Published online Apr 15, 2024. doi: 10.4251/wjgo.v16.i4.1547
Peer-review started: December 8, 2023
First decision: December 22, 2023
Revised: January 8, 2024
Accepted: February 7, 2024
Article in press: February 7, 2024
Published online: April 15, 2024
Processing time: 124 Days and 19.9 Hours
Increasing data indicated that long noncoding RNAs (lncRNAs) were directly or indirectly involved in the occurrence and development of tumors, including hepatocellular carcinoma (HCC). Recent studies had found that the expression of lncRNA HAND2-AS1 was downregulated in HCC tissues, but its role in HCC progression is unclear. Ultrasound targeted microbubble destruction mediated gene transfection is a new method to overexpress genes.
To study the role of ultrasound microbubbles (UTMBs) mediated HAND2-AS1 in the progression of HCC, in order to provide a new reference for the treatment of HCC.
In vitro, we transfected HAND2-AS1 siRNA into HepG2 cells by UTMBs, and detected cell proliferation, apoptosis, invasion and epithelial-mesenchymal transition (EMT) by cell counting kit-8 assay, flow cytometry, Transwell invasion assay and Western blotting, respectively. In addition, we transfected miR-837-5p mimic into UTMBs treated cells and observed the changes of cell behavior. Next, the UTMBs treated HepG2 cells were transfected together with miR-837-5p mimic and tissue inhibitor of matrix metalloproteinase-2 (TIMP2) overexpression vector, and we detected cell proliferation, apoptosis, invasion and EMT. In vivo, we established a mouse model of subcutaneous transplantation of HepG2 cells and observed the effect of HAND2-AS1 silencing on tumor formation ability.
We found that UTMBs carrying HAND2-AS1 restricted cell proliferation, invasion, and EMT, encouraged apoptosis, and HAND2-AS1 silencing eliminated the effect of UTMBs. Additionally, miR-873-5p targets the gene HAND2-AS1, which also targets the 3’UTR of TIMP2. And miR-873-5p mimic counteracted the impact of HAND2-AS1. Further, miR-873-5p mimic solely or in combination with pcDNA-TIMP2 had been transformed into HepG2 cells exposed to UTMBs. We discovered that TIMP2 reversed the effect of miR-873-5p mimic caused by the blocked signalling cascade for matrix metalloproteinase (MMP) 2/MMP9. In vivo results showed that HAND2-AS1 silencing significantly inhibited tumor formation in mice.
LncRNA HAND2-AS1 promotes TIMP2 expression by targeting miR-873-5p to inhibit HepG2 cell growth and delay HCC progression.
Core Tip: In this study, we found that ultrasound microbubbles loaded with long noncoding RNA HAND2-AS1 inhibited the growth of hepatocellular carcinoma cells and tumor formation in mice in vivo and in vitro by downregulating miR-873-5p to promote tissue inhibitor of matrix metalloproteinase-2 expression.
- Citation: Zou Q, Wang HW, Di XL, Li Y, Gao H. Long noncoding RNAs HAND2-AS1 ultrasound microbubbles suppress hepatocellular carcinoma progression by regulating the miR-873-5p/tissue inhibitor of matrix metalloproteinase-2 axis. World J Gastrointest Oncol 2024; 16(4): 1547-1563
- URL: https://www.wjgnet.com/1948-5204/full/v16/i4/1547.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i4.1547
Hepatocellular carcinoma (HCC) is a malignant tumor with a high incidence. A previous study reported more than 40000 new patients with HCC in a year, accounting for 50% of the global number of new patients[1]. Since patients with early-stage HCC do not show indicative clinical symptoms, once diagnosed, patients are primarily at advanced stages, and the prognosis of HCC is often unsatisfactory due to high postoperative recurrence and intrahepatic or extrahepatic meta
Studies have confirmed that long noncoding RNAs (lncRNAs) play pivotal roles in biological activities such as epigenetic inheritance, cell cycle progression and cell differentiation[2,3]. In addition, several scholars have shown that lncRNAs are directly or indirectly involved in the tumorigenesis, development and regulation of numerous biological functions. A study showed that the lncRNA HAND2-AS1 inhibited HCC progression by regulating the Janus kinase-signal transducer and activator of transcription signaling pathway[4]. Moreover, it has been reported that HAND2-AS1 inhibits HCC cell viability[5,6]. MiRNAs represent a novel epigenetic mechanism for regulating gene expression in various cells and are regulatory transcripts approximately 19-22 nucleotides in length. Dysfunction of these proteins is associated with many diseases. miR-873-5p expression is upregulated in samples from patients with cholestasis and cirrhosis, and the miR-873-5p inhibitor suppressed hepatocyte apoptosis[7].
Recently, ultrasound-targeted microbubble destruction-mediated gene transfection has been found to be a promising new method. Studies have shown that the cavitating effect of ultrasound may be the primary mechanism by which it enhances gene transfection. Microbubbles are ruptured by vibration under ultrasound, releasing high energy to promote the formation of reversible micropores on the cell membrane, increasing cell membrane permeability and thus helping exogenous genes of interest to enter the cell. Several studies have confirmed that ultrasound-targeted microbubble destruction-mediated plasmid DNA transfection can improve gene transfection and the number of local tissues and cells, and this approach is expected to be a highly efficient, safe, and somewhat effective method for targeted gene transfection and gene therapy[8,9]. Liu et al transfected shCD133 into CD133+ cells isolated from HCC cell lines using both ultrasound microbubbles (UTMBs) and liposomes, and the results indicated that the transfection efficiency was significantly greater in the UTMB group than in the liposome group[10]. Another study revealed that after the transfection of shEZH2 with UTMBs or liposomes, the survival and proliferation of CD133–overexpressing Huh7 cells were inhibited[11]. As a result, in the present study, we investigated the role of UTMB-mediated HAND2-AS1 in HCC progression to provide an addi
HepG2, Huh-7, SMMC-7721, MHCC97 (Human HCC cell line), and L-02 cells were obtained from ATCC and cultured in RPMI 1640 medium supplemented with foetal bovine Sserum, penicillin and streptomycin in an incubator with 5% CO2. RiboBio Co., Ltd., provided the overexpression vectors for HAND2-AS1 and tissue inhibitor of matrix metalloproteinase-2 (TIMP2) and the negative control. Santa Cruz Biotechnology supplied the si-HAND2-AS1 and scrambled the RNA.
