Published online Apr 15, 2024. doi: 10.4251/wjgo.v16.i4.1421
Peer-review started: December 8, 2023
First decision: December 21, 2023
Revised: January 4, 2024
Accepted: February 7, 2024
Article in press: February 7, 2024
Published online: April 15, 2024
Processing time: 124 Days and 12.9 Hours
Metabolic reprogramming plays a key role in cancer progression and clinical outcomes; however, the patterns and primary regulators of metabolic repro
To explore the role of nicotinamide adenine dinucleotide phosphate oxidase 4 (NOX4) in promoting progression of CRC.
We evaluated the expression and function of dysregulated and survival-related metabolic genes using Gene Ontology and Kyoto Encyclopedia of Genes and Genomes. Consensus clustering was used to cluster CRC based on dysregulated metabolic genes. A prediction model was constructed based on survival-related metabolic genes. Sphere formation, migration, invasion, proliferation, apoptosis and clone formation was used to evaluate the biological function of NOX4 in CRC. mRNA sequencing was utilized to explore the alterations of gene expression NOX4 over-expression tumor cells. In vivo subcutaneous and lung metastasis mouse tumor model was used to explore the effect of NOX4 on tumor growth.
We comprehensively analyzed 3341 metabolic genes in CRC and identified three clusters based on dysregulated metabolic genes. Among these genes, NOX4 was highly expressed in tumor tissues and correlated with worse survival. In vitro, NOX4 overexpression induced clone formation, migration, invasion, and stemness in CRC cells. Furthermore, RNA-sequencing analysis revealed that NOX4 overexpression activated the mitogen-activated protein kinase-MEK1/2-ERK1/2 signaling pathway. Trametinib, a MEK1/2 inhibitor, abolished the NOX4-mediated tumor progression. In vivo, NOX4 overexpression promoted subcutaneous tumor growth and lung metastasis, whereas trametinib treatment can reversed the metastasis.
Our study comprehensively analyzed metabolic gene expression and highlighted the importance of NOX4 in promoting CRC metastasis, suggesting that trametinib could be a potential therapeutic drugs of CRC clinical therapy targeting NOX4.
Core Tip: We first identified three clusters with different survival status based on dysregulated metabolic genes in colorectal cancer (CRC). In addition, based on differentially expressed survival-related metabolic genes, we constructed and validated a prediction model using different cohorts. Among these genes, nicotinamide adenine dinucleotide phosphate oxidase 4 (NOX4) overexpression activated the mitogen-activated protein kinase-MEK1/2-ERK1/2 signaling pathway to promote cancer metastasis, whereas trametinib (a MEK1/2 inhibitor) treatment reversed this effect. Our study analyzed metabolic gene expression and highlighted the importance of NOX4 in promoting CRC metastasis, suggesting that NOX4 could be a new therapeutic target and modulating the response to clinical therapy.
- Citation: Xu YJ, Huo YC, Zhao QT, Liu JY, Tian YJ, Yang LL, Zhang Y. NOX4 promotes tumor progression through the MAPK-MEK1/2-ERK1/2 axis in colorectal cancer. World J Gastrointest Oncol 2024; 16(4): 1421-1436
- URL: https://www.wjgnet.com/1948-5204/full/v16/i4/1421.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i4.1421
Colorectal cancer (CRC) is the third most common cause of cancer-related death, accounting for approximately 10% of all cancers diagnosed worldwide[1]. For early CRC, treatment options include endoscopic and surgical approaches, while for patients with advanced stage CRC, such as fluoropyrimidine-based chemotherapy, radiotherapy, have been used to improve the survival rate[2-5]. In addition, targeted therapies and immunotherapies have been used to treat advanced and metastatic CRC[6,7]. However, the majority of patients do not respond to these therapies[8]. This outcome is partially due to the tumor metabolic reprogramming.
Metabolic reprogramming is a hallmark of cancer and is responsible for the ability of tumor cells to evade drug therapy[9,10]. The first recognized concept of metabolic reprogramming was called Warburg effect[11]. Warburg effect is the enhanced conversion of glucose to pyruvate even in the presence of abundant oxygen[12]. Additionally, in KRAS-mutant CRC, solute carrier family 7, member 5 maintains intracellular amino acid levels required for tumor growth[13]. V-9302, an antagonist of SLC1A5, was reported to reduce tumor growth in the mouse models of breast and liver cancers[14,15]. To date, numerous metabolic genes have been identified as potential drug candidates; however, off-target side effects and metabolic plasticity add to the challenges of drug development. Thus, a comprehensive understanding of the metabolic patterns and identification of key molecules in CRC are essential for targeting cancer metabolism.
Nicotinamide adenine dinucleotide phosphate oxidase 4 (NOX4) is an aberrantly-expressed metabolic gene that correlated with tumor progression. NOX4 belongs to the NOX family, and is the major oxidase involved in the production of reactive oxygen species (ROS)[16]. NOX4 is widely expressed and is involved in the regulation of numerous cell functions[17,18]. Furthermore, it has been proposed that an equilibrium between the production and elimination of ROS is required for the initiation and progression of cancer[16], suggesting that the expression of NOX4 is linked to tumor progression.
Here, we performed a comprehensive analysis of aberrant expression of metabolic gene and signaling pathways involved in CRC, and further classified the patients into three clusters based on different metabolic gene patterns. A prediction model was constructed to predict survival of patients with CRC based on survival-related genes. We found that NOX4 overexpression promoted CRC progression and metastasis via mitogen-activated protein kinase (MAPK)-MEK1/2-ERK1/2 signaling.
The RNA sequencing data and corresponding clinical information of CRC samples from The Cancer Genome Atlas (TCGA) Program were downloaded from the online database UCSC Xena (xena.ucsc.edu) with log2(RSEM + 1) format. Metabolic genes were selected by integrating two previously published datasets[19,20]. The colon cancer microarray expression data and corresponding clinical information were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) with accession number GSE17536.
The R package “limma” was used to identify the metabolic differentially expressed genes (DEGs) between CRC and normal tissues, and the genes with an adjusted P-value < 0.05 and log2-fold change |log2 FC)| > 1 were visualized using a volcano map. The top ten dysregulated metabolic genes (ranked by adjusted P-value) were analyzed using the R package “ggplot2”. The R packages “clusterProfiler”, “org.Hs.eg.db”, and “enrichplot” were used to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) of DEGs, and significantly enrichment GO and KEGG categories (P-value < 0.05 and q-value < 0.05) were selected.
The R package “ConsensusClusterPlus” was used to cluster CRC based on metabolic DEGs. Clusters were visualized using “pheatmap”. Pie and bar plots were used to display patient clinical parameters and the R package “survival” was used to analyze differences of overall survival (OS) and progression free interval (PFI) across the three clusters.
