Published online Nov 15, 2024. doi: 10.4251/wjgo.v16.i11.4300
Revised: June 13, 2024
Accepted: July 2, 2024
Published online: November 15, 2024
Processing time: 242 Days and 21.5 Hours
This editorial reviews the molecular mechanisms underlying the roles of the long non-coding RNA (lncRNA) small nucleolar RNA host gene 16 (SNHG16) in digestive system cancers based on two recent studies on lncRNAs in digestive system tumors. The first study, by Zhao et al, explored how hBD-1 affects colon cancer, via the lncRNA TCONS_00014506, by inhibiting mTOR and promoting autophagy. The second one, by Li et al, identified the lncRNA prion protein testis specific (PRNT) as a factor in oxaliplatin resistance by sponging ZNF184 to regulate HIPK2 and influence colorectal cancer progression and chemoresistance, suggesting PRNT as a potential therapeutic target for colorectal cancer. Both of these two articles discuss the mechanisms by which lncRNAs contribute to the development and progression of digestive system cancers. As a recent research hotspot, SNHG16 is a typical lncRNA that has been extensively studied for its association with digestive system cancers. The prevailing hypothesis is that SNHG16 participates in the development and progression of digestive system tumors by acting as a competing endogenous RNA, interacting with other pro
Core Tip: The long non-coding RNA (lncRNA) small nucleolar RNA host gene 16 (SNHG16) plays a significant role in the development and progression of various digestive system cancers, including esophageal, liver, pancreatic, gastric, and colorectal cancers. It is involved in processes such as cell proliferation, migration, invasion, apoptosis, and chemoresistance. SNHG16 acts as a competing endogenous RNA, interacting with microRNAs and regulating target genes, and is associated with a poor prognosis in digestive system cancers. Its expression is influenced by transcription factors and its polymorphisms are linked to cancer susceptibility. SNHG16 has potential as a biomarker and therapeutic target for digestive system cancers.
- Citation: Yang TF, Li XR, Kong MW. Molecular mechanisms underlying roles of long non-coding RNA small nucleolar RNA host gene 16 in digestive system cancers. World J Gastrointest Oncol 2024; 16(11): 4300-4308
- URL: https://www.wjgnet.com/1948-5204/full/v16/i11/4300.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i11.4300
This editorial reviews the molecular mechanisms underlying the roles of the long non-coding RNA (lncRNA) small nucleolar RNA host gene 16 (SNHG16) in digestive system cancers based on two recent studies on long lncRNAs in colorectal cancer (CRC), one is by Zhao et al[1] and the other by Li et al[2]. The study of Zhao et al[1] linked hBD-1 to mTOR pathway regulation and autophagy via the lncRNA TCONS_00014506, highlighting the potential of hBD-1 in cancer cell destruction. The study of Li et al[2] identified the lncRNA prion protein testis specific (PRNT) as a regulator of oxaliplatin resistance, showing its upregulation in resistant CRC cells and its role in HIPK2 expression, suggesting PRNT as a therapeutic target for CRC treatment. Both of these two articles discuss the mechanisms by which lncRNAs con
Digestive system tumors remain a major cause of global mortality, with increasing incidence and mortality rates[1,2]. In 2018, there were approximately 18.1 million new cancer cases and 9.6 million cancer-related deaths worldwide, with digestive system tumors accounting for a significant proportion[3]. Despite significant advances in understanding the potential molecular mechanisms of digestive system tumors[4-7] and substantial progress in their treatment[8,9], the recurrence and mortality rates of digestive system tumors remain dismal[1]. Therefore, it is crucial to seek novel effective biomarkers and therapeutic targets for these tumors.
Recent studies have shown that non-coding RNAs (ncRNAs) play a role in the development of digestive system tumors[10]. ncRNAs are generally divided into two categories based on their length: Small ncRNAs with less than 200 nuc
This paper aims to systematically review the recent findings on the biological functions, related mechanisms, and potential clinical significance of SNHG16 in various digestive system cancers, and to explore the close relationship between SNHG16 and digestive system cancers.
