Published online Apr 15, 2024. doi: 10.4251/wjgo.v16.i4.1344
Peer-review started: December 20, 2023
First decision: December 27, 2023
Revised: January 9, 2024
Accepted: February 25, 2024
Article in press: February 25, 2024
Published online: April 15, 2024
Processing time: 112 Days and 19 Hours
Cholangiocarcinoma (CCA) is a highly malignant cancer, characterized by frequent mucin overexpression. MUC1 has been identified as a critical oncogene in the progression of CCA. However, the comprehensive understanding of how the mucin family influences CCA progression and prognosis is still incomplete.
To investigate the functions of mucins on the progression of CCA and to establish a risk evaluation formula for stratifying CCA patients.
Single-cell RNA sequencing data from 14 CCA samples were employed for elucidating the roles of mucins, complemented by bioinformatic analyses. Subse
CCA tumor cells with elevated levels of MUC1 and MUC4 showed activated nucleotide metabolic pathways and increased invasiveness. MUC5AC-high cells were found to promote CCA progression through WNT signaling. MUC5B-high cells exhibited robust cellular oxidation activities, leading to resistance against antitumoral treatments. MUC13-high cells were observed to secret chemokines, recruiting and transforming macrophages into the M2-polarized state, thereby suppressing antitumor immunity. MUC16-high cells were found to promote tumor progression through interleukin-1/nuclear factor kappa-light-chain-enhancer of activated B cells signaling upon interaction with neutrophils. Utilizing the expression levels of these mucins, a risk factor evaluation formula for CCA was developed and validated across multiple cohorts. CCA samples with higher risk factors exhibited stronger metastatic potential, chemotherapy resistance, and poorer prognosis.
Our study elucidates the functional mechanisms through which mucins contribute to CCA development, and provides tools for risk stratification in CCA.
Core Tip: In this study, we have conducted a comprehensive investigation of mucins in cholangiocarcinoma (CCA) using a combination of bioinformatics analysis, including single-cell RNA sequencing and spatial transcriptomics, along with experimental validations. Our findings highlight the significant roles of MUC1, MUC4, and MUC5B in CCA metabolism, contributing to tumor progression and therapy resistance. Additionally, MUC5AC has been identified as a regulator of CCA invasiveness through the WNT signaling. MUC13 and MUC16 are found to play critical roles in tumor-immune interactions, regulating antitumoral immune defense. The collect impact of these mucins enables the development of a CCA prognosis evaluation model that effectively predicts tumor malignancy, treatment effectiveness, and prognosis in CCA cases.
- Citation: Yang CY, Guo LM, Li Y, Wang GX, Tang XW, Zhang QL, Zhang LF, Luo JY. Establishment of a cholangiocarcinoma risk evaluation model based on mucin expression levels. World J Gastrointest Oncol 2024; 16(4): 1344-1360
- URL: https://www.wjgnet.com/1948-5204/full/v16/i4/1344.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i4.1344
Cholangiocarcinoma (CCA) is the second most common malignant liver cancer and has exhibited a rising incidence and mortality trend over the past four decades[1,2]. CCA is categorized into intrahepatic CCA (iCCA), perihilar CCA (pCCA) and distal CCA based on the primary anatomic region, among which the former two subtypes constitute approximately 70%-80% of cases[3]. The majority of patients are diagnosed at an advanced stage, limiting treatment options[4]. For patients who are ineligible for surgical resection, chemotherapy, particularly gemcitabine and cisplatin combination, becomes their primary choice, however, the median survival period remains less than one year[5]. While molecularly targeted therapies show promising antitumor effects[6], patients suitable for these targeted therapies are relatively scarce.
Histopathologically, CCA is classified into two primary subgroups: the mucin-producing subgroup and the nonmucin-producing subgroup[7]. Consequently, mucins play pivotal roles in CCA development. Mucins, a family of proteins widely distributed on the epithelial surface of various organs, serve essential functions in lubrication and defense against toxins and infections[8]. In addition to their physiological roles, mucins play important roles in pathological conditions, including cancer. Abnormal expression and distribution of mucins are found in various cancer types, such as lung cancer, breast cancer, pancreatic cancer, colorectal cancer, gastric cancer, liver cancer, and ovarian cancer[9,10]. Numerous studies have explored the mechanisms of MUC1 in tumorigenesis, implicating multiple signaling pathways, including the Ras, β-catenin, TP53, nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), transforming growth factor β, and vascular endothelial growth factor[10]. However, clinical trials investigating MUC1 have not demonstrated significant treatment effects compared to those in control groups[11,12], highlighting the need to systematically target multiple mucins.
In recent decades, single-cell RNA sequencing (scRNA-seq) has revolutionized cancer research, leading to remarkable milestones. The application of scRNA-seq to CCA has revealed intratumor heterogeneity at the single-cell level[13], identified new markers for different CCA subtypes[14], characterized sub-clusters of cancer-associated fibroblasts[15], and elucidated intercellular crosstalk within the tumor microenvironment (TME)[16]. Nevertheless, mucin functions and the characteristics of mucin-expressing CCA cells have not been previously explored at single-cell resolution. In this study, we comprehensively analyzed the scRNA-seq data from 14 human CCA samples, including 13 iCCA patients and one pCCA patient, to determine the mechanisms of various mucins, specifically MUC1, MUC4, MUC5AC, MUC5B, MUC13, and MUC16, in promoting CCA progression. The mucin-based CCA patient stratification system provides a new dimension for CCA prognosis prediction, and offers potential strategies for CCA treatment.
Our scRNA-seq data of CCA samples were sourced from the gene expression omnibus database (GSE154170, GSE138709, GSE142784, and GSE125449). After selecting samples from treatment-naïve CCA patients, we integrated these data into a unified dataset using R package Harmony. Following normalization, nonlinear dimensionality reduction was applied to reduce the dimensionality of data. Subsequently, cell clustering was performed using the standard methods suggested by satijalab (https://satijalab.org/seurat/). This resulted in the identification of six cell types. Cells from normal tissues and hepatocytes were filtered out, leaving only CCA tumor cells for the functional evaluation of mucins. Myeloid cells were performed under the same methodology.
The R package SCENIC (http://scenic.aertslab.org) was utilized to identify putative transcription factors and regulons of MUC1-high and MUC1-low cells[17]. Marker regulons of MUC1-high cells were identified by comparing different regulons between MUC1-high and MUC1-low cells using FindAllMarker function.
Our gene set enrichment analysis (GSEA) was conducted using the R package fgsea. The oncogenic and metabolic pathways involved in our study were obtained from MSigDB (http://www.gseamsigdb.org/gsea/msigdb). The normalized enrichment score value was calculated by the difference enrichment score between MUC1-high and MUC1-low cells, MUC5AC-high vs MUC5AC-low cells, MUC5B-high vs MUC5B-low cells, MUC13-high cells vs MUC13-low cells, and MUC16-high cells vs MUC16-low cells.
Comprehensive analyses of 70 metabolic signaling pathways were performed using gene set variation analysis (GSVA) with the R package GSVA. The 70 metabolic pathways were collected based on a previous article[18]. After calculation the rank values of each gene in each geneset, the rank value was constrained from -3 to 3 to avoid extreme values. Subsequently, the rank values of each cluster of cells were normalized to their mean value to generate the heatmap.
Spatial transcriptomics (ST) data were obtained from a published liver cancer dataset (cohort 9, Supplementary Table 1), which included hepatocellular carcinoma samples, CCA samples, and combined hepatocellular-CCA[19]. We selected the cHC-1T sample, exhibiting clear CCA histological features, for functional detection of MUC1 and MUC5B. ST data analysis followed the guidelines provided by satijala (https://satijalab.org/seurat/articles/spatial_vignette).
Expression data of mucins and WNT7B, ALDH1A1, UCP2, CSF3R, NFKB1, NFKB2, RELA were obtained from a previously reported dataset[20]. Correlations between mucins and these genes was evaluated using the R package ggplot2. The significance of these correlations was determined by Pearson’s correlation analysis (P ≤ 0.05).
Cellular interaction network analysis was performed using the R package CellChat (http://www.cellchat.org/)[21]. When analyzing interactions between CCA sub-cluster and other cell types in TME, we randomly selected 1000 cells in each TME cell partitions, including lymphocytes, fibroblasts, endothelial cells, and myeloid cells.
To assess CCA prognosis based on mucins, we applied the least absolute shrinkage and selection operator (LASSO) algorithm on the RNA levels of MUC1, MUC13, MUC16, MUC4, MUC5AC, and MUC5B by R package glmnet. RNA levels were normalized using the trans per million method. The CCA risk score was then established as follows: risk score = sum (each MUC gene expression × corresponding coefficient). The coefficient for MUC1, MUC13, MUC16, MUC4, MUC5AC, and MUC5B are 0.0297, 0.0460, 0.1217, 0.0574, 0.0501, and 0.0657, respectively. Subsequently, the CCA patients were stratified into high-risk and low-risk groups.
