Published online Mar 15, 2024. doi: 10.4251/wjgo.v16.i3.1019
Peer-review started: October 30, 2023
First decision: December 6, 2023
Revised: December 16, 2023
Accepted: January 17, 2024
Article in press: January 17, 2024
Published online: March 15, 2024
Processing time: 133 Days and 23.8 Hours
Through experimental research on the biological function of GATA6-AS1, it was confirmed that GATA6-AS1 can inhibit the proliferation, invasion, and migration of gastric cancer cells, suggesting that GATA6-AS1 plays a role as an anti-oncogene in the occurrence and development of gastric cancer. Further experi
To investigate the effects of GATA6-AS1 on the proliferation, invasion and migration of gastric cancer cells and its mechanism of action.
We used bioinformatics methods to analyze the Cancer Genome Atlas (https://portal.gdc.cancer.gov/. The Cancer Genome Atlas) and download expression data for GATA6-AS1 in gastric cancer tissue and normal tissue. We also constructed a GATA6-AS1 lentivirus overexpression vector which was transfected into gastric cancer cells to investigate its effects on proliferation, migration and invasion, and thereby clarify the expression of GATA6-AS1 in gastric cancer and its biological role in the genesis and development of gastric cancer. Next, we used a database (http://starbase.sysu.edu.cn/starbase2/) to analysis GATA6-AS1 whether by m6A methylation modify regulation and predict the methyltransferases that may methylate GATA6-AS1. Furthermore, RNA immunoprecipitation experiments confirmed that GATA6-AS1 was able to bind to the m6A methylation modification enzyme. These data allowed us to clarify the ability of m6A methylase to influence the action of GATA6-AS1 and its role in the occurrence and development of gastric cancer.
Low expression levels of GATA6-AS1 were detected in gastric cancer. We also determined the effects of GATA6-AS1 overexpression on the biological function of gastric cancer cells. GATA6-AS1 had strong binding ability with the m6A demethylase FTO, which was expressed at high levels in gastric cancer and negatively correlated with the expression of GATA6-AS1. Following transfection with siRNA to knock down the expression of FTO, the expression levels of GATA6-AS1 were up-regulated. Finally, the proliferation, migration and invasion of gastric cancer cells were all inhibited following the knockdown of FTO expression.
During the occurrence and development of gastric cancer, the overexpression of FTO may inhibit the expression of GATA6-AS1, thus promoting the proliferation and metastasis of gastric cancer.
Core Tip: Long non-coding RNA GATA6-AS1 is down-regulated in gastric malignant tumor, and capable to inhibit the proliferation, migration and invasion of tumor cells, acting as a tumor suppressor gene in gastric cancer cells. Fat mass and obesity-associated protein (FTO) is highly expressed in gastric cancer, and the down-regulation of GATA6-AS1 in gastric cancer is regulated by the N6-methyladenine demethylase FTO. Therefore, during the occurrence and development of gastric cancer, overexpressed FTO may inhibit the expression of GATA6-AS1, thus promoting the proliferation and metastasis of gastric cancer.
- Citation: Shen JJ, Li MC, Tian SQ, Chen WM. Long non-coding RNA GATA6-AS1 is mediated by N6-methyladenosine methylation and inhibits the proliferation and metastasis of gastric cancer. World J Gastrointest Oncol 2024; 16(3): 1019-1028
- URL: https://www.wjgnet.com/1948-5204/full/v16/i3/1019.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i3.1019
According to the latest data, of the top 10 malignant tumors in the world, the incidence of gastric cancer ranks fifth and the mortality rate ranks fourth[1]. Of the 10 most common forms of cancer in China, the incidence and mortality of gastric cancer both rank third. Thus, it is evident that stomach cancer remains very common in malignant tumors and it poses a great threat to the lives of patients affected by this disease. The primary causes of death in patients with gastric cancer are tumor metastasis and recurrence[2]. Therefore, there is an urgent need to investigate the regulatory factors and pathways involved in the recurrence and metastasis of cancer, and to develop interventions with which to block these mechanisms. The formation of gastric cancer is a complex process involving multiple factors, steps, and stages[3]. Previous studies have reported that abnormal expression levels of long non-coding RNA (lncRNA) appear to influence the progression of gastric cancer. It is now well accepted that lncRNAs are transcribed by RNA polymerase II and are greater than 200 nucleotides in length. Furthermore, lncRNAs do not encode proteins[4]; rather, they play an indispensable role in multiple cellular processes, including the cell cycle, differentiation, metabolism and disease. In addition, lncRNAs are capable of acting at the epigenetic, transcriptional and post-transcriptional levels, and play key roles in the regulation of gene expression via RNA[5]. With the continuous in-depth exploration of lncRNA, an increasing body of evidence now supports the fact that lncRNA also plays an important role in the metabolic recombination, occurrence and development of tumors[6]. In addition, previous reports have demonstrated that various lncRNAs can influence the occurrence and development of gastric cancer. For example, HOXC-AS3 was shown to inhibit the proliferation and metastasis of gastric cancer cells by regulating Y-box-binding protein 1[7]. In addition, HOXA11-AS was shown to promote the proliferation, invasion and metastasis of gastric cancer by inhibiting the expression of PRSS8 and Kruppel-like factor 2[8]. Other research showed that the miR-708-5p/ upstream stimulatory factor 1 pathway is regulated by LOXL1-AS1 to promote the progression of gastric cancer[9] and that FEZF1-AS1 promotes the proliferation of gastric cancer cells by inhibiting the expression of p21[10]. Therefore, it is highly evident that lncRNA is closely related to the occurrence and development of gastric cancer. Subsequently, our group analyzed chip data from the GSE13911 dataset in the gene expression omnibus database and found that the expression levels of lncRNA GATA6-AS1 in gastric cancer tissues were significantly down-regulated when compared with normal tissues; moreover, the prognosis of patients with high expression levels of GATA6-AS1 was significantly better than that of patients with low expression levels. lncRNA GATA6-AS1 was first discovered by Sigova et al[11] in 2013; however, its specific biological function and role in the occurrence and development of gastric cancer have yet to be fully elucidated. The clear downregulation of GATA6-AS1 expression in gastric cancer tissues subsequently led to the discovery of five N6-methyladenine (m6A) modification sites in the database m6Avar and RMBase v2.0 databases by us. Therefore, we hypothesized that the downregulation of GATA6-AS1 may be modified by m6A methylation. m6A is the most common form of internal RNA modification in eukaryotic cells. Research has shown that m6A is enriched near the stop codon and the 3’-untranslated terminal region, and is translated in a cap-independent manner near the 5'-untranslated terminal region, thus affecting the transcription, processing, translation, and metabolism of RNA[12-14]. m6A methylation modification is a dynamic, reversible epigenetic regulatory process that is installed by m6A methyltransferase, reversed by demethylase, and recognized by m6A binding proteins, and plays an important role in a variety of diseases, such as obesity, infertility, and tumors[15]. Moreover, lncRNA can mediate the promotion of cell proliferation and development as well as tumor development and migration[16]. Since GATA6-AS1 is expressed at low levels in gastric cancer tissues, we first constructed an overexpression vector and used lentivirus transfection to investigate how GATA6-AS1 affects the progression of gastric cancer. We also verified whether the downregulation of GATA6-AS1 in gastric cancer is affected by m6A methylation modification and identified the methylation modification enzyme involved. Our findings provide an experimental and theoretical basis for identifying new diagnostic markers and therapeutic targets for gastric cancer.
