in the studies may lead to limited comparability between results, hindering the use of these biomarkers in clinical practice. The combined biomarker approach requires standardization of the testing process to reduce testing costs, ensure reproducibility, and enhance early diagnosis, which would further improve the outcomes in high-risk populations.
GC represents a major global disease burden, with H. pylori infection being a major risk factor. Prevention strategies that focus on controlling H. pylori infection, improving environmental conditions, and identifying individuals at risk to cancer due to H. pylori infection are the most promising approaches for reducing cancer risk. Serologic index testing, a simple and cost-effective noninvasive method, facilitates early identification of high-risk individuals, enhancing the accuracy and efficiency of screening and aiding in early diagnosis. In the future, advancements in technologies such as high-throughput sequencing and artificial intelligence will lead to the discovery of more sensitive serum biomarkers for detecting H. pylori-related GC. The integration of serum indicator tests with other screening tools, optimization of detection methods, and improvements in their clinical application will be key areas of research. In this review, the relationship between H. pylori and GC is discussed in detail, focusing on the unique value of H. pylori-related serum indicators and the applications of multiple serum indicators in GC screening. H. pylori-related serum indicators are clinically significant as they offer a noninvasive, cost-effective approach for the early detection and screening of GC, enhancing diagnostic accuracy and enabling timely intervention in high-risk populations. In recent years, novel molecular biomarkers, such as noncoding genes, have shown promise in improving the accuracy of early screening. These indicators can reveal potential cancer risks at an early stage of infection, leading to earlier intervention and diagnosis. The new strategy of combined biomarker testing aims to improve the accuracy of the identification of high-risk populations, thereby supporting personalized treatment approaches and providing new ideas for developing more accurate and reliable screening tools. The current studies have certain limitations, including insufficient sample size, complex interpretation of indicators, and large individual differences. To enhance the effectiveness and practicality of GC screening, future research should focus on multicenter clinical studies with larger sample sizes to validate and ensure the reliability of new serum biomarkers, thereby facilitating their integration into clinical practice. Additionally, a comprehensive assessment combining multiple serum indicators with other screening tools, such as endoscopy and imaging, is essential to improve screening accuracy and efficiency. These efforts will enhance the early diagnosis of GC, reduce the disease burden associated with H. pylori infection and GC, and ultimately improve the health outcomes of high-risk populations.
The author would like to thank the associate editor and the reviewers for their useful feedback that improved this paper.
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 68593] [Article Influence: 13718.6] [Reference Citation Analysis (201)] |
| 2. | Jin G, Lv J, Yang M, Wang M, Zhu M, Wang T, Yan C, Yu C, Ding Y, Li G, Ren C, Ni J, Zhang R, Guo Y, Bian Z, Zheng Y, Zhang N, Jiang Y, Chen J, Wang Y, Xu D, Zheng H, Yang L, Chen Y, Walters R, Millwood IY, Dai J, Ma H, Chen K, Chen Z, Hu Z, Wei Q, Shen H, Li L. Genetic risk, incident gastric cancer, and healthy lifestyle: a meta-analysis of genome-wide association studies and prospective cohort study. Lancet Oncol. 2020;21:1378-1386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 185] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 3. | Kashyap S, Pal S, Chandan G, Saini V, Chakrabarti S, Saini NK, Mittal A, Thakur VK, Saini AK, Saini RV. Understanding the cross-talk between human microbiota and gastrointestinal cancer for developing potential diagnostic and prognostic biomarkers. Semin Cancer Biol. 2022;86:643-651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 4. | Romańczyk M, Osmola M, Link A, Druet A, Hémont C, Martin J, Chapelle N, Matysiak-Budnik T. Non-Invasive Markers for the Detection of Gastric Precancerous Conditions. Cancers (Basel). 2024;16:2254. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 5. | Yang C, Rodriguez Y Baena A, Manso BA, Hu S, Lopez-Magaña R, Ohanyan M, Ottemann KM. Helicobacter pylori luxS mutants cause hyperinflammatory responses during chronic infection. Microbiol Spectr. 2024;e0107324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 6. | Hooi JKY, Lai WY, Ng WK, Suen MMY, Underwood FE, Tanyingoh D, Malfertheiner P, Graham DY, Wong VWS, Wu JCY, Chan FKL, Sung JJY, Kaplan GG, Ng SC. Global Prevalence of Helicobacter pylori Infection: Systematic Review and Meta-Analysis. Gastroenterology. 2017;153:420-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1361] [Cited by in RCA: 2211] [Article Influence: 245.7] [Reference Citation Analysis (3)] |
| 7. | Aniekwe O, Jolaiya T, Ajayi A, Adeleye IA, Gerhard M, Smith SI. Co-infection of Helicobacter pylori and intestinal parasites in children of selected low-income communities in Lagos State, Nigeria. Parasitol Int. 2024;101:102896. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 8. | Dincă AL, Meliț LE, Mărginean CO. Old and New Aspects of H. pylori-Associated Inflammation and Gastric Cancer. Children (Basel). 2022;9:1083. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 9. | Maleki Kakelar H, Barzegari A, Dehghani J, Hanifian S, Saeedi N, Barar J, Omidi Y. Pathogenicity of Helicobacter pylori in cancer development and impacts of vaccination. Gastric Cancer. 2019;22:23-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 50] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 10. | Lee YC, Chiang TH, Chou CK, Tu YK, Liao WC, Wu MS, Graham DY. Association Between Helicobacter pylori Eradication and Gastric Cancer Incidence: A Systematic Review and Meta-analysis. Gastroenterology. 2016;150:1113-1124.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 737] [Cited by in RCA: 722] [Article Influence: 72.2] [Reference Citation Analysis (0)] |