LncRNAs are single-stranded RNA molecules transcribed by RNA polymerase II, with a length of more than 200 nts and lacking protein-coding ability[22]. Overwhelming evidence from numerous studies indicates that lncRNAs play a crucial role in the development, proliferation, migration, and prognosis of various cancers by regulating a series of biological processes, such as interacting with target genes at the transcriptional level, regulating histone modifications and chromatin remodeling, and interacting with miRNAs of approximately 22 nts in length (also known as competing endogenous RNAs [ceRNAs])[22-25]. For example, the lncRNA BC069792 acts as a ceRNA to sponge miR-658 and miR-4739, increasing the expression of the target gene KCNQ4, leading to AKT phosphorylation, and thus inhibiting the proliferation and invasion of breast cancer[24,25].
SNHG16 is a member of the SNHG family, located on human chromosome 17q25.1, and consists of four exons. Initially, it was identified as a potent carcinogenic factor that promotes the progression of neuroblastoma[26]. Therefore, SNHG16 is also known as non-coding RNA expressed in aggressive neuroblastoma. Subsequent studies further revealed the widespread involvement of SNHG16 in the complex molecular regulatory networks in different human cancers[27-29]. For instance, by regulating the miR-32-5p/SPN axis, the silencing of SNHG16 inhibits the proliferation and radioresistance of nasopharyngeal carcinoma cells[28]. SNHG16 may act as an oncogene by binding and recruiting EZH2 to the p21 promoter, silencing the expression of p21, thereby promoting cell proliferation and reducing apoptosis in bladder cancer cells[29]. SNHG16 plays a key role in the progression, distant metastasis, and prognosis of ovarian cancer by increasing the expression of MMP9[30]. In addition, in oral squamous cell carcinoma, the expression of SNHG16 is regulated by a transcription factor called c-Myc, which recruits histone acetyltransferases and induces the clearance of RNA polymerase II[31]. These functions indicate that SNHG16 plays an important role in the progression, invasion, and carcinogenesis of human cancers through various molecular mechanisms.
Esophageal cancer (EC) is one of the major cancer types worldwide, ranking 7th in incidence (3.1%, 604100 new cases) and 6th in mortality (5.5%, 544076 deaths) among all cancers[2]. There are differences in the incidence and mortality rates of EC across geographic regions[2]. For example, due to economic underdevelopment and dietary habits, the burden of EC in East Asia is heavier, with esophageal squamous cell carcinoma (ESCC) being the predominant histological type[2]. Studies have shown that SNHG16 is upregulated in EC and closely related to tumor stage, lymph node metastasis, and clinical stage. Silencing of SNHG16 inhibits the proliferation and invasion of EC-1 and Eca-109 cells by reducing the expression of β-catenin, cyclin D1, and c-Myc proteins, and promotes apoptosis[32]. This study also showed that SNHG16 is upregulated in ESCC tissue and cell lines, and disrupting SNHG16 expression promotes apoptosis and inhibits epithelial-mesenchymal transition (EMT) through the miR-140-5p/ZEB1 axis. Another study found that increased expression of SNHG16 also promotes the growth and metastasis of ESCC, and is related to tumor differentiation and T stage, with the mechanism being that SNHG16 can bind and recruit EIF4A3 to regulate the expression of RhoU by enhancing the stability of RhoU mRNA[33]. These results indicate that the upregulation of SNHG16 is closely related to the development of ESCC, and SNHG16 is expected to serve as a marker for ESCC, providing new clues for its clinical treatment and the development of related drugs.