The patient survival information, along with corresponding RNA-seq data and protein expression levels of tumor samples, were obtained from cohort 2 and cohort 10. The correlation between CCA patient survival, individual mucin expression (including MUC1, MUC4, MUC5AC, MUC5B, MUC13, and MUC16), and risk factors was assessed using the log-rank (Mantel-Cox) test.
Validation of mucin functions was conducted using primary CCA samples resected at Peking University Third Hospital. Our study was approved by the Ethics Committee of Peking University Third Hospital. All research was conducted in accordance with both the Declarations of Helsinki and Istanbul. All patients were informed and kept anonymous.
Formalin-fixed and paraffin-embedded 4-µm tissue sections were used for immunohistochemistry (IHC) staining. Briefly, sections were dehydrated with graded concentrations of ethanol and immersed in 3% hydrogen peroxide for 15 min. Antigen retrieval was performed by heating for 2 min in a pressure cooker using 0.01 M citrate buffer (pH 6.0). Sections were then incubated with primary antibodies against MUC13 (Abcam, Cat ab235450), MUC16 (Origene, Cat ZM-0019), CD163 (Origene, Cat ZM-0428), and CD66b (Abcam, Cat ab300122) at 4 ℃ overnight. The GTVisionTM Double Staining Detection System (Dako, Cat GK700110) was used for the secondary antibody and 3,3-diaminobenzidine/hydrogen peroxide was used as the chromogen. Substitution of the primary antibody with phosphate-buffered saline was used as a negative control.
Comparisons of gene expression levels between tumor and non-tumoral normal tissue were performed using paired or unpaired student’s t-test. The signaling pathway enrichment analysis was performed using Fisher’s exact test. The GSEA was performed using Wilcoxon-Mann-Whitney test. The GSVA was performed using Kolmogorov-Smirnov like random walk test. Patient survival analysis was performed using the log-rank (Mantel-Cox) test. Correlation analyses of mucin expression levels and CCA pathological phenotypes were subjected to Chi-square analysis. All analyses were performed by R or GraphPad Prism. Differences were regarded significant when P < 0.05.
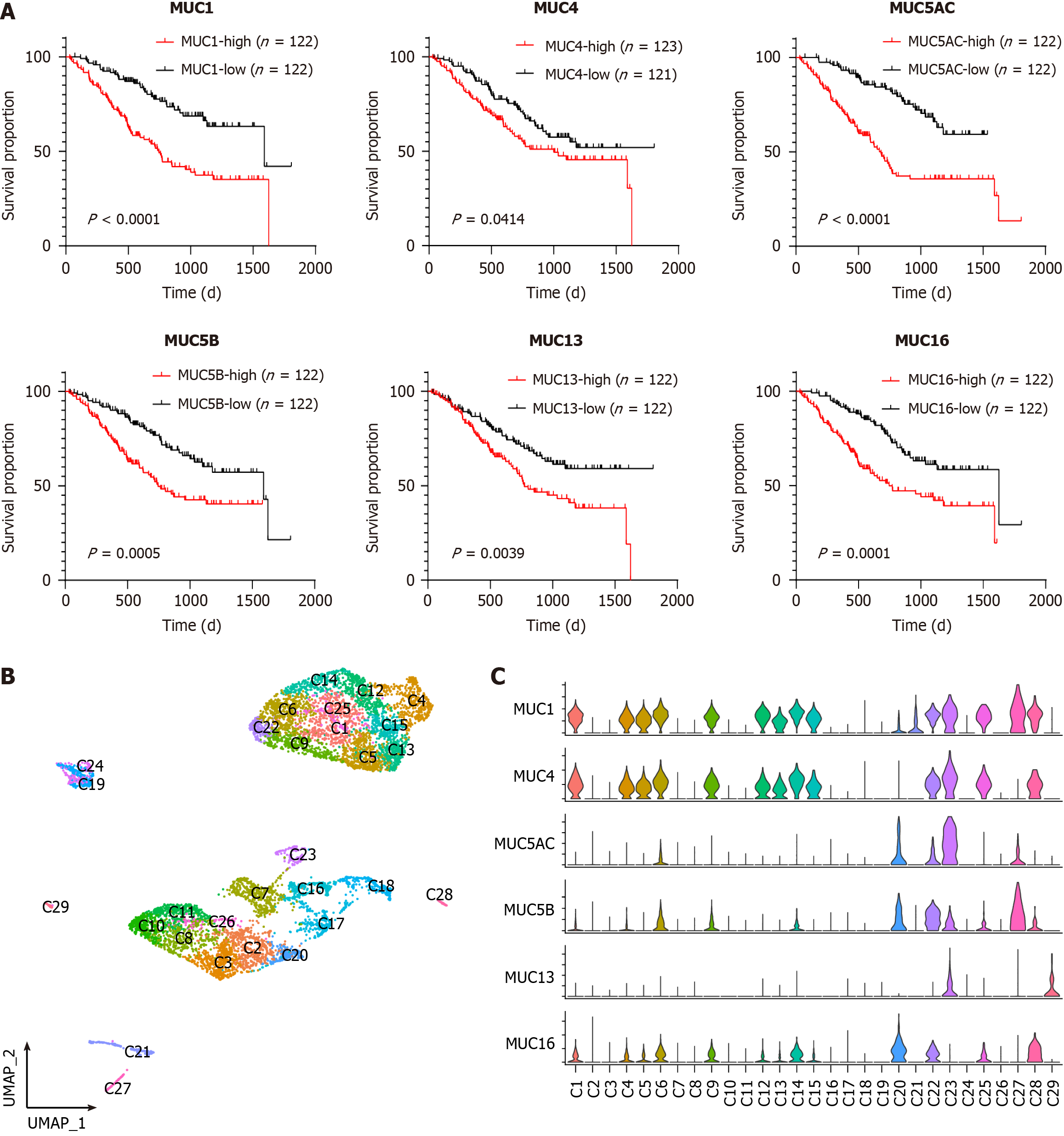
Despite previous reports suggesting the potential diagnostic roles of mucins in CCA[10,22,23], systematic evaluation of all mucins in CCA is lacking. This study introduced multiple clinical cohorts to assess the clinical relevance of mucins in CCA. Cohort 1 (Supplementary Table 1) was sourced from the CCA dataset in The Cancer Genome Atlas database. Among the 20 mucin-encoding genes, many mucins exhibited significantly higher expression levels in tumor tissues than in normal tissues (Supplementary Figure 1A), emphasizing the critical functions of mucins in CCA development. Notably, elevated expression levels of MUC1, MUC4, MUC5AC, MUC5B, MUC13, and MUC16 were found to predict poor CCA prognosis in cohort 2 (Supplementary Table 1)[24] (Figure 1A). Therefore, we focused on the functional evaluation of these mucins in this study. Two additional independent CCA cohorts (cohorts 3 and 4, Supplementary Table 1) further validated the upregulation of the abovementioned mucins in both unpaired[25] (Supplementary Figure 1B) and paired[26] (Supplementary Figure 1C) CCA samples. However, given the heterogeneity of CCA, a detailed investigation of mucins at the single-cell level is imperative.
To evaluate mucin heterogeneity at the single-cell level, we combined the scRNA-seq data from four independent datasets (cohorts 5, 6, 7, and 8 in Supplementary Table 1)[15,16,27], and reanalyzed the integrated data. The integrated dataset comprised 51810 single cells from 14 patients (Supplementary Table 2), and was classified into six cell types, namely cycling cells, dying cells, epithelial cells, lymphocytes, myeloid cells, and stromal cells (Supplementary Figure 2A). The marker gene expression levels and proportions further confirmed the annotation of these cell types (Supplementary Figure 2B). For quality control, we examined the batch effect of our data integration by evaluating the distribution features of cells from different patients. The result indicated a fine mixture of cells from different patients, suggesting low batch effects (Supplementary Figure 2C). Additionally, the observed differences in gene expression between cells from normal regions and tumor regions (Supplementary Figure 2D) suggested the occurrence of transcriptomic remodeling during CCA development.
To evaluate the functions of mucins (MUC1, MUC4, MUC5AC, MUC5B, MUC13, and MUC16) in CCA at the single-cell level, we extracted tumor cells from the integrated data. Re-clustering of the tumor cells resulted in the formation of 29 sub-clusters, designated C1 to C29 (Figure 1B). The marker gene expression for each sub-cluster is shown in Supplementary Figure 3. Notably, the detection of mucin expression revealed many sub-clusters expressing high levels of the MUC1 and MUC4 genes, including C1, C4, C5, C6, C9, C12, C13, C14, C15, C22, C23, C25, C27 and C28 (Figure 1C). Therefore, we examined MUC1 and MUC4 collectively afterward. In contrast, MUC5AC, MUC5B, MUC13 and MUC16 exhibited relatively unique distribution patterns (Figure 1C). MUC5AC was enriched in C23. MUC5B was enriched in C22 and C27. MUC13 was enriched in C23 and C29. MUC16 was enriched in C20 and C28. Subsequently, functional analyses were performed on MUC1, MUC5AC, MUC5B, MUC13, and MUC16, independently.