| 11. | Usui Y, Taniyama Y, Endo M, Koyanagi YN, Kasugai Y, Oze I, Ito H, Imoto I, Tanaka T, Tajika M, Niwa Y, Iwasaki Y, Aoi T, Hakozaki N, Takata S, Suzuki K, Terao C, Hatakeyama M, Hirata M, Sugano K, Yoshida T, Kamatani Y, Nakagawa H, Matsuda K, Murakami Y, Spurdle AB, Matsuo K, Momozawa Y. Helicobacter pylori, Homologous-Recombination Genes, and Gastric Cancer. N Engl J Med. 2023;388:1181-1190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 161] [Article Influence: 53.7] [Reference Citation Analysis (1)] |
| 12. | Choi IJ, Kim CG, Lee JY, Kim YI, Kook MC, Park B, Joo J. Family History of Gastric Cancer and Helicobacter pylori Treatment. N Engl J Med. 2020;382:427-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 287] [Article Influence: 47.8] [Reference Citation Analysis (0)] |
| 13. | Kowada A. A Population-Based Helicobacter pylori Eradication Strategy Is More Cost-Effective than Endoscopic Screening. Dig Dis Sci. 2023;68:1735-1746. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 23] [Reference Citation Analysis (0)] |
| 14. | Slomski A. H pylori Treatment May Reduce Long-term Gastric Cancer Risk. JAMA. 2019;322:1948. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 15. | Liou JM, Malfertheiner P, Lee YC, Sheu BS, Sugano K, Cheng HC, Yeoh KG, Hsu PI, Goh KL, Mahachai V, Gotoda T, Chang WL, Chen MJ, Chiang TH, Chen CC, Wu CY, Leow AH, Wu JY, Wu DC, Hong TC, Lu H, Yamaoka Y, Megraud F, Chan FKL, Sung JJ, Lin JT, Graham DY, Wu MS, El-Omar EM; Asian Pacific Alliance on Helicobacter and Microbiota (APAHAM). Screening and eradication of Helicobacter pylori for gastric cancer prevention: the Taipei global consensus. Gut. 2020;69:2093-2112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 335] [Cited by in RCA: 346] [Article Influence: 57.7] [Reference Citation Analysis (0)] |
| 16. | Kowada A. Cost-Effectiveness of Population-Based Helicobacter pylori Screening With Eradication for Optimal Age of Implementation. Helicobacter. 2024;29:e13120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 17. | Thrift AP, Wenker TN, El-Serag HB. Global burden of gastric cancer: epidemiological trends, risk factors, screening and prevention. Nat Rev Clin Oncol. 2023;20:338-349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 484] [Reference Citation Analysis (1)] |
| 18. | Sarmasti M, Khoshbaten M, Khalili F, Yousefi M. Cost-Effectiveness of Screening Helicobacter pylori for Gastric Cancer Prevention: a Systematic Review. J Gastrointest Cancer. 2022;53:1093-1103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (1)] |
| 19. | Bansil R, Constantino MA, Su-Arcaro C, Liao W, Shen Z, Fox JG. Motility of Different Gastric Helicobacter spp. Microorganisms. 2023;11:634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 20. | Cunha ES, Chen X, Sanz-Gaitero M, Mills DJ, Luecke H. Cryo-EM structure of Helicobacter pylori urease with an inhibitor in the active site at 2.0 Å resolution. Nat Commun. 2021;12:230. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 49] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 21. | Ruch TR, Engel JN. Targeting the Mucosal Barrier: How Pathogens Modulate the Cellular Polarity Network. Cold Spring Harb Perspect Biol. 2017;9:a027953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 22. | Fagoonee S, Pellicano R. Helicobacter pylori: molecular basis for colonization and survival in gastric environment and resistance to antibiotics. A short review. Infect Dis (Lond). 2019;51:399-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 72] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 23. | Salvatori S, Marafini I, Laudisi F, Monteleone G, Stolfi C. Helicobacter pylori and Gastric Cancer: Pathogenetic Mechanisms. Int J Mol Sci. 2023;24:2895. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 164] [Reference Citation Analysis (1)] |
| 24. | Murata-Kamiya N, Hatakeyama M. Helicobacter pylori-induced DNA double-stranded break in the development of gastric cancer. Cancer Sci. 2022;113:1909-1918. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 50] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 25. | Kuo YC, Ko HJ, Yu LY, Shih SC, Wang HY, Lin YC, Hu KC. Kill Two Birds with One Stone? The Effect of Helicobacter pylori Eradication in Decreased Prevalence of Gastric Cancer and Colorectal Cancer. Cancers (Basel). 2024;16:3881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 26. | Shimizu T, Chiba T, Marusawa H. Helicobacter pylori-Mediated Genetic Instability and Gastric Carcinogenesis. Curr Top Microbiol Immunol. 2017;400:305-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 27. | Nabavi-Rad A, Yadegar A, Sadeghi A, Aghdaei HA, Zali MR, Klionsky DJ, Yamaoka Y. The interaction between autophagy, Helicobacter pylori, and gut microbiota in gastric carcinogenesis. Trends Microbiol. 2023;31:1024-1043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 30] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 28. | Lim NR, Chung WC. Helicobacter pylori-associated Chronic Atrophic Gastritis and Progression of Gastric Carcinogenesis. Korean J Gastroenterol. 2023;82:171-179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 31] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 29. | Khazaaleh S, Alomari M, Rashid MU, Castaneda D, Castro FJ. Gastric intestinal metaplasia and gastric cancer prevention: Watchful waiting. Cleve Clin J Med. 2024;91:33-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 18] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 30. | Lopes C, Almeida TC, Pimentel-Nunes P, Dinis-Ribeiro M, Pereira C. Linking dysbiosis to precancerous stomach through inflammation: Deeper than and beyond imaging. Front Immunol. 2023;14:1134785. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 31. | Butcher LD, den Hartog G, Ernst PB, Crowe SE. Oxidative Stress Resulting From Helicobacter pylori Infection Contributes to Gastric Carcinogenesis. Cell Mol Gastroenterol Hepatol. 2017;3:316-322. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 180] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 32. | Smirnova OV, Sinyakov AA, Kasparov EV. The Role of Immunoglobulin G (IgG), IgA and IgE-Antibodies against Helicobacter pylori in the Development of Oxidative Stress in Patients with Chronic Gastritis. Biomedicines. 2022;10:2053. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 33. | Li Y, Choi H, Leung K, Jiang F, Graham DY, Leung WK. Global prevalence of Helicobacter pylori infection between 1980 and 2022: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2023;8:553-564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 290] [Reference Citation Analysis (0)] |