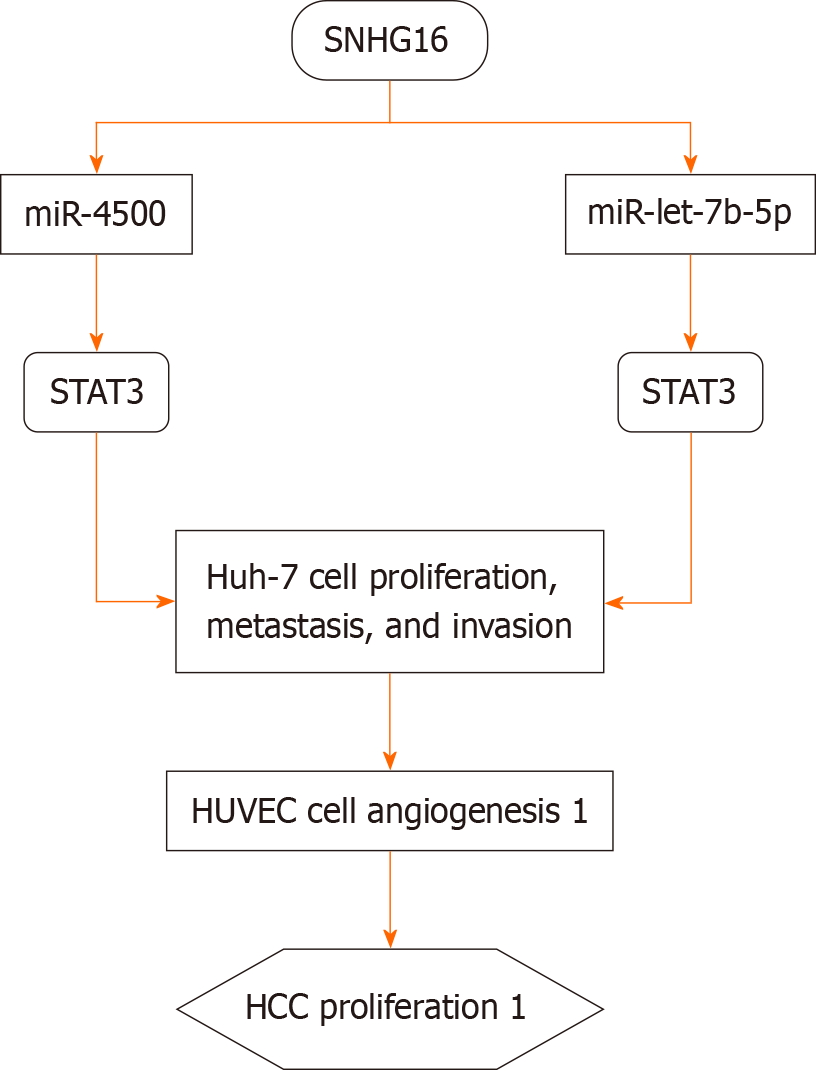
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors worldwide, accounting for 4.7% of all new cancer cases (906000 cases) and 8.3% of cancer-related deaths (830000 cases), ranking sixth and third among all malignancies, respectively[2]. In most studies, SNHG16 is considered an oncogene for HCC, and the upregulation of SNHG16 expression is closely related to the malignant prognosis and tumor stage of HCC. The expression of SNHG16 in late-stage HCC is significantly higher than that in early-stage HCC[34,35]. Additionally, high expression of SNHG16 is also associated with tumor size, TNM stage, and vascular invasion in HCC patients[36]. SNHG16, as a ceRNA, targets STAT3 and GALNT1 through miR-4500 and miR-let-7b-5p in Huh-7 and HUVEC cells, respectively, promoting the proliferation, metastasis, and angiogenesis of Huh-7 cells and enhancing vascular formation in HUVEC cells[34,36] (Figure 1).