Human gastric mucosal epithelial cells and a human gastric cancer cell line (HGC-27) were acquired from the Beijing Bena culture collection (Beijing, China). In addition, a human gastric cancer cell line (AGS) was purchased from Procell Life Science and Technology Co., Ltd (Wuhan, China). The authenticity of all cell lines was confirmed by short tandem repeat DNA profiling analysis. Cells were cultured in 1640 medium supplemented with 20% fetal bovine serum (FBS) (Gibco, Thermo Fisher Scientific, Inc., Waltham, MA, United States) and Ham’s F-12 medium (Procell Life Science and Technology Co., Ltd, Wuhan, China) supplemented with 10% FBS in a humidified atmosphere at 37 °C with 5% CO2. Cancer cells that had been sub-cultured less than 5-6 times were used in all experiments.
A lncRNA GATA6-AS1 (oe-GATA6-AS1) overexpression lentivirus vector and its negative control were obtained from Genechem Medical Technology Co. Ltd. (Shanghai, China). The small interfering RNA of fat mass and obesity-associated protein (Si-FTO) and its negative control were obtained from Guangzhou Ruibo Biological Technology Co. Ltd. (Guangzhou, China). These constructs were transfected into cells with Infection enhancer fluid A/P (Shanghai Jikai, Shanghai, China) and Lipofectamin 3000 (Invitrogen, Carlsbad, CA).
Total RNA was extracted with a MicroElute Total RNA Kit R6831 (Omega, MA, United States) or TRIzol Reagent (Thermo Fisher Scientific, MA, United States). cDNA was then synthesized from the total RNA with a PrimeScript RT Reagent Kit (Takara, Kusatsu, Japan). The stem-loop RT primer method was applied for miRNA reverse transcription and quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was performed using TB Green Premix Ex Taq II (Takara) in a CFX96 touch system (Bio-Rad, Hercules, CA, United States). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an endogenous control for mRNA or miRNA detection, and fold change was calculated by the relative quantification method (2-ΔΔCt). The primers used in the study were as follows (5′-3′): LncRNA GATA6-AS1; (F): TTTGGCTCAGTTTGTGTCCA; LncRNA GATA6-AS1 (R): TCCACGCAGACATCCTTGTA; FTO (F): ATTGGTAATCCAGGCTGCAC; FTO (R): GCAGCAAGTTCTTCCAAAGC; GAPDH (F): CCAGGTGGTCTCCTCTGAT; GAPDH (R): GCTGT
A total of 2000 transfected cells were seeded into 96-well plates. At the indicated time points, 10 µL of cell counting kit-8 reagent (CCK8, Dojindo, Kumamoto, Japan) was added to each well in accordance with the manufacturer’s instructions. Absorbance at 450 nm (A450) was then measured on a Multifunctional Enzyme Marker (Thermo Fisher Scientific, MA, United States).
In brief, 4 × 104 cells in 200 µL of serum-free 1640 or F12 media were reseeded into the top of the insert of a Boyden chamber (Corning, NY, United States) with 300 µg/mL Matrigel (BD Bioscience, San Jose, CA, United States), while 600 µL of medium with 10% FBS was loaded into the well below. After 20-24 h of incubation, invasive cells that had passed through the filter were fixed with 0.1% paraformaldehyde (Beijing Solarbio Technology Co. Ltd., Beijing, China) and stained with 0.1% crystal violet solution (Beijing Solarbio Technology Co. Ltd.). For the Trans-well migration assay, we used the same procedure but without incubation with Matrigel. The cells that had passed through the filter were imaged at 100 × magnification in six random fields, and measured using ImageJ software (National Institutes of Health and the Laboratory, Bethesda, MD, United States).
Gastric cancer cells, with or without transfection, were harvested and lysed. Protein concentration was then determined with a bicinchoninicacid Protein Assay Kit (Thermo Fisher Scientific). Equal amounts of protein were separated by 10% or 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes (Thermo Fisher Scientific). Membranes were incubated with primary antibodies at 4 °C overnight, followed by secondary antibodies at room temperature for 2 h. After washing, signals were detected using a ChemiDo imaging system (Bio-Rad). The following antibodies were used: anti-FTO (1:5000, 31687, Cell Signaling Technology, United States), anti-YTH domain-containing family protein 1 (1: 5000, 86463, Cell Signaling Technology), anti- methyltransferase-like 3 (1:5000, 86132, Cell Signaling Technology), and anti-β-actin (1:5000, AF7018, Affinity, United States). The secondary antibody was anti-rabbit IgG (1:5000, S0001, Affinity, United States).
