Published online Apr 15, 2024. doi: 10.4251/wjgo.v16.i4.1514
Peer-review started: November 24, 2023
First decision: January 12, 2024
Revised: January 16, 2024
Accepted: February 4, 2024
Article in press: February 4, 2024
Published online: April 15, 2024
Processing time: 138 Days and 11 Hours
Competitive endogenous RNA (ceRNA) is an innovative way of gene expression modulation, which plays a crucial part in neoplasia. However, the intricacy and behavioral characteristics of the ceRNA network in hepatocellular carcinoma (HCC) remain dismal.
To establish a cyclin dependent kinase inhibitor 2A (CDKN2A)-related ceRNA network and recognize potential prognostic indicators for HCC.
The mutation landscape of CDKN2A in HCC was first explored using the cBioPortal database. Differential expression analysis was implemented between CDKN2Ahigh and CDKN2Alow expression HCC samples. The targeted microRNAs were predicted by lncBasev3.0, and the targeted mRNAs were predicted by miRDB, and Targetscan database. The univariate and multivariate analysis were utilized to identify independent prognostic indicators.
CDKN2A was frequently mutated and deleted in HCC. The single-cell RNA-sequencing analysis revealed that CDKN2A participated in cell cycle pathways. The CDKN2A-related ceRNA network-growth arrest specific 5 (GAS5)/miR-25-3p/SRY-box transcription factor 11 (SOX11) was successfully established. GAS5 was recognized as an independent prognostic biomarker, whose overexpression was correlated with a poor prognosis in HCC patients. The association between GAS5 expression and methylation, immune infiltration was explored. Besides, traditional Chinese medicine effective components targeting GAS5 were obtained.
This CDKN2A-related ceRNA network provides innovative insights into the molecular mechanism of HCC formation and progression. Moreover, GAS5 might be a significant prognostic biomarker and therapeutic target in HCC.
Core Tip: In this study, the mutation landscape, and molecular biological mechanisms of cyclin dependent kinase inhibitor 2A (CDKN2A) in hepatocellular carcinoma (HCC) was first explored, and a CDKN2A-related competitive endogenous RNA axis was constructed. We identified growth arrest specific 5 as an independent prognostic biomarker and established a prognostic nomogram model for HCC. Moreover, we analyzed its methylation level, immune infiltration, and targeted agents, which might be an independent prognostic biomarker and therapeutic target in HCC.
- Citation: Pan Y, Zhang YR, Wang LY, Wu LN, Ma YQ, Fang Z, Li SB. Construction of CDKN2A-related competitive endogenous RNA network and identification of GAS5 as a prognostic indicator for hepatocellular carcinoma. World J Gastrointest Oncol 2024; 16(4): 1514-1531
- URL: https://www.wjgnet.com/1948-5204/full/v16/i4/1514.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i4.1514
Hepatocellular carcinoma (HCC) takes up 75%-85% of primary liver cancer and is also the third leading cause of cancer deaths globally[1,2]. The development of HCC involves many genetic alterations and interactions, which is the molecular basis for its recurrence, metastasis, and drug resistance[3-5]. Although there have been some advances in HCC treatment, the 5-year survival rate for HCC patients is less than 17%[6]. The tumor recurrence rate is still up to 70% within five years after surgery or radiofrequency ablation for early HCC[7]. Therefore, seeking valuable prognostic predictors and thera
Competitive endogenous RNA (ceRNA) has received much scholarly attention in recent years, representing a novel model of gene expression regulation. ceRNA regulatory networks are more elaborate and complex than microRNA (miRNA) regulatory networks, involving many RNA molecules, including mRNAs, long noncoding RNAs (lncRNAs), and pseudogenes encoding genes[8-10]. ceRNA molecules increase the expression of target genes by competitively linking the same miRNA with mRNA, thereby diluting the concentration of free miRNA in the cell and reducing the inhibition of mRNA by miRNA[11,12]. Besides, Salmena et al[8] proposed that lncRNAs mostly functioned as endogenous competitors in the cytoplasm. Regarding HCC, the DLEU2L/TAOK1 axis was recognized as a valuable prognostic factor engaging in HCC[13]. Li et al[14] proposed that TRHDE-AS1 influenced the prognosis of hepatitis B virus-infected HCC via miR-23b/PKIA axis. According to Hao et al[15], the transcript produced from the pseudogene UBE2MP1 acted as a molecular sponge for miR-145-5p, promoting the proliferation and resistance to apoptosis of HCC cells in vitro.
Cyclin dependent kinase inhibitor 2A (CDKN2A) is frequently mutated or deleted in various tumors, including HCC, and is known to be an essential tumor suppressor gene[16]. Inactivation of CDKN2A may result in uncontrolled cell growth and proliferation, leading to cancer development. Notably, CDKN2A was one of the cuproptosis (copper-induced cell death) genes discovered by Tsvetkov et al[17]. Prior research has revealed that deleted CDKN2A was a common activity in HCC and was connected to invasion, metastasis, recurrence, and prognosis[18-20]. For example, Desjonqueres et al[21] mentioned that aberrant methylation changes of CDKN2A at early stages of malignant transformation in HCC correlated with increased cell proliferation.
In our study, we established a CDKN2A-related ceRNA network-GAS5/miR-25-3p/SOX11 axis in HCC (Figure 1). First, we sought differentially expressed RNAs (DEGs) between CDKN2Ahigh and CDKN2Alow expression HCC patients and obtained CDKN2A-related lncRNA-miRNA-mRNA triple regulatory networks. Subsequently, a hub ceRNA network was selected through differential expression and survival analysis, and GAS5 was considered an independent prognostic factor. Finally, we carried out methylation analysis, immune infiltration analysis and targeted drug analysis to explore potential biological roles of GAS5 in HCC. In conclusion, our study constructed GAS5/miR-25-3p/SOX11 axis and identified GAS5 as a significant prognostic indicator and therapeutic target in HCC.
The RNA-Seq expression profiles of 374 HCC samples and 50 normal samples were provided by the The Cancer Genome Atlas (TCGA) database (http://portal.gdc.com), together with corresponding clinical information of HCC patients. The raw read counts of gene expression were converted into transcripts per million. The clinical information of patients in the TCGA cohort is illustrated in Supplementary Table 1.
Besides, the mutation landscape of CDKN2A in HCC was investigated via the cBioPortal for Cancer Genomics (http://www.cbioportal.org/).