Tumor samples from TCGA were randomly divided into training and test groups in a 1:1 ratio based on survival status. Univariate survival analysis was performed for each gene to select candidate metabolic genes related to OS in the training datasets. The least absolute shrinkage and selection operator (Lasso) algorithm was used to optimize the gene signatures, and multivariate Cox regression analysis was used to calculate the formula for the prediction model. The R packages “survival”, “survminer”, and “survival ROC” were used to analyze the differences of survival and receiver operating characteristic (ROC) curve between high- and low-risk groups. Only gene signatures from prediction model were used for validating in our clinical samples and GEO datasets.
Pairs of freshly frozen samples of primary CRC and adjacent tumor tissues (n = 82) were obtained from patients who underwent surgical resection at the First Affiliated Hospital of Zhengzhou University. None of the patients received preoperative chemotherapy, radiotherapy, or other tumor-specific treatments. Total of 82 patients were followed up through an interview at the clinic or by telephone. The clinicopathological parameters of patients with CRC are summarized in Table 1.
| Factor | Category | Total | High NOX4 (N = 52) | Low NOX4 (N = 30) | P value |
| Age (yr) | 64.46 ± 12.6 | 64.79 ± 12.38 | 63.9 ± 13.18 | 0.765 | |
| Gender | Female | 49 (60) | 31 (60) | 18 (60) | 1.000 |
| Male | 33 (40) | 21 (40) | 12 (40) | ||
| Tumor location | Left colon or rectum | 51 (62) | 35 (67) | 16 (53) | 0.307 |
| Right colon | 31 (38) | 17 (33) | 14 (47) | ||
| Histopathology | Well differentiated | 31 (38) | 17 (33) | 14 (47) | 0.373 |
| Moderately differentiated | 32 (39) | 23 (44) | 9 (30) | ||
| Poorly differentiated | 19 (23) | 12 (23) | 7 (23) | ||
| pT | T1 | 17 (21) | 11 (21) | 6 (20) | 0.357 |
| T2 | 26 (32) | 13 (25) | 13 (43) | ||
| T3 | 24 (29) | 17 (33) | 7 (23) | ||
| T4 | 15 (18) | 11 (21) | 4 (13) | ||
| pN | N0 | 45 (55) | 25 (48) | 20 (67) | 0.233 |
| N1 | 26 (32) | 18 (35) | 8 (27) | ||
| N2 | 11 (13) | 9 (17) | 2 (7) | ||
| pM | M0 | 82 (100) | 52 (100) | 30 (100) | 1.000 |
| M1 | 0 | 0 | 0 | ||
| pStage | Stage I | 19 (23) | 5 (10) | 14 (47) | < 0.001 |
| Stage II | 24 (29) | 19 (37) | 5 (17) | ||
| Stage III | 39 (48) | 28 (54) | 11 (37) |
CRC cell line RKO was cultured in the DMEM (Dulbecco’s Modified Eagle’s Medium-high glucose) (Sigma, United States) supplemented with 10% fetal bovine serum, 100 U/mL penicillin and 100 μg/mL streptomycin in a 5% CO2 atmosphere at 37 °C. To overexpress NOX4 in RKO cells, the target gene fragment was amplified using the primers listed in Supplementary Table 1. The fragment was cloned into the lentiviral vector Ubi-MCS-3FLAG-CBh-gcGFP-IRES-puromycin (GeneChem Co., Ltd., Shanghai, China). Next, lentiviruses containing the target fragment or negative control were transfected into RKO cells. After 72 h, NOX4-overexpressing RKO cells were selected with puromycin (2 μg/mL; Santa Cruz).
Total RNA was extracted using the Trizol reagent, and 1 μg of total RNA was reverse-transcribed using the PrimeScript RT-PCR kit (Takara, Japan). Real-time quantitative polymerase chain reaction (RT-qPCR, Applied Biosystems, Grand Island, NY, United States) was performed using the SYBR Green method (Takara, Japan). The 2-ΔΔCt method was used to quantify the relative NOX4 expression levels. The primers used for RT-PCR were listed in Supplementary Table 2.
Whole cell lysates were prepared using RIPA lysis buffer containing a protease inhibitor cocktail (Sigma-Aldrich) and phosphatase inhibitor cocktail 2 (Sigma-Aldrich). Equal amounts of protein were loaded onto a 10% sodium-dodecyl sulfate gel electrophoresis gel and then transferred to a polyvinylidene fluoride membrane (Bio-Rad, Hercules, CA, United States). Membranes were blocked in 5% nonfat milk and incubated with primary antibodies overnight at 4 °C, followed by treatment with secondary antibody for 2 h at 37 °C. The primary antibodies were NOX4 (1:1000, ab133303, Abcam), phospho-p44/42 MAPK (Erk1/2) (1:2000, 4370T, Cell Signaling Technology), p44/42 MAPK (Erk1/2) (1:1000, 4695T, Cell Signaling Technology), MEK1/2 (1:1000, 8727T, Cell Signaling Technology), phospho-MEK1/2 (1:1000, 9154T, Cell Signaling Technology), β-actin (1:5000, 4967S, Cell Signaling Technology). Blots were imaged using enhanced chemiluminescence (Thermo Fisher, United States).
NOX4-overexpressing and negative control RKO cells were permeabilized with 0.3% Triton X-100 for 10 min and then washed. The sections were incubated with primary antibodies anti-NOX4 (1:500, Abcam) and anti-p-ERK (1:200, Abcam) overnight at 4 °C. Fluorescein isothiocyanate and Cy3-conjugated secondary antibodies (1:500, BioLegend) were used to detect the primary antibodies. The stained cells were counterstained with 4¢6-diamidino-2-phenylindole (1:1000; Roche, Basel, Switzerland) and analyzed using an IX71 inverted fluorescence microscope (model 1003; Olympus).
CRC tissues and corresponding adjacent tissues were evaluated for NOX4 and p-Erk expression using immunohistochemical (IHC) staining. Sections were treated with 3% hydrogen peroxide and 5% BSA, and then incubated with primary antibodies (NOX4, 1:100, Abcam, Cambridge; p-Erk, 1:200, Abcam) overnight at 4 °C. Next, the sections were incubated with horseradish peroxidase-conjugated secondary antibody for 1 h at 37 °C, counterstained with hematoxylin, and then visualized under a microscope (Olympus, Tokyo, Japan). NOX4 staining was semi-quantitatively evaluated using a grading system based on staining intensity (scored as follows: 0, no staining; 1, weak staining; 2, moderate staining; 3, strong staining) and the percentage of positive tumor cells (scored as follows: 0, none; 1, 1%-29%; 2, 30%-69%; 3, > 70%). The total IHC score for each tissue sample was calculated by multiplying the staining intensity score by the positive tumor cell score. The final score, which ranged from 0 to 9 was defined as follows: 0, negative (-); 1-3 (+), weak; 4-6 (++), moderate; and > 6, strong (+++).