The Magna RIP Kit (Millipore, United States) and qRT-PCR were utilized to analyze the direct interactions between the lncRNA GATA6-AS1 transcript and indicated proteins. All procedures were carried out in accordance with the manufacturer’s instructions. Cells were broken up with RIP lysis buffer containing protease inhibitor cocktail and RNase inhibitors. Next, the cell lysates and magnetic beads conjugated with specific antibodies or IgG were incubated in RIP immunoprecipitation buffer at 4 °C for 3 h or overnight with rotation. After digestion with protease K, the immunoprecipitated RNA samples were purified by phenol/chloroform/isoamyl alcohol and 100% ethanol, and finally detected by qRT-PCR.
All statistical tests were performed using data acquired from three independent experiments, and were each performed in triplicate. SPSS version 21.0 (IBM Corporation, Armonk, NY, United States) and GraphPad Prism version 8.0 (GraphPad, La Jolla, CA, United States) were used to analyze and present experimental data. Statistical comparisons were performed between the two groups with unpaired t-tests. A P value < 0.05 was considered statistically significant.
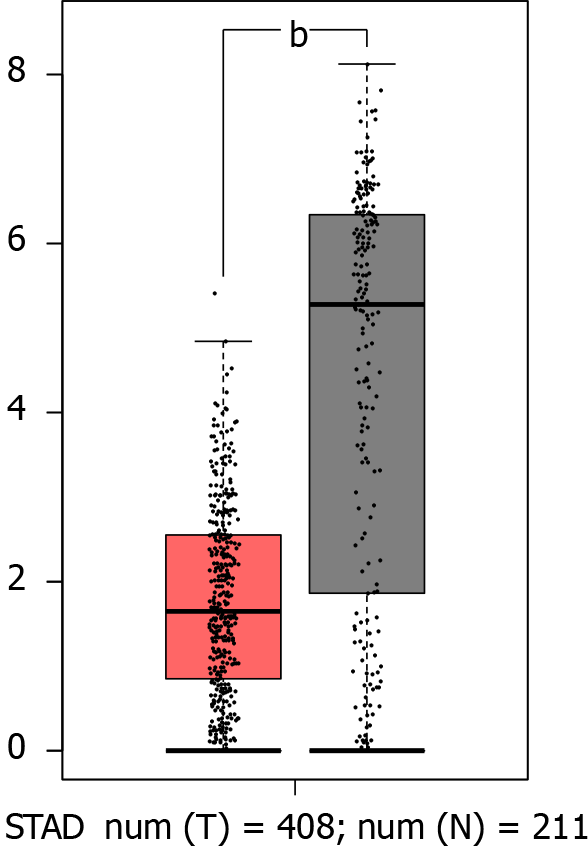
GATA6-AS1 was expressed at low levels in gastric cancer. We got that the expression levels of GATA6-AS1 in gastric cancer tissues were significantly down-regulated when compared with normal tissues (aP < 0.05) through the bioinformatics database, as shown in Figure 1.
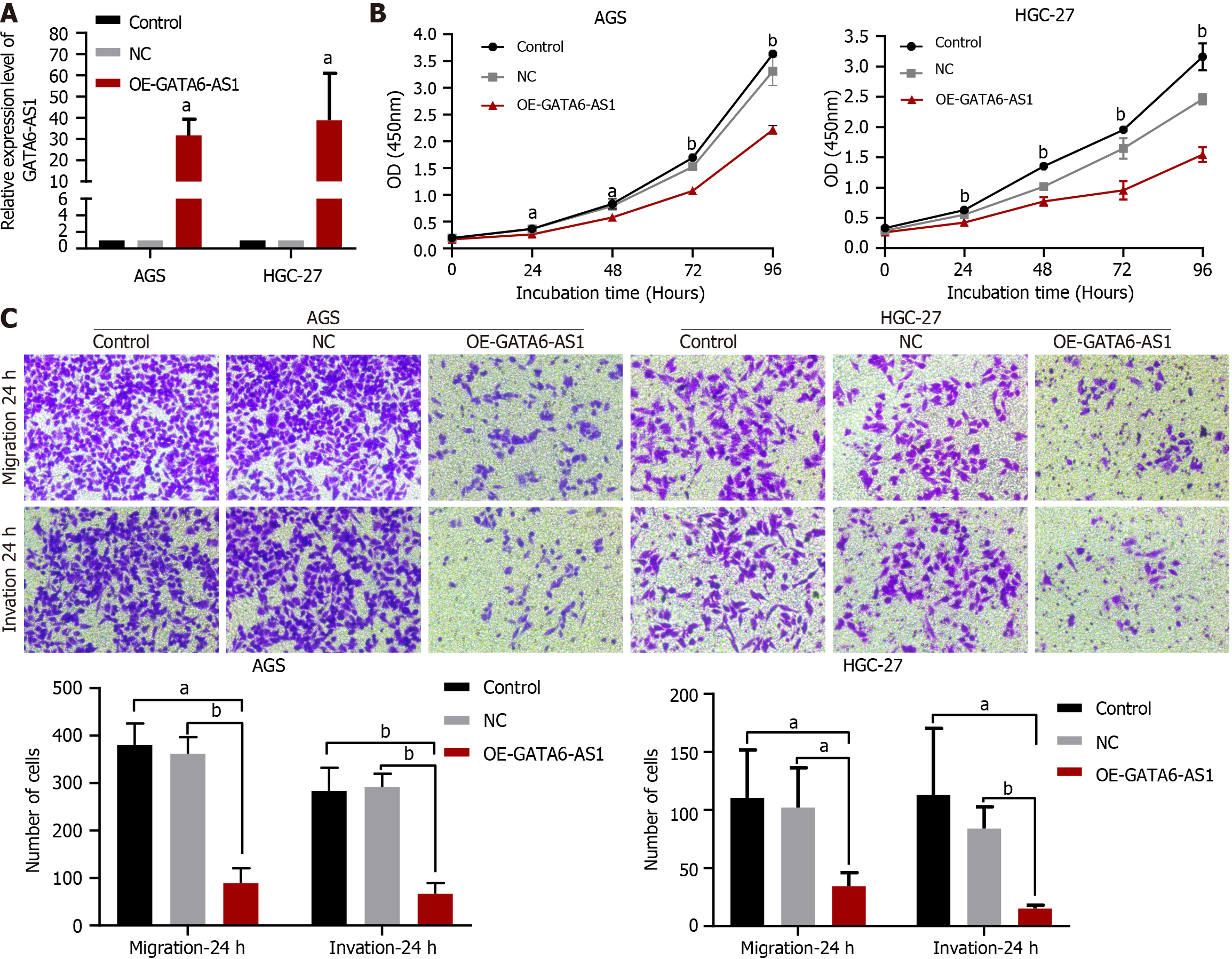
The effects of GATA6-AS1 overexpression on biological function in gastric cancer cells. The overexpression vector was constructed by lentivirus transfection, and the transfection efficiency was verified by RT-PCR (P < 0.05), as shown in Figure 2A. CCK8 assays showed that the overexpression of GATA6-AS1 could reduce the proliferation ability of HGC-27 and AGS gastric cancer cell lines when compared with blank and negative controls (P < 0.01), as shown in Figure 2B. The result that when compared with the negative control and blank control groups, the migration and invasion ability of AGS and HGC-27 cells in the GATA6-AS1 overexpression group were decreased (P < 0.05) is obtained from trans-well assays, as shown in Figure 2C.