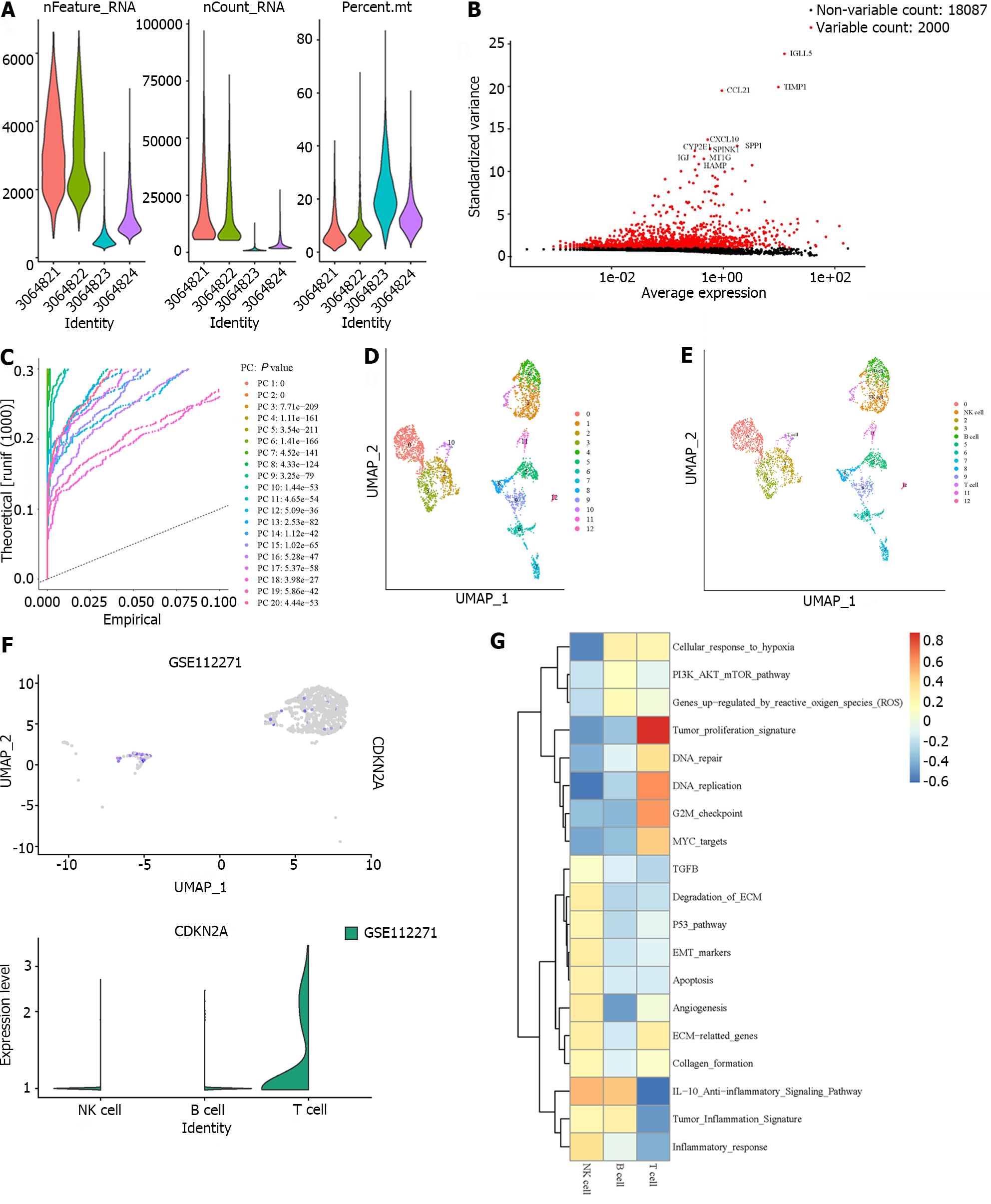
The Single-cell RNA-seq (scRNA-seq) dataset (GSE112271) of HCC was acquired from the GEO database (https://ncbi.nlm.nih.gov/geo/). The “Seurat” package was used to preprocess before dimensionality reduction. Then the principal component analysis (PCA) was performed to identify the top 20 PC based on the top 2000 highly variable genes. Cluster visualization was performed by uniform manifold approximation and projection reduction, and the marker genes for each cluster were selected using the FindAllMarkers function with the adjusted P value < 0.01 and absolute log2 (fold change) value > 1. Finally, each cluster was annotated based on the corresponding canonical marker gene.
For metabolism analysis, the mean expression levels of the cells contained in various clusters were initially determined by the Average Expression function. For every cluster, the scores of the relevant pathways were determined using the “GSVA” R package[22], which were finally visualized by the “pheatmap” R package.
For DEGs in CDKN2Ahigh and CDKN2Alow HCC samples, we selected differentially expressed long non-coding RNAs (DElncRNAs), differentially expressed microRNAs (DEmiRNAs), and differentially expressed messenger RNAs (DEmRNAs) with limits of all P < 0.05 and |logFC| > 0.5, |logFC| > 0.3, |logFC| > 0.5, respectively. For DEGs between HCC samples and adjacent non-tumor samples, we set P < 0.05 and |logFC| > 0.5 as the cut-off value of DElncRNAs, P < 0.05 and |logFC|> 0.3 as the cut-off value of DEmiRNAs, P < 0.05 and |logFC| > 0.7 as the cut-off value of DEmRNAs.
The following traits of ceRNA are based on its notion and mechanism: (1) The presence of same miRNA binding sites for ceRNAs; (2) ceRNA are modulated by miRNAs; (3) the mutual regulatory relationship between ceRNAs and the consistent trend in expression levels; and (4) lncRNAs mainly function as ceRNAs in the cytoplasm. Hence, the ceRNA network was constructed based on the following procedures: (1) LncBasev.3 (https://diana.ece.uth.gr/Lncbase-v3/interactions) was utilized to forecast potential miRNAs targeted by DElncRNAs and lncRNA-miRNA matched bindings; (2) miRDB (http://www.mirdb.org/) and Targetscan (version 7.2, http://www.targetscan.org/) were applied to predict target genes of the DEmiRNAs and search for miRNA-mRNA matched bindings; (3) “VennDiagram” package was used to screen the target genes that intersected with DEmRNAs in this study; (4) LncATLAS (https://Lncatlas.crg.eu/) database[23] was utilized to select cytoplasmic lncRNAs for the subsequent analysis; (5) Cytoscape software (version 3.9.1) was used to construct and draw the lnRNA-miRNA-mRNA triple regulatory network; and (6) The Cytoscape plug-in CytoHubba was applied to determine the hub triple regulatory network which ranked top 15 by maximum clique centrality (MCC) algorithm.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) research of DEmRNAs was performed using “clusterProfiler” package[24]. To investigate the potential interplay of 15 hub genes, we applied an interactive network analysis through the GENEMANIA website[25].
The univariate and multivariate Cox regression was utilized to recognize independent prognostic indicators for HCC. Using “rms” package, the nomogram was established to incorporate every predictive variable in the multivariate Cox regression model. The model’s predictive ability was assessed using the calibration curve.
For validation, four independent datasets (GSE45267, GSE45436, GSE55092, and GSE64041) were collected from the GEO database. The data of GSE45267, GSE45436, GSE55092 were generated using the platform GPL570, while the data of GSE64041 was produced by the platform GPL6244. We adopted a column bar graph to present differential expression of GAS5 in a variety of datasets using GraphPad Prism v9.3.1.
To enhance the statistical credibility, we implemented a meta-analysis of differential expression results of diverse datasets by inputting mean and standard deviation. We took Q test (I2 statistics) result to measure the heterogeneity between different data. In addition, a forest plot was created to display the mean difference and correlated 95% confidence interval of GAS5. All procedures were accomplished by the software Review Manager 5.4.