To establish HCC xenografts, a total of 12 BALB/c nude mice (4-6 wk; 18-22 g) were injected subcutaneously with 5 × 105 HepG2 cells suspended in 0.2 mL of 0.9% NaCl. The mice were housed in a clean and well-ventilated animal environment at 20 ± 2 °C, with a relative humidity of 60%-70% and a day/night cycle of 12/12 h. The mice were fed with free diet and water intake for more than 3 d. The mice were randomly divided into following groups (n = 6 per group): the control group (mice were given only 0.9% NaCl at 1 MHz) and the UTMB group (mice were injected with HAND2-AS1 microbubbles; 200 μg; 1 MHz). At days 35, we used the method of cervical dislocation to euthanize the mice in all experimental group. The Tianjin Medical University’s animal ethics committee approved this investigation.
A PrimeScript Reagent Kit was used to create single-stranded cDNA after total RNA was extracted from the cells using the TRIzol method. Quantitative polymerase chain reaction (qPCR) was performed using the SYBR Premix Ex TaqTM Kit. The study primers used were created and dyed by Sangon Biotech. The 2-ΔΔCt method was used to standardize the relative expression levels.
Total protein was extracted from the RIPA lysis buffer and then transferred to polyvinylidene fluoride membranes. The membranes were incubated with primary antibodies for 12 hours after being blocked at room temperature for 2 h. Snail (1:1000, ab216347), Glyceraldehyde-3-phosphate dehydrogenase (1:1500, ab8245), E-cadherin (1:10000, ab40772), vimentin (1:2000, ab92547), TIMP2 (1:500, ab180630), matrix metalloproteinase (MMP) 2 (1:2500, ab92536), and MMP9 (1:3000, ab76003). The membranes were then incubated for 1 h with a secondary antibody (1:2500, ab6721). After that, liquid exposure was applied, and the visible protein bands were recorded.
Before exposure to H2O2, tissue from the tumor pieces was trypsinized for 10 min. The sections were then treated with the appropriate antibodies for 12 h at 4 °C. The slices were then treated with secondary antibodies following a phosphate buffered saline (PBS) rinse. The sections were washed once more before being exposed to diaminobenzidine substrate for two minutes.
After adding the media to the bottom chamber, the cells were grown for 48 h at room temperature after being sown in the chamber on top coated with Matrigel. The cells in the top compartment of the chamber were then removed, 70% ethanol was added to the bottom compartment of the chamber, the cells were stained with 0.1% crystal violet, and cell invasion was examined using a microscope.
After the cells were digested with 0.25% trypsin, the cell suspension (3 × 104/mL) was incubated with Dulbecco’s modified Eagle’s medium once more, seeded into 96-well plates (100 mL), and incubated for 4 h (37 °C; 5% CO2). A microplate reader (450 nm) was used to measure the absorbance.
PBS was used to prepare the cell suspension (100 mL, 1 × 105 cells/mL), which was subsequently added to the culture tube. The culture tube was then incubated in the dark for 20 min with annexin V-FITC and propidium iodide. Finally, flow cytometry was used to identify apoptotic cells.
In brief, probe-miR-873-5p, probe-NC, and the positive control (Input) were generated from the scratch and added to the HepG2 cells. Real-time qPCR (RT-qPCR) was used to identify the enriched RNA after the cells had been lysed at low temperatures for 10 minutes and rinsed with PBS after 48 h.
The SPSS program (version 21.0; SPSS, Chicago, IL) was used for all the statistical analyses. P < 0.05 was regarded as a statistically significant value, and the quantitative results collected from three independent experiments are expressed as the mean ± SD.
Prediction results from an online bioinformatics database revealed that HAND2-AS1 was strongly downregulated in HCC tissue samples (Figure 1A and B). Additionally, we obtained tumor and paracancerous tissue samples from 35 HCC patients and discovered that the expression of HAND2-AS1 was downregulated in tumor tissues compared to paracancerous tissues (Figure 1C). In addition, the HepG2, Huh-7, SMMC-7721, and MHCC97 human HCC cell lines presented decreased HAND2-AS1 expression (Figure 1D).
The UTMBs were round-shaped microbubbles with a white powder shape, a smooth surface and good dispersibility. We found that UTMB treatment alone increased HAND2-AS1 expression, while si-HAND2-AS1 decreased HAND2-AS1 expression (Figure 2A). In addition, UTMB treatment alone inhibited cell growth, while si-HAND2-AS1 enhanced cell proliferation, inhibited apoptosis and promoted invasion (Figure 2B-D). Western blotting showed that cell epithelial-mesenchymal transition (EMT) was reduced after UTMB treatment alone, and si-HAND2-AS1 abolished the effects of UTMBs (Figure 2E).