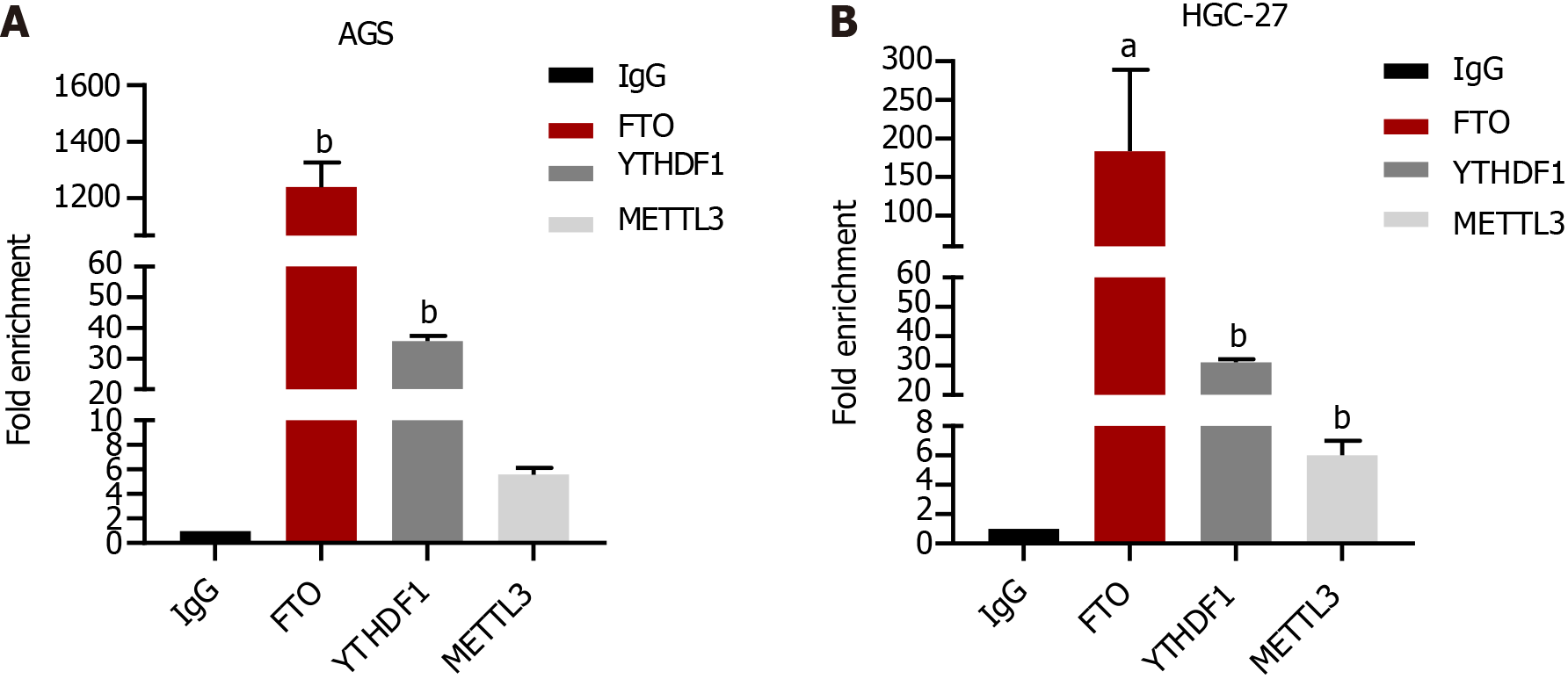
GATA6-AS1 exhibited the highest binding ability with m6A demethylase FTO. RIP experiments confirmed that GATA6-AS1 bound most strongly with the m6A demethylase FTO in gastric cancer cells (P < 0.05), as shown in Figure 3.