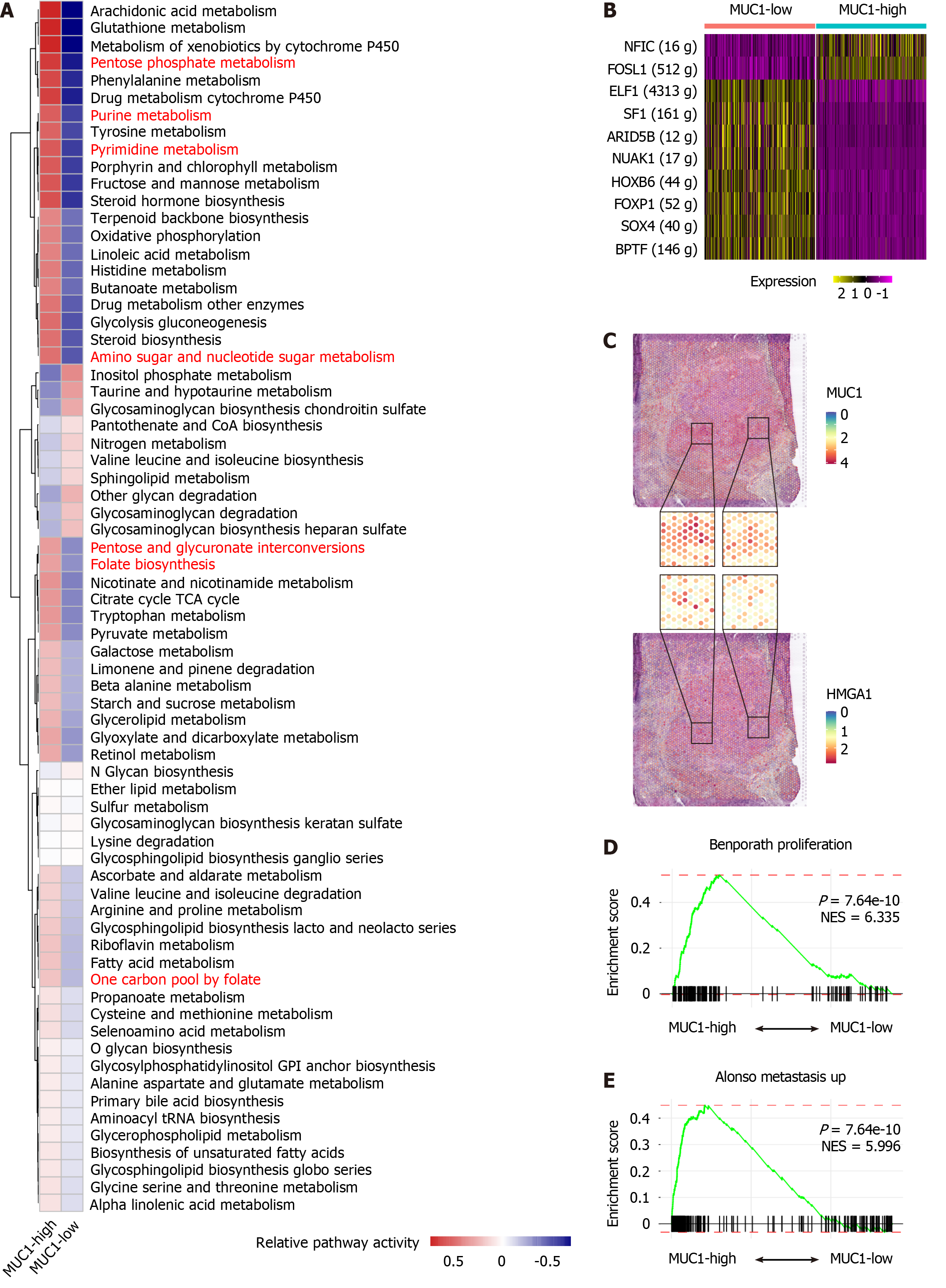
To elucidate the functions of MUC1 in CCA, we integrated sub-clusters C1, C4, C5, C6, C9, C12, C13, C14, C15, C22, C23, C25, C27, and C28 into MUC1-high subgroup, and integrated the remaining sub-clusters into MUC1-low subgroup. Next, we compared the gene expression profiles between MUC1-high cells and MUC1-low cells, and identified differentially expressed genes (DEGs). Pathway enrichment analysis of these DEGs revealed pronounced activation of metabolic pathways in MUC1-high cells (Supplementary Figure 4A). To determine the metabolic characteristics of MUC1-high cells, we employed a 70-pathway metabolic analysis panel, which demonstrated heightened activation of pathways related to nucleotide, energy, amino acid, and detoxification metabolism in MUC1-high cells (Figure 2A). Among these metabolic processes, nucleotide metabolism exhibited a prominent role, as indicated by the identification of numerous pathways (Figure 2A, highlighted in red). Independent analyses of nucleotide metabolic pathways (Supplementary Figure 4B-D) and nucleotide excision repair (Supplementary Figure 4E and F) further validated the distinctive features of MUC1-high cells. Moreover, compared with MUC1-low cells, MUC1-high cells displayed elevated expression levels of RNA polymerase components and DNA repair regulators (Supplementary Figure 4G).
To unravel the mechanisms underlying the observed metabolic characteristics of MUC1-high cells, we analyzed the transcription factor expression in these cells. Compared with MUC1-low cells, MUC1-high cells exhibited uniquely higher expression levels of NFIC and FOSL1 (Figure 2B). FOSL1 is known to regulate the expression of another transcription factor, HMGA1[28], which was found to play important roles in nucleotide metabolism[29,30]. Examination of the co-expression patterns of FOSL1, HMGA1, and MUC1 in both scRNA-seq (Supplementary Figure 4H) and ST (cohort 9, Supplementary Table 1) (Figure 2C) data indicated significant co-expression. Importantly, both FOSL1 and HMGA1 are implicated tumor progression[30,31]. GSEA confirmed the enhanced proliferation and metastasis characteristics of MUC1-high cells compared to MUC1-low cells (Figure 2D and E). Taken together, our analyses reveal the activation of nucleotide metabolism and high invasion status of MUC1-high cells.
Given the substantial expression of MUC5AC in cluster C23 (Figure 1C), we defined C23 cells as MUC5AC-high cells, and the remaining sub-clusters as MUC5AC-low cells. Analysis of DEGs in the MUC5AC-high cluster vs the MUC5AC-low cluster revealed enrichment of cell-cell junctions and actin-related cellular mobility pathways (Figure 3A), implicating MUC5AC in CCA metastasis. Cellular interaction analysis using CellChat demonstrated activation of the WNT signaling pathway in MUC5AC-high cells (Figure 3B). Given the tumor-promoting functions of the WNT pathway[32], we examined the expression of WNT ligands, including WNT7A and WNT7B, as well as the WNT target gene MMP7[33], in MUC5AC-high and MUC5AC-low cells (Figure 3C). Co-expression analysis of MUC5B and WNT7B in an independent CCA cohort (cohort 10, Supplementary Table 1) confirmed a significant correlation (Figure 3D), supporting the activation of the WNT signaling pathway in MUC5AC-high CCA cells. Subsequent GSEA further verified the metastasis-promoting role of MUC5AC-high cells (Figure 3E and Supplementary Figure 5).