| 34. | Zahir SO, El Khadir M, Boukhris SA, Benajah DA, Ibrahimi SA, Chbani L, El Abkari M, Bennani B. Helicobacter pylori vacA Allelic Combination, dupA, cagE and cagA Genotypes and Their Associations with Gastric Diseases in the Moroccan Population. Jpn J Infect Dis. 2024;77:325-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 35. | Li F, Si YT, Tang JW, Umar Z, Xiong XS, Wang JT, Yuan Q, Tay ACY, Chua EG, Zhang L, Marshall BJ, Yang WX, Gu B, Wang L. Rapid profiling of carcinogenic types of Helicobacter pylori infection via deep learning analysis of label-free SERS spectra of human serum. Comput Struct Biotechnol J. 2024;23:3379-3390. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 36. | Tran SC, Bryant KN, Cover TL. The Helicobacter pylori cag pathogenicity island as a determinant of gastric cancer risk. Gut Microbes. 2024;16:2314201. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 42] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 37. | Gou L, Yang X, Yun J, Ma Z, Zheng X, Du H, Zhang D. Roles of the components of the cag-pathogenicity island encoded type IV secretion system in Helicobacter pylori. Future Microbiol. 2024;19:1253-1267. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 38. | Zhu X, Ma C, Sa R, Wang Y, Zhu C, Zhao Y, Luo J, Liu X. CagA 3' region polymorphism of Helicobacter pylori and its association with chronic gastritis in the Chinese population. J Med Microbiol. 2024;73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 39. | Linz B, Sticht H, Tegtmeyer N, Backert S. Cancer-associated SNPs in bacteria: lessons from Helicobacter pylori. Trends Microbiol. 2024;32:847-857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 11] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 40. | Wang F, Meng W, Wang B, Qiao L. Helicobacter pylori-induced gastric inflammation and gastric cancer. Cancer Lett. 2014;345:196-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 607] [Cited by in RCA: 606] [Article Influence: 50.5] [Reference Citation Analysis (1)] |
| 41. | He X, Huang T, Wang Q, Bao L, Wang Z, Song H, Li Y, Zhou J, Zhao Y, Xie Y. A prominent role of LncRNA H19 in H. pylori CagA induced DNA damage response and cell malignancy. Sci Rep. 2024;14:14185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 42. | Palrasu M, Zaika E, El-Rifai W, Garcia-Buitrago M, Piazuelo MB, Wilson KT, Peek RM Jr, Zaika AI. Bacterial CagA protein compromises tumor suppressor mechanisms in gastric epithelial cells. J Clin Invest. 2020;130:2422-2434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 43. | Baj J, Forma A, Sitarz M, Portincasa P, Garruti G, Krasowska D, Maciejewski R. Helicobacter pylori Virulence Factors-Mechanisms of Bacterial Pathogenicity in the Gastric Microenvironment. Cells. 2020;10:27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 235] [Cited by in RCA: 246] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 44. | Qiao Z, Wang E, Bao B, Tan X, Yuan L, Wang D. Association of Helicobacter pylori CagA seropositivity with gastric precancerous lesions: a systematic review and meta-analysis. Eur J Gastroenterol Hepatol. 2024;36:687-694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Reference Citation Analysis (0)] |
| 45. | Naing C, Aung HH, Aye SN, Poovorawan Y, Whittaker MA. CagA toxin and risk of Helicobacter pylori-infected gastric phenotype: A meta-analysis of observational studies. PLoS One. 2024;19:e0307172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 46. | Foegeding NJ, Caston RR, McClain MS, Ohi MD, Cover TL. An Overview of Helicobacter pylori VacA Toxin Biology. Toxins (Basel). 2016;8:173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 169] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 47. | Sharafutdinov I, Knorr J, Soltan Esmaeili D, Backert S, Tegtmeyer N. Cortactin Promotes Effective AGS Cell Scattering by Helicobacter pylori CagA, but Not Cellular Vacuolization and Apoptosis Induced by the Vacuolating Cytotoxin VacA. Pathogens. 2021;11:3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 48. | Chauhan N, Tay ACY, Marshall BJ, Jain U. Helicobacter pylori VacA, a distinct toxin exerts diverse functionalities in numerous cells: An overview. Helicobacter. 2019;24:e12544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 102] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 49. | Shirani M, Shariati S, Bazdar M, Sojoudi Ghamnak F, Moradi M, Shams Khozani R, Taki E, Arabsorkhi Z, Heidary M, Eskandari DB. The immunopathogenesis of Helicobacter pylori-induced gastric cancer: a narrative review. Front Microbiol. 2024;15:1395403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 50. | Utsch C, Haas R. VacA's Induction of VacA-Containing Vacuoles (VCVs) and Their Immunomodulatory Activities on Human T Cells. Toxins (Basel). 2016;8:190. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 51. | Sedarat Z, Taylor-Robinson AW. Helicobacter pylori Outer Membrane Proteins and Virulence Factors: Potential Targets for Novel Therapies and Vaccines. Pathogens. 2024;13:392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 22] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 52. | Matsuo Y, Kido Y, Yamaoka Y. Helicobacter pylori Outer Membrane Protein-Related Pathogenesis. Toxins (Basel). 2017;9:101. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 73] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 53. | Kim JH, Han KD, Lee JK, Kim HS, Cha JM, Park S, Kim JS, Kim WH; Big Data Research Group (BDRG) of the Korean Society of Gastroenterology (KSG). Association between the National Cancer Screening Programme (NSCP) for gastric cancer and oesophageal cancer mortality. Br J Cancer. 2020;123:480-486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 54. | Mabe K, Inoue K, Kamada T, Kato K, Kato M, Haruma K. Endoscopic screening for gastric cancer in Japan: Current status and future perspectives. Dig Endosc. 2022;34:412-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 84] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 55. | Lomba-Viana R, Dinis-Ribeiro M, Fonseca F, Vieira AS, Bento MJ, Lomba-Viana H. Serum pepsinogen test for early detection of gastric cancer in a European country. Eur J Gastroenterol Hepatol. 