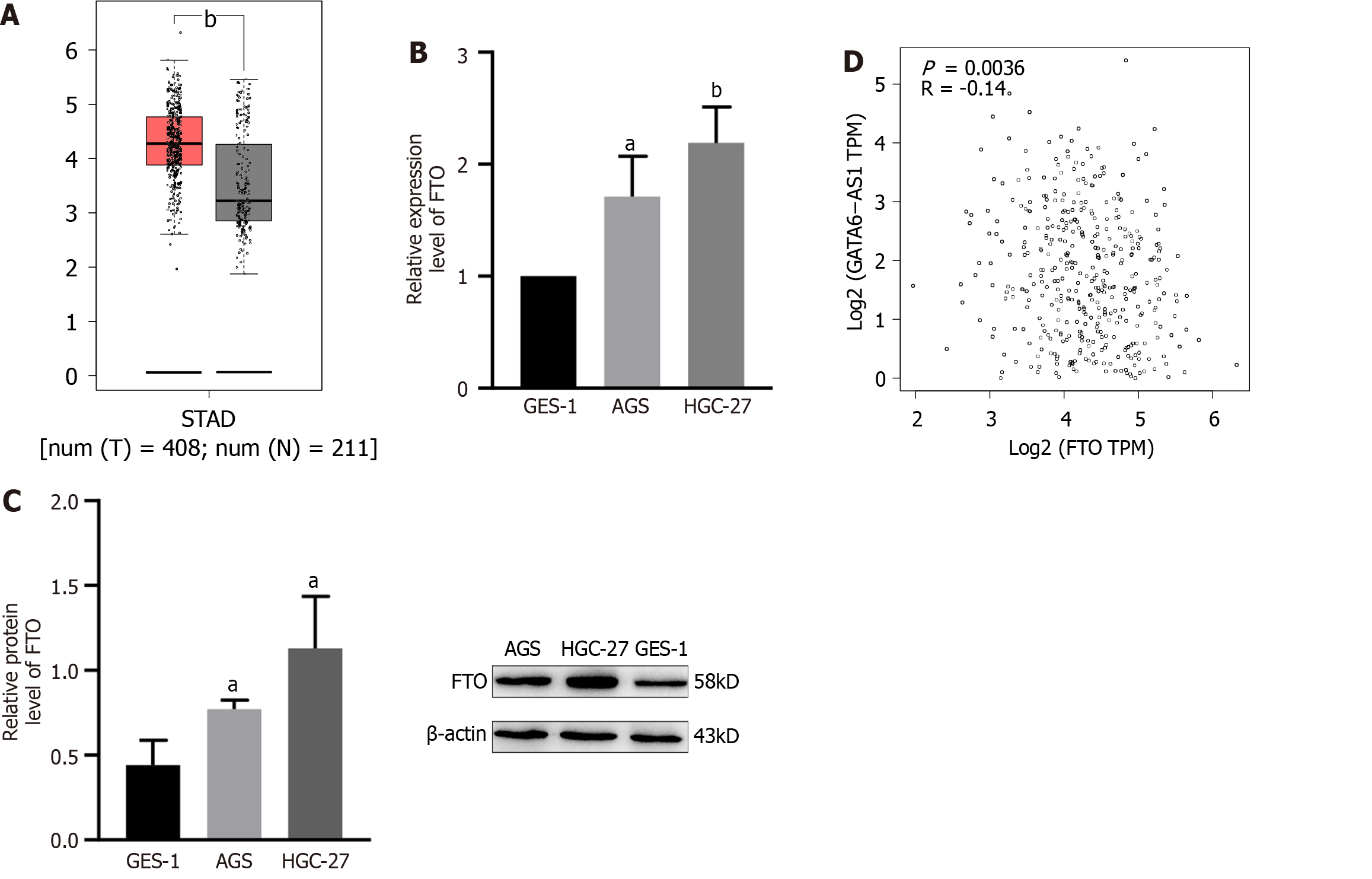
The expression level of FTO was higher in stomach cancer, and negatively correlated with the expression of GATA6-AS1. A founding that the expression levels of FTO in gastric cancer tissues were significantly higher than those in normal tissues (P < 0.05) was got by bioinformatics analysis, as shown in Figure 4A. RT-PCR and western blotting experiments confirmed that the expression levels of FTO in gastric cancer cells were higher than GES-1 in gastric mucosa cells (P < 0.05), as shown in Figure 4B and C. Moreover, there was a negative correlation with the expression levels of GATA6-AS1 (P < 0.01), as shown in Figure 4D.
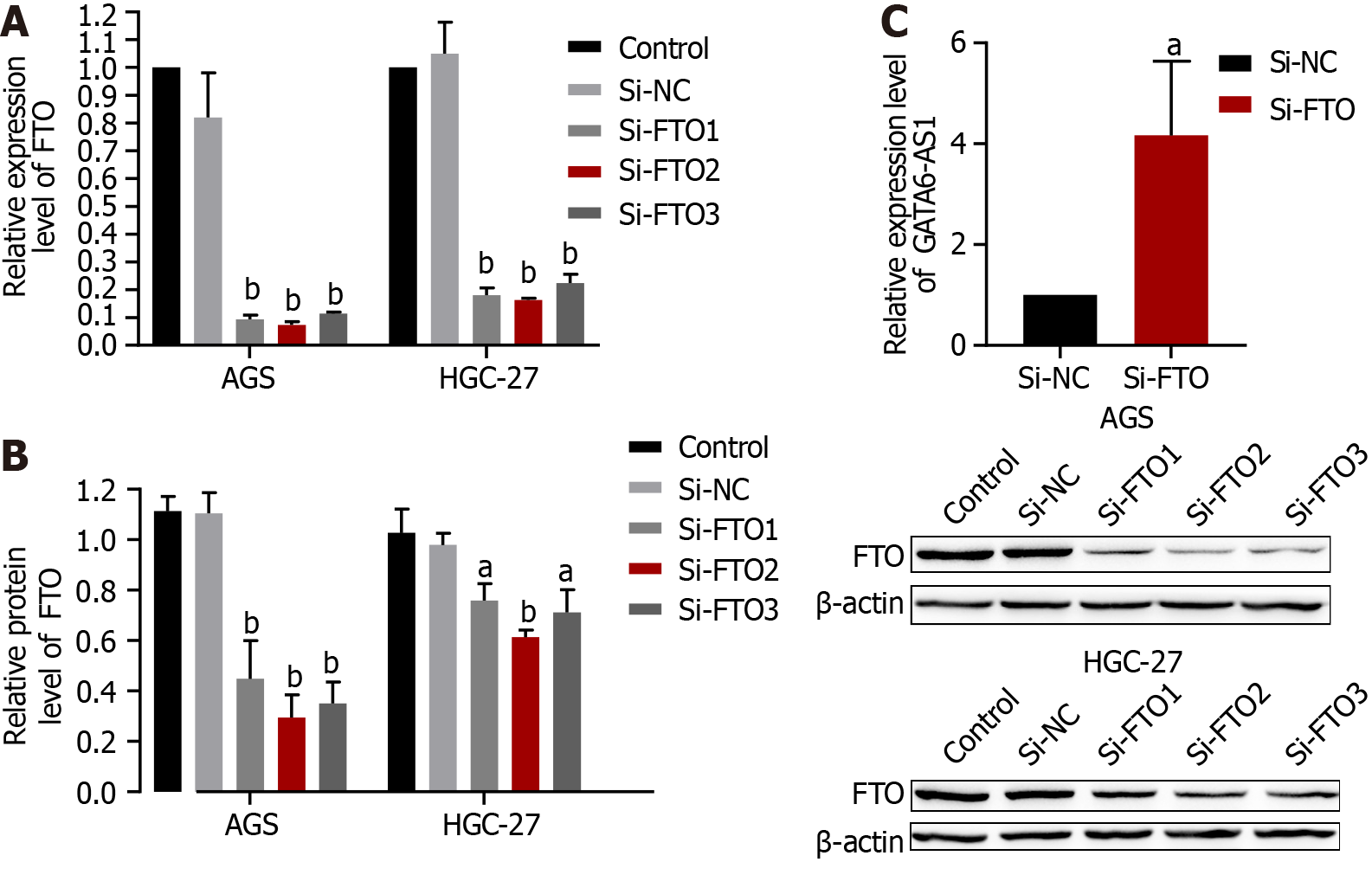
The expression of GATA6-AS1 was up-regulated after transfection with the siRNA knockdown of FTO expression. After transfection with Si-FTO, the interference efficiency was verified by RT-PCR and western blotting (P < 0.01), respectively. As shown in Figure 5A and B, Si-FTO2 exhibited the best interference efficiency. Subsequently, RT-PCR was used to detect changes in the expression levels of GATA6-AS1 after Si-FTO. Following the knockdown of FTO expression, the expression of GATA6-AS1 in AGS cells was up-regulated (P < 0.05) (Figure 5C).