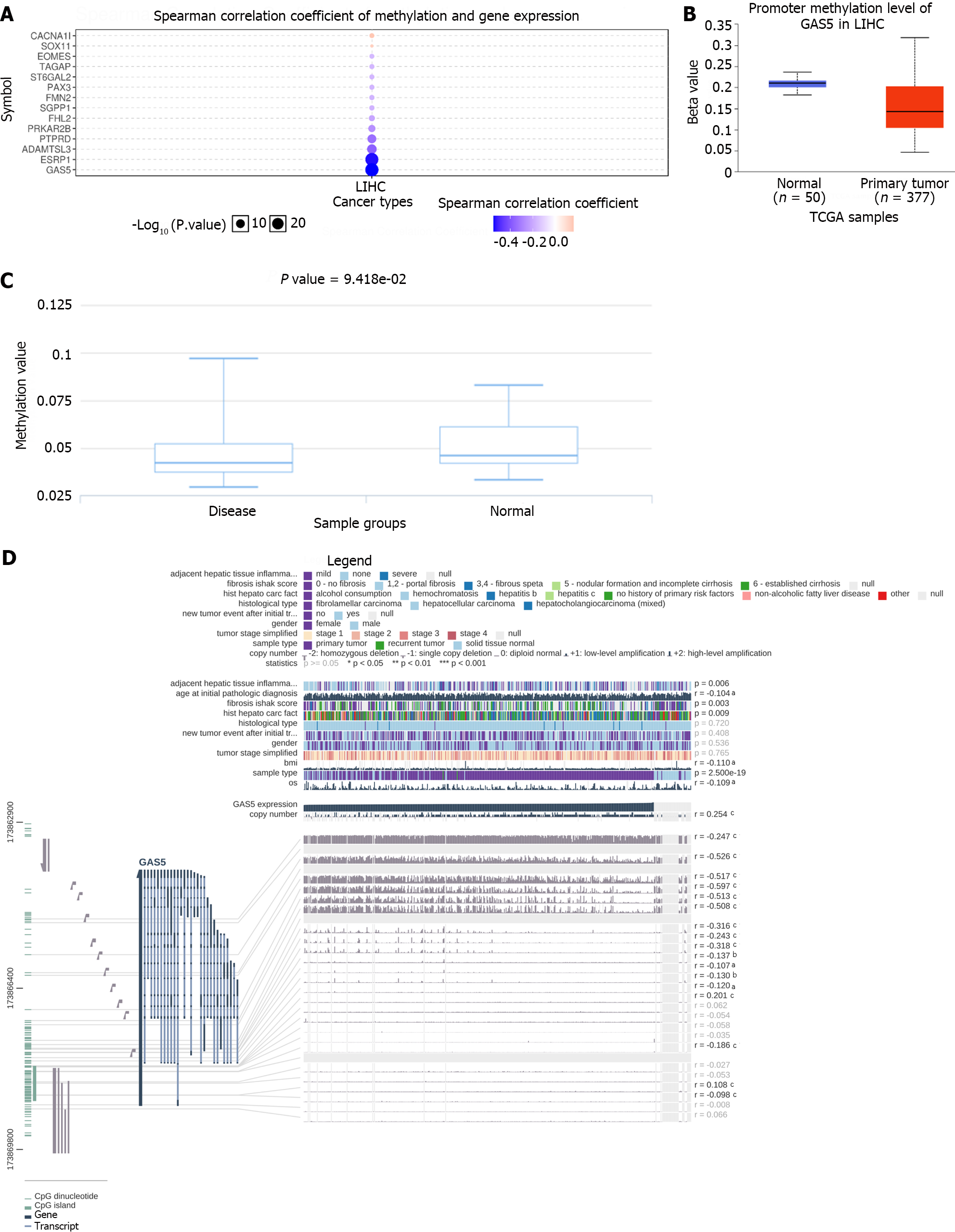
DNA methylation, a common epigenetic mechanism, could modulate gene expression. We first used the GSCALite database[26] to investigate the correlation between methylation and GAS5 expression in LIHC. Then we separately utilized UALCAN and DiseaseMeth version 2.0 to evaluate the methylation degree of GAS5. Besides, the methylation site of GAS5 DNA sequence correlation with gene expression was visualized through MEXPRESS.
The TIMER database[27] was utilized to evaluate the abundance degree of different immune cell types in HCC tumor microenvironment. Additionally, we explored the association between three significant immune checkpoints (ICKs), including PDCD1, CD274, HAVCR2, and GAS5 in HCC using the Spearman correlation coefficient, since the expression of ICK-related genes influences the immunotherapy effect of ICK blockades[28].
The CTD database (http://ctdbase.org/) was utilized to recognize traditional Chinese medicine (TCM) effective components targeting GAS5, and PubChem database (pubchem.ncbi.nlm.nih.gov) was applied to visualize their structures.
As for statistical description, numerical variables were depicted as mean ± SD, while categorical variables were depicted as frequency and proportion. The student’s t test and nonparametric tests were utilized to make a comparison between two groups of data. All analysis methods and R packages were conducted by R (3.6.3) software. A difference of P < 0.05 was defined as statistically significant.
We first explored genomic alterations, copy number, and mRNA expression of CDKN2A using the cBioPortal database. As shown in Figure 2A, CDKN2A had an 8% genomic alteration rate, among which deep depletion was the most common type. Furthermore, HCC samples harboring CDKN2A deletion exhibited the highest mRNA expression, while HCC samples containing CDKN2A deep deletion showed the lowest mRNA expression (Figure 2B). There was a positive association between mRNA expression and CDKN2A copy number values in HCC samples in terms of both Spearman correlation coefficient and Pearson correlation coefficient (Figure 2C). As shown in Figure 2D, there were nine mutations occurring at different positions in the CDKN2A protein for HCC patients, including eight putative drivers (five missense mutations, two truncating mutations, and one splice mutation) and one variant of uncertain significance (one missense mutation). In addition, we found that CDKN2A participated in cell cycle pathways in HCC using pathway sources from the cBioPortal database (Figure 2E).
Given that genetic mutation can drive tumorigenesis and cause aberrant cell proliferation, CDKN2A was very likely to participate in HCC progression.
One scRNA-seq dataset (GSE112271) was collected to further explore CDKN2A expression in the cells from HCC tissues. As shown in Figure 3A, three quality measures were applied. After the top 2000 highly variable genes were screened for clustering (Figure 3B), we utilized PCA to identify 20 PCs with significant difference (Figure 3C). Thirteen clusters were identified (Figure 3D), and three immune cell types (NK cell, B cell, and T cell) were annotated (Figure 3E). Notably, almost only T cells expressed CDKN2A (Figure 3F), More interestingly, we found that the many pathways enriched in T cells were associated with cell cycle, including “tumor proliferation signature”, “DNA repair”, “DNA replication”, and “G2M checkpoint” (Figure 3G), which re-indicated that CDKN2A may participate in cell cycle modulation.
To construct a ceRNA network related to CDKN2A, we first screened out the DElncRNAs, DEmiRNAs, and DEmRNAs in HCC samples with CDKN2Ahigh and CDKN2Alow expression groups as well as in HCC and adjacent non-tumor tissues using TCGA database. The P value and each |logFC| threshold was listed in the material and method part.
Totally, 2032 DElncRNAs, 23 DEmiRNAs, and 2055 DEmRNAs were selected from HCC samples with CDKN2Ahigh and CDKN2Alow expression groups. Meanwhile, 4544 DElncRNAs, 69 DEmiRNAs, and 6560 DEmRNAs were determined between HCC samples and normal liver tissue samples.
We employed volcano plots to visualize the distribution of DElncRNAs, DEmiRNAs, and DEmRNAs (Figure 4A-C; Supplementary Figure 1A-C), and the heatmaps to exhibit the expression of 15 significant variable genes in CDKN2Ahigh expression and CDKN2Alow expression HCC samples, as well as in HCC and normal samples (Figure 4D-F;Supplementary Figure 1D-F).
We then put 4544 DElncRNAs into lncBasev3 to find targeted miRNAs that overlapped with DEmiRNAs between CDKN2Ahigh HCC and CDKN2Alow HCC group and 3 targeted miRNAs (miR-25-3p, miR-210-3p, and miR-148a-3p) and their corresponding lncRNAs (MALAT1, NEAT1, SNHG7, and GAS5) were sorted out. We analyzed the localization of 4 LncRNAs through the lncATLAS database and found that only GAS5 was mainly located in the cytoplasm (Figure 5A). So, we put GAS5 targeting miRNA-miR-25-3p into the miRDB database and ENCORI database, respectively and acquired 63 shared targeted mRNAs. Then we utilized cytoscape software to construct the lncRNA-miRNA-mRNA triple regulatory network in HCC (Figure 5B) and the MCC algorithm to select top 15 score genes as hub genes and visualized them by Cytoscape plug-in CytoHubba (Figure 5C). Additionally, we implemented GO and KEGG analysis of the DEmRNAs in the network. We discovered that they were enriched in the “inclusion body”, “apical dendrite”, “actin-based cell projection”, and “positive regulation of neuron differentiation” (Figure 5D), indicating that they possibly played a crucial role in the growth and differentiation of cells. Finally, we built a gene network to analyze the interaction of hub genes by GENEMANIA website and revealed that these genes took part in positive regulation of cell development (Figure 5E).