2012;24:37-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 56] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 56. | Budzyń K, Pelczar M, Romańczyk M, Barański K, Hartleb M. Willingness to undergo screening gastroscopy in a population with low-to-moderate prevalence rate of esophageal and gastric cancer. Pol Arch Intern Med. 2024;134:16671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 57. | Romańczyk M, Ostrowski B, Budzyń K, Koziej M, Wdowiak M, Romańczyk T, Błaszczyńska M, Kajor M, Januszewski K, Zajęcki W, Hartleb M. The role of endoscopic and demographic features in the diagnosis of gastric precancerous conditions. Pol Arch Intern Med. 2022;132:16200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 58. | Kim SY, Park JM. Quality indicators in esophagogastroduodenoscopy. Clin Endosc. 2022;55:319-331. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 59. | Xia JY, Aadam AA. Advances in screening and detection of gastric cancer. J Surg Oncol. 2022;125:1104-1109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 106] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 60. | Matsuoka T, Yashiro M. Biomarkers of gastric cancer: Current topics and future perspective. World J Gastroenterol. 2018;24:2818-2832. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 230] [Cited by in RCA: 343] [Article Influence: 42.9] [Reference Citation Analysis (7)] |
| 61. | Jing R, Cui M, Ju S, Pan S. The Changes and Clinical Significance of Preoperative and Postoperative Serum CEA and CA19-9 in Gastric Cancer. Clin Lab. 2020;66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 62. | Sekiguchi M, Matsuda T. Limited usefulness of serum carcinoembryonic antigen and carbohydrate antigen 19-9 levels for gastrointestinal and whole-body cancer screening. Sci Rep. 2020;10:18202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (11)] |
| 63. | Chen XZ, Zhang WK, Yang K, Wang LL, Liu J, Wang L, Hu JK, Zhang B, Chen ZX, Chen JP, Zhou ZG, Mo XM. Correlation between serum CA724 and gastric cancer: multiple analyses based on Chinese population. Mol Biol Rep. 2012;39:9031-9039. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 86] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 64. | Zhang Y, Zhang M, Bai X, Li C, Zhang L. Increased serum CA724 levels in patients suffering gout vs cancers. Prog Mol Biol Transl Sci. 2019;162:177-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 65. | Tsai CY, Liu KH, Chiu CT, Hsueh SW, Hung CY, Hsu JT, Tsang NM, Hung YS, Chou WC. Alpha-fetoprotein for Gastric Cancer Staging: An Essential or Redundant Tumor Marker? Anticancer Res. 2021;41:2711-2718. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 66. | Wang Z, Mo TM, Tian L, Chen JQ. Gastrin-17 Combined with CEA, CA12-5 and CA19-9 Improves the Sensitivity for the Diagnosis of Gastric Cancer. Int J Gen Med. 2021;14:8087-8095. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 67. | Chen C, Chen Q, Zhao Q, Liu M, Guo J. Value of Combined Detection of Serum CEA, CA72-4, CA19-9, CA15-3 and CA12-5 in the Diagnosis of Gastric Cancer. Ann Clin Lab Sci. 2017;47:260-263. [PubMed] |
| 68. | Huang YK, Yu JC, Kang WM, Ma ZQ, Ye X, Tian SB, Yan C. Significance of Serum Pepsinogens as a Biomarker for Gastric Cancer and Atrophic Gastritis Screening: A Systematic Review and Meta-Analysis. PLoS One. 2015;10:e0142080. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 135] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 69. | Qin Y, Geng JX, Huang B. Clinical value of serum pepsinogen in the diagnosis and treatment of gastric diseases. World J Gastrointest Oncol. 2023;15:1174-1181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (2)] |
| 70. | Kishikawa H, Kimura K, Takarabe S, Kaida S, Nishida J. Helicobacter pylori Antibody Titer and Gastric Cancer Screening. Dis Markers. 2015;2015:156719. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 47] [Article Influence: 4.3] [Reference Citation Analysis (1)] |
| 71. | Lin JT. Screening of gastric cancer: who, when, and how. Clin Gastroenterol Hepatol. 2014;12:135-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 52] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 72. | Liu L, Lang J, Jin Y, Chen Y, Chang W, Yao Y, Yu J. The Value of Pepsinogen in GC Screening: A Systematic Review and Meta-Analysis. Gastroenterol Res Pract. 2019;2019:7087232. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 73. | Pasechnikov V, Chukov S, Fedorov E, Kikuste I, Leja M. Gastric cancer: prevention, screening and early diagnosis. World J Gastroenterol. 2014;20:13842-13862. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 247] [Cited by in RCA: 309] [Article Influence: 25.8] [Reference Citation Analysis (2)] |
| 74. | Trivanovic D, Plestina S, Honovic L, Dobrila-Dintinjana R, Vlasic Tanaskovic J, Vrbanec D. Gastric cancer detection using the serum pepsinogen test method. Tumori. 2022;108:386-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 75. | Meliț LE, Mărginean CO, Borka Balas R. The Most Recent Insights into the Roots of Gastric Cancer. Life (Basel). 2024;14:95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Reference Citation Analysis (0)] |
| 76. | Wang F. Diagnostic value of combined detection of three gastric functions and Helicobacter pylori typing in chronic gastritis and gastric cancer. SLAS Technol. 2024;29:100141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 7] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 77. | Guo X, Schreurs MWJ, Marijnissen FE, Mommersteeg MC, Nieuwenburg SAV, Doukas M, Erler NS, Capelle LG, Bruno MJ, Peppelenbosch MP, Spaander MCW; Proregal study group, Fuhler GM. Increased Prevalence of Autoimmune Gastritis in Patients with a Gastric Precancerous Lesion. J Clin Med. 2023;12:6152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 78. | Leja M, Kupcinskas L, Funka K, Sudraba A, Jonaitis L, Ivanauskas A, Janciauskas D, Kuidelis G, Chiu HM, Lin JT. Value of gastrin-17 in detecting antral atrophy. Adv Med Sci. 2011;56:145-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 79. | Wang X, Ling L, Li S, Qin G, Cui W, Li X, Ni H. The Diagnostic Value of Gastrin-17 Detection in Atrophic Gastritis: A Meta-Analysis. Medicine (Baltimore). 2016;95:e3599. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 80. | Shigeyasu K, Toden S, Zumwalt TJ, Okugawa Y, Goel A. Emerging Role of MicroRNAs as Liquid Biopsy Biomarkers in Gastrointestinal Cancers. Clin Cancer Res. 2017;23:2391-2399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 112] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 81. | Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, Douville C, Javed AA, Wong F, Mattox A, Hruban RH, Wolfgang CL, Goggins MG, Dal Molin M, Wang TL, Roden R, Klein AP, Ptak J, Dobbyn L, Schaefer J, Silliman N, Popoli M, Vogelstein JT, Browne JD, Schoen RE, Brand RE, Tie J, Gibbs P, Wong HL, Mansfield AS, Jen J, Hanash SM, Falconi M, Allen PJ, Zhou S, Bettegowda C, Diaz LA Jr, Tomasetti C, Kinzler KW, Vogelstein B, Lennon AM, Papadopoulos N. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926-930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1752] [Cited by in RCA: 2031] [Article Influence: 253.9] [Reference Citation Analysis (0)] |