The migration and invasion of NOX4-overexpressing and negative control RKO cells were evaluated using transwell assays. Briefly, 2 × 106 cells/well were cultured in foetal bovine serum-free DMEM in the upper chambers of a 8.0 μm pore transwell insert (Corning, United States) and treated with either MEK inhibitor (MEKi, trametinib, 100 nM) or vehicle control. For the invasion assay, the upper chambers of transwell plates were pre-coated with 40 μL Matrigel (1:8 dilution, BD Biosciences) at 37 °C for 30 min for gelling. Cells were cultured for 36 h, fixed with 4% paraformaldehyde (Sigma-Aldrich; Merck KGaA) at 37 °C for 20 min, and then stained with 0.5% crystal violet (Sigma-Aldrich; Merck KGaA) at 37 °C for 15 min. The number of invaded cells was counted under an optical microscope at 100 × magnification (five random fields per sample).
Cell viability of NOX4-overexpressing and negative-control RKO cells was evaluated using the cell counting kit-8 (CCK8, Dojindo Molecular Technologies, Japan). Briefly, the RKO cells were seeded into 96-well plates at a density of 5 × 103 cells/well. CCK8 solution was added to each well at 24, 48, and 72 h, and cells were incubated for 30 min at 37 °C. Absorbance at 450 nm was measured using a microplate reader (spectraMax iD3, PerkinElmer, United States). The cell cycle was examined using propidium iodide (PI) (BD Biosciences) staining. Cells were washed with cold phosphate buffered saline (PBS), fixed in 75% ethanol at -20 °C overnight, incubated with PI for 15 min, and then analyzed using flow cytometry.
NOX4-overexpressing and negative control RKO cells were seeded into 6-well plates at a density of 1 × 106 cells/well. Next, three evenly-spaced vertical lines were made in each well using a 200 μL enzyme-free yellow pipette tip. The culture medium was then removed, wells were washed three times with PBS, and 2 mL of serum-free DMEM was slowly added. Images at 0, 24, and 48 h time points were obtained using an optical microscope at 100 × magnification.
NOX4-overexpressing and negative control RKO cells were seeded into the 6-well plates at a density of 1 × 103 cells/mL and incubated in a 5% CO2 atmosphere at 37 °C. When the cell clones in the wells were visible by eye, the cells were fixed with 4% paraformaldehyde (Sigma-Aldrich; Merck KGaA) at 37 °C for 20 min, stained with 0.5% crystal violet (Sigma-Aldrich; Merck KGaA) at 37 °C for 15 min, and the number of invaded cells was counted under an optical microscope.
NOX4-overexpressing and negative-control-RKO cells were seeded into low adhesion 6-well plates at a density of 2 × 103 cells/mL and then cultured in tumor sphere formation medium [DMEM/F12 + basic fibroblast growth factor (20 ng/mL) + epidermal growth factor (20 ng/mL) + B27 (1:50) + Heparin (4 mg/mL)] in a 5% CO2 atmosphere at 37 °C for 5-7 d. The number of tumor spheres was counted using an optical microscope.
RNA-sequencing analysis were conducted by SeqHealth Technology Co., LTD (Wuhan, China). Principal component analysis was performed using the R packages “FactoMineR” and “ggplot2”. “Limma” was used to identify DEGs and the results were visualized using volcano and pheatmap plots. “ClusterProfiler” was used to perform GO and KEGG analyses of the DEGs.
Female BALB/c nude mice (5-6 wk-old) were purchased from Vital River Laboratory Animal Technology Co. Ltd. (Beijing, China). The mice were housed under pathogen-free conditions. To generate a xenograft mouse model mice were randomly divided into two groups (n = 6/group). NOX4-overexpressing and negative control RKO cells (5 × 106) were injected subcutaneously and intravenously. Tumor volume was evaluated every 3 d using digital calipers. Next, the mice were intravenously treated with: (1) trametinib (1 mg/kg, daily; n = 3); or (2) Vehicle control (n = 3) at day 14. At day 28, mice were sacrificed, and the tumors and lungs were harvested for further analysis. All animal experiments were approved by the Institutional Animal Care and Use Committee of the First Affiliated Hospital of Zhengzhou University, China.
Statistical analysis was performed using the R software (version 4.0.1) and GraphPad Prism (version 9). Two-tailed unpaired or paired t-tests were used to compare the differences between two groups in vitro experiments, the Wilcoxon test was used to compare the differences between two groups in the bioinformatic analysis, and analysis of variance (ANOVA) was used to compare the differences between more than two groups, data were showed by mean ± SEM. P < 0.05 was considered statistically significant.
To evaluate the expression of metabolic genes in CRC, we integrated two published metabolic datasets and performed a multistep analysis (Figure 1A). Total of 3341 metabolic genes were selected, of which 942 DEGs were identified dysregulated in tumor and adjacent normal tissues (726 genes were upregulated in tumor tissues and 216 genes were downregulated) (Figure 1B). The top ten DEGs, as shown in a boxplot, were all upregulated in tumor tissues (Figure 1C). The top five KEGG signaling pathways for the upregulated DEGs enrichment were: Biosynthesis of amino acids; GABAergic synapse; alanine, aspartate, and glutamate metabolism; steroid biosynthesis and bile secretion. The downregulated DEGs were enriched in chemical carcinogenesis, drug metabolism-cytochrome P450, bile secretion, xenobiotic metabolism by cytochrome P450, morphine addiction, and retinol metabolism (Figure 1D). The GO annotation results revealed that the upregulated DEGs were primarily enriched in the transmembrane transporter activity of anions and metabolites, whereas the downregulated genes were involved in the channel activity of ions, as well as transmembrane transporter activity (Supplementary Figure 1A). Consensus clustering of CRC samples using these dysregulated DEGs identified three clusters (Figure 1E), and each cluster had a unique gene expression pattern of dysregulated DEGs (Supplementary Figure 1B). Cluster 3 (C3) had a higher proportion of patients with a smaller tumor size (T1 + T2), lower stage (stage I + stage II), less metastasis (M0 and N0), and higher microsatellite instability (MSI) than C1 and C2 (Supplementary Figure 1C-G). In line with these results, C3 had longer OS and PFI (Figure 1F and G).