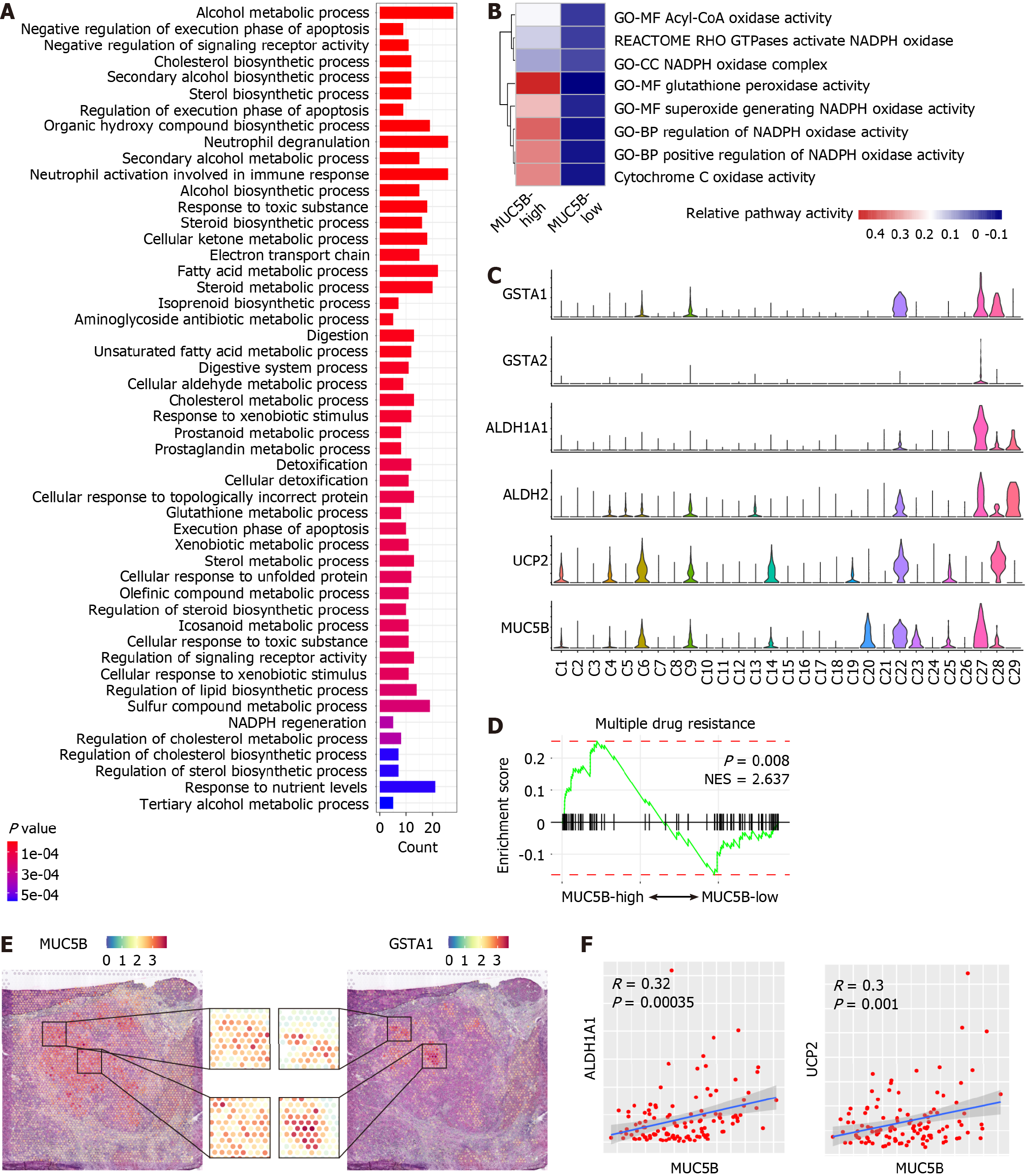
To unravel the functions of MUC5B in CCA, we stratified CCA cells based on MUC5B expression level in the scRNA-seq data. Combining sub-clusters C22 and C27 as the MUC5B-high subgroup (Figure 1C), the remaining sub-clusters formed the MUC5B-low subgroup. Comparative analysis revealed that the DEGs of MUC5B-high cells were highly enriched in cellular oxidation and detoxification pathways (Figure 4A). Additional cellular oxidation pathways from independent datasets further confirmed the active oxidation status of MUC5B-high cells (Figure 4B). Furthermore, the scRNA-seq data revealed that GSTA1, GSTA2, ALDH1A1, ALDH2, and UCP2 were highly co-expressed with MUC5B (Figure 4C). These genes not only participate in detoxification processes, but also indicate antitumor treatment resistance and poor prognosis in cancer patients[34-37]. Chemotherapy sensitivity analysis of MUC5B-high cells demonstrated that MUC5B-high cells exhibited significantly greater resistance to antitumor drugs than MUC5B-low cells did (Figure 4D, and Supplementary Figure 6). Furthermore, in-situ co-expression of MUC5B and GSTA1 was observed in the ST data (Figure 4E). Co-expression of MUC5B with ALDH1A1 or UCP2 was further confirmed in cohort 10 (Figure 4F). Collectively, these data reveal the active oxidation state and chemotherapy resistance characteristics of MUC5B-high cells.
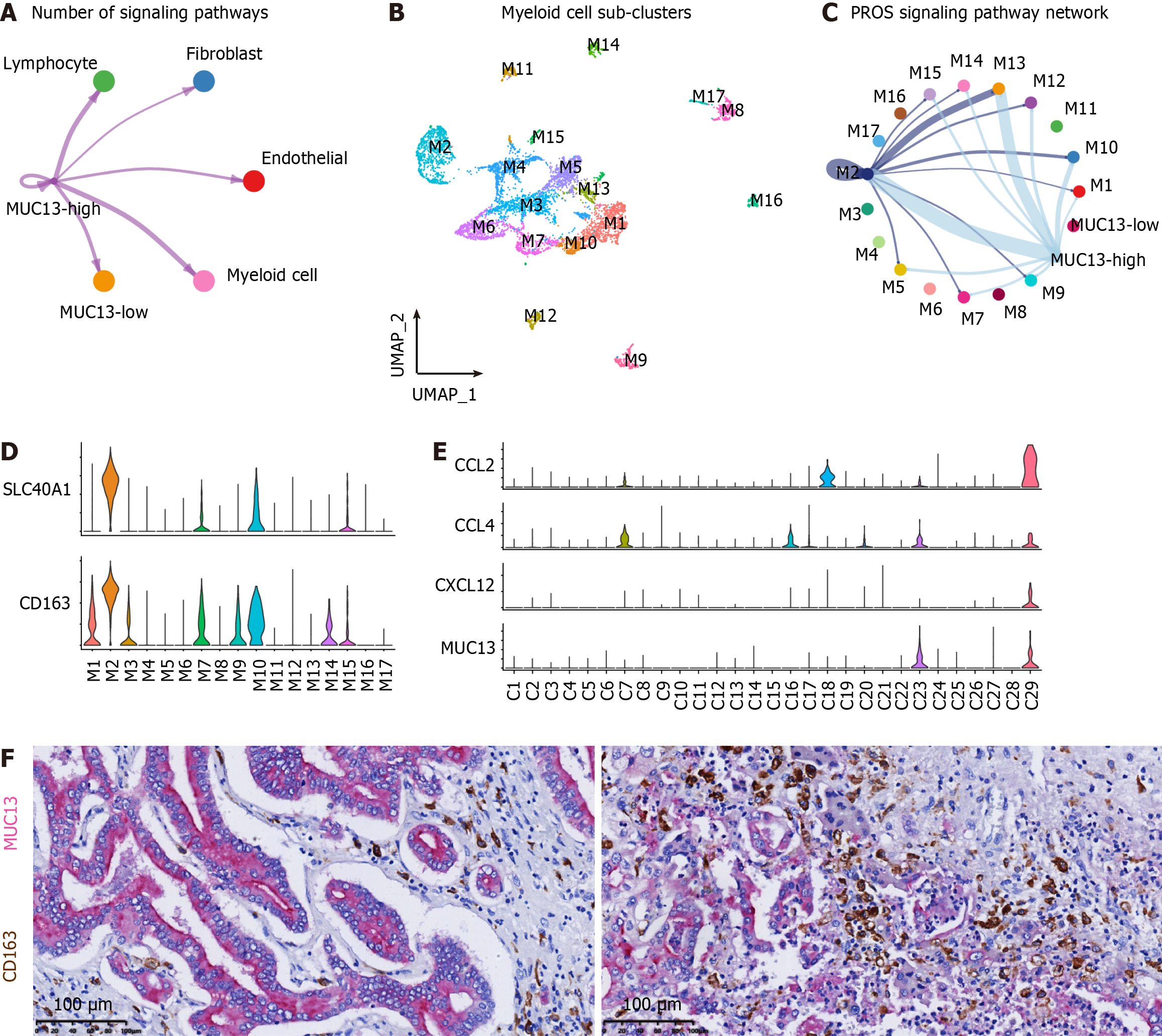
Although MUC13 has been reported to play important roles in the progression of various tumors[38], the interaction between MUC13-high cells and the TME has not been fully elucidated. Considering the distinctive expression of MUC13 in C23 and C29 (Figure 1C), we integrated C23 and C29 to generate MUC13-high CCA cells, while the remaining sub-clusters were categorized as MUC13-low cells. Interaction analyses between MUC13-high cells and TME components, namely MUC13-low cells, myeloid cells, endothelial cells, fibroblasts, and lymphocytes, revealed significant interactions between MUC13-high cells and myeloid cells (Figure 5A). To comprehensively evaluate the transcriptomic characteristics of the myeloid cell sub-populations interacting with MUC13-high cells, we isolated myeloid cell components and re-clustered them into 17 sub-populations (Figure 5B). In comparison with MUC13-low cells, MUC13-high cells exhibited a specific interaction with the M2 sub-cluster of myeloid cells in the PROS1-AXL signaling pathway (Figure 5C). The PROS signaling pathway, particularly the AXL receptor on the macrophage surface, has been shown to induce macrophage M2-polarization and tumor progression[39]. Therefore, the communication between MUC13-high cells and the M2 sub-cluster suggested that M2-polarization was induced by MUC13-high cells. Moreover, the M2 sub-cluster was characterized by high expression of SLC40A1 (Supplementary Figure 7, red frame), which was reported to be a marker gene of macrophage M2-polarization[40]. Subsequently, we examined the expression of SLC40A1 and another M2-polarization marker, CD163, in the myeloid sub-clusters of our data. The results indicated significantly elevated expression levels of both SLC40A1 and CD163 in the M2 sub-cluster (Figure 5D), confirming the M2-polarization of these cells. We next explored the mechanisms of these interactions, and identified uniquely expressed CCL2, CCL4, and CXCL12 in MUC13-high clusters (Figure 5E). The chemokine activities of CCL2 and CCL4 in macrophage recruitment and CXCL12 in macrophage M2-polarization suggest that MUC13-high cells recruit macrophages into the TME and induce M2 polarization[41]. Additionally, double staining IHC was used to confirm the histologically adjacent distribution between MUC13-high cells and M2-polarized macrophages in multiple CCA samples (Figure 5F). Taken together, these findings suggest that MUC13-high cells promote CCA progression by inducing M2-polarization of macrophages through the PROS1-AXL signaling pathway.