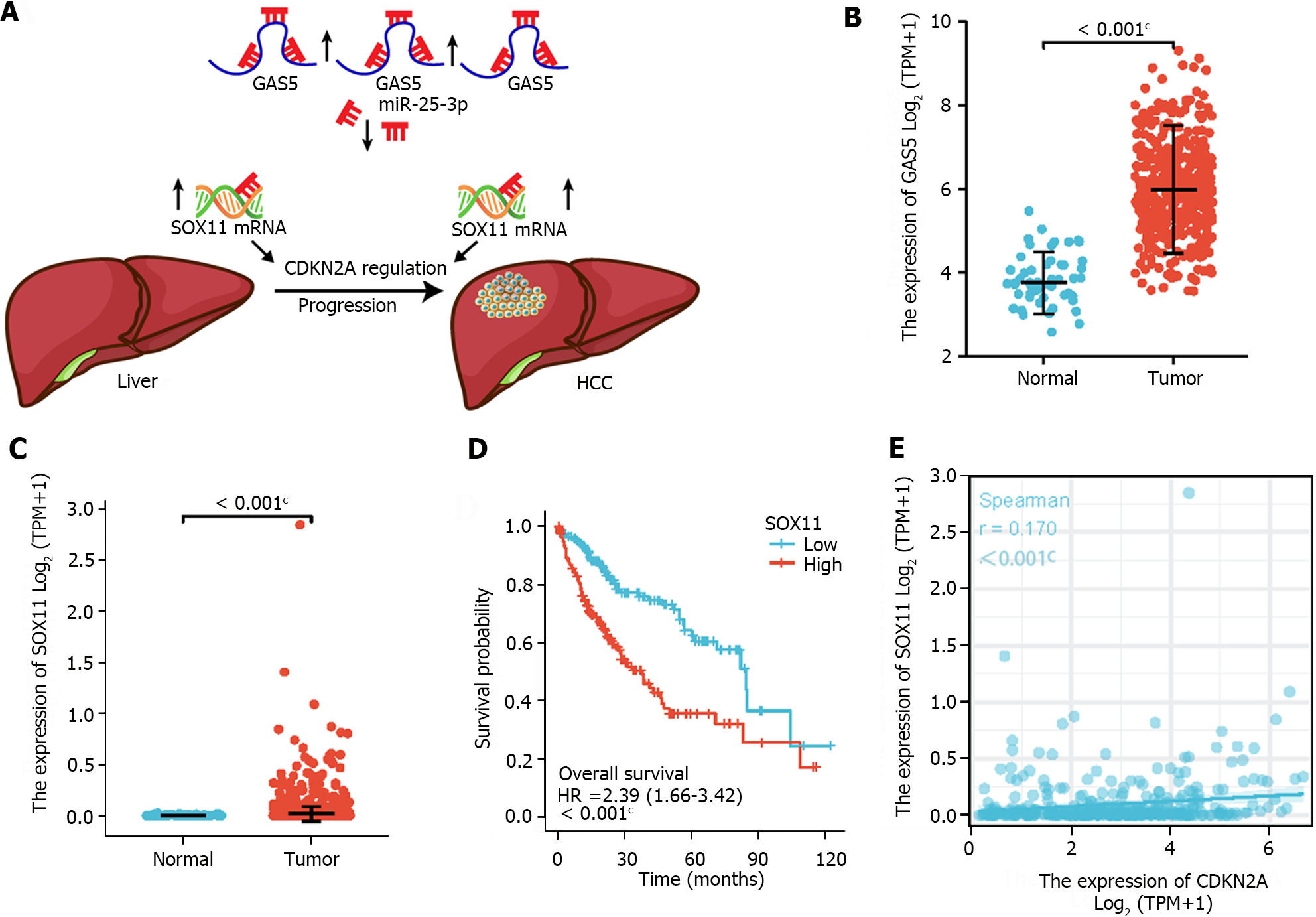
To establish a ceRNA regulatory network that truly influenced the progression of HCC, we analyzed whether the 15 hub genes were differentially expressed and the relationship between 15 hub genes expression and OS in HCC patients. The results showed that 3 mRNAs (CACNA1, SOX11, and PDZD2) were differentially expressed, and only SOX11 was significantly linked to OS in HCC patients. Therefore, a CDKN2A-related ceRNA network-GAS5/miR-25-3p/SOX11 axis was constructed (Figure 6A). To validate whether the regulatory relationship is truly established, we explored the expression level of GAS5 and SOX11 in HCC patients and discovered that both GAS5 and SOX11 were remarkably upregulated in HCC patients (Figure 6B and C). Then, we examined whether SOX11 expression influenced the prognosis of HCC patients, and Kaplan-Meier analysis revealed that SOX11 overexpression was associated with poor OS in HCC patients (Figure 6D). Moreover, spearman correlation analysis revealed that CDKN2A expression was positively related to SOX11 expression (Figure 6E).
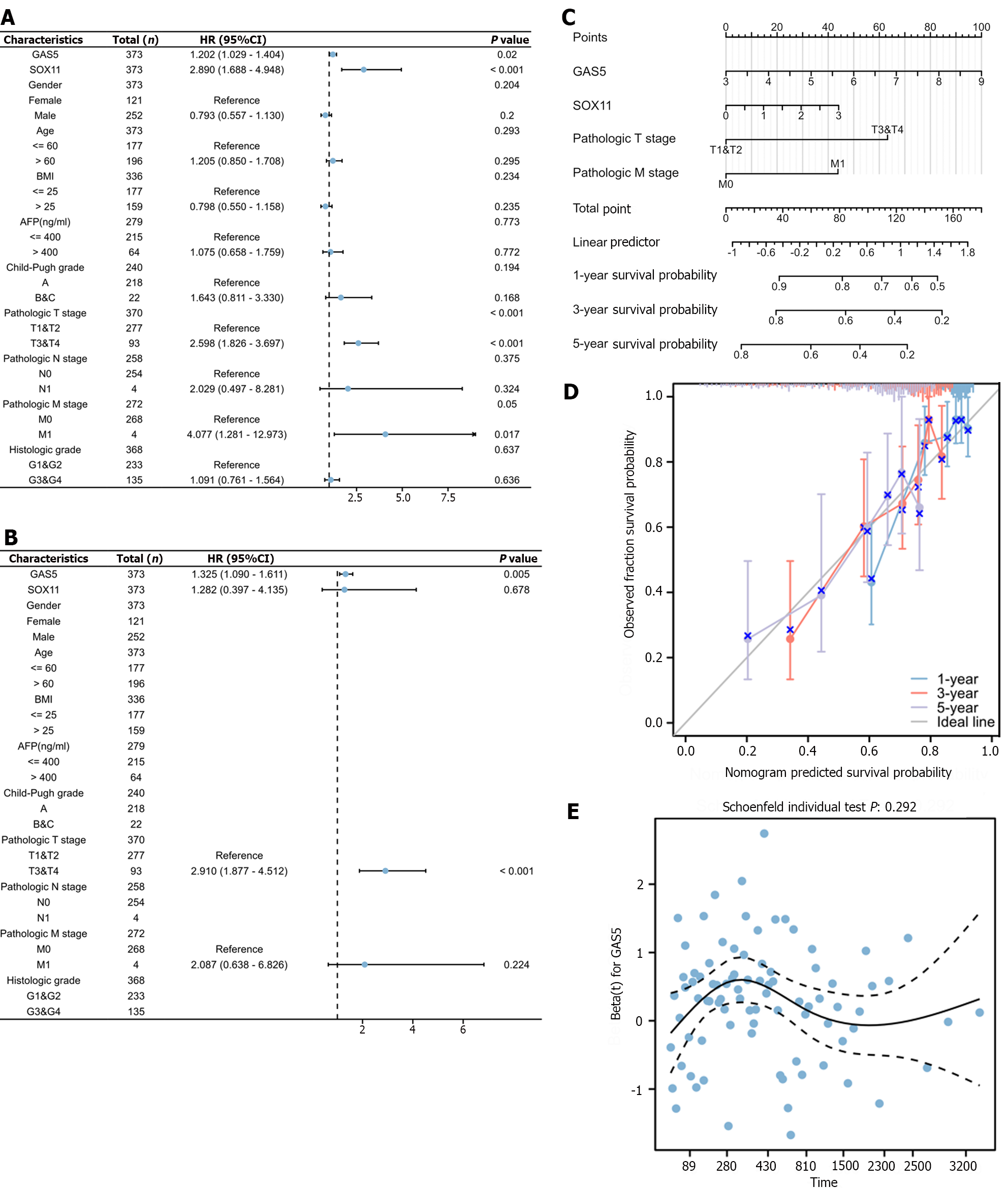
GAS5 expression, SOX11 expression, and clinical characteristics were subjected to univariate and multivariate Cox analyses to identify independent prognostic factors. The univariate Cox regression analysis (Figure 7A) revealed that tumor diameter, distant metastasis, GAS5 expression, and SOX11 expression were tightly connected with a poor OS in HCC patients. The multivariate Cox regression analysis (Figure 7B) suggested that the only factors still associated with OS were tumor diameter and GAS5 expression. All variables in the multivariate Cox regression model were incorporated to establish a nomogram (Figure 7C). The calibration curve (Figure 7D) demonstrated the nomogram’s superior prediction ability. In addition, the hazard proportional curve (Figure 7E) showed Cox regression coefficients for SNHG3 at various prognostic times.