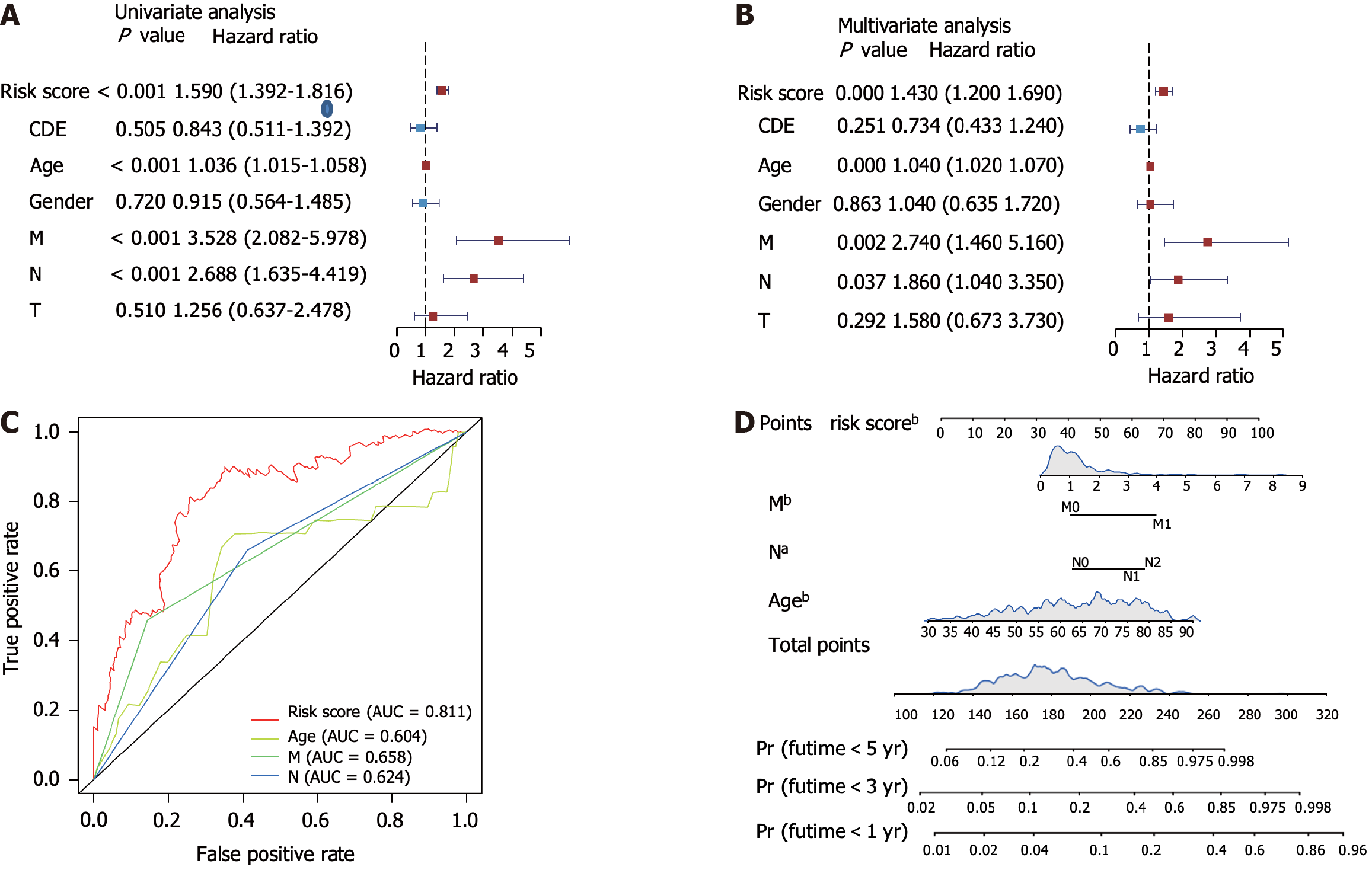
We next performed a unicox test and found that 536 metabolic genes correlated with OS. LASSO regression analysis identified six gene signatures that could predict OS (Supplementary Figure 2A and B). Using multivariate Cox regression analysis, we constructed the risk score model formula as follows: -0.38 × CPT2 expression - 0.64 × PGM2 expression - 0.06 × CLCA1 expression + 0.20 × RYR2 expression - 0.54 × ARSK expression + 0.16 × SLC6A1 expression (Supplementary Figure 2C). Next, we calculated the risk score of each patient using this formula and found that patients with high-risk scores had worse survival times in the training, test, and GEO datasets (Figure 2A and B, Supplementary Figure 2D). ROC analysis revealed that this prediction model had a high predictive efficacy across three datasets (Figure 2D and E, Supplementary Figure 2E). Next, our clinical cohort was used to validate the prediction model and clinical information was summarized in Table 1. In line with our findings, patients with high risk showed shorter survival time (Figure 2C). The prediction model showed high ROC values (Figure 2F). Univariate and multivariate analyses of the risk scores combined with several other clinical parameters revealed that risk score, age, M-stage, and N-stage were correlated with OS and could serve as independent survival risk factors in CRC (Figure 3A and B). In addition, the ROC analysis showed that the risk score had the highest sensitivity and specificity (Figure 3C). Finally, we constructed a nomogram using these independent risk scores to predict the survival of patients with CRC (Figure 3D).
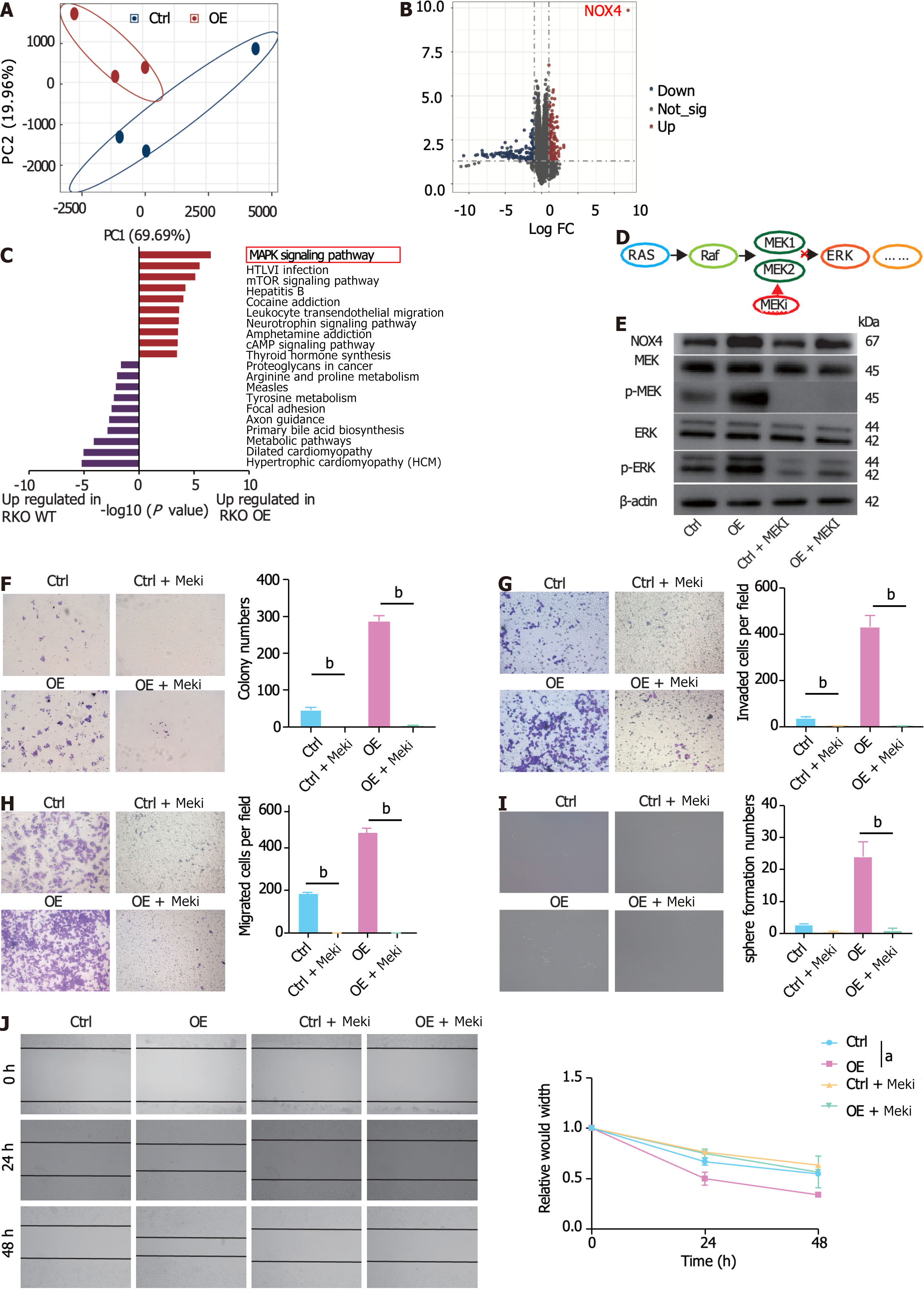
To identify potential metabolic targets, the upregulated DEGs (logFC > 2.5, adjusted P value < 0.05) in tumor tissues were overlapped with unfavorable survival-related genes (P < 0.01) (Figure 4A). Three genes were identified: NOX4, Biglycan, and phosphatidic acid phosphatase type 2 domain containing 1A. Between these three genes, NOX4 had the highest expression levels in tumor tissues (Table 2, Supplementary Figure 3A and B), indicating its potent role in promoting tumor progression. In line with these results, our clinical data demonstrated that NOX4 was highly expressed in tumor tissues at both mRNA and protein levels (Figure 4B-D). Furthermore, high NOX4 expression levels predicted a shorter OS and a higher stage of CRC (Figure 4E, Table 2). We next overexpressed NOX4 in the CRC cell line RKO (Supplementary Figure 3C-E). and found that the overexpression of NOX4 induced clone and tumorsphere formation, as well as cell migration and invasion of tumor cells (Figure 4F-J), while it had no effect on cell proliferation, cell cycle, and apoptosis (Supplementary Figure 3F-H).