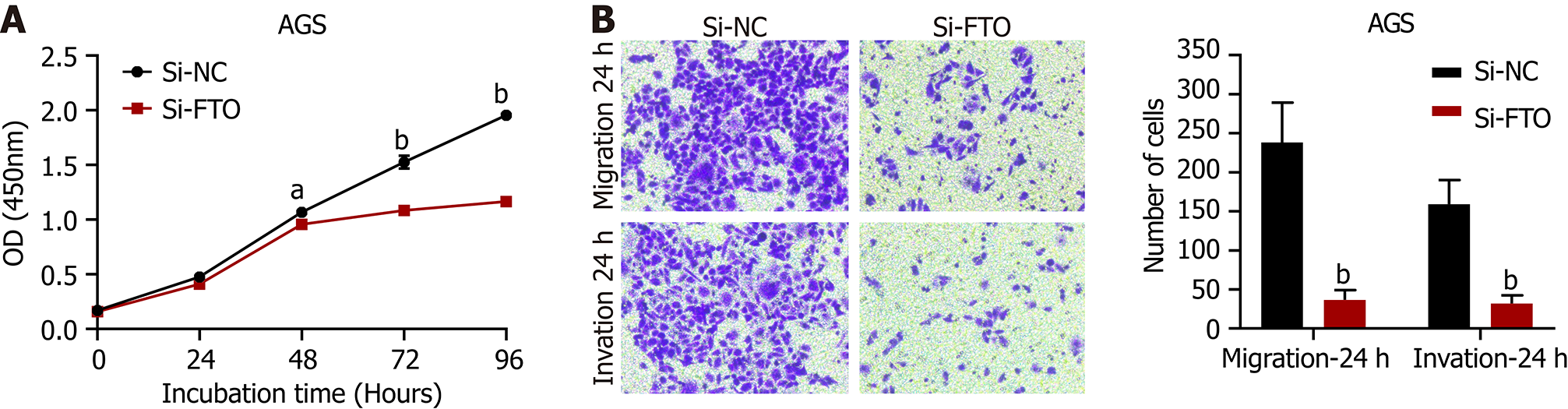
The proliferation, migration and invasion of gastric cancer cells were inhibited after the knockdown of FTO expression. The results of CCK8 proliferation assays showed that when compared with the negative control group, the proliferation ability of AGS cells in the FTO interference group was decreased (P < 0.05), as shown in Figure 6A. Trans-well migration and invasion assays showed that the migration and invasion ability of AGS cells in the FTO interference group were decreased when compared with the negative control group (P < 0.01), as shown in Figure 6B.
Gastric cancer is a global disease, with more than 1 million new cases every year. As most cases have reached the advanced stage when they are diagnosed and treated, gastric cancer has a high mortality rate and is now the third most common cause of death related to malignant tumors[17]. The onset of gastric cancer is also a progressive process, involving normal mucosa, chronic gastritis, multifocal atrophic gastritis, gastrointestinal metaplasia, dysplasia (uncertain, low grade, high grade), and finally, adenocarcinoma. This process underlies the complex diversity of the etiology and pathogenesis of gastric cancer. More and more studies have proven that lncRNAs exert a certain influence on the occurrence and development of gastric cancer[18]; however, the impact of lncRNA GATA6-AS1 on the biological function of gastric cancer remains unclear.