Therefore, GAS5 might develop into an independent prognostic factor for HCC patients.
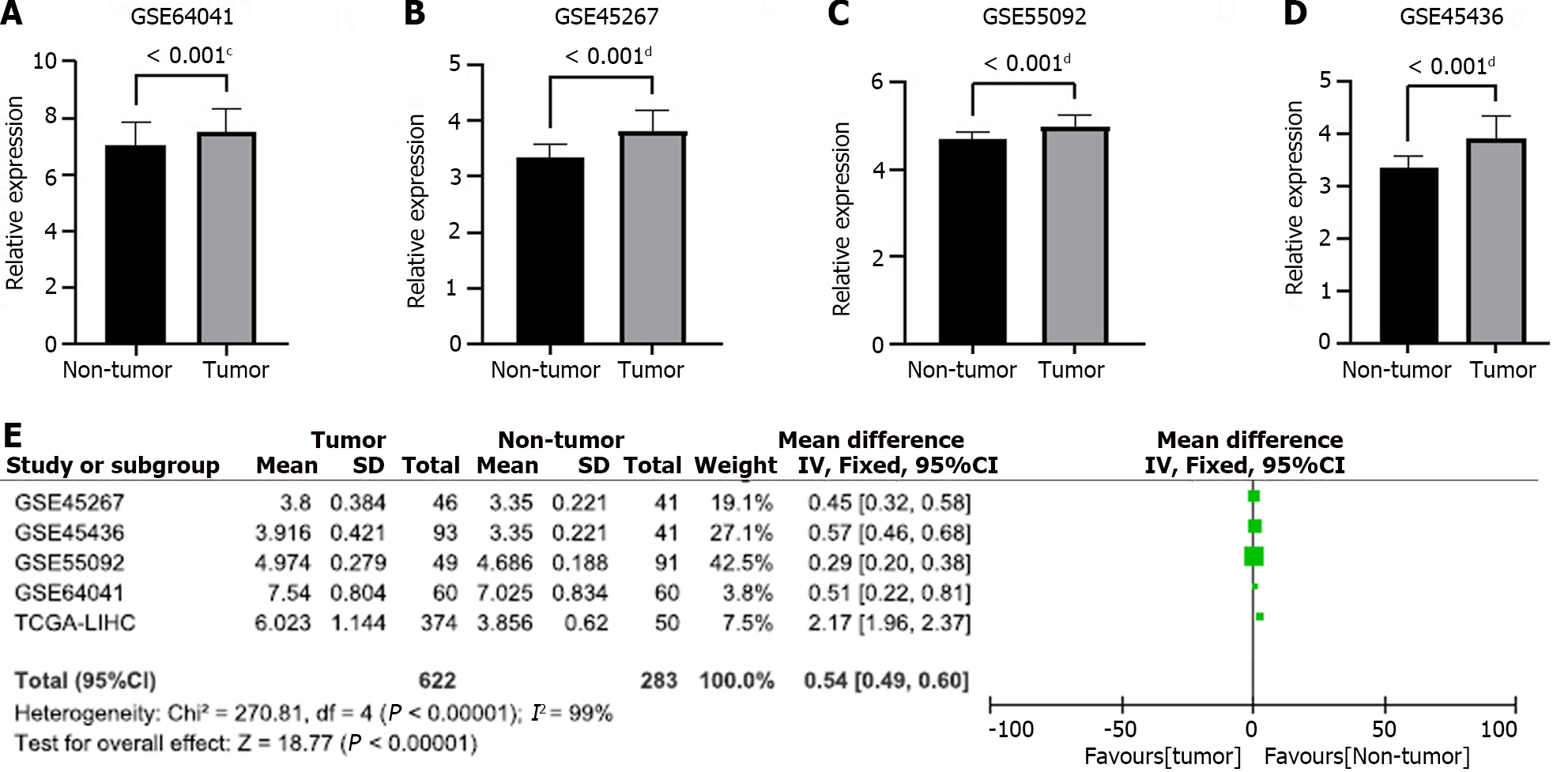
To verify that GAS5 was overexpressed in HCC tumor, we collected four independent GEO datasets (GSE64041, GSE45267, GSE55092, and GSE45436) and carried out a meta-analysis to aggregate research results. In GEO datasets, GAS5 exhibited a significant overexpression trend in HCC tumor tissues (GSE64041: t = 3.443, P = 0.0008; GSE45267: t = 6.596, P < 0.0001; GSE55092: t = 7.257, P < 0.0001; GSE45436: t = 4.220, P < 0.0001; Figure 8A-D). Because several datasets (including the TCGA-LIHC cohort) differ from one another, we used a random effects model in our meta-analysis. After combining all outcomes, we concluded that GAS5 was significantly overexpressed in HCC tissues (Figure 8E), which revealed the role of GAS5 as an oncogene.
To better understand the methylation level of GAS5 in HCC, we assessed the methylation of all hub genes through the GSCALite database. It is revealed that GAS5 expression was most correlated with methylation level among all hub genes (Figure 9A). We also discovered that the methylation level of GAS5 in normal samples was higher than that in HCC tumor samples both in the UALCAN database (Figure 9B) and the DiseaseMeth database (Figure 9C), which proved differential expression of GAS5 in HCC and normal samples from another point of view since methylation could inhibit gene expression. Furthermore, we detected ten methylation sites (cg08947084, cg06644515, cg03044573, cg16290996, cg17025683, cg07177756, cg05396044, cg03584783, cg06704773, and cg17868533) in the DNA sequences of GAS5 had a negative correlation with their expression at a standard of P value < 0.001 (Figure 9D).