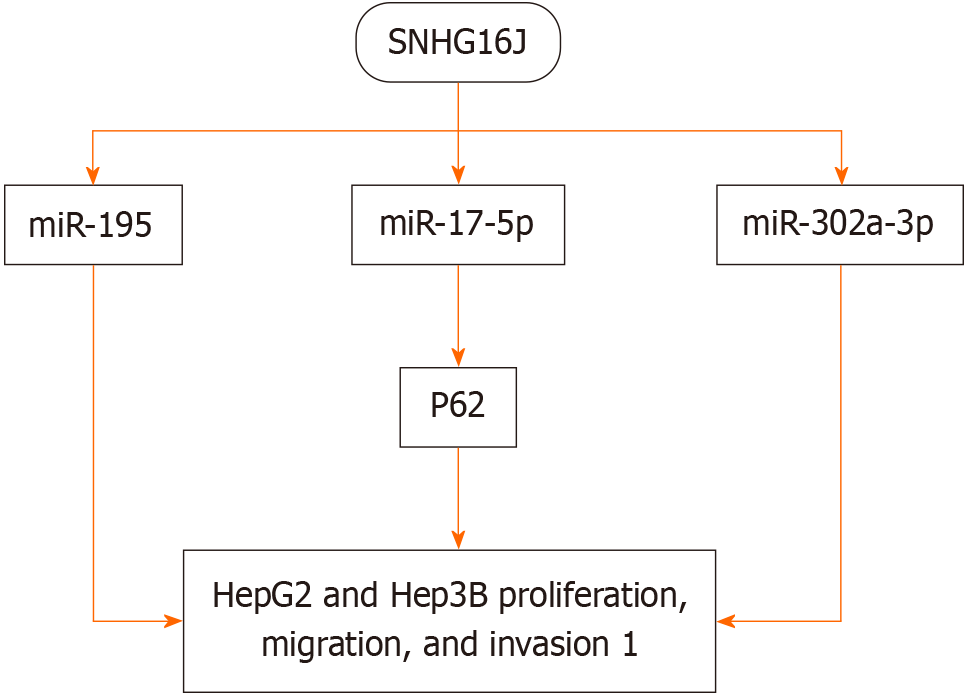
Other studies have shown that downregulating SNHG16 expression affects the SNHG16/miR-195, SNHG16/miR-17-5p/P62, and SNHG16/miR-302a-3p/FGF19 axes, inhibiting the proliferation, migration, and invasion of HepG2 and Hep3B cells[34,37-39] (Figure 2).
The overexpression of SNHG16 affects the G2/M transition of HCC cells by sponging miR-let-7b-5p[40]. SNHG16 is overexpressed in sorafenib-resistant HCC tumor tissues and cells, enhancing the resistance of HCC cells to sorafenib[36]. Conversely, when the expression of SNHG16 is inhibited, the resistance to sorafenib disappears[41]. Assarzadegan et al[42] proposed that SNHG16 enhances the autophaic function of HCC cells to protect HCC from sorafenib resistance through the miR-23b-3p/EGR1 axis. Moreover, SNHG16 exosomes can be engulfed by microglia, and through the miR-942-3p/MMP9 axis, they mediate microglia to promote the metastasis of HCC cells[43]. In addition, Hu et al[44] found that the overexpression of SNHG16 promotes the expression of TRAF6 by sponging miR-605-3p, activating NF-κB and thus promoting the development of HCC. Conversely, activated NF-κB can enhance the activity of the SNHG16 promoter, forming a positive SNHG16-NF-κB feedback loop, further exacerbating HCC.
Studies have shown that SNHG16 regulates a large lncRNA-miRNA-mRNA network in HCC and is closely related to immune cell infiltration, the release of immune regulatory factors, and the expression of chemokines in tumor tissues[45-47]. In addition, researchers have pointed out that the progression of HCC promoted by UBE4B and SEMA3F is regulated by its upstream SNHG16/miR-22-3p and SNHG16/Let-7c-5p axes[47,48]. Furthermore, Liu et al[49] revealed that SNHG16 can serve as a potential biomarker for poor prognosis in HCC patients. In summary, SNHG16 is upregulated in HCC and promotes the development of HCC. However, a recent paper presented the opposite view, suggesting that compared to normal liver tissue, the expression of SNHG16 is reduced in HCC, and the overexpression of SNHG16 reduces the proliferation of Hep3B and Huh-7 cells by sponging miR-93, inhibiting the development and chemoresistance of HCC[50]. Further research is needed to address the above-mentioned inconsistent findings.