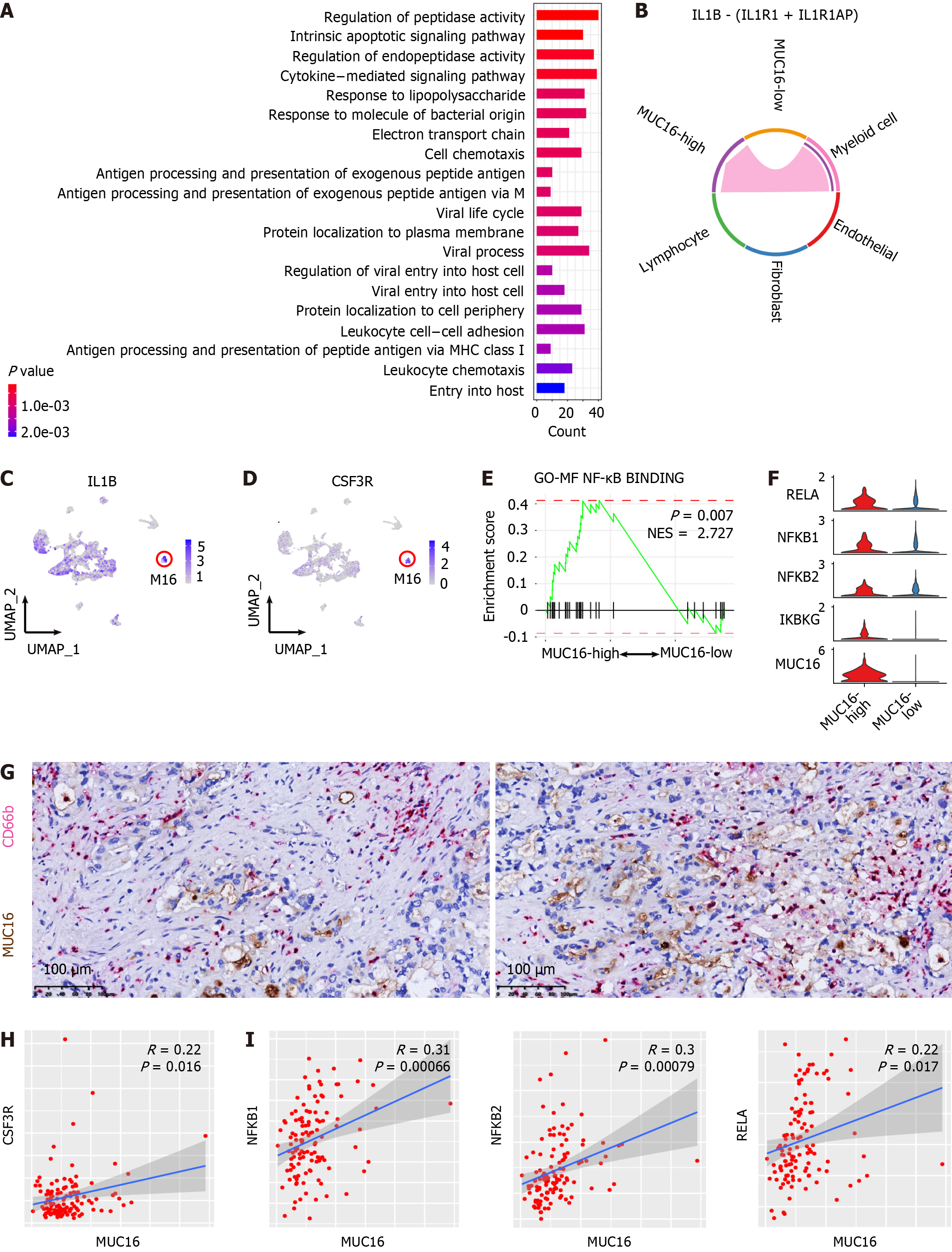
MUC16, also known as CA125, serves as a widely used biomarker for ovarian cancer[42]. Nevertheless, the impact of MUC16 on CCA progression and the underlying mechanisms remain elusive. Given the abundant and relatively unique expression of MUC16 in the C20 cluster (Figure 1C), we designated the C20 cluster as MUC16-high cells, and the remaining sub-clusters were termed MUC16-low cells. To unravel the functions of MUC16, we first compared the transcriptomics of MUC16-high and MUC16-low cells. The enrichment of DEGs in MUC16-high cells revealed multiple pathways involved in immunoregulatory processes (Figure 6A). Therefore, we hypothesized interactions between MUC16-high cells and immune cells. Through an analysis of the interaction network of MUC16-high cells with cells in the TME, we identified unique interactions between myeloid cells and MUC16-high cells in the IL-1 signaling pathway, whereas MUC16-low cells did not show such interactions (Figure 6B). To further specify the sub-cluster of myeloid cells interacting with MUC16-high cells in the IL-1 signaling pathway, we examined the expression level of IL-1B in myeloid sub-clusters and found that the highest expression of IL-1B was in the M16 sub-cluster (Figure 6C). Intriguingly, M16 was characterized by high expression levels of CXCL8, FCGR3B, NAMPT, and PTGS2 (Supplementary Figure 7, blue frame), indicating that M16 cells are a cluster of neutrophils. Consistently, CSF3R, a neutrophil marker[43], was found to be uniquely highly expressed in the M16 cluster (Figure 6D), supporting the interaction between MUC16-high cells and neutrophils through the IL1 signaling pathway. The activity of the IL-1 signaling pathway has been reported to be one of the most potent triggers for NF-κB signaling[44]. Therefore, we analyzed the activity of the NF-κB signaling in MUC16-high cells, and found significant enrichment of NF-κB signaling pathway genes in MUC16-high cells compared with MUC16-low cells (Figure 6E). To further confirm the activation of the NF-κB pathway in MUC16-high cells, we compared the expression levels of critical molecules in the NF-κB pathway, including RELA, NFKB1, NFKB2, and IKBKG, and found that the expression levels of these molecules were greater in MUC16-high cells than in MUC16-low cells (Figure 6F). Double staining IHC was further employed to confirm the histological adjacent distribution between MUC16-high cells and neutrophils in multiple CCA samples (Figure 6G). Moreover, gene co-expression pattern detection in cohort 10 further validated the interaction between MUC16-high cells and neutrophils (Figure 6H) and the activation of NF-κB signaling in MUC16-high cells (Figure 6I). Taken together, these findings suggest that MUC16-high cells interact with neutrophils, and induce CCA progression through activation of the NF-κB signaling pathway.
Given the intricate yet pivotal roles of mucins in CCA progression, we aimed to establish a comprehensive prognostic assessment strategy for CCA using mucins systematically. The RNA levels of mucins in different CCA patients exhibited mosaic distribution features (Figure 7A). Therefore, we used the LASSO-Cox regression strategy to construct a CCA risk factor evaluation system based on the RNA levels of MUC1, MUC4, MUC5AC, MUC5B, MUC13, and MUC16. This resulted in the following formula: Risk factor = 0.0297 × ExprMUC1 + 0.0574 × ExprMUC4 + 0.0501 × ExprMUC5AC + 0.0657 × ExprMUC5B + 0.0460 × ExprMUC13 + 0.1217 × ExprMUC16. Survival analysis of high-risk CCA patients and low-risk CCA patients revealed a significant difference (Figure 7B), suggesting the efficacy of our CCA risk assessment system. The superior performance of the risk factor compared with that of the individual mucins was further confirmed by area under curve analysis (Supplementary Figure 8A). Notably, compared to low-risk CCA patients, high-risk CCA patients exhibited markedly greater cell proliferation (Figure 7C), tumor metastasis (Figure 7D), and antitumor drug resistance (Figure 7E and F), thereby providing additional confirmation of the mechanisms involving mucins as we established. Intriguingly, we then tested the applicability of the risk factor evaluation formula to other types of carcinoma, and found that this formula could effectively predict the prognosis of pancreatic carcinoma and pulmonary adenocarcinoma (Supplementary Figure 8B).