As shown in Figure 10A, infiltration abundance of CD8+ T cells, macrophages, neutrophils, and dendritic cells were possibly associated with GAS5 gene copy number. In addition, we detected that GAS5 expression was remarkably positively associated with tumor purity and infiltration abundance of B cells, CD4+ T cells, CD8+ T cells, macrophages, neutrophils, and dendritic cells (Figure 10B).
Besides, we explored the relationship between GAS5 expression and the expression of PDCD1, CD274, and HAVCR2 in HCC patients, respectively. The GAS5 expression showed a positive correlation with PDCD1 expression and a negative correlation with CD274 expression, while GAS5 expression bore no notable association with HAVCR2 expression (Figure 10C).
To find out potential TCM active ingredients targeting GAS5, we acquired chemical components of TCM that may correlate with GAS5 using the CTD database. The top 10 chemicals were listed as follow: Valproic acid, bisphenol A, carbon tetrachloride, 1-methyl-4-phenylpyridinium, acetaminophen, tetrachlorodibenzodioxin, trichostatin A, 4-(5-benzo (1,3) dioxol-5-yl-4-pyridin-2-yl-1H-imidazol-2-yl) benzamide, aflatoxin B, and dorsomorphin. The structure of Valproic Acid, bisphenol A, 1-Methyl-4-phenylpyridinium, and Acetaminophen was visualized by the PubChem database (Supplementary Figure 2).
HCC pathophysiology is a multistep, intricate process, which entails gene mutation and signaling pathway abnormalities. Although substantial advancements have been made in diagnosing and managing HCC, additional studies are still required to understand the disease’s potential molecular mechanism fully and to enhance the prognosis for HCC patients. Herein, considering that CDKN2A was frequently mutated or deleted in HCC, we established a CKND2A-related ceRNA network-GAS5/miR-25-3p/SOX11, which might help to illuminate the molecular process of HCC carcinogenesis and identified GAS5 as an independent prognostic biomarker for HCC.
We first discovered that CDKN2A exhibited a relatively high mutation frequency in HCC patients. Cancer is featured in aberrant and limitless cell proliferation caused by genetic mutations, which are called “drivers” for they drive tumorigenesis, and their mutation form affects the homeostasis of a collection of crucial cell functions[29]. The single-cell analysis re-indicated that CDKN2A participated in cell cycle pathways in HCC. Therefore, CDKN2A was most likely to engage in the progression of HCC and was chosen for further study. Interestingly, CDKN2A, also called multiple tumor suppressor 1, has been proven to be the most common somatic mutated gene in metastatic tumors. If the CDKN2A gene is mutated or missing, the inhibition of the cyclinD-CDK4 complex is lifted, allowing it to function as an inhibitor of retinoblastoma protein and removing barriers in the cell cycle, which provides the cell unlimited proliferative capacity.
The significance of ceRNA network in the development and spread of malignancies has been increasingly clear in recent years. For example, several studies have found that the expression of key oncogene PTEN was downregulated by ceRNAs (e.g., PTEN pseudogene, VAPA, BCL11B, etc.), thus leading to the development of lung adenocarcinoma[30], renal clear cell carcinoma[31], and glioma[32]. Nevertheless, the predictive significance of ceRNA for HCC remains incompletely understood, and scant research has pinpointed ceRNA regulatory networks associated with CDKN2A that influence patient survival in HCC patients. Accordingly, we intended to construct a CDKN2A-related ceRNA network that likely predicts the prognosis of HCC and provides a potential target for the therapy of HCC.
First, after excluding lncRNAs whose subcellular localization was the nucleus, the lncRNA-miRNA-mRNA triple regulatory network containing one lncRNA (GAS5), one miRNA (miR-25-3p), and 63 mRNAs was acquired by bioinformatic analysis. Next, we identified the hub ceRNA network using the cytoHubba plug-in based on the MCC score. Functional analysis and gene network revealed that 15 hub genes might participate in the positive regulation of cell development. After differential expression analysis and survival analysis of 15 hub genes in HCC, we discovered that only differential expression of SOX11 was significantly linked to the prognosis of HCC patients. Furthermore, we demonstrated consistent expression trends of GAS5 and SOX11 in HCC patients, which lived up to the features of ceRNAs. Thus, CDKN2A-related ceRNA regulatory network- GAS5/miR-25-3p/SOX11 axis was established.
Based on published data, we observed that studies into the roles of GAS5, miR-25a-3p, and SOX11 in cancer or their connections to cancer have taken place. Hsieh et al[33] evaluated the influences of lncRNA GAS5 genetic variants on the clinicopathological characteristics of lung adenocarcinoma. Similarly, miR-25-3p was also identified as a stable tumor biomarker for its knockdown suppressed breast cancer cell proliferation and invasion[34]. As for SOX11, many studies have demonstrated that SOX11 participated in various cancer progression, such as lung adenocarcinoma[35], breast cancer[36], bladder cancer[37], neuroblastoma[38]. However, few studies have explored the role of ceRNA regulatory networks containing GAS5, miR-25-3p, or SOX11 in HCC.
In this study, we performed a clinical correlation analysis of the GAS5/miR-25-3p/SOX11 axis in HCC patients. GAS5 and SOX11 were markedly correlated with the OS of HCC patients in univariate Cox regression, and only GAS5 was remarkably linked to the OS of HCC patients in multivariate Cox regression. Hence, GAS5 was more likely to be an independent prognostic indicator for HCC patients. Besides, we established a nomogram containing all variables in the multivariate Cox regression model, exhibiting an excellent predictive capacity. We also demonstrated GAS5 abnormally high expression in four independent GEO datasets to improve the credibility of the conclusion. To further investigate the potential clinical significance of GAS5 for HCC treatment, we carried out methylation analysis, immune infiltration analysis and identified TCM effective drugs targeting GAS5. We validated the abnormal methylation level of GAS5 in HCC and detected ten methylation sites in the DNA sequences of GAS5. Both copy number and expression of GAS5 were tightly associated with the abundance of some immune cells, such as CD8+ cells, macrophages, neutrophils, and dendritic cells. Moreover, we discovered that GAS5 expression was positively linked to PDCD1 expression and negatively correlated with CD274 expression. These findings indicated that these differences induced by the GAS5/miR-25-3p/SOX11 axis might influence the tumor immune microenvironment, thus promoting the progression of HCC. Since Chinese herbal medicine was considered an essential source for discovering innovational medications[39,40], we identified ten potential TCM chemical components, which promisingly become potent methods for the treatment of HCC patients.
GAS5 has been widely considered as a potential cancer suppressor gene, with its downregulation associated with cancer in multiple different tissues[41-43]. In our study, however, we discovered that GAS5 was overexpressed in HCC tumor tissues, and its overexpression led to a poor prognosis of HCC patients. There was also some contradictory information about GAS5’s correlation with HCC. Two previous studies pointed out that downregulation of GAS5 promoted tumor progression in HCC[44,45]. In contrast, a study using bioinformatic tools revealed that GAS5 overexpression was significantly linked to shorter relapse free survival in the TCGA-LIHC cohort[46]. Similarly, Li et al[47] suggested that gain of GAS5 was an independent indicator of poor prognosis in HCC from the TCGA-LIHC dataset. Hence, we predicted that GAS5 might have its distinctive function and involve in the occurrence and progression of LIHC. Besides, GAS5 might exhibit different roles based on different genetic backgrounds.
Of course, these findings need to be demonstrated by detailed experiments in vivo and in vitro in the future, and there are still some defects in our study. First, although we validated the robustness of the predictive effect of GAS5 in the TCGA cohort, more prospective studies with a suitably high sample size are needed to confirm each other as this study was solely retrospective. Second, we are supposed to detect the function and mechanism of the GAS5/miR-25-3p/SOX11 axis in HCC by in vivo and in vitro experiments.