Pancreatic cancer (PC) is one of the most severe malignant tumors in the digestive system. Due to its poor prognosis, the number of PC-related deaths (466000 cases) is almost equivalent to the number of cases (496000 cases), making it the seventh leading cause of cancer death in both genders[2]. Similar to EC, the incidence of PC in high human development index (HDI) countries is 4 to 5 times higher than that in low HDI countries[2]. Studies have shown that SNHG16 exp
Gastric cancer (GC) remains a significant malignancy worldwide, being the fifth most common malignant tumor (5.6%, over 1000000 new cases) and the fourth leading cause of cancer-related deaths (7.7%, about 769000 deaths)[2]. GC has different characteristics in different parts of the world. The highest age-standardized incidence rate was observed in East Asia, followed by Central and Eastern Europe[2]. Many studies have shown that the expression of SNHG16 is signi
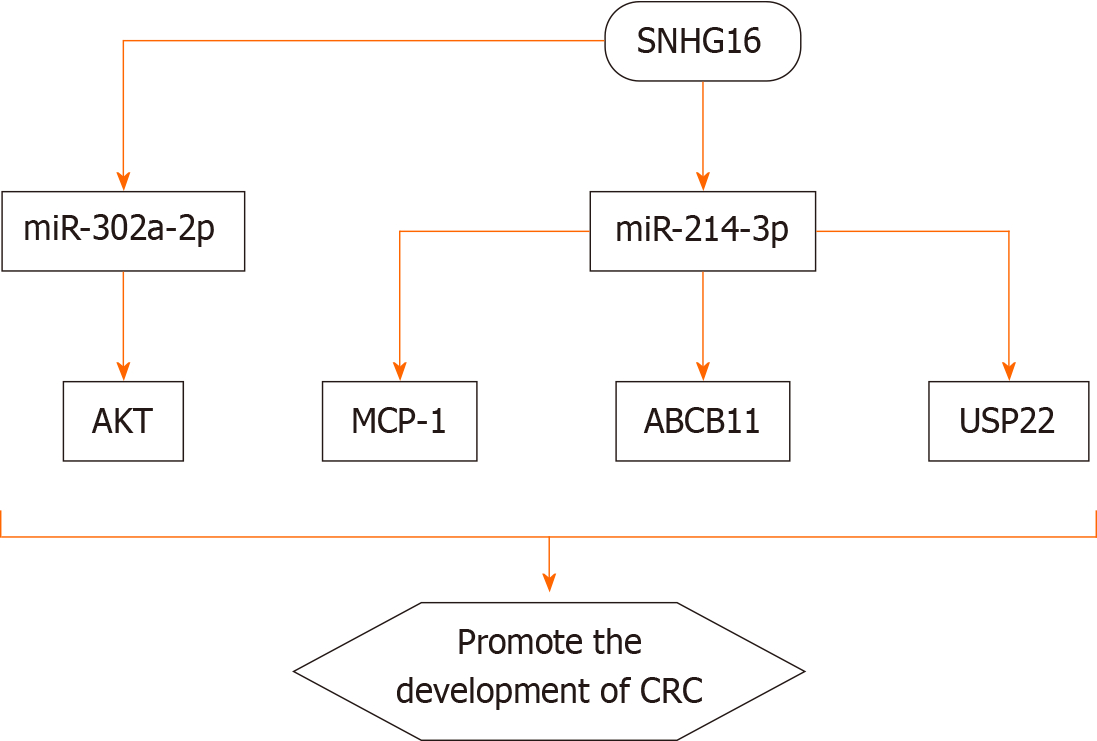
In 2020, there were over 1.9 million new cases of CRC worldwide, with 935000 deaths, ranking third in incidence after breast cancer in women and lung cancer, and second in mortality, close to lung cancer[2]. Increasing evidence indicates that the expression level of SNHG16 is positively correlated with advanced TNM stage, distant metastasis, and shortened overall survival time in CRC[61-63]. SNHG16 is mainly present in the cytoplasm, suggesting that SNHG16 functions as a ceRNA to regulate the activity of multiple miRNAs and target genes. Li et al[62] revealed that SNHG16 is associated with the malignancy and poor prognosis of CRC by sponging miR-200a-3p. Tan et al[64] demonstrated that SNHG16 promotes the proliferation of CRC cells by upregulating its target gene ABCB1 through interaction with miR-214-3p. It has also been found that SNHG16 promotes the progression of CRC by activating the expression of USP22 via sponging miR-132-3p[65,66]. Ke et al[67] discovered that SNHG16 supports the growth of colon cancer cells by targeting the miR-302a-2p/AKT axis. Chen et al[47] showed that the expression level of SNHG16 in cancerous tissue is higher than that in matched normal tissue and is positively correlated with CRC grade. Moreover, SNHG16 promotes the proliferation, migration, and EMT of CRC cells via the miR-124-3p/MCP-1 axis[48]. Bioinformatics analysis also led to the same conclusion, indicating that SNHG16 plays an important role in CRC[50,53]. Recent studies have further established the close relationship between SNHG16 and autophagy in CRC[68,69].