| Gene symbol | Log FC | HR | Function |
| NOX4 | 2.868242 | 1.197865 | Constitutive NADPH oxidase which generates superoxide intracellularly |
| BGN | 2.368592 | 1.259598 | May be involved in collagen fiber assembly |
| PPAPDC1A | 2.345055 | 1.126713 | Magnesium-independent phospholipid phosphatase with broad substrate specificity |
To elucidate the NOX4-mediated signaling pathway, we performed RNA-sequencing analysis of wild-type (RKO-Ctrl) and NOX4-overexpressing RKO (RKO-OE) cells. Our results showed distinct transcriptional expression patterns in RKO-Ctrl and -OE cells (Figure 5A). Of the DEGs between two groups, NOX4 was the most DEG among the upregulated genes, validating our transfection efficiency (Figure 5B, Supplementary Figure 4A). GO analysis of the upregulated DEGs revealed that these genes were involved in RNA polymerase II transcription factor activity, DNA and ion binding. Furthermore, GO analysis of the downregulated genes demonstrated that DEGs were enriched in ‘protein binding’ (Supplementary Figure 4B). KEGG analysis showed that the upregulated DEGs were mostly enriched in the MAPK signal transduction pathway (Figure 5C), indicating that NOX4 could be involved in MAPK signaling. The RAS-RAF-MEK1/2-ERK1/2 axis is one of MAPK classic activation pathways[21] and our results showed that the overexpression of NOX4 increased the levels of phosphorylated MEK and its downstream target phosphorylated ERK; however, these effects were reversed by the MEK inhibitor, trametinib (Figure 5D and E). In addition, the inhibition of MAPK signaling by trametinib decreased clone formation, migration, invasion, and sphere formation in CRC cells (Figure 5F-J).
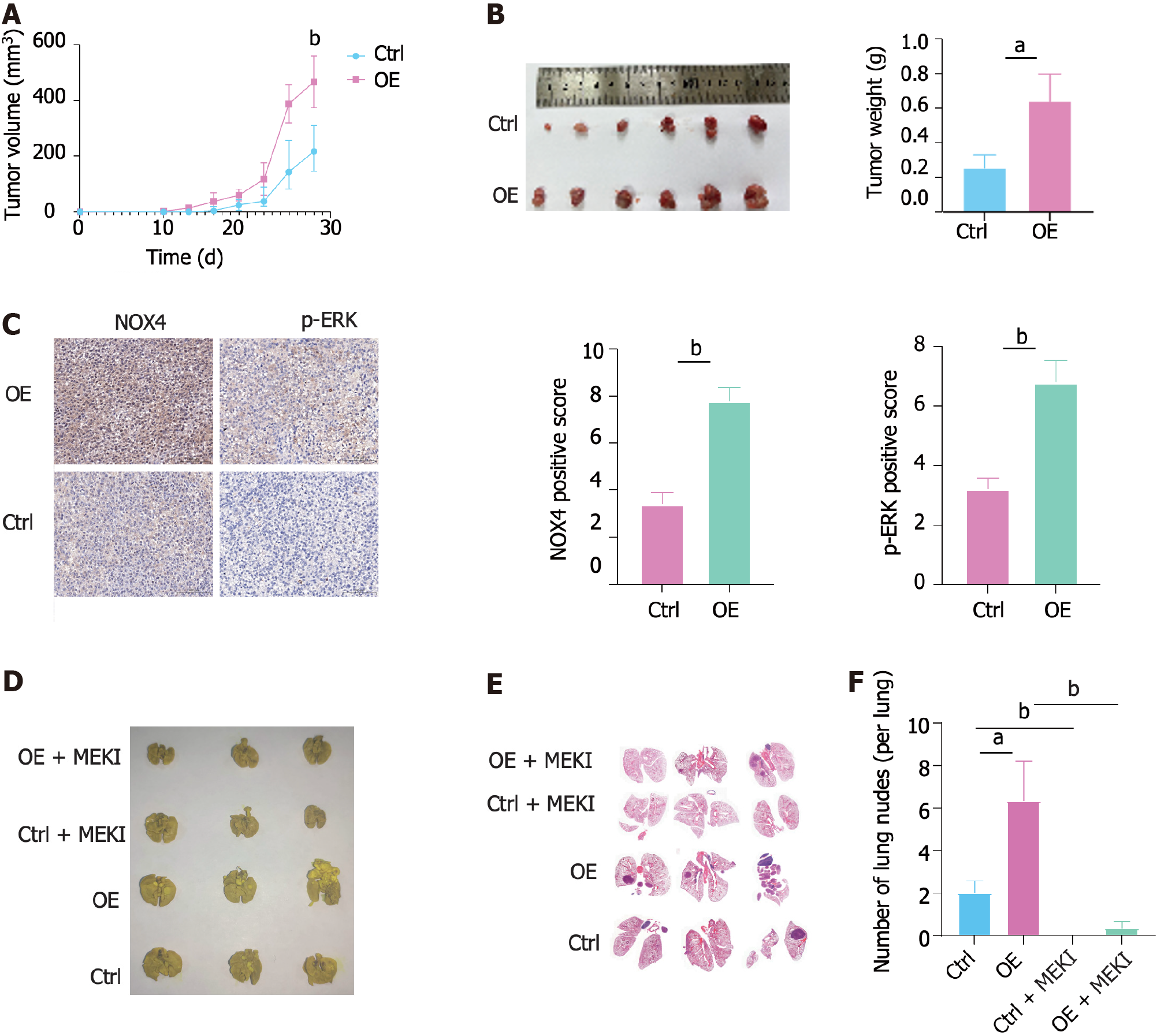
We next constructed a subcutaneous CRC xenograft model in nude mice using RKO-Ctrl and RKO-OE cells. The results showed that NOX4 overexpression promoted subcutaneous tumor growth (Figure 6A and B). In addition, IHC analysis indicated that RKO-OE cells induced NOX4 overexpression and ERK phosphorylation in tumor tissues (Figure 6C). Meanwhile, we established a lung metastasis tumor model using a tail vein injection to evaluate the role of NOX4 in metastasis in vivo. Our results demonstrated that a higher number of pulmonary nodules was observed in mice injected with RKO-OE cells compared to RKO-Ctrl injected mice, whereas trametinib treatment reduced the number of NOX4-mediated metastasis (Figure 6D-F).