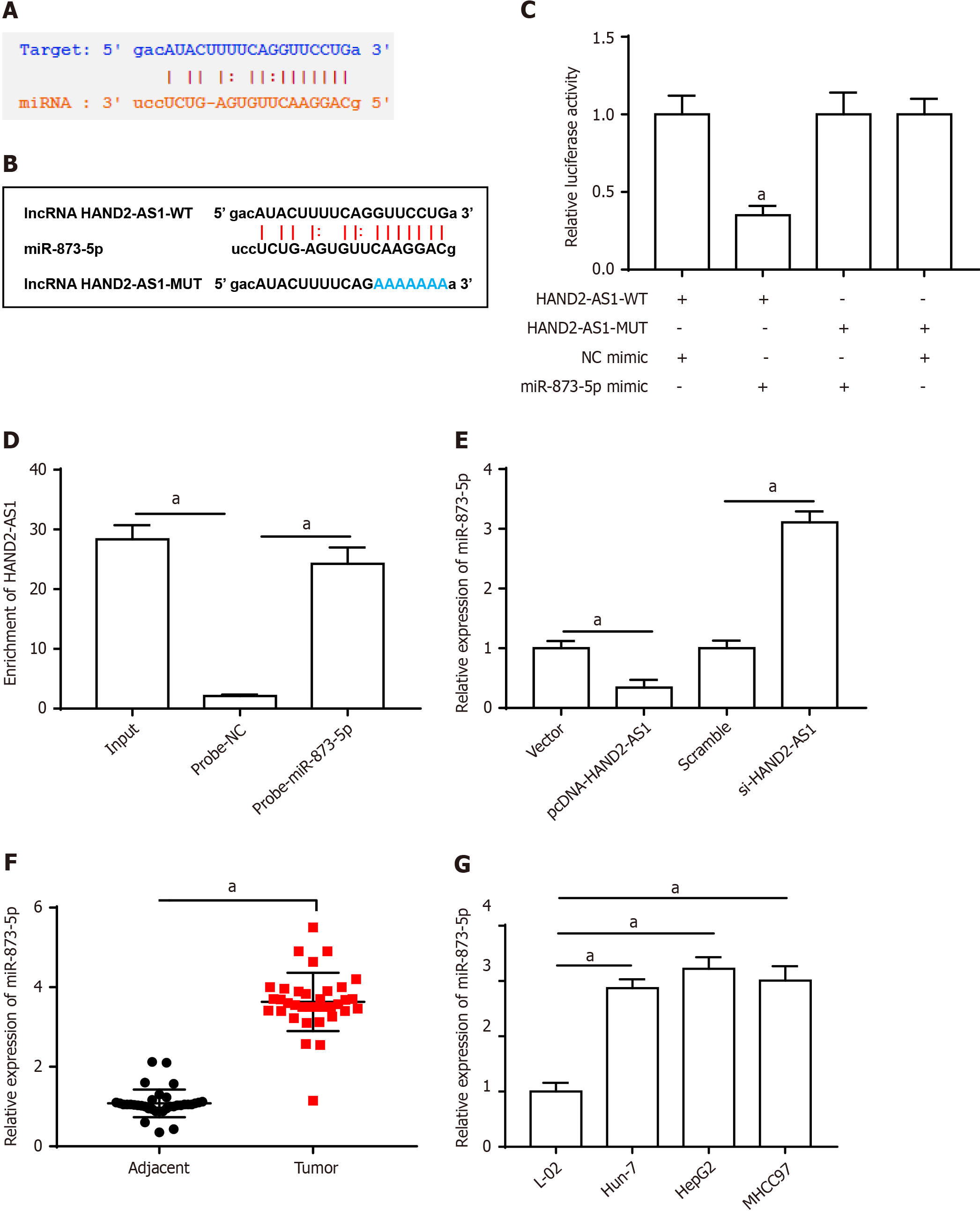
Figure 3A shows the binding site between HAND2-AS1 and miR-873-5p, and Figure 3B shows the altered bases of HAND2-AS1. Furthermore, the luciferase activity of the HAND2-AS1 wild-type reporter was decreased by the miR-873-5p mimic, whereas the HAND2-AS1 mutant reporter was unaffected (Figure 3C). Additionally, HAND2-AS1 was enriched on the miR-873-5p probe according to the results of the RNA pull-down method (Figure 3D). Furthermore, HAND2-AS1 silencing increased miR-873-5p expression, while HAND2-AS1 overexpression blocked this expression (Figure 3E). MiR-873-5p expression was downregulated in HCC tissues (Figure 3F) and cell populations (Figure 3G).
The effects of UTMBs were reversed by the miR-873-5p mimic. These compounds increased HAND2-AS1 expression while inhibiting miR-873-5p expression (Figure 4A). Furthermore, UTMB administration suppressed cell growth, whereas the miR-873-5p mimic increased cell proliferation, decreased apoptosis, and promoted cell invasion (Figure 4B-D). Furthermore, after UTMB treatment, the expression of EMT-related proteins was reduced, and the miR-873-5p mimic eliminated the impact of UTMBs (Figure 4E).