To assess the universality of the CCA risk evaluation system, we expanded the input of the formula from RNA levels to protein levels. The protein levels of mucins in CCA patients showed mosaic distribution features similar to those of the RNA levels (Supplementary Figure 8C). By applying mucin proteins to the risk factor formula, we stratified the CCA patients into two groups, namely the high-risk group and low-risk group. As a result, high-risk CCA patients showed significantly shorter survival than low-risk CCA patients (Figure 7G), consistent with the result obtained from mucin RNA. Additionally, CCA patients in the high-risk group showed a higher vascular invasion rate (Figure 7H), regional lymph node metastasis rate (Figure 7I), and TNM stage (Figure 7J), further verifying the worse prognosis of high-risk patients. Notably, we further confirmed the reliability of our risk factor evaluation system using data from cohort 10, which included 83 iCCA patients and 29 pCCA patients, and found that patients with a higher risk factor showed a significantly worse prognosis than those with a lower risk factor (Figure 7K). In conclusion, we construct a CCA risk assessment tool that can assess both the RNA and protein levels of mucins.
Mucins, a group of secretory proteins, play important roles in both physiological and pathological conditions. Elevated mucin levels have been observed in various tumor types, including CCA[10]. However, previous investigations into mucin functions in CCA have relied primarily on techniques such as IHC or enzyme-linked immunosorbent assay, limiting the discovery of mucin functions to specific molecules. In contrast, in our study, we innovatively employed scRNA-seq for mucin investigation, which offers significant advantages in the following aspects: (1) Comprehensive analysis of the entire mucin family. In human, 21 mucins have been identified, with MUC1, MUC4, MUC5AC, and MUC16 (CA125) being the most studied mucins in CCA. However, the functions of the remaining mucins in CCA are largely unknown. Our study considered the clinical significance and expression abundance of all mucins in CCA, leading to the identification of potential prognostic mucins, namely MUC1, MUC4, MUC5AC, MUC5B, MUC13, and MUC16; (2) Metabolic profiling of mucins. Metabolic reprogramming is a hallmark of cancer, and has drawn increased amounts of attention in CCA[45]. However, the metabolic states of mucin-positive cells in CCA have rarely been investigated. Our study is the first to report the metabolic characteristics of critical mucins in CCA. FOSL1 and HMGA1 were identified as critical transcription factors regulating active nucleotide metabolism in MUC1-high cells. Additionally, GSTA1, GSTA2, ALDH1A1, ALDH2, and UCP2 were found to be overexpressed in MUC5B-high cells, regulating cellular oxidation processes to resist chemotherapies. These findings shed light on the mechanisms of CCA progression and treatment resistance in an unprecedented way; and (3) interplay between mucin-positive cells and the TME. Despite numerous studies on MUC1-activated signaling pathways in tumor cells[10], the cellular interaction network between mucin-positive cells and TME components has seldom been illustrated. Using scRNA-seq data, we identified interactions between mucin-positive cells and myeloid cell sub-clusters. MUC13-high cells were found to induce macrophage infiltration into the TME, leading to subsequent M2-polarization through PROS1/AXL signaling. Additionally, NF-κB signaling pathway activation was observed in MUC16-high cells following interaction with neutrophils. These interactions elucidate the immunosuppressive status in CCA TME and offer potential targets for CCA immunotherapy. Although the number of patients used for functional mechanism investigations is limited, the CCA risk evaluation model based on mucin expression levels are applicable in large CCA cohorts, indicating the reliability of our conclusions.
Given its wide overexpression pattern and oncogenic characteristics, MUC1 is anticipated to be a potential antitumor treatment target. However, the outcomes of clinical trials investigating MUC1 have proven unsatisfactory[11,12]. Previous reports attributed this discrepancy to the scarcity of major histocompatibility complex class I epitopes within the MUC1 protein, resulting in insufficient immune reactions[46]. Nevertheless, our data offer new possible explanations. Although MUC1 is one of the most widely expressed mucins, there are numerous mucins, including MUC4, MUC5AC, MUC5B, MUC13, and MUC16, which share oncogenic and metabolic characteristics with MUC1. Therefore, targeting MUC1 alone may be insufficient for effective tumor therapy. Our study developed a mucin-based CCA risk evaluation system that links risk factors to multiple CCA pathological indicators, such as tumor cell proliferation, metastasis, and resistance to antitumor drugs.
In fact, the risk factor formula, which includes MUC1, MUC4, MC5AC, MUC5B, MUC13, and MUC16, implies the involvement and necessity of all six mucins in CCA development. Consequently, targeting all mucins together may yield more effective therapeutic outcomes than focusing solely on MUC1. Notably, although the functional mechanisms of mucin-positive cells may not be directly facilitated by mucins, mucin-directed immune therapy or chemotherapy still shows strong clinical application potential. Recently, MUC1-directed chimeric antigen receptor (CAR) T cell therapies have shown promising killing effects both in-vitro and in-vivo[47,48]. Our data reveal attractive targets for immunotherapy, suggesting that anti-MUC4, anti-MUC5AC, anti-MUC5B, anti-MUC13 or anti-MUC16 CAR T cells may also show promising treatment effects in the future.
Through a comprehensive approach involving both bioinformatic analysis strategies and experimental validation, our study successfully unveils the functional mechanisms of mucin-positive cells in CCA progression, through MUC1, MUC4, MUC5AC, MUC5B, MUC13, and MUC16. We subsequently construct a CCA risk factor evaluation system based on the expression levels of these six mucins. The risk factor evaluation model effectively predicts CCA patient prognosis, providing a novel patient stratification method. The results obtained in our study not only enrich the current understanding of mucins in CCA progression, but also identify new targets for precision treatments of CCA.
In conclusion, we analyzed ten clinical CCA cohorts to investigate the functions of mucins on CCA progression and prognosis. The mucin family, includes MUC1, MUC4, MUC5AC, MUC5B, MUC13, and MUC16, regulates tumor metabolism, invasiveness, chemotherapy resistance, and cellular interaction with immune cells in microenvironments, comprehensively promoting CCA progression. We also construct a CCA risk evaluation model based on the expression levels of these mucins, applicable at both RNA and protein levels. Given the critical roles of these mucins on CCA development, our model serves as a promising tool for evaluating tumor malignancy and stratifying CCA patients.
Cholangiocarcinoma (CCA) is the second most common type of liver cancer and exhibits a high mortality rate. Mucins are a family of protein that are elevated in various tumor types, including CCA. However, the comprehensive functional mechanisms and prognosis evaluation significance of mucins in CCA progression remain largely unknown.
MUC1 has been identified as an oncogene that induce CCA progression through multiple signaling pathways. Nevertheless, how the mucin family regulate CCA is still elusive.
To investigate the functional mechanisms of mucins in CCA and to conduct a CCA risk evaluation model based on mucin expression levels.
For the detection of mucin functions in CCA, single-cell RNA sequencing data from 14 CCA samples were employed, supported by comprehensive bioinformatic analyses. Validations were pursued through spatial transcriptomics and immunohistochemistry. The establishment of a CCA risk evaluation model based on mucin expression levels employed the least absolute shrinkage and selection operator regression algorithm. The risk evaluation model was constructed using RNA level of mucins, and subsequently validated by both RNA and protein levels of mucins, as well as multiple independent cohorts.
Elevated levels of MUC1 and MUC4 in CCA tumor cells were associated with activated nucleotide metabolic pathways and higher invasiveness. CCA tumor cells with heightened MUC5AC expression were found to induce tumor progression through the WNT signaling pathway. Robust cellular oxidation activities in MUC5B-high CCA tumor cells facilitated antitumoral treatment resistance. MUC13-high cells transformed macrophage into M2-polarization state through the PROS signaling and chemokines, including CCL2, CCL4, and CXCR12. Neutrophils induced the activation of nuclear factor kappa-light-chain-enhancer of activated B cells signaling in MUC16-high cells through the IL1B signaling, thereby promoting CCA development. Utilizing the expression levels of these mucins, a CCA prognosis evaluation model was developed and validated across multiple cohorts, which simultaneously exhibited predictive functions on the evaluation of CCA malignancy, metastasis potential, and chemotherapy sensitivity.
Our study unveils the functional mechanisms by which mucins contribute to CCA progression, and offers a potential tool for CCA risk stratification.
The discovery of mucin functions in CCA development and prognosis prediction indicate that mucins may be promising treatment targets for CCA.