The expression of SNHG16 is also activated by other proteins such as c-Myc. Christensen et al[70] found that in CRC, the expression of SNHG16 is determined by Wnt-regulated transcription factors, and the inhibition of β-catenin reduces the expression of SNHG16 and c-Myc. Additionally, the inhibition or overexpression of c-Myc can respectively decrease or increase the expression of SNHG16. In another study, Xiang et al[71] for the first time discovered a positive feedback loop of SNHG16-YAP1/TEAD1 in CRC cells. SNHG16, as a ceRNA, physically binds to miR-195-5p, further regulating the expression of YAP1 and promoting tumor progression. YAP1 binds to TEAD1 to form a YAP1/TEAD1 complex, which in turn binds to two sites on the SNHG16 promoter, activating the transcription of SNHG16[71]. The mechanism of
The polymorphisms of SNHG16 are significantly associated with susceptibility to CRC. The A>G variant at the rs7353 locus of the SNHG16 gene is associated with a reduced susceptibility to CRC. However, the G>A variant at the rs8038 locus and the A>G and G>A variants at the rs15278 locus may increase the susceptibility to CRC[72].
An increasing number of studies indicate that the occurrence of tumors is caused by a combination of genetic and environmental factors. The focus of this editorial is on the molecular mechanisms underlying the roles of the lncRNA SNHG16 in the development and progression of digestive system cancers (Figure 3). Existing data suggest that SNHG16 is closely related to the proliferation, migration, invasion, apoptosis, and poor prognosis of EC, HCC, PC, GC, and CRC. Moreover, SNHG16 is positively correlated with clinical stage and lymph node metastasis of digestive system cancers. In terms of mechanisms of action of SNHG16 in digestive system cancers, there are mainly four aspects: (1) Many transcription factors, including CTCF, c-Myc, NF-κB, STAT3, and TEAD1, are positively correlated with SNHG16; (2) SNHG16 directly controls the expression of downstream target genes such as DKK3; (3) SNHG16 can bind and recruit EIF4A3 to regulate the expression of RhoU by enhancing the stability of RhoU mRNA, and SNHG16 can also bind to EZH2 and recruit EZH2 to the promoter of Smad4, thereby inhibiting the expression of Smad4; and (4) SNHG16 can compete with miRNAs, regulate the expression of downstream target genes, and activate different signaling pathways. However, these studies are only preliminary, and the expression levels of SNHG16 in body fluids and chemical stability have not yet been clarified. In addition, the clinical application of SNHG16 as a biomarker in digestive system cancers needs further research in the future.