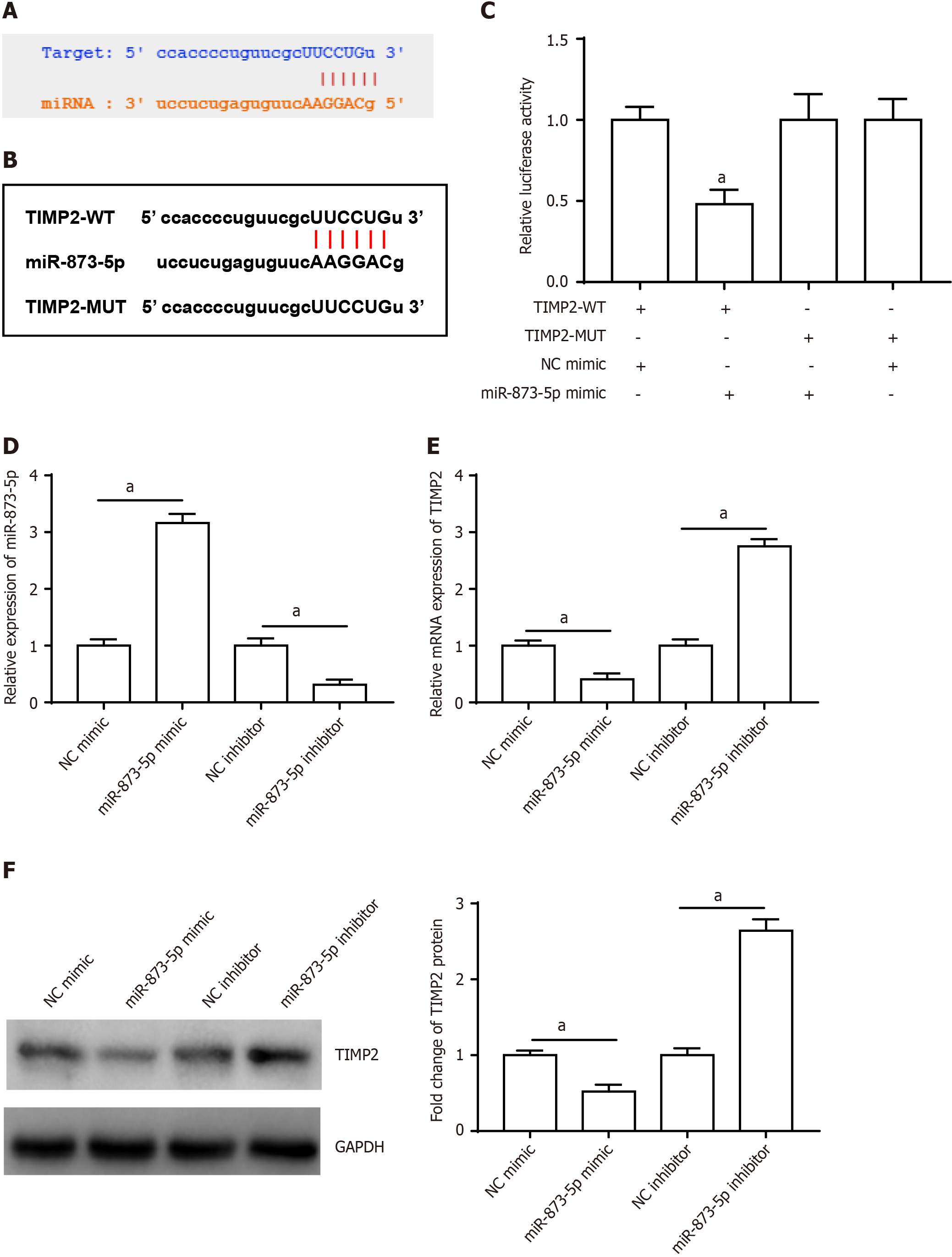
Furthermore, miR-873-5p targeted the 3’UTR of TIMP2 (Figure 5A), and the TIMP2 sequences are displayed in Figure 5B. Furthermore, luciferase reporter gene analysis validated the connection between miR-873-5p and TIMP2 (Figure 5C). Figure 5D shows the transfection efficiency of the miR-873-5p mimic and the miR-873-5p inhibitor. The miR-873-5p mimic reduced TIMP2 mRNA (Figure 5E) and protein expression (Figure 5F), and the miR-873-5p inhibitor increased TIMP2 expression.
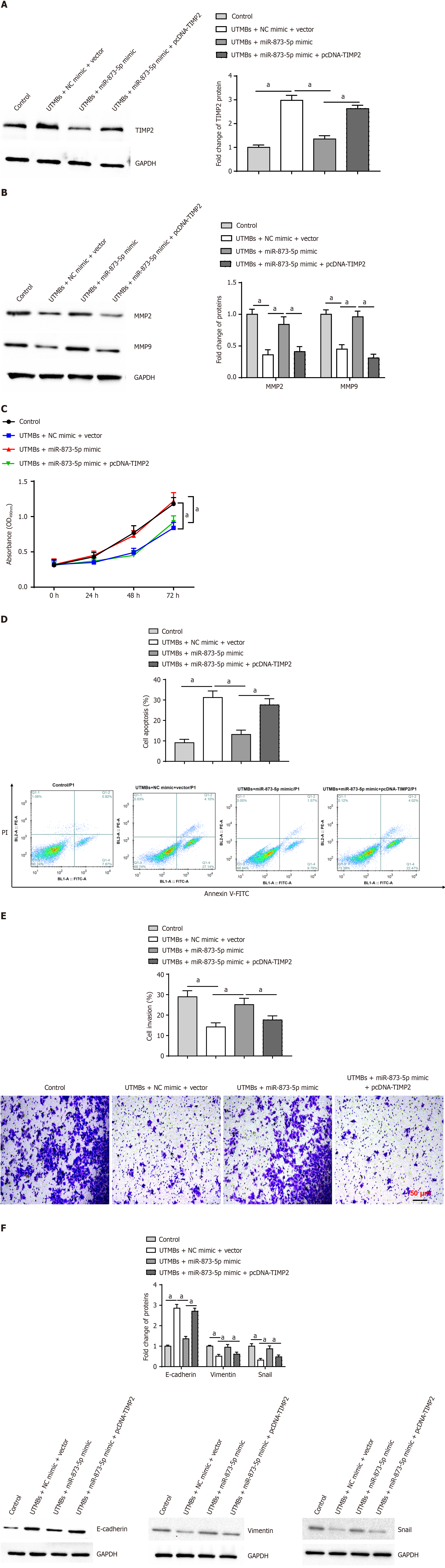
Western blotting revealed that UTMB administration increased the expression of the TIMP2 protein. The miR-873-5p mimic reversed this effect, although the impact of the miR-873-5p mimic was again blocked by pcDNA-TIMP2 (Figure 6A). Furthermore, UTMBs inhibited MMP2 and MMP9 protein expression, and the miR-873-5p mimic promoted MMP2 and MMP9 protein expression, which was reversed by pcDNA-TIMP2 (Figure 6B). We also observed that the miR-873-5p mimic promoted proliferation (Figure 6C), inhibited apoptosis (Figure 6D), promoted invasion (Figure 6E) and accelerated EMT (Figure 6F) in UTMB-treated HepG2 cells, but these effects were reversed after transfection with pcDNA-TIMP2.