In the present study, bioanalysis of a database found that GATA6-AS1 have a significantly lower expression in gastric cancer tissues than normal tissues. Then, by constructing a lentivirus overexpression vector, we demonstrated that the expression levels of GATA6-AS1 were up-regulated; subsequently, we detected changes in the biological function of gastric cancer cell lines. The lentivirus infection of AGS and HGC-27 increased led to an increase in GATA6-AS1 expression by 30- and 40-fold, respectively; subsequent experiments were conducted with transfected cells. The proliferation, migration and invasion ability of HGC-27 and AGS cells in the GATA6-AS1 overexpression group were decreased when compared with the negative control and blank control groups (aP < 0.05). These data suggested that GATA6-AS1 acts as a tumor suppressor gene in gastric cancer. Next, we investigated the reasons responsible for the downregulation of GATA6-AS1 in gastric cancer and identified m6A modification sites on GATA6-AS1 which could modified by m6A demethylase FTO, as determined by database screening (http://starbase.sysu.edu.cn/starbase2/). Next, we confirmed the binding of GATA6-AS1 and FTO by performing RIP experiments. m6A modification is the most common modification in human RNA, and the modification site is mainly located in the DRACH sequence (D = A/G R = A/G, H = A/C/U). Modification does not change the coding ability of the transcript; rather, it only plays a certain regulatory role[19]. m6A is widely present in mRNA, microRNA, lncRNA, circular RNA, transfer RNA and so on, and is currently a study focus in the field of epigenetics[20]. With the increasing number of new technologies and new methods, such as second-generation sequencing, have been used in m6A research[21]; these studies proved that m6A abnormal modification has an important relationship with the occurrence , metastasis, progression, drug resistance and prognosis of various cancer[22]. Of these, FTO was the first demethylation enzyme to be discovered[23]; the m6A demethylation of FTO can lead to changes in protein levels by regulating mRNA stability, degradation and translation efficiency[24]. Over recent years, many studies have proven that FTO is involved in the progression of tumors. For example, FTO regulates the MALAT/miR-384/MAL2 axis via m6A modification, promotes the occurrence, and influences the prognosis of bladder cancer[25]. In addition, FTO can accelerate the progression of breast cancer and play a carcinogenic role by inhibiting the up-regulation of ARL5B by miR-181b-3p[26]. Furthermore, FTO promotes the growth of esophageal squamous cell carcinoma via the demethylation of lncRNA LINC00022[27]. In order to investigate the relationship between FTO and gastric cancer, we verified that the expression of FTO in gastric cancer cells was higher than GES-1 in gastric mucosa cells by RT-PCR and western blotting experiments; these data were consistent with our analysis of the bioinformatics database. Subsequently, the levels of FTO in gastric cancer cells were knocked down by the transfection of siRNA, and the interference efficiency was verified by RT-PCR and western blotting. The interference efficiency of the Si-FTO2 interference sequence was identified as the most efficient and could eliminate 93% and 84% of FTO in AGS and HGC-27 gastric cancer cells, respectively. Moreover, the expression levels of GATA6-AS1 in AGS cells after FTO knockout were detected by RT-PCR. We found that the expression levels of GATA6-AS1 in AGS cells were up-regulated after FTO knockdown. Finally, we conducted functional experiments with the cells affected by FTO interference and found that the proliferation, migration and invasion of AGS cells in the FTO interference group were decreased when compared with the negative control group. In conclusion, the effects of FTO interference and GATA6-AS1 overexpression on the biological characteristics of gastric cancer cells were consistent, thus further validating the functional combination of FTO and GATA6-AS1. In summary, we found that FTO was highly expressed in gastric cancer, and by down-regulating the expression level of GATA6-AS1 in gastric cancer, we were capable to boost the growth, migration and invasion ability of stomach cancer cells. However, we did not identify whether the methylation level of GATA6-AS1 can be increased after FTO knockdown. Furthermore, the downstream target molecules that GATA6-AS1 acts upon to inhibit the proliferation, migration and invasion of gastric cancer cells still need to be investigated. However, our findings provide new concepts for investigating the effect of FTO on gastric cancer via the regulation of GATA6-AS1.
In the present study, we found that lncRNA GATA6-AS1 was down-regulated in gastric cancer, and inhibited the proliferation, migration and invasion of gastric cancer cells, acting as a tumor suppressor gene in gastric cancer cells. FTO is highly expressed in gastric cancer, and the down-regulation of GATA6-AS1 in gastric cancer is regulated by the m6A demethylase FTO. Therefore, during the occurrence and development of gastric cancer, the overexpression of FTO may inhibit the expression of GATA6-AS1, thus promoting the proliferation and metastasis of gastric cancer. Furthermore, GATA6-AS1 is expected to become a new indicator for diagnosing gastric cancer and predicting tumor recurrence, which has important clinical significance.
Long non-coding RNA (lncRNA) GATA6-AS1 is known to be closely associated with tumorigenesis and development; however, its expression in gastric cancer and its role in the development of gastric cancer has not been fully explained. This study aimed to discuss the effects of GATA6-AS1 on the invasion, migration and proliferation of gastric malignant tumor cells by cell biology experiments, and investigate its mechanism of action.
Previous studies have reported that abnormal expression levels of lncRNA appear to influence the progression of gastric cancer, this paper first used bioinformatics analysis to screen out the lncRNA GATA6-AS1 that is abnormally expressed in gastric cancer tissues.
This study aimed to investigate the effects of GATA6-AS1 on the proliferation, invasion and migration of gastric cancer cells by cell biology experiments, and investigate its mechanism of action, so as to provide a basis for the early diagnosis and precise treatment of gastric cancer.
By constructing a lentivirus overexpression vector, we demonstrated that GATA6-AS1 acts as a tumor suppressor gene in gastric cancer. Next, we confirmed the binding of GATA6-AS1 and fat mass and obesity-associated protein (FTO) by performing RIP experiments.
This paper first used bioinformatics analysis to screen out the lncRNA GATA6-AS1 that is abnormally expressed in gastric cancer tissues. Then, through experimental research on the biological function of GATA6-AS1, it was confirmed that GATA6-AS1 can inhibit the proliferation, invasion, and migration of gastric tumor cells, suggesting that GATA6-AS1 plays a role as an anti-oncogene in the occurrence and development of gastric cancer. Further analysis of the reasons for the downregulation of GATA6-AS1 expression confirmed that it was regulated by the m6A demethylase FTO, which is upregulated in gastric cancer.
During the occurrence and development of gastric cancer, the overexpression of FTO may inhibit the expression of GATA6-AS1, thus promoting the proliferation and metastasis of gastric cancer.
GATA6-AS1 is expected to become a new indicator for diagnosing gastric cancer and predicting tumor recurrence, which has important clinical significance.
The authors would like to acknowledge Dao-Ping Sun, MD, Shu-Long Jiang, MD, Dong-Mei Shi, MD, for skillful technical assistance.