We thank Guangze Zhang and Xin Zhang in Peking University Health Science Center for assistance in data analysis. We thank Prof. Jin Gu in Tsinghua University for ST data sharing.
| 1. | Saha SK, Zhu AX, Fuchs CS, Brooks GA. Forty-Year Trends in Cholangiocarcinoma Incidence in the U.S.: Intrahepatic Disease on the Rise. Oncologist. 2016;21:594-599. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 364] [Cited by in RCA: 595] [Article Influence: 59.5] [Reference Citation Analysis (0)] |
| 2. | Khan SA, Taylor-Robinson SD, Toledano MB, Beck A, Elliott P, Thomas HC. Changing international trends in mortality rates for liver, biliary and pancreatic tumours. J Hepatol. 2002;37:806-813. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 408] [Cited by in RCA: 424] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 3. | Rizvi S, Khan SA, Hallemeier CL, Kelley RK, Gores GJ. Cholangiocarcinoma - evolving concepts and therapeutic strategies. Nat Rev Clin Oncol. 2018;15:95-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1052] [Cited by in RCA: 1206] [Article Influence: 150.8] [Reference Citation Analysis (0)] |
| 4. | Jarnagin WR, Fong Y, DeMatteo RP, Gonen M, Burke EC, Bodniewicz BS J, Youssef BA M, Klimstra D, Blumgart LH. Staging, resectability, and outcome in 225 patients with hilar cholangiocarcinoma. Ann Surg. 2001;234:507-17; discussion 517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 973] [Cited by in RCA: 982] [Article Influence: 39.3] [Reference Citation Analysis (0)] |
| 5. | Valle J, Wasan H, Palmer DH, Cunningham D, Anthoney A, Maraveyas A, Madhusudan S, Iveson T, Hughes S, Pereira SP, Roughton M, Bridgewater J; ABC-02 Trial Investigators. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med. 2010;362:1273-1281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2617] [Cited by in RCA: 3332] [Article Influence: 208.3] [Reference Citation Analysis (15)] |
| 6. | Abou-Alfa GK, Sahai V, Hollebecque A, Vaccaro G, Melisi D, Al-Rajabi R, Paulson AS, Borad MJ, Gallinson D, Murphy AG, Oh DY, Dotan E, Catenacci DV, Van Cutsem E, Ji T, Lihou CF, Zhen H, Féliz L, Vogel A. Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: a multicentre, open-label, phase 2 study. Lancet Oncol. 2020;21:671-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 809] [Cited by in RCA: 1178] [Article Influence: 196.3] [Reference Citation Analysis (0)] |
| 7. | Kendall T, Verheij J, Gaudio E, Evert M, Guido M, Goeppert B, Carpino G. Anatomical, histomorphological and molecular classification of cholangiocarcinoma. Liver Int. 2019;39 Suppl 1:7-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 254] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 8. | Leal J, Smyth HDC, Ghosh D. Physicochemical properties of mucus and their impact on transmucosal drug delivery. Int J Pharm. 2017;532:555-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 264] [Cited by in RCA: 359] [Article Influence: 39.9] [Reference Citation Analysis (0)] |
| 9. | Dhanisha SS, Guruvayoorappan C, Drishya S, Abeesh P. Mucins: Structural diversity, biosynthesis, its role in pathogenesis and as possible therapeutic targets. Crit Rev Oncol Hematol. 2018;122:98-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 139] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 10. | Kasprzak A, Adamek A. Mucins: the Old, the New and the Promising Factors in Hepatobiliary Carcinogenesis. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 62] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 11. | Rossmann E, Österborg A, Löfvenberg E, Choudhury A, Forssmann U, von Heydebreck A, Schröder A, Mellstedt H. Mucin 1-specific active cancer immunotherapy with tecemotide (L-BLP25) in patients with multiple myeloma: an exploratory study. Hum Vaccin Immunother. 2014;10:3394-3408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 12. | Butts C, Socinski MA, Mitchell PL, Thatcher N, Havel L, Krzakowski M, Nawrocki S, Ciuleanu TE, Bosquée L, Trigo JM, Spira A, Tremblay L, Nyman J, Ramlau R, Wickart-Johansson G, Ellis P, Gladkov O, Pereira JR, Eberhardt WE, Helwig C, Schröder A, Shepherd FA; START trial team. Tecemotide (L-BLP25) versus placebo after chemoradiotherapy for stage III non-small-cell lung cancer (START): a randomised, double-blind, phase 3 trial. Lancet Oncol. 2014;15:59-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 406] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 13. | Wang T, Xu C, Zhang Z, Wu H, Li X, Zhang Y, Deng N, Dang N, Tang G, Yang X, Shi B, Li Z, Li L, Ye K. Cellular heterogeneity and transcriptomic profiles during intrahepatic cholangiocarcinoma initiation and progression. Hepatology. 2022;76:1302-1317. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 40] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 14. | Song G, Shi Y, Meng L, Ma J, Huang S, Zhang J, Wu Y, Li J, Lin Y, Yang S, Rao D, Cheng Y, Lin J, Ji S, Liu Y, Jiang S, Wang X, Zhang S, Ke A, Cao Y, Ji Y, Zhou J, Fan J, Zhang X, Xi R, Gao Q. Single-cell transcriptomic analysis suggests two molecularly subtypes of intrahepatic cholangiocarcinoma. Nat Commun. 2022;13:1642. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 107] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 15. | Affo S, Nair A, Brundu F, Ravichandra A, Bhattacharjee S, Matsuda M, Chin L, Filliol A, Wen W, Song X, Decker A, Worley J, Caviglia JM, Yu L, Yin D, Saito Y, Savage T, Wells RG, Mack M, Zender L, Arpaia N, Remotti HE, Rabadan R, Sims P, Leblond AL, Weber A, Riener MO, Stockwell BR, Gaublomme J, Llovet JM, Kalluri R, Michalopoulos GK, Seki E, Sia D, Chen X, Califano A, Schwabe RF. Promotion of cholangiocarcinoma growth by diverse cancer-associated fibroblast subpopulations. Cancer Cell. 2021;39:866-882.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 266] [Article Influence: 53.2] [Reference Citation Analysis (0)] |
| 16. | Zhang M, Yang H, Wan L, Wang Z, Wang H, Ge C, Liu Y, Hao Y, Zhang D, Shi G, Gong Y, Ni Y, Wang C, Zhang Y, Xi J, Wang S, Shi L, Zhang L, Yue W, Pei X, Liu B, Yan X. Single-cell transcriptomic architecture and intercellular crosstalk of human intrahepatic cholangiocarcinoma. J Hepatol. 2020;73:1118-1130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 412] [Article Influence: 68.7] [Reference Citation Analysis (0)] |
| 17. | Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine JC, Geurts P, Aerts J, van den Oord J, Atak ZK, Wouters J, Aerts S. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14:1083-1086. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1585] [Cited by in RCA: 4154] [Article Influence: 461.6] [Reference Citation Analysis (0)] |
| 18. | Gaude E, Frezza C. Tissue-specific and convergent metabolic transformation of cancer correlates with metastatic potential and patient survival. Nat Commun. 2016;7:13041. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 182] [Cited by in RCA: 288] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 19. | Wu R, Guo W, Qiu X, Wang S, Sui C, Lian Q, Wu J, Shan Y, Yang Z, Yang S, Wu T, Wang K, Zhu Y, Liu C, Zhang Y, Zheng B, Li Z, Shen S, Zhao Y, Wang W, Bao J, Hu J, Wu X, Jiang X, Wang H, Gu J, Chen L. Comprehensive analysis of spatial architecture in primary liver cancer. Sci Adv. 2021;7:eabg3750. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 252] [Article Influence: 50.4] [Reference Citation Analysis (0)] |
| 20. | Jusakul A, Cutcutache I, Yong CH, Lim JQ, Huang MN, Padmanabhan N, Nellore V, Kongpetch S, Ng AWT, Ng LM, Choo SP, Myint SS, Thanan R, Nagarajan S, Lim WK, Ng CCY, Boot A, Liu M, Ong CK, Rajasegaran V, Lie S, Lim AST, Lim TH, Tan J, Loh JL, McPherson JR, Khuntikeo N, Bhudhisawasdi V, Yongvanit P, Wongkham S, Totoki Y, Nakamura H, Arai Y, Yamasaki S, Chow PK, Chung AYF, Ooi LLPJ, Lim KH, Dima S, Duda DG, Popescu I, Broet P, Hsieh SY, Yu MC, Scarpa A, Lai J, Luo DX, Carvalho AL, Vettore AL, Rhee H, Park YN, Alexandrov LB, Gordân R, Rozen SG, Shibata T, Pairojkul C, Teh BT, Tan P. Whole-Genome and Epigenomic Landscapes of Etiologically Distinct Subtypes of Cholangiocarcinoma. Cancer Discov. 2017;7:1116-1135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 415] [Cited by in RCA: 723] [Article Influence: 80.3] [Reference Citation Analysis (0)] |
| 21. | Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, Myung P, Plikus MV, Nie Q. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021;12:1088. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3052] [Cited by in RCA: 5212] [Article Influence: 1042.4] [Reference Citation Analysis (0)] |