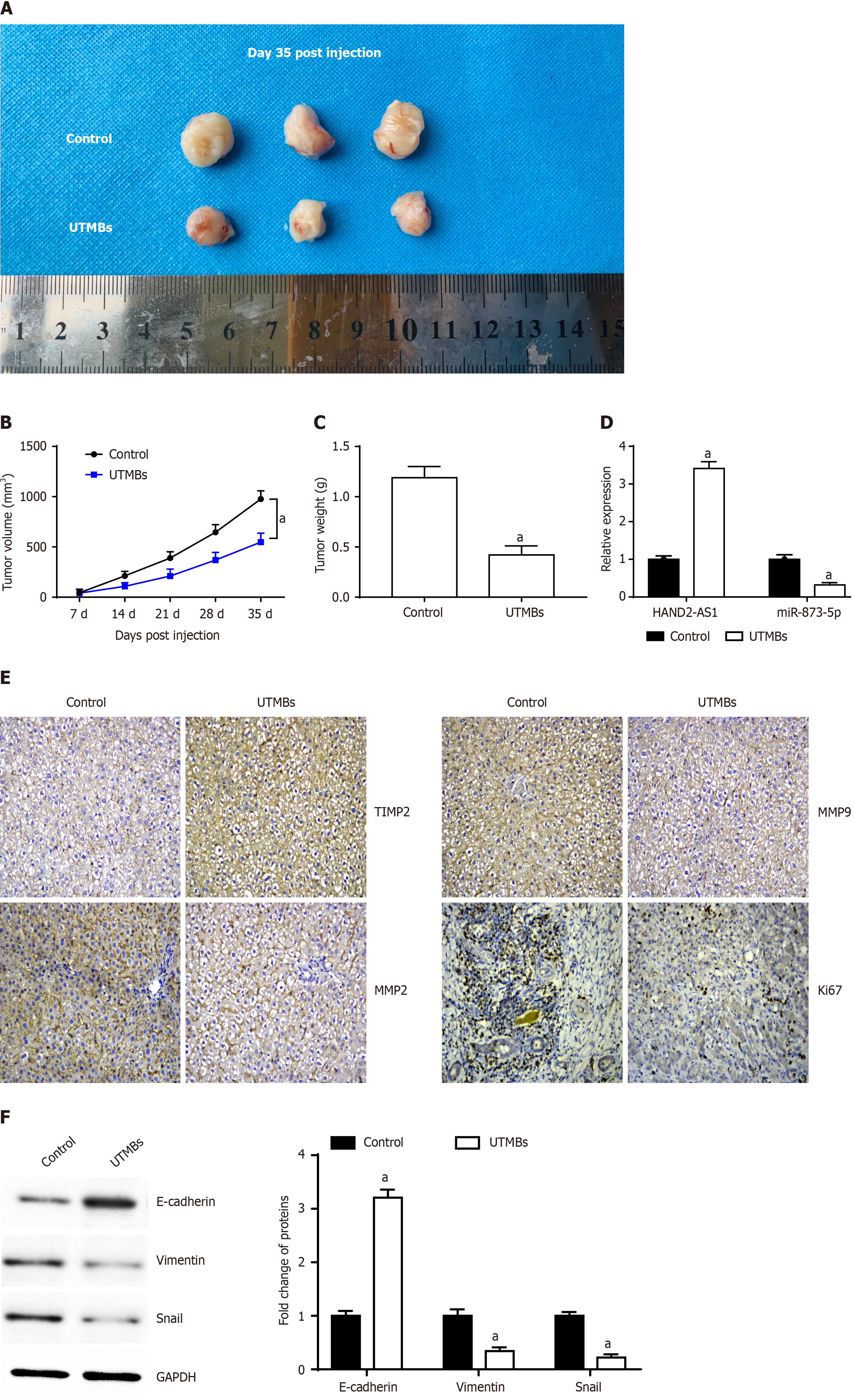
A representative image of the tumor is shown in Figure 7A. We observed that, compared with those of the control mice, the tumor volume and weight (Figure 7B and C) of the nude mice injected with the UTMBs harboring HAND2-AS1 were significantly lower. RT-qPCR indicated that injection of UTMBs promoted HAND2-AS1 expression and inhibited miR-873-5p expression (Figure 7D). Immunohistochemistry revealed that UTMBs promoted TIMP2 expression and inhibited MMP2, MMP9 and Ki67 expression (Figure 7E). Furthermore, UTMBs increased E-cadherin expression while decreasing vimentin and snail expression (Figure 7F).