Metabolic reprogramming is a feature of cancer, numerous studies have shown that dysregulated metabolic process promotes CRC progression[22,23]. Herein, we identified all dysregulated metabolic genes in CRC and comprehensively analyzed their function.
CRC is heterogeneous and can be divided into several subgroups which correlate with disease prognosis and treatment efficiency. Chromosomal instability accounts for 85% of CRC and is characterized by the activation of proliferation pathways[24]. MSI involves changes in the number of short repeated sequences called microsatellites that are spread across the genome, and patients with MSI-high have longer survival rates and would benefit from immunotherapy[7]. The CpG island methylator phenotype is characterized by high levels of CpG island hypermethylation at the promoters of several tumor suppressor genes[25]. Guinney et al[26] used a large gene expression dataset to identify four molecular subtypes called consensus molecular subtypes. Herein, we observed remarkable differences in prognosis of patients with CRC who were subgrouped by metabolic genes, indicating that metabolic classification is an important method for defining heterogeneity. For example, Xiao et al[27] generated a large metabolomic atlas of triple-negative breast cancers and classified these triple-negative breast cancers into three distinct metabolomic subgroups, while Liu et al[28] identified two CRC immune phenotypes using consensus clustering of immune-related gene expression profiles. Song et al[29] used survival-related N6-methyladenosine (m6A)-modified Long non-coding RNAs to classify CRC and to identify two subtypes. These results, combined with our findings, revealed a different perspective on CRC heterogeneity.
NOX4 plays a central role in generating ROS to facilitate tumor progression in several types of cancers. NOX4 has been identified as a modulator of two oncogenic signaling pathways, hypoxia inducible factor 1 signaling and AMP-activated protein kinase signaling[30,31]. In this study, we found that NOX4 was overexpressed in tumor tissues. Similar findings describing the role of NOX4 in CRC have been previously reported[32,33]. NOX4 overexpression in CRC cells upregulated clone formation, stemness, migration, and invasion in vitro, but had no effects on cell proliferation and apoptosis. The overexpression of NOX4 increased subcutaneous tumor volume and weight in vivo, which could have been caused by the enhanced ability to form clones and augment stemness. We also observed that NOX4 overexpression increased lung metastasis in vivo. Shen et al[34] reported that angiopoietin-like 4 induced the expression of NOX4 to promote CRC metastasis, while Cho et al[35] analyzed the expression of the NOX family and found that NOX4 expression levels were related to the degree of tumor malignancy. Mechanistically, we found that NOX4 activates the MAPK/MEK1/2/ERK1/2 signaling pathway. MAPKs are well-known oncogenic signaling molecules involved in tumor progression[36]. Mutations and the overexpression of genes involved in MAPK signaling have been frequently observed in CRC[37]. Previous studies have shown that NOX4 can activate MAPK signaling in pancreatic carcinoma and breast cancer[38,39]. In our study, RNA-sequencing analysis results showed that the overexpression of NOX4 activated MAPK signaling, and in vitro and in vivo analyses demonstrated that the inhibition of MAPK signaling (trametinib) abolished NOX4-mediated tumor growth and metastasis. Our findings further demonstrated the correlation between NOX4 expression and MAPK signaling and highlighted trametinib targeting NOX4 to inhibit MAPK signaling in CRC.
In summary, our in vitro and in vivo studies have explored the tumorigenic functions of NOX4 and downstream MAPK signaling. Our results suggest that NOX4 induces tumor progression and, therefore, could be used as a potential metabolic target in CRC.
In this study, we identified metabolic DEGs and comprehensively explored their expression and function in CRC. The CRC clusters based on dysregulated metabolic genes provided a novel insight into the heterogeneity of CRC. A prediction model generated using metabolic gene signatures could be used to predict patient prognosis. In summary, our in vitro and in vivo studies have explored the tumorigenic functions of NOX4 and downstream MAPK signaling. Our results suggest that NOX4 induces tumor progression and, therefore, could be used as a potential metabolic target in CRC.
Colorectal cancer (CRC) is the leading cause of cancer-related death. Although conventional therapy has improved the prognosis of CRC, majority of patient do not well respond to these therapies. Therefore, identification of new targets is urgent for CRC.
Metabolic reprogramming is the major cause of therapy failure. In this study, we aim to explore the metabolic alteration in CRC tumor tissue and identify the key metabolic genes mediated this process.
We aim to explore the differently expressed metabolic genes between tumor and normal tissue as well as correlated with survival. In addition, we want to construct a prediction model based on this metabolic gene and identify key metabolic genes.
We used bioinformatics analysis to identify differently expressed genes and survival related genes. Cox regression model was used to construct prediction model. Tumor biology experiments and in vivo mouse model were used to validate the role of nicotinamide adenine dinucleotide phosphate oxidase 4 (NOX4) in mediating CRC progression.
We found that most of metabolic genes were dys-regulated between tumor and normal tissue. The NOX4 was the key metabolic gene that promoted proliferation, migration invasion and stemness of tumor cell through mitogen-activated protein kinase signaling pathway. In vivo mouse models, over-expression of NOX4 increased tumor growth and metastasis.
NOX4 is a key metabolic gene that promote CRC progression.