In conclusion, we constructed a CDKN2A-related ceRNA network-GAS5/miR-25-3p/SOX11 for HCC patients, which could possibly serve as a prognostic indicator for clinical outcome and therapeutic responsiveness, provide innovative insights into the molecular basis of HCC initiation and development. Besides, regulating GAS5 expression might be an effective therapeutic strategy in inducing immune infiltration and immunotherapy for HCC.
Hepatocellular carcinoma (HCC) is a tumor with multi-etiology and multi-pathway involvement, and it is characterized by a low 5- year survival rate. Competitive endogenous RNA (ceRNA) is an innovative way of gene expression modulation, which plays a crucial part in HCC. Nevertheless, the complexity and behavioral features of the ceRNA network in HCC are not fully elucidated.
To construct a cyclin dependent kinase inhibitor 2A (CDKN2A)-related ceRNA network in HCC.
To establish a CDKN2A-related ceRNA network in HCC and investigate the effect of the ceRNA on the prognosis of HCC.
TCGA database, GEO database, cBioPortal database, lncBasev.3 database, miRDB database, Targetscan database, lncATLAS database, principal component analysis, The Gene Ontology enrichment, Kyoto Encyclopedia of Genes and Genomes pathway, meta-analysis, Kaplan-Meier plotter, nomogram, calibration curve, univariate and multivariate Cox regression, Spearman correlation coefficient, GSCALite database, UALCAN database, DiseaseMeth version 2.0 database, MEXPRESS database, TIMER database, CTD database, and PubChem database were used in this study.
CDKN2A was frequently mutated and deleted in and participated in cell cycle pathways in HCC. The CDKN2A-related ceRNA network- Growth arrest specific 5 (GAS5)/miR-25-3p/SRY-box transcription factor 11 (SOX11) was successfully established. GAS5 was recognized as an independent prognostic biomarker for HCC patients. GAS5 expression was correlated with methylation level, immune infiltration, and the expression of three immune checkpoint genes in HCC.
The CDKN2A-related ceRNA network-GAS5/miR-25-3p/SOX11 was successfully established and GAS5 was an independent prognostic indicator for HCC patients.
The CDKN2A-related ceRNA network provides innovative insights into the molecular mechanism of HCC formation and progression. Moreover, regulating GAS5 expression might be an effective therapeutic strategy in inducing immune infiltration and immunotherapy for HCC.
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 68628] [Article Influence: 13725.6] [Reference Citation Analysis (201)] |
| 2. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 56675] [Article Influence: 7084.4] [Reference Citation Analysis (135)] |
| 3. | Huang W, Skanderup AJ, Lee CG. Advances in genomic hepatocellular carcinoma research. Gigascience. 2018;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 4. | Dominguez DA, Wang XW. Impact of Next-Generation Sequencing on Outcomes in Hepatocellular Carcinoma: How Precise Are We Really? J Hepatocell Carcinoma. 2020;7:33-37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 5. | Caruso S, O'Brien DR, Cleary SP, Roberts LR, Zucman-Rossi J. Genetics of Hepatocellular Carcinoma: Approaches to Explore Molecular Diversity. Hepatology. 2021;73 Suppl 1:14-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 65] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 6. | Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18694] [Cited by in RCA: 21466] [Article Influence: 1951.5] [Reference Citation Analysis (6)] |
| 7. | Brown ZJ, Greten TF, Heinrich B. Adjuvant Treatment of Hepatocellular Carcinoma: Prospect of Immunotherapy. Hepatology. 2019;70:1437-1442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 84] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 8. | Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4127] [Cited by in RCA: 5734] [Article Influence: 382.3] [Reference Citation Analysis (0)] |
| 9. | Karreth FA, Reschke M, Ruocco A, Ng C, Chapuy B, Léopold V, Sjoberg M, Keane TM, Verma A, Ala U, Tay Y, Wu D, Seitzer N, Velasco-Herrera Mdel C, Bothmer A, Fung J, Langellotto F, Rodig SJ, Elemento O, Shipp MA, Adams DJ, Chiarle R, Pandolfi PP. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell. 2015;161:319-332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 269] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 10. | Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2305] [Cited by in RCA: 3189] [Article Influence: 265.8] [Reference Citation Analysis (0)] |
| 11. | Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272-283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1116] [Cited by in RCA: 1638] [Article Influence: 163.8] [Reference Citation Analysis (0)] |
| 12. | Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3:1113-1121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 504] [Cited by in RCA: 694] [Article Influence: 53.4] [Reference Citation Analysis (0)] |
| 13. | Shi Y, Zhang DD, Liu JB, Yang XL, Xin R, Jia CY, Wang HM, Lu GX, Wang PY, Liu Y, Li ZJ, Deng J, Lin QL, Ma L, Feng SS, Chen XQ, Zheng XM, Zhou YF, Hu YJ, Yin HQ, Tian LL, Gu LP, Lv ZW, Yu F, Li W, Ma YS, Da F. Comprehensive analysis to identify DLEU2L/TAOK1 axis as a prognostic biomarker in hepatocellular carcinoma. Mol Ther Nucleic Acids. 2021;23:702-718. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 14. | Li G, Wang Z, Chen D, Yin J, Mo Z, Sun B, Yang T, Zhang X, Zhai Z, Li Y, Chen P, Dai Y, Ma J. Comprehensive analysis of a TPX2-related TRHDE-AS1/PKIA ceRNA network involving prognostic signatures in Hepatitis B virus-infected hepatocellular carcinoma. Front Cell Infect Microbiol. 2022;12:1025900. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 15. | Hao F, Wang N, Gui H, Zhang Y, Wu Z, Wang J. Pseudogene UBE2MP1 derived transcript enhances in vitro cell proliferation and apoptosis resistance of hepatocellular carcinoma cells through miR-145-5p/RGS3 axis. Aging (Albany NY). 2022;14:7906-7925. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 16. | Liggett WH, Sidransky D. Role of the p16 tumor suppressor gene in cancer. J Clin Oncol. 1998;16:1197-1206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 460] [Cited by in RCA: 488] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 17. | Tsvetkov P, Coy S, Petrova B, Dreishpoon M, Verma A, Abdusamad M, Rossen J, Joesch-Cohen L, Humeidi R, Spangler RD, Eaton JK, Frenkel E, Kocak M, Corsello SM, Lutsenko S, Kanarek N, Santagata S, Golub TR. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science. 2022;375:1254-1261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 282] [Cited by in RCA: 3060] [Article Influence: 765.0] [Reference Citation Analysis (1)] |
| 18. | Zhu C, Soto-Feliciano YM, Morris JP, Huang CH, Koche RP, Ho YJ, Banito A, Chen CW, Shroff A, Tian S, Livshits G, Chen CC, Fennell M, Armstrong SA, Allis CD, Tschaharganeh DF, Lowe SW. MLL3 regulates the CDKN2A tumor suppressor locus in liver cancer. Elife. 2023;12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 19. | Liu H, Jia S, Guo K, Li R. INK4 cyclin-dependent kinase inhibitors as potential prognostic biomarkers and therapeutic targets in hepatocellular carcinoma. Biosci Rep. 2022;42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 20. | Fu Y, Yang Z, Hu Z, Pan Y, Chen J, Wang J, Hu D, Zhou Z, Xu L, Chen M, Zhang Y. Preoperative serum ctDNA predicts early hepatocellular carcinoma recurrence and response to systemic therapies. Hepatol Int. 2022;16:868-878. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 32] [Reference Citation Analysis (0)] |