HCC is a malignant tumor characterized by invasion and metastasis with an extremely high mortality rate, and its malignant transformation behavior is orchestrated by multiple factors. Tumorigenesis occurs through disruption of the balance of various biochemical factor metabolites around cancerous tissue and in the microenvironment where cancer cells are located far from normal tissue; this process involves growth factors, tissue suppressors, tumor factors, endocrine factors, and proteolytic enzymes[12]. In recent years, the importance of MMPs in tumor progression has been recognized. Generally, cancerous tissues first release sufficient amounts of MMPs to degrade the basement membrane and matrix, after which the defect site and stromal gap that break through the basement membrane grow peripherally. At the same time cancer cells' degradation edge spreads, the vascular endothelial cells in the tissue also metastasize and reconstruct, at which point the new blood vessels provide nutrition for the growth and propagation of the tumor[13]. A considerable body of research suggests that there is a significant link between MMP expression and tumor differentiation degree, suggesting that MMP expression has the potential to be a biological marker for predicting tumor invasion and metastasis, as well as determining the risk of recurrence[14,15]. Wen et al[16] showed that citrus reticulate peel black tea could inhibit liver cancer progression by inhibiting the phosphorylation of the Phosphoinositide 3-kinase and protein kinase B proteins, upregulating the ratio of Bax/B cell lymphoma-2, and inhibiting the protein expression of MMP2, MMP9, N-cadherin, and vimentin. Additional studies have shown that alternative splice variants of CXCR3 that mediate CXCL9 caused significant changes in the phosphorylation levels of extracellular signal-regulated kinase 1/2 in the mitogen-activated protein kinases signaling pathway, thereby upregulating MMP2 and MMP9 expression and promoting CD133+ hepatoma cell invasion and metastasis[17].
TIMP2 is a specific inhibitor of MMP2 that inhibits the degradation of the extracellular matrix by the MMP-2 protein and antagonizes neovascularization to control the spread and metastasis of cancer cells[18]. At present, this pair of anta
The EMT is a crucial factor for tumors to acquire the ability to metastasize and invade. After EMT occurs, the tumor cell's epithelial cell characteristics disappear while exhibiting the properties of mesenchymal cells; thus, the tumor cells have stronger invasive and metastatic abilities. The primary molecular mechanism involved is the reduced expression of the tumor epithelial marker E-cadherin[21,22]. E-cadherin acts as a suppressor of cancer cell metastasis, and its expression is correlated with the degree of differentiation, invasion, and malignant tumor metastasis. According to previous reports, overexpression of TIMP2 can upregulate E-cadherin expression, which helps to maintain cell-cell adhesion and inhibit EMT progression induced by epithelial growth factor[23]. At present, additional studies have focused on systemic treatment of HCC; for example, patients with improved responses to tyrosine kinase inhibitors (TKIs), such as sorafenib, lenvatinib, regorafenib, cabozantinib and immune checkpoint inhibitors (ICIs), including anti-PD1, anti-PDL1 and anti-CTLA-4 drugs, have been found. A recent comprehensive evaluation showed that the efficacy of TKI-ICI combination therapy for HCC was more significant than that of other therapies[24]. In addition, studies have shown that HCC patients who fail first-line treatment with sorafenib also have good tolerance and safety of metronomic capecitabine[25,26]. The results of this study suggested that HAND2-AS1 may be used as a prognostic marker for HCC. Therefore, our subsequent studies will further explore the relationship between HAND2-AS1 and TKI or ICI therapy to determine the best systemic treatment for HCC.
In brief, in this study, we delivered HAND2-AS1 into HepG2 HCC cells via UTMBs. We found that UTMBs harboring HAND2-AS1 suppressed cell invasion, proliferation and EMT, and the mechanistic findings indicated that HAND2-AS1 suppressed the MMP2/MMP9 signaling pathway and subsequently suppressed tumor progression by upregulating TIMP2 by targeting miR-873-5p. Furthermore, in vivo results demonstrated that tumor formation was inhibited in xenograft mice injected with HAND2-AS1-bearing UTMBs.
Hepatocellular carcinoma (HCC) is a malignant tumor with high incidence and poor prognosis. Studies have confirmed that long noncoding RNAs (lncRNAs) are directly or indirectly involved in the occurrence and development of tumors and the regulation of various biological functions, including HCC, in which the expression of lncRNA HAND2-AS1 is downregulated in HCC tissues, but the specific mechanism of its involvement in HCC progression still needs to be further explored. In addition, ultrasound targeted microbubble destruction mediated gene transfection is a promising new method in recent years. Therefore, studying the role of ultrasound microbubbles (UTMBs) mediated HAND2-AS1 in HCC progression can provide a new reference for the treatment of HCC.
lncRNA HAND2-AS1 expression was downregulated in HCC tissues and cells, which may be involved in tumor progression. We tried to transfect lncRNA HAND2-AS1 into HCC cell line HepG2 by ultrasound targeted microbubble destruction mediated gene transfection technology to detect the effect of HAND2-AS1 on the proliferation, invasion, epithelial mesenchymal transition and apoptosis of HepG2 cells, and further explore the specific regulatory mechanism. In addition, we established a subcutaneous tumor xenograft mouse model to observe the effect of lncRNA HAND2-AS1 on the tumor forming ability of mice. We aimed to clarify the role of lncRNA HAND2-AS1 in HCC progression through in vivo and in vitro studies, in order to provide new ideas for the treatment of HCC.
We transfected lncRNA HAND2-AS1 into HepG2 cells through ultrasound targeted microbubble destruction mediated gene transfection technology, and detected the effect of lncRNA HAND2-AS1 on the biological behavior of HepG2 cells through a series of experiments in vitro, and found the downstream target genes of lncRNA HAND2-AS1 through online bioinformatics data retrieval, and further clarified the specific mechanism of lncRNA HAND2-AS1 participating in HCC cell growth. In addition, we successfully established a subcutaneous tumor xenograft mouse model and verified the inhibitory effect of lncRNA HAND2-AS1 on tumor formation in vivo in mice. Our results clarify the feasibility of ultrasound targeted microbubble destruction mediated gene transfection technology, and provide a new idea for finding gene therapy for HCC.