Combined therapies with targeting NOX4 may be a promising strategy for CRC treatment.
| 1. | Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394:1467-1480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1570] [Cited by in RCA: 3413] [Article Influence: 487.6] [Reference Citation Analysis (4)] |
| 2. | Hayashi N, Tanaka S, Hewett DG, Kaltenbach TR, Sano Y, Ponchon T, Saunders BP, Rex DK, Soetikno RM. Endoscopic prediction of deep submucosal invasive carcinoma: validation of the narrow-band imaging international colorectal endoscopic (NICE) classification. Gastrointest Endosc. 2013;78:625-632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 282] [Cited by in RCA: 326] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 3. | Jayanna M, Burgess NG, Singh R, Hourigan LF, Brown GJ, Zanati SA, Moss A, Lim J, Sonson R, Williams SJ, Bourke MJ. Cost Analysis of Endoscopic Mucosal Resection vs Surgery for Large Laterally Spreading Colorectal Lesions. Clin Gastroenterol Hepatol. 2016;14:271-8.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 165] [Article Influence: 16.5] [Reference Citation Analysis (2)] |
| 4. | Taylor FG, Quirke P, Heald RJ, Moran B, Blomqvist L, Swift I, Sebag-Montefiore DJ, Tekkis P, Brown G; MERCURY study group. Preoperative high-resolution magnetic resonance imaging can identify good prognosis stage I, II, and III rectal cancer best managed by surgery alone: a prospective, multicenter, European study. Ann Surg. 2011;253:711-719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 429] [Cited by in RCA: 474] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 5. | Schmoll HJ, Tabernero J, Maroun J, de Braud F, Price T, Van Cutsem E, Hill M, Hoersch S, Rittweger K, Haller DG. Capecitabine Plus Oxaliplatin Compared With Fluorouracil/Folinic Acid As Adjuvant Therapy for Stage III Colon Cancer: Final Results of the NO16968 Randomized Controlled Phase III Trial. J Clin Oncol. 2015;33:3733-3740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 208] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 6. | Cunningham D, Lang I, Marcuello E, Lorusso V, Ocvirk J, Shin DB, Jonker D, Osborne S, Andre N, Waterkamp D, Saunders MP; AVEX study investigators. Bevacizumab plus capecitabine versus capecitabine alone in elderly patients with previously untreated metastatic colorectal cancer (AVEX): an open-label, randomised phase 3 trial. Lancet Oncol. 2013;14:1077-1085. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 422] [Cited by in RCA: 486] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 7. | Ganesh K, Stadler ZK, Cercek A, Mendelsohn RB, Shia J, Segal NH, Diaz LA Jr. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol. 2019;16:361-375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1114] [Cited by in RCA: 1368] [Article Influence: 195.4] [Reference Citation Analysis (0)] |
| 8. | Luo M, Yang X, Chen HN, Nice EC, Huang C. Drug resistance in colorectal cancer: An epigenetic overview. Biochim Biophys Acta Rev Cancer. 2021;1876:188623. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 63] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 9. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51728] [Cited by in RCA: 48731] [Article Influence: 3248.7] [Reference Citation Analysis (12)] |
| 10. | Martínez-Reyes I, Chandel NS. Cancer metabolism: looking forward. Nat Rev Cancer. 2021;21:669-680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 290] [Cited by in RCA: 1191] [Article Influence: 238.2] [Reference Citation Analysis (0)] |
| 12. | Vaupel P, Schmidberger H, Mayer A. The Warburg effect: essential part of metabolic reprogramming and central contributor to cancer progression. Int J Radiat Biol. 2019;95:912-919. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 636] [Article Influence: 90.9] [Reference Citation Analysis (0)] |
| 13. | Najumudeen AK, Ceteci F, Fey SK, Hamm G, Steven RT, Hall H, Nikula CJ, Dexter A, Murta T, Race AM, Sumpton D, Vlahov N, Gay DM, Knight JRP, Jackstadt R, Leach JDG, Ridgway RA, Johnson ER, Nixon C, Hedley A, Gilroy K, Clark W, Malla SB, Dunne PD, Rodriguez-Blanco G, Critchlow SE, Mrowinska A, Malviya G, Solovyev D, Brown G, Lewis DY, Mackay GM, Strathdee D, Tardito S, Gottlieb E; CRUK Rosetta Grand Challenge Consortium, Takats Z, Barry ST, Goodwin RJA, Bunch J, Bushell M, Campbell AD, Sansom OJ. The amino acid transporter SLC7A5 is required for efficient growth of KRAS-mutant colorectal cancer. Nat Genet. 2021;53:16-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 184] [Article Influence: 36.8] [Reference Citation Analysis (0)] |
| 14. | Schulte ML, Fu A, Zhao P, Li J, Geng L, Smith ST, Kondo J, Coffey RJ, Johnson MO, Rathmell JC, Sharick JT, Skala MC, Smith JA, Berlin J, Washington MK, Nickels ML, Manning HC. Pharmacological blockade of ASCT2-dependent glutamine transport leads to antitumor efficacy in preclinical models. Nat Med. 2018;24:194-202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 267] [Cited by in RCA: 403] [Article Influence: 50.4] [Reference Citation Analysis (0)] |
| 15. | Edwards DN, Ngwa VM, Raybuck AL, Wang S, Hwang Y, Kim LC, Cho SH, Paik Y, Wang Q, Zhang S, Manning HC, Rathmell JC, Cook RS, Boothby MR, Chen J. Selective glutamine metabolism inhibition in tumor cells improves antitumor T lymphocyte activity in triple-negative breast cancer. J Clin Invest. 2021;131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 255] [Article Influence: 51.0] [Reference Citation Analysis (0)] |
| 16. | Szanto I. NADPH Oxidase 4 (NOX4) in Cancer: Linking Redox Signals to Oncogenic Metabolic Adaptation. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 17. | Tang CT, Gao YJ, Ge ZZ. NOX4, a new genetic target for anti-cancer therapy in digestive system cancer. J Dig Dis. 2018;19:578-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 18. | Vermot A, Petit-Härtlein I, Smith SME, Fieschi F. NADPH Oxidases (NOX): An Overview from Discovery, Molecular Mechanisms to Physiology and Pathology. Antioxidants (Basel). 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 240] [Cited by in RCA: 471] [Article Influence: 94.2] [Reference Citation Analysis (0)] |