| 21. | Desjonqueres E, Campani C, Marra F, Zucman-Rossi J, Nault JC. Preneoplastic lesions in the liver: Molecular insights and relevance for clinical practice. Liver Int. 2022;42:492-506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 22. | Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7222] [Cited by in RCA: 10484] [Article Influence: 806.5] [Reference Citation Analysis (4)] |
| 23. | Mas-Ponte D, Carlevaro-Fita J, Palumbo E, Hermoso Pulido T, Guigo R, Johnson R. LncATLAS database for subcellular localization of long noncoding RNAs. RNA. 2017;23:1080-1087. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 203] [Cited by in RCA: 254] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 24. | Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11591] [Cited by in RCA: 24970] [Article Influence: 1783.6] [Reference Citation Analysis (2)] |
| 25. | Franz M, Rodriguez H, Lopes C, Zuberi K, Montojo J, Bader GD, Morris Q. GeneMANIA update 2018. Nucleic Acids Res. 2018;46:W60-W64. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 341] [Cited by in RCA: 1023] [Article Influence: 146.1] [Reference Citation Analysis (1)] |
| 26. | Liu CJ, Hu FF, Xia MX, Han L, Zhang Q, Guo AY. GSCALite: a web server for gene set cancer analysis. Bioinformatics. 2018;34:3771-3772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 307] [Cited by in RCA: 361] [Article Influence: 45.1] [Reference Citation Analysis (0)] |
| 27. | Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017;77:e108-e110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2728] [Cited by in RCA: 4315] [Article Influence: 479.4] [Reference Citation Analysis (0)] |
| 28. | He X, Xu C. Immune checkpoint signaling and cancer immunotherapy. Cell Res. 2020;30:660-669. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 231] [Cited by in RCA: 972] [Article Influence: 162.0] [Reference Citation Analysis (0)] |
| 29. | Martínez-Jiménez F, Muiños F, Sentís I, Deu-Pons J, Reyes-Salazar I, Arnedo-Pac C, Mularoni L, Pich O, Bonet J, Kranas H, Gonzalez-Perez A, Lopez-Bigas N. A compendium of mutational cancer driver genes. Nat Rev Cancer. 2020;20:555-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 385] [Cited by in RCA: 843] [Article Influence: 140.5] [Reference Citation Analysis (0)] |
| 30. | Xin R, Shen B, Jiang YJ, Liu JB, Li S, Hou LK, Wu W, Jia CY, Wu CY, Fu D, Ma YS, Jiang GX. Comprehensive analysis to identify a novel PTEN-associated ceRNA regulatory network as a prognostic biomarker for lung adenocarcinoma. Front Oncol. 2022;12:923026. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 31. | Zhou X, Liu G, Xu M, Ying X, Li B, Cao F, Cheng S, Xiao B, Cheng M, Liang L, Jia M, Li W, Liu J, Li Z. Comprehensive analysis of PTEN-related ceRNA network revealing the key pathways WDFY3-AS2 - miR-21-5p/miR-221-3p/miR-222-3p - TIMP3 as potential biomarker in tumorigenesis and prognosis of kidney renal clear cell carcinoma. Mol Carcinog. 2022;61:508-523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 32. | Liu Y, Chen S, Peng G, Liao Y, Fan X, Zhang Z, Shen C. CircRNA NALCN acts as an miR-493-3p sponge to regulate PTEN expression and inhibit glioma progression. Cancer Cell Int. 2021;21:307. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 33. | Hsieh MH, Wu YL, Tsao TC, Huang YW, Lin JC, Lee CY, Hsieh MJ, Yang SF. Impact of LncRNA GAS5 Genetic Variants and the Epidermal Growth Factor Receptor Phenotypes on the Clinicopathological Characteristics of Lung Adenocarcinoma Patients. Int J Environ Res Public Health. 2022;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 34. | Zhao T, Meng W, Chin Y, Gao L, Yang X, Sun S, Pan X, He L. Identification of miR253p as a tumor biomarker: Regulation of cellular functions via TOB1 in breast cancer. Mol Med Rep. 2021;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 35. | Ma Z, Chen G, Chen Y, Guo Z, Chai H, Tang Y, Zheng L, Wei K, Pan C, Ma Z, Xia Y, Zhang A. MiR-937-3p promotes metastasis and angiogenesis and is activated by MYC in lung adenocarcinoma. Cancer Cell Int. 2022;22:31. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 36. | Pan JK, Lin CH, Kuo YL, Ger LP, Cheng HC, Yao YC, Hsiao M, Lu PJ. MiR-211 determines brain metastasis specificity through SOX11/NGN2 axis in triple-negative breast cancer. Oncogene. 2021;40:1737-1751. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 37. | Wu Z, Huang W, Wang X, Wang T, Chen Y, Chen B, Liu R, Bai P, Xing J. Circular RNA CEP128 acts as a sponge of miR-145-5p in promoting the bladder cancer progression via regulating SOX11. Mol Med. 2018;24:40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 75] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 38. | Wang J, Wang Z, Lin W, Han Q, Yan H, Yao W, Dong R, Jia D, Dong K, Li K. LINC01296 promotes neuroblastoma tumorigenesis via the NCL-SOX11 regulatory complex. Mol Ther Oncolytics. 2022;24:834-848. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 39. | Luo H, Vong CT, Chen H, Gao Y, Lyu P, Qiu L, Zhao M, Liu Q, Cheng Z, Zou J, Yao P, Gao C, Wei J, Ung COL, Wang S, Zhong Z, Wang Y. Naturally occurring anti-cancer compounds: shining from Chinese herbal medicine. Chin Med. 2019;14:48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 296] [Cited by in RCA: 389] [Article Influence: 55.6] [Reference Citation Analysis (0)] |
| 40. | Ma X, Hu M, Wang H, Li J. Discovery of traditional Chinese medicine monomers and their synthetic intermediates, analogs or derivatives for battling P-gp-mediated multi-drug resistance. Eur J Med Chem. 2018;159:381-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 38] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 41. | Wu S, Ren K, Zhao J, Li J, Jia B, Wu X, Dou Y, Fei X, Huan Y, He X, Wang T, Lv W, Wang L, Wang Y, Fei Z, Li S. LncRNA GAS5 represses stemness and malignancy of gliomas via elevating the SPACA6-miR-125a/let-7e Axis. Front Oncol. 2022;12:803652. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 42. | Xie J, Wang JJ, Li YJ, Wu J, Gu XJ, Yang XR. LncRNA GAS5 Suppresses Colorectal Cancer Progress by Target miR-21/LIFR Axis. Evid Based Complement Alternat Med. 2022;2022:3298939. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 43. | Tu J, Tan X, Chen Y, Li Z, Zhang Y, Chen X, Yang H, Chen H, Yu Z. Growth arrest-specific transcript 5 represses endometrial cancer development by promoting antitumor function of tumor-associated macrophages. Cancer Sci. 2022;113:2496-2512. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 44. | Zhang F, Yang C, Xing Z, Liu P, Zhang B, Ma X, Huang L, Zhuang L. LncRNA GAS5-mediated miR-1323 promotes tumor progression by targeting TP53INP1 in hepatocellular carcinoma. Onco Targets Ther. 2019;12:4013-4023. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 45. | Wang C, Ke S, Li M, Lin C, Liu X, Pan Q. Downregulation of LncRNA GAS5 promotes liver cancer proliferation and drug resistance by decreasing PTEN expression. Mol Genet Genomics. 2020;295:251-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 46. | Zhu Q, Yang H, Cheng P, Han Q. Bioinformatic analysis of the prognostic value of the lncRNAs encoding snoRNAs in hepatocellular carcinoma. Biofactors. 2019;45:244-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 47. | Li J, Li Y, He X, Zhao Q. Gain of GAS5 reveals worse prognosis in kidney renal clear cell carcinoma and liver hepatocellular carcinoma from the Cancer Genome Atlas dataset. Transl Cancer Res. 2021;10:223-232. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Gupta T, India S-Editor: Chen YL L-Editor: A P-Editor: Cai YX