We detected the expression levels of lncRNA HAND2-AS1 and miR-873-5p in tumor cells and tumor tissues by real-time quantitative polymerase chain reaction. The proliferation, apoptosis and invasion of HepG2 cells were detected by cell counting kit-8 assay, flow cytometry and Transwell cell invasion assay, respectively. Western botting was used to detect the protein expression levels of tissue inhibitor of matrix metalloproteinase-2 (TIMP2), matrix metalloproteinase (MMP)-2, MMP9 and epithelial mesenchymal transition related proteins in tumor cells and tumor tissues. In addition, immunohistochemistry was used to detect the expression of TIMP2, MMP2, MMP9 and Ki67 in tumor tissues. Luciferase reporter gene analysis was used to verify the targeting relationship of lncRNA HAND2-AS1 and miR-873-5p, as well as miR-873-5p and TIMP2. The SPSS program (version 21.0; SPSS, Chicago, IL) was used for all statistical analyses.
UTMBs loaded with lncRNA HAND2-AS1 inhibited the proliferation, invasion, and epithelial mesenchymal transition of HepG2 cells, and promoted apoptosis. We found that miR-873-5p was a target gene of lncRNA HAND2-AS1, and overexpression of miR-873-5p abolished the inhibitory effect of lncRNA HAND2-AS1 on tumor cell growth. In addition, miR-873-5p targeted the 3'UTR of TIMP2, and TIMP2 again reversed the promoting effect of miR-873-5p on tumor cell growth, and the mechanism study showed that this was mediated by blocking the MMP2/MMP9 signaling pathway. In the subcutaneous tumor xenograft mouse model, we observed that UTMBs carrying lncRNA HAND2-AS1 inhibited tumor formation in mice. Our results provide a new idea for gene therapy of HCC. Considering that this study only uses HepG2 cells, we will verify the results of this study in a variety of HCC cell lines later.
We delivered lncRNA HAND2-AS1 into HeGp2 cells by UTMBs, and found that UTMBs carrying lncRNA HAND2-AS1 suppressed the cell invasion, proliferation and epithelial-mesenchymal transition, and the mechanistic findings indicated that lncRNA HAND2-AS1 suppressed the MMP2/MMP9 signaling pathway and then suppressed tumor progression by upregulating TIMP2 via targeting miR-873-5p. Furthermore, in vivo results demonstrated that tumor formation was inhibited in xenograft mice injected with lncRNA HAND2-AS1-bearing UTMBs.
We identified the regulatory role of lncRNA HAND2-AS1/miR-873-5p/TIMP2 axis in HCC progression, which is a classic ceRNA pattern. Subsequently, we will take lncRNA HAND2-AS1 as a starting point to explore whether it is involved in tumor immune evasion microenvironment, or its relationship with tyrosine kinase inhibitors and immune checkpoint inhibitors, which were mentioned in the discussion section of the manuscript.
| 1. | Liu C, Wu J, Chang Z. Trends and Age-Period-Cohort Effects on the Prevalence, Incidence and Mortality of Hepatocellular Carcinoma from 2008 to 2017 in Tianjin, China. Int J Environ Res Public Health. 2021;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 2. | Ma W, Chen X, Wu X, Li J, Mei C, Jing W, Teng L, Tu H, Jiang X, Wang G, Chen Y, Wang K, Wang H, Wei Y, Liu Z, Yuan Y. Long noncoding RNA SPRY4-IT1 promotes proliferation and metastasis of hepatocellular carcinoma via mediating TNF signaling pathway. J Cell Physiol. 2020;235:7849-7862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 3. | Wu M, Yang LZ, Chen LL. Long noncoding RNA and protein abundance in lncRNPs. RNA. 2021;27:1427-1440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 4. | Yan D, Jin F, Lin Y. lncRNA HAND2-AS1 Inhibits Liver Cancer Cell Proliferation and Migration by Upregulating SOCS5 to Inactivate the JAK-STAT Pathway. Cancer Biother Radiopharm. 2020;35:143-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 5. | Bi HQ, Li ZH, Zhang H. Long noncoding RNA HAND2-AS1 reduced the viability of hepatocellular carcinoma via targeting microRNA-300/SOCS5 axis. Hepatobiliary Pancreat Dis Int. 2020;19:567-574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 6. | Jing GY, Zheng XZ, Ji XX. lncRNA HAND2-AS1 overexpression inhibits cancer cell proliferation in hepatocellular carcinoma by downregulating RUNX2 expression. J Clin Lab Anal. 2021;35:e23717. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 7. | Fernández-Ramos D, Fernández-Tussy P, Lopitz-Otsoa F, Gutiérrez-de-Juan V, Navasa N, Barbier-Torres L, Zubiete-Franco I, Simón J, Fernández AF, Arbelaiz A, Aransay AM, Lavín JL, Beraza N, Perugorria MJ, Banales JM, Villa E, Fraga MF, Anguita J, Avila MA, Berasain C, Iruzibieta P, Crespo J, Lu SC, Varela-Rey M, Mato JM, Delgado TC, Martínez-Chantar ML. MiR-873-5p acts as an epigenetic regulator in early stages of liver fibrosis and cirrhosis. Cell Death Dis. 2018;9:958. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 8. | Wang J, Li Y, Ma Q, Huang J. miR‑378 in combination with ultrasonic irradiation and SonoVue microbubbles transfection inhibits hepatoma cell growth. Mol Med Rep. 2020;21:2493-2501. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 9. | Zhao Z, Lin X, Zhang L, Liu X, Wang Q, Shi Y, Cui G, Cai H, Chen Y, Li Y, Hu A, Zhang Z, Liu J, Xie H, Zheng T, Liang X, Shuai X, Sun D. Lipidated Methotrexate Microbubbles: A Promising Rheumatoid Arthritis Theranostic Medicine Manipulated via Ultrasonic Irradiation. J Biomed Nanotechnol. 