| 82. | Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ, Cheang M, Osin P, Nerurkar A, Kozarewa I, Garrido JA, Dowsett M, Reis-Filho JS, Smith IE, Turner NC. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. 2015;7:302ra133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 796] [Cited by in RCA: 885] [Article Influence: 80.5] [Reference Citation Analysis (0)] |
| 83. | Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, Silliman N, Tacey M, Wong HL, Christie M, Kosmider S, Skinner I, Wong R, Steel M, Tran B, Desai J, Jones I, Haydon A, Hayes T, Price TJ, Strausberg RL, Diaz LA Jr, Papadopoulos N, Kinzler KW, Vogelstein B, Gibbs P. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8:346ra92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 729] [Cited by in RCA: 1097] [Article Influence: 121.9] [Reference Citation Analysis (0)] |
| 84. | Park CK, Cho HJ, Choi YD, Oh IJ, Kim YC. A Phase II Trial of Osimertinib in the Second-Line Treatment of Non-small Cell Lung Cancer with the EGFR T790M Mutation, Detected from Circulating Tumor DNA: LiquidLung-O-Cohort 2. Cancer Res Treat. 2019;51:777-787. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 85. | Kim ST, Cristescu R, Bass AJ, Kim KM, Odegaard JI, Kim K, Liu XQ, Sher X, Jung H, Lee M, Lee S, Park SH, Park JO, Park YS, Lim HY, Lee H, Choi M, Talasaz A, Kang PS, Cheng J, Loboda A, Lee J, Kang WK. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med. 2018;24:1449-1458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 643] [Cited by in RCA: 1235] [Article Influence: 154.4] [Reference Citation Analysis (0)] |
| 86. | Gao Y, Zhang K, Xi H, Cai A, Wu X, Cui J, Li J, Qiao Z, Wei B, Chen L. Diagnostic and prognostic value of circulating tumor DNA in gastric cancer: a meta-analysis. Oncotarget. 2017;8:6330-6340. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 87. | Kato S, Okamura R, Baumgartner JM, Patel H, Leichman L, Kelly K, Sicklick JK, Fanta PT, Lippman SM, Kurzrock R. Analysis of Circulating Tumor DNA and Clinical Correlates in Patients with Esophageal, Gastroesophageal Junction, and Gastric Adenocarcinoma. Clin Cancer Res. 2018;24:6248-6256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 102] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 88. | Abbosh C, Swanton C, Birkbak NJ. Clonal haematopoiesis: a source of biological noise in cell-free DNA analyses. Ann Oncol. 2019;30:358-359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 89. | Ashrafizadeh M, Rafiei H, Mohammadinejad R, Farkhondeh T, Samarghandian S. Wnt-regulating microRNAs role in gastric cancer malignancy. Life Sci. 2020;250:117547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 90. | Shaker F, Razi S, Rezaei N. Circulating miRNA and circulating tumor DNA application as liquid biopsy markers in gastric cancer. Clin Biochem. 2024;129:110767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 91. | Yu L, Wu D, Gao H, Balic JJ, Tsykin A, Han TS, Liu YD, Kennedy CL, Li JK, Mao JQ, Tan P, Oshima M, Goodall GJ, Jenkins BJ. Clinical Utility of a STAT3-Regulated miRNA-200 Family Signature with Prognostic Potential in Early Gastric Cancer. Clin Cancer Res. 2018;24:1459-1472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 92. | Shin VY, Ng EK, Chan VW, Kwong A, Chu KM. A three-miRNA signature as promising non-invasive diagnostic marker for gastric cancer. Mol Cancer. 2015;14:202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 90] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 93. | So JBY, Kapoor R, Zhu F, Koh C, Zhou L, Zou R, Tang YC, Goo PCK, Rha SY, Chung HC, Yoong J, Yap CT, Rao J, Chia CK, Tsao S, Shabbir A, Lee J, Lam KP, Hartman M, Yong WP, Too HP, Yeoh KG. Development and validation of a serum microRNA biomarker panel for detecting gastric cancer in a high-risk population. Gut. 2021;70:829-837. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 149] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 94. | Tsai MM, Wang CS, Tsai CY, Huang CG, Lee KF, Huang HW, Lin YH, Chi HC, Kuo LM, Lu PH, Lin KH. Circulating microRNA-196a/b are novel biomarkers associated with metastatic gastric cancer. Eur J Cancer. 2016;64:137-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 52] [Article Influence: 5.2] [Reference Citation Analysis (1)] |
| 95. | Wei L, Sun J, Zhang N, Zheng Y, Wang X, Lv L, Liu J, Xu Y, Shen Y, Yang M. Noncoding RNAs in gastric cancer: implications for drug resistance. Mol Cancer. 2020;19:62. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 279] [Cited by in RCA: 366] [Article Influence: 61.0] [Reference Citation Analysis (0)] |
| 96. | Zhen-Hua W, Yi-Wei G, Li-Qin Z, Jie-Yun Z, Zhe G, Wei-Jian G. Silencing of LncRNA C1RL-AS1 Suppresses the Malignant Phenotype in Gastric Cancer Cells via the AKT/β-Catenin/c-Myc Pathway. Front Oncol. 2020;10:1508. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 97. | Fang D, Ou X, Sun K, Zhou X, Li Y, Shi P, Zhao Z, He Y, Peng J, Xu J. m6A modification-mediated lncRNA TP53TG1 inhibits gastric cancer progression by regulating CIP2A stability. Cancer Sci. 2022;113:4135-4150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 89] [Reference Citation Analysis (0)] |
| 98. | Yang H, Hu Y, Weng M, Liu X, Wan P, Hu Y, Ma M, Zhang Y, Xia H, Lv K. Hypoxia inducible lncRNA-CBSLR modulates ferroptosis through m6A-YTHDF2-dependent modulation of CBS in gastric cancer. J Adv Res. 2022;37:91-106. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 169] [Cited by in RCA: 204] [Article Influence: 51.0] [Reference Citation Analysis (0)] |