| 1. | Zhao YX, Cui Y, Li XH, Yang WH, An SX, Cui JX, Zhang MY, Lu JK, Zhang X, Wang XM, Bao LL, Zhao PW. Human β-defensin-1 affects the mammalian target of rapamycin pathway and autophagy in colon cancer cells through long non-coding RNA TCONS_00014506. World J Gastrointest Oncol. 2024;16:1465-1478. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 2. | Li SN, Yang S, Wang HQ, Hui TL, Cheng M, Zhang X, Li BK, Wang GY. Upregulated lncRNA PRNT promotes progression and oxaliplatin resistance of colorectal cancer cells by regulating HIPK2 transcription. World J Gastrointest Oncol. 2024;16:1564-1577. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 3. | Xu W, Li B, Xu M, Yang T, Hao X. Traditional Chinese medicine for precancerous lesions of gastric cancer: A review. Biomed Pharmacother. 2022;146:112542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 106] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 4. | Zhang X, Qiu H, Li C, Cai P, Qi F. The positive role of traditional Chinese medicine as an adjunctive therapy for cancer. Biosci Trends. 2021;15:283-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 254] [Article Influence: 50.8] [Reference Citation Analysis (0)] |
| 5. | Hojman P, Gehl J, Christensen JF, Pedersen BK. Molecular Mechanisms Linking Exercise to Cancer Prevention and Treatment. Cell Metab. 2018;27:10-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 418] [Article Influence: 52.3] [Reference Citation Analysis (0)] |
| 6. | Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017;77:3965-3981. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2088] [Cited by in RCA: 2229] [Article Influence: 247.7] [Reference Citation Analysis (0)] |
| 7. | Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4127] [Cited by in RCA: 5722] [Article Influence: 381.5] [Reference Citation Analysis (0)] |
| 8. | Lorenzi L, Avila Cobos F, Decock A, Everaert C, Helsmoortel H, Lefever S, Verboom K, Volders PJ, Speleman F, Vandesompele J, Mestdagh P. Long noncoding RNA expression profiling in cancer: Challenges and opportunities. Genes Chromosomes Cancer. 2019;58:191-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 110] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 9. | Zhang Y, Dong X, Guo X, Li C, Fan Y, Liu P, Yuan D, Ma X, Wang J, Zheng J, Li H, Gao P. LncRNA-BC069792 suppresses tumor progression by targeting KCNQ4 in breast cancer. Mol Cancer. 2023;22:41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 61] [Reference Citation Analysis (0)] |
| 10. | Yu M, Ohira M, Li Y, Niizuma H, Oo ML, Zhu Y, Ozaki T, Isogai E, Nakamura Y, Koda T, Oba S, Yu B, Nakagawara A. High expression of ncRAN, a novel non-coding RNA mapped to chromosome 17q25.1, is associated with poor prognosis in neuroblastoma. Int J Oncol. 2009;34:931-938. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 56] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 11. | Gong CY, Tang R, Nan W, Zhou KS, Zhang HH. Role of SNHG16 in human cancer. Clin Chim Acta. 2020;503:175-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 12. | Ghafouri-Fard S, Khoshbakht T, Taheri M, Shojaei S. A Review on the Role of Small Nucleolar RNA Host Gene 6 Long Non-coding RNAs in the Carcinogenic Processes. Front Cell Dev Biol. 2021;9:741684. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 13. | Yang M, Wei W. SNHG16: A Novel Long-Non Coding RNA in Human Cancers. Onco Targets Ther. 2019;12:11679-11690. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 14. | Zhang W, Zhou X, Tang Z, Fu L, Zou S, Tang S. Knockdown of lncRNA SNHG16 Attenuates the Proliferation and Radioresistance of Nasopharyngeal Carcinoma Cells by Mediating miR-31-5p/SFN Axis. Radiat Res. 2023;199:124-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 15. | Cao X, Xu J, Yue D. LncRNA-SNHG16 predicts poor prognosis and promotes tumor proliferation through epigenetically silencing p21 in bladder cancer. Cancer Gene Ther. 2018;25:10-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 86] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 16. | Yang XS, Wang GX, Luo L. Long non-coding RNA SNHG16 promotes cell growth and metastasis in ovarian cancer. Eur Rev Med Pharmacol Sci. 2018;22:616-622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 21] [Reference Citation Analysis (0)] |