| 19. | Haider S, McIntyre A, van Stiphout RG, Winchester LM, Wigfield S, Harris AL, Buffa FM. Genomic alterations underlie a pan-cancer metabolic shift associated with tumour hypoxia. Genome Biol. 2016;17:140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 71] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 20. | Peng X, Chen Z, Farshidfar F, Xu X, Lorenzi PL, Wang Y, Cheng F, Tan L, Mojumdar K, Du D, Ge Z, Li J, Thomas GV, Birsoy K, Liu L, Zhang H, Zhao Z, Marchand C, Weinstein JN; Cancer Genome Atlas Research Network, Bathe OF, Liang H. Molecular Characterization and Clinical Relevance of Metabolic Expression Subtypes in Human Cancers. Cell Rep. 2018;23:255-269.e4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 229] [Cited by in RCA: 227] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 21. | Tsai HC, Tsou HH, Lin CC, Chen SC, Cheng HW, Liu TY, Chen WS, Jiang JK, Yang SH, Chang SC, Teng HW, Wang HT. Acrolein contributes to human colorectal tumorigenesis through the activation of RAS-MAPK pathway. Sci Rep. 2021;11:12590. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 22. | Coleman O, Ecker M, Haller D. Dysregulated lipid metabolism in colorectal cancer. Curr Opin Gastroenterol. 2022;38:162-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 23. | Mika A, Duzowska K, Halinski LP, Pakiet A, Czumaj A, Rostkowska O, Dobrzycka M, Kobiela J, Sledzinski T. Rearrangements of Blood and Tissue Fatty Acid Profile in Colorectal Cancer - Molecular Mechanism and Diagnostic Potential. Front Oncol. 2021;11:689701. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 24. | Geigl JB, Obenauf AC, Schwarzbraun T, Speicher MR. Defining 'chromosomal instability'. Trends Genet. 2008;24:64-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 248] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 25. | Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, Koh H, Simms L, Barker M, Leggett B, Levine J, Kim M, French AJ, Thibodeau SN, Jass J, Haile R, Laird PW. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787-793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1423] [Cited by in RCA: 1520] [Article Influence: 76.0] [Reference Citation Analysis (0)] |
| 26. | Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda G, Angelino P, Bot BM, Morris JS, Simon IM, Gerster S, Fessler E, De Sousa E Melo F, Missiaglia E, Ramay H, Barras D, Homicsko K, Maru D, Manyam GC, Broom B, Boige V, Perez-Villamil B, Laderas T, Salazar R, Gray JW, Hanahan D, Tabernero J, Bernards R, Friend SH, Laurent-Puig P, Medema JP, Sadanandam A, Wessels L, Delorenzi M, Kopetz S, Vermeulen L, Tejpar S. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350-1356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3408] [Cited by in RCA: 3821] [Article Influence: 347.4] [Reference Citation Analysis (8)] |
| 27. | Xiao Y, Ma D, Yang YS, Yang F, Ding JH, Gong Y, Jiang L, Ge LP, Wu SY, Yu Q, Zhang Q, Bertucci F, Sun Q, Hu X, Li DQ, Shao ZM, Jiang YZ. Comprehensive metabolomics expands precision medicine for triple-negative breast cancer. Cell Res. 2022;32:477-490. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 222] [Article Influence: 55.5] [Reference Citation Analysis (0)] |
| 28. | Liu Z, Guo Y, Yang X, Chen C, Fan D, Wu X, Si C, Xu Y, Shao B, Chen Z, Dang Q, Cui W, Han X, Ji Z, Sun Z. Immune Landscape Refines the Classification of Colorectal Cancer With Heterogeneous Prognosis, Tumor Microenvironment and Distinct Sensitivity to Frontline Therapies. Front Cell Dev Biol. 2021;9:784199. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 29. | Song W, Ren J, Yuan W, Xiang R, Ge Y, Fu T. N6-Methyladenosine-Related lncRNA Signature Predicts the Overall Survival of Colorectal Cancer Patients. Genes (Basel). 2021;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 30. | Semenza GL. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J Clin Invest. 2013;123:3664-3671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 828] [Cited by in RCA: 1052] [Article Influence: 80.9] [Reference Citation Analysis (0)] |
| 31. | Faubert B, Vincent EE, Poffenberger MC, Jones RG. The AMP-activated protein kinase (AMPK) and cancer: many faces of a metabolic regulator. Cancer Lett. 2015;356:165-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 241] [Cited by in RCA: 280] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 32. | Ford K, Hanley CJ, Mellone M, Szyndralewiez C, Heitz F, Wiesel P, Wood O, Machado M, Lopez MA, Ganesan AP, Wang C, Chakravarthy A, Fenton TR, King EV, Vijayanand P, Ottensmeier CH, Al-Shamkhani A, Savelyeva N, Thomas GJ. NOX4 Inhibition Potentiates Immunotherapy by Overcoming Cancer-Associated Fibroblast-Mediated CD8 T-cell Exclusion from Tumors. Cancer Res. 2020;80:1846-1860. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 269] [Article Influence: 44.8] [Reference Citation Analysis (0)] |
| 33. | Lin XL, Yang L, Fu SW, Lin WF, Gao YJ, Chen HY, Ge ZZ. Overexpression of NOX4 predicts poor prognosis and promotes tumor progression in human colorectal cancer. Oncotarget. 2017;8:33586-33600. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 34. | Shen CJ, Chang KY, Lin BW, Lin WT, Su CM, Tsai JP, Liao YH, Hung LY, Chang WC, Chen BK. Oleic acid-induced NOX4 is dependent on ANGPTL4 expression to promote human colorectal cancer metastasis. Theranostics. 2020;10:7083-7099. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 54] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 35. | Cho SY, Kim JS, Eun HS, Kang SH, Lee ES, Kim SH, Sung JK, Lee BS, Jeong HY, Moon HS. Expression of NOX Family Genes and Their Clinical Significance in Colorectal Cancer. Dig Dis Sci. 2018;63:2332-2340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 36. | Guo YJ, Pan WW, Liu SB, Shen ZF, Xu Y, Hu LL. ERK/MAPK signalling pathway and tumorigenesis. Exp Ther Med. 2020;19:1997-2007. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 239] [Cited by in RCA: 856] [Article Influence: 142.7] [Reference Citation Analysis (0)] |
| 37. | Stefani C, Miricescu D, Stanescu-Spinu II, Nica RI, Greabu M, Totan AR, Jinga M. Growth Factors, PI3K/AKT/mTOR and MAPK Signaling Pathways in Colorectal Cancer Pathogenesis: Where Are We Now? Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 217] [Cited by in RCA: 269] [Article Influence: 53.8] [Reference Citation Analysis (0)] |
| 38. | Witte D, Bartscht T, Kaufmann R, Pries R, Settmacher U, Lehnert H, Ungefroren H. TGF-β1-induced cell migration in pancreatic carcinoma cells is RAC1 and NOX4-dependent and requires RAC1 and NOX4-dependent activation of p38 MAPK. Oncol Rep. 2017;38:3693-3701. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 39. | Chiou JT, Shi YJ, Lee YC, Wang LJ, Chen YJ, Chang LS. Carboxyl group-modified α-lactalbumin induces TNF-α-mediated apoptosis in leukemia and breast cancer cells through the NOX4/p38 MAPK/PP2A axis. Int J Biol Macromol. 2021;187:513-527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Tchilikidi KY, Russia S-Editor: Wang JJ L-Editor: A P-Editor: Zhang XD