| 99. | Zheng P, Zhang H, Gao H, Sun J, Li J, Zhang X, Gao L, Ma P, Li S. Plasma Exosomal Long Noncoding RNA lnc-SLC2A12-10:1 as a Novel Diagnostic Biomarker for Gastric Cancer. Onco Targets Ther. 2020;13:4009-4018. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 100. | Li R, Jiang J, Shi H, Qian H, Zhang X, Xu W. CircRNA: a rising star in gastric cancer. Cell Mol Life Sci. 2020;77:1661-1680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 249] [Cited by in RCA: 296] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 101. | Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH, Pandolfi PP. Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell. 2016;165:289-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 344] [Cited by in RCA: 484] [Article Influence: 48.4] [Reference Citation Analysis (0)] |
| 102. | Hossain MT, Li S, Reza MS, Feng S, Zhang X, Jin Z, Wei Y, Peng Y. Corrigendum: Identification of circRNA biomarker for gastric cancer through integrated analysis. Front Mol Biosci. 2023;10:1249019. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 103. | Sun H, Tang W, Rong D, Jin H, Fu K, Zhang W, Liu Z, Cao H, Cao X. Hsa_circ_0000520, a potential new circular RNA biomarker, is involved in gastric carcinoma. Cancer Biomark. 2018;21:299-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 124] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 104. | Li T, Shao Y, Fu L, Xie Y, Zhu L, Sun W, Yu R, Xiao B, Guo J. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J Mol Med (Berl). 2018;96:85-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 216] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 105. | Kustanovich A, Schwartz R, Peretz T, Grinshpun A. Life and death of circulating cell-free DNA. Cancer Biol Ther. 2019;20:1057-1067. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 339] [Cited by in RCA: 446] [Article Influence: 63.7] [Reference Citation Analysis (0)] |
| 106. | Herrera-Pariente C, Montori S, Llach J, Bofill A, Albeniz E, Moreira L. Biomarkers for Gastric Cancer Screening and Early Diagnosis. Biomedicines. 2021;9:1448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 107. | Merker JD, Oxnard GR, Compton C, Diehn M, Hurley P, Lazar AJ, Lindeman N, Lockwood CM, Rai AJ, Schilsky RL, Tsimberidou AM, Vasalos P, Billman BL, Oliver TK, Bruinooge SS, Hayes DF, Turner NC. Circulating Tumor DNA Analysis in Patients With Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol. 2018;36:1631-1641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 494] [Cited by in RCA: 675] [Article Influence: 84.4] [Reference Citation Analysis (0)] |
| 108. | Hensel KO, Boland V, Postberg J, Zilbauer M, Heuschkel R, Vogel S, Gödde D, Wirth S, Jenke AC. Differential expression of mucosal trefoil factors and mucins in pediatric inflammatory bowel diseases. Sci Rep. 2014;4:7343. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 109. | Xiao P, Ling H, Lan G, Liu J, Hu H, Yang R. Trefoil factors: Gastrointestinal-specific proteins associated with gastric cancer. Clin Chim Acta. 2015;450:127-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 41] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 110. | Huang Z, Zhang X, Lu H, Wu L, Wang D, Zhang Q, Ding H. Serum trefoil factor 3 is a promising non-invasive biomarker for gastric cancer screening: a monocentric cohort study in China. BMC Gastroenterol. 2014;14:74. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 111. | Im S, Yoo C, Jung JH, Choi HJ, Yoo J, Kang CS. Reduced expression of TFF1 and increased expression of TFF3 in gastric cancer: correlation with clinicopathological parameters and prognosis. Int J Med Sci. 2013;10:133-140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 112. | Meng JR, Tang HZ, Zhou KZ, Shen WH, Guo HY. TFF3 and survivin expressions associate with a lower survival rate in gastric cancer. Clin Exp Med. 2013;13:297-303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 113. | Mora Barthelmess R, Stijlemans B, Van Ginderachter JA. Hallmarks of Cancer Affected by the MIF Cytokine Family. Cancers (Basel). 2023;15:395. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 40] [Reference Citation Analysis (0)] |
| 114. | Mondal D, Shinde S, Sinha V, Dixit V, Paul S, Gupta RK, Thakur S, Vishvakarma NK, Shukla D. Prospects of liquid biopsy in the prognosis and clinical management of gastrointestinal cancers. Front Mol Biosci. 2024;11:1385238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 115. | Wang X, Huang J, Chen W, Li G, Li Z, Lei J. The updated role of exosomal proteins in the diagnosis, prognosis, and treatment of cancer. Exp Mol Med. 2022;54:1390-1400. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 90] [Reference Citation Analysis (0)] |
| 116. | Nikanjam M, Kato S, Kurzrock R. Liquid biopsy: current technology and clinical applications. J Hematol Oncol. 2022;15:131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 589] [Reference Citation Analysis (0)] |
| 117. | Shirani M, Pakzad R, Haddadi MH, Akrami S, Asadi A, Kazemian H, Moradi M, Kaviar VH, Zomorodi AR, Khoshnood S, Shafieian M, Tavasolian R, Heidary M, Saki M. The global prevalence of gastric cancer in Helicobacter pylori-infected individuals: a systematic review and meta-analysis. BMC Infect Dis. 2023;23:543. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 53] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 118. | El Hafa F, Wang T, Ndifor VM, Jin G. Association between Helicobacter pylori antibodies determined by multiplex serology and gastric cancer risk: A meta-analysis. Helicobacter. 2022;27:e12881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 119. | Liao EC, Yu CH, Lai JH, Lin CC, Chen CJ, Chang WH, Chien DK. A pilot study of non-invasive diagnostic tools to detect Helicobacter pylori infection and peptic ulcer disease. Sci Rep. 2023;13:22800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 120. | Tonkic A, Tonkic M, Lehours P, Mégraud F. Epidemiology and diagnosis of Helicobacter pylori infection. Helicobacter. 2012;17 Suppl 1:1-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 96] [Article Influence: 6.9] [Reference Citation Analysis (1)] |