2021;17:1293-1304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 10. | Liu YM, Li XF, Liu H, Wu XL. Ultrasound-targeted microbubble destruction-mediated downregulation of CD133 inhibits epithelial-mesenchymal transition, stemness and migratory ability of liver cancer stem cells. Oncol Rep. 2015;34:2977-2986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 11. | Wu J, Sun L, Liu T, Dong G. Ultrasound-Targeted Microbubble Destruction-Mediated Downregulation of EZH2 Inhibits Stemness and Epithelial-Mesenchymal Transition of Liver Cancer Stem Cells. Onco Targets Ther. 2021;14:221-237. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 12. | Mughees M, Sengupta A, Khowal S, Wajid S. Mechanism of tumour microenvironment in the progression and development of oral cancer. Mol Biol Rep. 2021;48:1773-1786. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 13. | Gonzalez-Avila G, Sommer B, García-Hernández AA, Ramos C. Matrix Metalloproteinases' Role in Tumor Microenvironment. Adv Exp Med Biol. 2020;1245:97-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 77] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 14. | Pang D, Yang C, Li C, Zou Y, Feng B, Li L, Liu W, Luo Q, Chen Z, Huang C. Polyphyllin II inhibits liver cancer cell proliferation, migration and invasion through downregulated cofilin activity and the AKT/NF-κB pathway. Biol Open. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 15. | Zhu W, Li W, Geng Q, Wang X, Sun W, Jiang H, Pu X. Silence of Stomatin-Like Protein 2 Represses Migration and Invasion Ability of Human Liver Cancer Cells via Inhibiting the Nuclear Factor Kappa B (NF-κB) Pathway. Med Sci Monit. 2018;24:7625-7632. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 16. | Wen S, Sun L, An R, Zhang W, Xiang L, Li Q, Lai X, Huo M, Li D, Sun S. A combination of Citrus reticulata peel and black tea inhibits migration and invasion of liver cancer via PI3K/AKT and MMPs signaling pathway. Mol Biol Rep. 2020;47:507-519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 17. | Ding Q, Xia Y, Ding S, Lu P, Sun L, Liu M. An alternatively spliced variant of CXCR3 mediates the metastasis of CD133+ liver cancer cells induced by CXCL9. Oncotarget. 2016;7:14405-14414. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 18. | Yu L, Wang S, Lin X, Lu Y, Gu P. MicroRNA-124a inhibits cell proliferation and migration in liver cancer by regulating interleukin-11. Mol Med Rep. 2018;17:3972-3978. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 19. | Azevedo Martins JM, Rabelo-Santos SH, do Amaral Westin MC, Zeferino LC. Tumoral and stromal expression of MMP-2, MMP-9, MMP-14, TIMP-1, TIMP-2, and VEGF-A in cervical cancer patient survival: a competing risk analysis. BMC Cancer. 2020;20:660. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 70] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 20. | Fang F, Song T, Zhang T, Cui Y, Zhang G, Xiong Q. MiR-425-5p promotes invasion and metastasis of hepatocellular carcinoma cells through SCAI-mediated dysregulation of multiple signaling pathways. Oncotarget. 2017;8:31745-31757. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 21. | Huang Q, Pu M, Zhao G, Dai B, Bian Z, Tang H, Chen C, Liu W, Qu X, Shen L, Tao K. Tg737 regulates epithelial-mesenchymal transition and cancer stem cell properties via a negative feedback circuit between Snail and HNF4α during liver stem cell malignant transformation. Cancer Lett. 2017;402:52-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 22. | Feng L, Zhang Y, Yang Q, Guo L, Yang F. MicroRNA-885 regulates the growth and epithelial mesenchymal transition of human liver cancer cells by suppressing tropomodulin 1 expression. Arch Biochem Biophys. 2020;693:108588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 23. | Navarini NF, De Araújo VC, Sperandio M, Napimoga MH, Teixeira LN, De Araújo NS, Martinez EF. Effect of epithelial growth factor on matrix metalloproteinase-2 and E-cadherin/β-catenin expression in an in situ model of tumorigenesis. Oncol Lett. 2017;14:3136-3140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 24. | Stefanini B, Ielasi L, Chen R, Abbati C, Tonnini M, Tovoli F, Granito A. TKIs in combination with immunotherapy for hepatocellular carcinoma. Expert Rev Anticancer Ther. 2023;23:279-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 86] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 25. | Granito A, Marinelli S, Terzi E, Piscaglia F, Renzulli M, Venerandi L, Benevento F, Bolondi L. Metronomic capecitabine as second-line treatment in hepatocellular carcinoma after sorafenib failure. Dig Liver Dis. 2015;47:518-522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 75] [Article Influence: 6.8] [Reference Citation Analysis (2)] |
| 26. | Trevisani F, Brandi G, Garuti F, Barbera MA, Tortora R, Casadei Gardini A, Granito A, Tovoli F, De Lorenzo S, Inghilesi AL, Foschi FG, Bernardi M, Marra F, Sacco R, Di Costanzo GG. Metronomic capecitabine as second-line treatment for hepatocellular carcinoma after sorafenib discontinuation. J Cancer Res Clin Oncol. 2018;144:403-414. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 67] [Article Influence: 8.4] [Reference Citation Analysis (1)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Beenet L, United States S-Editor: Qu XL L-Editor: A P-Editor: Zhao YQ