| 22. | Abe T, Amano H, Shimamoto F, Hattori M, Kuroda S, Kobayashi T, Tashiro H, Ohdan H. Prognostic evaluation of mucin-5AC expression in intrahepatic cholangiocarcinoma, mass-forming type, following hepatectomy. Eur J Surg Oncol. 2015;41:1515-1521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 23. | Pabalan N, Sukcharoensin S, Butthongkomvong K, Jarjanazi H, Thitapakorn V. Expression and Serum Levels of Mucin 5AC (MUC5AC) as a Biomarker for Cholangiocarcinoma: a Meta-analysis. J Gastrointest Cancer. 2019;50:54-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 24. | Dong L, Lu D, Chen R, Lin Y, Zhu H, Zhang Z, Cai S, Cui P, Song G, Rao D, Yi X, Wu Y, Song N, Liu F, Zou Y, Zhang S, Zhang X, Wang X, Qiu S, Zhou J, Wang S, Shi Y, Figeys D, Ding L, Wang P, Zhang B, Rodriguez H, Gao Q, Gao D, Zhou H, Fan J. Proteogenomic characterization identifies clinically relevant subgroups of intrahepatic cholangiocarcinoma. Cancer Cell. 2022;40:70-87.e15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 245] [Article Influence: 61.3] [Reference Citation Analysis (0)] |
| 25. | Andersen JB, Spee B, Blechacz BR, Avital I, Komuta M, Barbour A, Conner EA, Gillen MC, Roskams T, Roberts LR, Factor VM, Thorgeirsson SS. Genomic and genetic characterization of cholangiocarcinoma identifies therapeutic targets for tyrosine kinase inhibitors. Gastroenterology. 2012;142:1021-1031.e15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 339] [Cited by in RCA: 440] [Article Influence: 31.4] [Reference Citation Analysis (0)] |
| 26. | Ahn KS, O'Brien D, Kang YN, Mounajjed T, Kim YH, Kim TS, Kocher JA, Allotey LK, Borad MJ, Roberts LR, Kang KJ. Prognostic subclass of intrahepatic cholangiocarcinoma by integrative molecular-clinical analysis and potential targeted approach. Hepatol Int. 2019;13:490-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 27. | Ma L, Hernandez MO, Zhao Y, Mehta M, Tran B, Kelly M, Rae Z, Hernandez JM, Davis JL, Martin SP, Kleiner DE, Hewitt SM, Ylaya K, Wood BJ, Greten TF, Wang XW. Tumor Cell Biodiversity Drives Microenvironmental Reprogramming in Liver Cancer. Cancer Cell. 2019;36:418-430.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 390] [Cited by in RCA: 616] [Article Influence: 88.0] [Reference Citation Analysis (13)] |
| 28. | Maurus K, Hufnagel A, Geiger F, Graf S, Berking C, Heinemann A, Paschen A, Kneitz S, Stigloher C, Geissinger E, Otto C, Bosserhoff A, Schartl M, Meierjohann S. The AP-1 transcription factor FOSL1 causes melanocyte reprogramming and transformation. Oncogene. 2017;36:5110-5121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 73] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 29. | Xu M, Sharma P, Pan S, Malik S, Roeder RG, Martinez E. Core promoter-selective function of HMGA1 and Mediator in Initiator-dependent transcription. Genes Dev. 2011;25:2513-2524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 30] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 30. | Adair JE, Maloney SC, Dement GA, Wertzler KJ, Smerdon MJ, Reeves R. High-mobility group A1 proteins inhibit expression of nucleotide excision repair factor xeroderma pigmentosum group A. Cancer Res. 2007;67:6044-6052. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 27] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 31. | Zhang M, Hoyle RG, Ma Z, Sun B, Cai W, Cai H, Xie N, Zhang Y, Hou J, Liu X, Chen D, Kellogg GE, Harada H, Sun Y, Wang C, Li J. FOSL1 promotes metastasis of head and neck squamous cell carcinoma through super-enhancer-driven transcription program. Mol Ther. 2021;29:2583-2600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 75] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 32. | Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2017;36:1461-1473. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1646] [Cited by in RCA: 1992] [Article Influence: 221.3] [Reference Citation Analysis (0)] |
| 33. | Han B, Zhou B, Qu Y, Gao B, Xu Y, Chung S, Tanaka H, Yang W, Giuliano AE, Cui X. FOXC1-induced non-canonical WNT5A-MMP7 signaling regulates invasiveness in triple-negative breast cancer. Oncogene. 2018;37:1399-1408. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 66] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 34. | Xu M, Wang Y, Duan W, Xia S, Wei S, Liu W, Wang Q. Proteomic Reveals Reasons for Acquired Drug Resistance in Lung Cancer Derived Brain Metastasis Based on a Newly Established Multi-Organ Microfluidic Chip Model. Front Bioeng Biotechnol. 2020;8:612091. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 35. | Liu H, Yang Z, Zang L, Wang G, Zhou S, Jin G, Pan X. Downregulation of Glutathione S-transferase A1 suppressed tumor growth and induced cell apoptosis in A549 cell line. Oncol Lett. 2018;16:467-474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 36. | Kleih M, Böpple K, Dong M, Gaißler A, Heine S, Olayioye MA, Aulitzky WE, Essmann F. Direct impact of cisplatin on mitochondria induces ROS production that dictates cell fate of ovarian cancer cells. Cell Death Dis. 2019;10:851. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 308] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 37. | Cubillos-Ruiz JR, Bettigole SE, Glimcher LH. Tumorigenic and Immunosuppressive Effects of Endoplasmic Reticulum Stress in Cancer. Cell. 2017;168:692-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 404] [Cited by in RCA: 714] [Article Influence: 79.3] [Reference Citation Analysis (0)] |
| 38. | Maher DM, Gupta BK, Nagata S, Jaggi M, Chauhan SC. Mucin 13: structure, function, and potential roles in cancer pathogenesis. Mol Cancer Res. 2011;9:531-537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 73] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 39. | Myers KV, Amend SR, Pienta KJ. Targeting Tyro3, Axl and MerTK (TAM receptors): implications for macrophages in the tumor microenvironment. Mol Cancer. 2019;18:94. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 145] [Cited by in RCA: 346] [Article Influence: 49.4] [Reference Citation Analysis (0)] |
| 40. | Zhang Q, He Y, Luo N, Patel SJ, Han Y, Gao R, Modak M, Carotta S, Haslinger C, Kind D, Peet GW, Zhong G, Lu S, Zhu W, Mao Y, Xiao M, Bergmann M, Hu X, Kerkar SP, Vogt AB, Pflanz S, Liu K, Peng J, Ren X, Zhang Z. Landscape and Dynamics of Single Immune Cells in Hepatocellular Carcinoma. Cell. 2019;179:829-845.e20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 763] [Cited by in RCA: 1112] [Article Influence: 158.9] [Reference Citation Analysis (0)] |
| 41. | Ruytinx P, Proost P, Van Damme J, Struyf S. Chemokine-Induced Macrophage Polarization in Inflammatory Conditions. Front Immunol. 2018;9:1930. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 148] [Cited by in RCA: 354] [Article Influence: 44.3] [Reference Citation Analysis (0)] |
| 42. | Dochez V, Caillon H, Vaucel E, Dimet J, Winer N, Ducarme G. Biomarkers and algorithms for diagnosis of ovarian cancer: CA125, HE4, RMI and ROMA, a review. J Ovarian Res. 2019;12:28. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 158] [Cited by in RCA: 349] [Article Influence: 49.9] [Reference Citation Analysis (0)] |
| 43. | Xue R, Zhang Q, Cao Q, Kong R, Xiang X, Liu H, Feng M, Wang F, Cheng J, Li Z, Zhan Q, Deng M, Zhu J, Zhang Z, Zhang N. Liver tumour immune microenvironment subtypes and neutrophil heterogeneity. Nature. 2022;612:141-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 593] [Article Influence: 148.3] [Reference Citation Analysis (0)] |
| 44. | Diep S, Maddukuri M, Yamauchi S, Geshow G, Delk NA. Interleukin-1 and Nuclear Factor Kappa B Signaling Promote Breast Cancer Progression and Treatment Resistance. Cells. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 45. | Raggi C, Taddei ML, Rae C, Braconi C, Marra F. Metabolic reprogramming in cholangiocarcinoma. J Hepatol. 2022;77:849-864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 113] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 46. | Taylor-Papadimitriou J, Burchell JM, Graham R, Beatson R. Latest developments in MUC1 immunotherapy. Biochem Soc Trans. 2018;46:659-668. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 109] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 47. | Supimon K, Sangsuwannukul T, Sujjitjoon J, Phanthaphol N, Chieochansin T, Poungvarin N, Wongkham S, Junking M, Yenchitsomanus PT. Anti-mucin 1 chimeric antigen receptor T cells for adoptive T cell therapy of cholangiocarcinoma. Sci Rep. 2021;11:6276. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 48. | Mao L, Su S, Li J, Yu S, Gong Y, Chen C, Hu Z, Huang X. Development of Engineered CAR T Cells Targeting Tumor-Associated Glycoforms of MUC1 for the Treatment of Intrahepatic Cholangiocarcinoma. J Immunother. 2023;46:89-95. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ueda H, Japan S-Editor: Qu XL L-Editor: A P-Editor: Zheng XM