| 121. | Song M, Camargo MC, Katki HA, Weinstein SJ, Männistö S, Albanes D, Surcel HM, Rabkin CS. Association of Antiparietal Cell and Anti-Intrinsic Factor Antibodies With Risk of Gastric Cancer. JAMA Oncol. 2022;8:268-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 122. | Deng D, Zhang Y, Zhang R, Yi J, Dong J, Sha L, Yan M. Circulating Proteins and Metabolite Biomarkers in Gastric Cancer: A Systematic Review and Meta-analysis. Arch Med Res. 2023;54:124-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 123. | Tatemichi M, Sasazuki S, Inoue M, Tsugane S; JPHC Study Group. Clinical significance of IgG antibody titer against Helicobacter pylori. Helicobacter. 2009;14:231-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 32] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 124. | Sun L, Tu H, Chen T, Yuan Q, Liu J, Dong N, Yuan Y. Three-dimensional combined biomarkers assay could improve diagnostic accuracy for gastric cancer. Sci Rep. 2017;7:11621. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 125. | Chen XZ, Huang CZ, Hu WX, Liu Y, Yao XQ. Gastric Cancer Screening by Combined Determination of Serum Helicobacter pylori Antibody and Pepsinogen Concentrations: ABC Method for Gastric Cancer Screening. Chin Med J (Engl). 2018;131:1232-1239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 126. | Ikeda F, Shikata K, Hata J, Fukuhara M, Hirakawa Y, Ohara T, Mukai N, Nagata M, Yoshida D, Yonemoto K, Esaki M, Kitazono T, Kiyohara Y, Ninomiya T. Combination of Helicobacter pylori Antibody and Serum Pepsinogen as a Good Predictive Tool of Gastric Cancer Incidence: 20-Year Prospective Data From the Hisayama Study. J Epidemiol. 2016;26:629-636. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 56] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 127. | Kurilovich S, Belkovets A, Reshetnikov O, Openko T, Malyutina S, Ragino Y, Scherbakova L, Leja M, Paloheimo L, Syrjänen K, Voevoda M. Stomach-specific Biomarkers (GastroPanel) Can Predict the Development of Gastric Cancer in a Caucasian Population: A Longitudinal Nested Case-Control Study in Siberia. Anticancer Res. 2016;36:247-253. [PubMed] |
| 128. | Syrjänen K. A Panel of Serum Biomarkers (GastroPanel®) in Non-invasive Diagnosis of Atrophic Gastritis. Systematic Review and Meta-analysis. Anticancer Res. 2016;36:5133-5144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 50] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 129. | Zagari RM, Rabitti S, Greenwood DC, Eusebi LH, Vestito A, Bazzoli F. Systematic review with meta-analysis: diagnostic performance of the combination of pepsinogen, gastrin-17 and anti-Helicobacter pylori antibodies serum assays for the diagnosis of atrophic gastritis. Aliment Pharmacol Ther. 2017;46:657-667. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 138] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 130. | Wu D, Zhang P, Ma J, Xu J, Yang L, Xu W, Que H, Chen M, Xu H. Serum biomarker panels for the diagnosis of gastric cancer. Cancer Med. 2019;8:1576-1583. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 114] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 131. | Xiao Y, Zhang B, Zhang H, Zhang Z, Meng F, Zhao X, Zhang J, Xiao D. Study of the relationships among known virulence genes, coccoid transformation and cytotoxicity of Helicobacter pylori in different clinical diseases. Virulence. 2024;15:2418407. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 132. | Pormohammad A, Ghotaslou R, Leylabadlo HE, Nasiri MJ, Dabiri H, Hashemi A. Risk of gastric cancer in association with Helicobacter pylori different virulence factors: A systematic review and meta-analysis. Microb Pathog. 2018;118:214-219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 58] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 133. | Soares CDRR, Silva LMVD, Almeida BR, Pereira JN, Santos MPD, Barbosa MS, Smith MAC, Payão SLM, Rasmussen LT. Decreased Expression of Microrna-629 In Gastric Cancer Samples Potentiated by The Virulence Marker of H. Pylori, Caga Gene. Arq Gastroenterol. 2024;61:e23139. [PubMed] [DOI] [Full Text] |
| 134. | Fernández de Larrea-Baz N, Pérez-Gómez B, Michel A, Romero B, Lope V, Pawlita M, Fernández-Villa T, Moreno V, Martín V, Willhauck-Fleckenstein M, López-Abente G, Castilla J, Fernández-Tardón G, Dierssen-Sotos T, Santibáñez M, Peiró R, Jiménez-Moleón JJ, Navarro C, Castaño-Vinyals G, Kogevinas M, Pollán M, de Sanjosé S, Del Campo R, Waterboer T, Aragonés N. Helicobacter pylori serological biomarkers of gastric cancer risk in the MCC-Spain case-control Study. Cancer Epidemiol. 2017;50:76-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 135. | Shakeri R, Malekzadeh R, Nasrollahzadeh D, Pawlita M, Murphy G, Islami F, Sotoudeh M, Michel A, Etemadi A, Waterboer T, Poustchi H, Brennan P, Boffetta P, Dawsey SM, Kamangar F, Abnet CC. Multiplex H. pylori Serology and Risk of Gastric Cardia and Noncardia Adenocarcinomas. Cancer Res. 2015;75:4876-4883. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 40] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 136. | Loh JT, Shaffer CL, Piazuelo MB, Bravo LE, McClain MS, Correa P, Cover TL. Analysis of cagA in Helicobacter pylori strains from Colombian populations with contrasting gastric cancer risk reveals a biomarker for disease severity. Cancer Epidemiol Biomarkers Prev. 2011;20:2237-2249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 43] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 137. | Zhao Z, Li Y, Liu S, Fu W. Serum Helicobacter pylori CagA antibody may not be used as a tumor marker for diagnosing gastric cancer in east Asian countries. Tumour Biol. 2014;35:12217-12224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (2)] |