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 68675] [Article Influence: 13735.0] [Reference Citation Analysis (201)] |
| 2. | van der Werf LR, Wijnhoven BPL, Fransen LFC, van Sandick JW, Nieuwenhuijzen GAP, Busweiler LAD, van Hillegersberg R, Wouters MWJM, Luyer MDP, van Berge Henegouwen MI. A National Cohort Study Evaluating the Association Between Short-term Outcomes and Long-term Survival After Esophageal and Gastric Cancer Surgery. Ann Surg. 2019;270:868-876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 83] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 3. | Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202-209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5015] [Cited by in RCA: 5095] [Article Influence: 424.6] [Reference Citation Analysis (4)] |
| 4. | Chen LL, Carmichael GG. Long noncoding RNAs in mammalian cells: what, where, and why? Wiley Interdiscip Rev RNA. 2010;1:2-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 94] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 5. | Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol. 2021;220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 528] [Cited by in RCA: 1114] [Article Influence: 222.8] [Reference Citation Analysis (0)] |
| 6. | Tan YT, Lin JF, Li T, Li JJ, Xu RH, Ju HQ. LncRNA-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun (Lond). 2021;41:109-120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 524] [Article Influence: 87.3] [Reference Citation Analysis (0)] |
| 7. | Zhang E, He X, Zhang C, Su J, Lu X, Si X, Chen J, Yin D, Han L, De W. A novel long noncoding RNA HOXC-AS3 mediates tumorigenesis of gastric cancer by binding to YBX1. Genome Biol. 2018;19:154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 167] [Cited by in RCA: 228] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 8. | Sun M, Nie F, Wang Y, Zhang Z, Hou J, He D, Xie M, Xu L, De W, Wang Z, Wang J. LncRNA HOXA11-AS Promotes Proliferation and Invasion of Gastric Cancer by Scaffolding the Chromatin Modification Factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76:6299-6310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 409] [Article Influence: 40.9] [Reference Citation Analysis (0)] |
| 9. | Sun Q, Li J, Li F, Li H, Bei S, Zhang X, Feng L. LncRNA LOXL1-AS1 facilitates the tumorigenesis and stemness of gastric carcinoma via regulation of miR-708-5p/USF1 pathway. Cell Prolif. 2019;52:e12687. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 65] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 10. | Liu YW, Xia R, Lu K, Xie M, Yang F, Sun M, De W, Wang C, Ji G. LincRNAFEZF1-AS1 represses p21 expression to promote gastric cancer proliferation through LSD1-Mediated H3K4me2 demethylation. Mol Cancer. 2017;16:39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 159] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 11. | Sigova AA, Mullen AC, Molinie B, Gupta S, Orlando DA, Guenther MG, Almada AE, Lin C, Sharp PA, Giallourakis CC, Young RA. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci U S A. 2013;110:2876-2881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 338] [Cited by in RCA: 370] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 12. | Chen XY, Zhang J, Zhu JS. The role of m(6)A RNA methylation in human cancer. Mol Cancer. 2019;18:103. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 400] [Cited by in RCA: 828] [Article Influence: 118.3] [Reference Citation Analysis (0)] |
| 13. | Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 5' UTR m(6)A Promotes Cap-Independent Translation. Cell. 2015;163:999-1010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1005] [Cited by in RCA: 1513] [Article Influence: 137.5] [Reference Citation Analysis (0)] |
| 14. | Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomed Pharmacother. 2019;112:108613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 631] [Article Influence: 90.1] [Reference Citation Analysis (0)] |
| 15. | Wei W, Ji X, Guo X, Ji S. Regulatory Role of N(6) -methyladenosine (m(6) A) Methylation in RNA Processing and Human Diseases. J Cell Biochem. 2017;118:2534-2543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 136] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 16. | Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou A, Liu J, Che L, Li J. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3. Mol Cancer. 2019;18:143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 220] [Cited by in RCA: 484] [Article Influence: 69.1] [Reference Citation Analysis (0)] |
| 17. | Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396:635-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1150] [Cited by in RCA: 3308] [Article Influence: 551.3] [Reference Citation Analysis (6)] |
| 18. | Zeng X, Shi H, Wang J, Cui S, Tang H, Zhang X. Long noncoding RNA aberrant expression profiles after cytoreductive surgery and hyperthermic intraperitoneal chemotherapy of AGC ascertained by microarray analysis. Tumour Biol. 2015;36:5021-5029. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 19. | Song P, Tayier S, Cai Z, Jia G. RNA methylation in mammalian development and cancer. Cell Biol Toxicol. 2021;37:811-831. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 89] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 20. | Xie F, Huang C, Liu F, Zhang H, Xiao X, Sun J, Zhang X, Jiang G. CircPTPRA blocks the recognition of RNA N(6)-methyladenosine through interacting with IGF2BP1 to suppress bladder cancer progression. Mol Cancer. 2021;20:68. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 111] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 21. | Nechay M, Kleiner RE. High-throughput approaches to profile RNA-protein interactions. Curr Opin Chem Biol. 2020;54:37-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 22. | Tsuchiya K, Yoshimura K, Inoue Y, Iwashita Y, Yamada H, Kawase A, Watanabe T, Tanahashi M, Ogawa H, Funai K, Shinmura K, Suda T, Sugimura H. YTHDF1 and YTHDF2 are associated with better patient survival and an inflamed tumor-immune microenvironment in non-small-cell lung cancer. Oncoimmunology. 2021;10:1962656. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 73] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 23. | Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, He C. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885-887. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2803] [Cited by in RCA: 3281] [Article Influence: 218.7] [Reference Citation Analysis (7)] |
| 24. | Azzam SK, Alsafar H, Sajini AA. FTO m6A Demethylase in Obesity and Cancer: Implications and Underlying Molecular Mechanisms. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 135] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 25. | Tao L, Mu X, Chen H, Jin D, Zhang R, Zhao Y, Fan J, Cao M, Zhou Z. FTO modifies the m6A level of MALAT and promotes bladder cancer progression. Clin Transl Med. 2021;11:e310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 97] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 26. | Xu Y, Ye S, Zhang N, Zheng S, Liu H, Zhou K, Wang L, Cao Y, Sun P, Wang T. The FTO/miR-181b-3p/ARL5B signaling pathway regulates cell migration and invasion in breast cancer. Cancer Commun (Lond). 2020;40:484-500. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 139] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 27. | Cui Y, Zhang C, Ma S, Li Z, Wang W, Li Y, Ma Y, Fang J, Wang Y, Cao W, Guan F. RNA m6A demethylase FTO-mediated epigenetic up-regulation of LINC00022 promotes tumorigenesis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2021;40:294. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 122] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Dilek ON, Turkey S-Editor: Qu XL L-Editor: A P-Editor: Zhao S