| 138. | Epplein M, Zheng W, Xiang YB, Peek RM Jr, Li H, Correa P, Gao J, Michel A, Pawlita M, Cai Q, Shu XO. Prospective study of Helicobacter pylori biomarkers for gastric cancer risk among Chinese men. Cancer Epidemiol Biomarkers Prev. 2012;21:2185-2192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 57] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 139. | Li H, Zhang B, Hu X, Dong Y, Fan Q, Guo F, Ren X, Zhou H, Tian W, Zhao Y. Serum Helicobacter pylori FliD antibody and the risk of gastric cancer. Oncotarget. 2016;7:22397-22408. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 140. | Nakamura K, Urabe Y, Kagemoto K, Yuge R, Hayashi R, Ono A, Hayes CN, Oka S, Ito M, Nishisaka T, Tanabe K, Arihiro K, Ohdan H, Tanaka S, Chayama K. Genomic Characterization of Non-Invasive Differentiated-Type Gastric Cancer in the Japanese Population. Cancers (Basel). 2020;12:510. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 141. | Choi HI, Choi JP, Seo J, Kim BJ, Rho M, Han JK, Kim JG. Helicobacter pylori-derived extracellular vesicles increased in the gastric juices of gastric adenocarcinoma patients and induced inflammation mainly via specific targeting of gastric epithelial cells. Exp Mol Med. 2017;49:e330. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 94] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 142. | Shiota S, Matsunari O, Watada M, Yamaoka Y. Serum Helicobacter pylori CagA antibody as a biomarker for gastric cancer in east-Asian countries. Future Microbiol. 2010;5:1885-1893. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 48] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 143. | Demiryas S, Caliskan R, Saribas S, Akkus S, Gareayaghi N, Kirmusaoglu S, Kepil N, Dinc H, Dag H, Dagdeviren E, Tokman HB, Kalayci F, Demirci M, Tasci I, Erzin Y, Bal K, Kocazeybek B. The association between cagL and cagA, vacAs-m, babA genes in patients with gastric cancer, duodenal ulcer, and non-ulcer dyspepsia related to Helicobacter pylori. Acta Gastroenterol Belg. 2020;83:385-392. [PubMed] |
| 144. | Li Q, Liu J, Gong Y, Yuan Y. Serum VacA antibody is associated with risks of peptic ulcer and gastric cancer: A meta-analysis. Microb Pathog. 2016;99:220-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 145. | Song H, Michel A, Nyrén O, Ekström AM, Pawlita M, Ye W. A CagA-independent cluster of antigens related to the risk of noncardia gastric cancer: associations between Helicobacter pylori antibodies and gastric adenocarcinoma explored by multiplex serology. Int J Cancer. 2014;134:2942-2950. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 48] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 146. | El Khadir M, Alaoui Boukhris S, Benajah DA, El Rhazi K, Ibrahimi SA, El Abkari M, Harmouch T, Nejjari C, Mahmoud M, Benlemlih M, Bennani B. VacA and CagA Status as Biomarker of Two Opposite End Outcomes of Helicobacter pylori Infection (Gastric Cancer and Duodenal Ulcer) in a Moroccan Population. PLoS One. 2017;12:e0170616. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 147. | Song L, Song M, Rabkin CS, Williams S, Chung Y, Van Duine J, Liao LM, Karthikeyan K, Gao W, Park JG, Tang Y, Lissowska J, Qiu J, LaBaer J, Camargo MC. Helicobacter pylori Immunoproteomic Profiles in Gastric Cancer. J Proteome Res. 2021;20:409-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 148. | Liu J, Liu H, Zhang T, Ren X, Nadolny C, Dong X, Huang L, Yuan K, Tian W, Jia Y. Serum Helicobacter pylori NapA antibody as a potential biomarker for gastric cancer. Sci Rep. 2014;4:4143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 149. | Su YL, Huang HL, Huang BS, Chen PC, Chen CS, Wang HL, Lin PH, Chieh MS, Wu JJ, Yang JC, Chow LP. Combination of OipA, BabA, and SabA as candidate biomarkers for predicting Helicobacter pylori-related gastric cancer. Sci Rep. 2016;6:36442. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 150. | Epplein M, Butt J, Zhang Y, Hendrix LH, Abnet CC, Murphy G, Zheng W, Shu XO, Tsugane S, Qiao YL, Taylor PR, Shimazu T, Yoo KY, Park SK, Kim J, Jee SH, Waterboer T, Pawlita M, You WC, Pan KF. Validation of a Blood Biomarker for Identification of Individuals at High Risk for Gastric Cancer. Cancer Epidemiol Biomarkers Prev. 2018;27:1472-1479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 151. | Shafaie E, Saberi S, Esmaeili M, Karimi Z, Najafi S, Tashakoripoor M, Abdirad A, Hosseini ME, Mohagheghi MA, Khalaj V, Mohammadi M. Multiplex serology of Helicobacter pylori antigens in detection of current infection and atrophic gastritis - A simple and cost-efficient method. Microb Pathog. 2018;119:137-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 152. | Bazin T, Nozeret K, Julié C, Lamarque D, Touati E. Protein Biomarkers of Gastric Preneoplasia and Cancer Lesions in Blood: A Comprehensive Review. Cancers (Basel). 2024;16:3019. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 153. | Boubrik F, Belmouden A, El Kadmiri N. Potential Non-invasive Biomarkers of Helicobacter pylori-Associated Gastric Cancer. J Gastrointest Cancer. 2022;53:1113-1120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 154. | Cai H, Ye F, Michel A, Murphy G, Sasazuki S, Taylor PR, Qiao YL, Park SK, Yoo KY, Jee SH, Cho ER, Kim J, Chen SC, Abnet CC, Tsugane S, Cai Q, Shu XO, Zheng W, Pawlita M, Epplein M. Helicobacter pylori blood biomarker for gastric cancer risk in East Asia. Int J Epidemiol. 2016;45:774-781. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 64] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 155. | Gao L, Michel A, Weck MN, Arndt V, Pawlita M, Brenner H. Helicobacter pylori infection and gastric cancer risk: evaluation of 15 H. pylori proteins determined by novel multiplex serology. Cancer Res. 2009;69:6164-6170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 76] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 156. | Chu LY, Wu FC, Guo HP, Xie JJ, Qu QQ, Li XH, Xu YW, Peng YH, Qiu B. Combined detection of serum EFNA1 and MMP13 as diagnostic biomarker for gastric cancer. Sci Rep. 2024;14:15957. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 157. | Li J, Zhao W, Yang J, Lu P, Sun H, Zhang Z, Gu J. Proteomic and serological markers for diagnosing cardia gastric cancer and precursor lesions in a Chinese population. Sci Rep. 2024;14:25309. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 158. | Pan KF, Formichella L, Zhang L, Zhang Y, Ma JL, Li ZX, Liu C, Wang YM, Goettner G, Ulm K, Classen M, You WC, Gerhard M. Helicobacter pylori antibody responses and evolution of precancerous gastric lesions in a Chinese population. Int J Cancer. 2014;134:2118-2125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 159. | Zhang C, Xu S, Xu D. Risk assessment of gastric cancer caused by Helicobacter pylori using CagA sequence markers. PLoS One. 2012;7:e36844. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 160. | Gu J, Xie S, Li X, Wu Z, Xue L, Wang S, Wei W. Identification of plasma proteomic signatures associated with the progression of cardia gastric cancer and precancerous lesions. J Natl Cancer Cent. 2023;3:286-294. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/