| 17. | Li S, Zhang S, Chen J. c-Myc induced upregulation of long non-coding RNA SNHG16 enhances progression and carcinogenesis in oral squamous cell carcinoma. Cancer Gene Ther. 2019;26:400-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 18. | Sealock T, Sharma S. Smoking Cessation. 2023 Jan 23. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2024 Jan-. [PubMed] |
| 19. | GBD 2019 Cancer Risk Factors Collaborators. The global burden of cancer attributable to risk factors, 2010-19: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. 2022;400:563-591. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 620] [Cited by in RCA: 538] [Article Influence: 134.5] [Reference Citation Analysis (0)] |
| 20. | Sheikh M, Roshandel G, McCormack V, Malekzadeh R. Current Status and Future Prospects for Esophageal Cancer. Cancers (Basel). 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 162] [Article Influence: 54.0] [Reference Citation Analysis (0)] |
| 21. | Cai C, Huo Q, Wang X, Chen B, Yang Q. SNHG16 contributes to breast cancer cell migration by competitively binding miR-98 with E2F5. Biochem Biophys Res Commun. 2017;485:272-278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 140] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 22. | Marti-Aguado D, Clemente-Sanchez A, Bataller R. Cigarette smoking and liver diseases. J Hepatol. 2022;77:191-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 119] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 23. | Lee YC, Cohet C, Yang YC, Stayner L, Hashibe M, Straif K. Meta-analysis of epidemiologic studies on cigarette smoking and liver cancer. Int J Epidemiol. 2009;38:1497-1511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 201] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 24. | Abdel-Rahman O, Helbling D, Schöb O, Eltobgy M, Mohamed H, Schmidt J, Giryes A, Mehrabi A, Iype S, John H, Tekbas A, Zidan A, Oweira H. Cigarette smoking as a risk factor for the development of and mortality from hepatocellular carcinoma: An updated systematic review of 81 epidemiological studies. J Evid Based Med. 2017;10:245-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 88] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 25. | Petrick JL, Campbell PT, Koshiol J, Thistle JE, Andreotti G, Beane-Freeman LE, Buring JE, Chan AT, Chong DQ, Doody MM, Gapstur SM, Gaziano JM, Giovannucci E, Graubard BI, Lee IM, Liao LM, Linet MS, Palmer JR, Poynter JN, Purdue MP, Robien K, Rosenberg L, Schairer C, Sesso HD, Sinha R, Stampfer MJ, Stefanick M, Wactawski-Wende J, Zhang X, Zeleniuch-Jacquotte A, Freedman ND, McGlynn KA. Tobacco, alcohol use and risk of hepatocellular carcinoma and intrahepatic cholangiocarcinoma: The Liver Cancer Pooling Project. Br J Cancer. 2018;118:1005-1012. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 155] [Cited by in RCA: 184] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 26. | Klein AP. Pancreatic cancer epidemiology: understanding the role of lifestyle and inherited risk factors. Nat Rev Gastroenterol Hepatol. 2021;18:493-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 854] [Article Influence: 170.8] [Reference Citation Analysis (0)] |
| 27. | Weissman S, Takakura K, Eibl G, Pandol SJ, Saruta M. The Diverse Involvement of Cigarette Smoking in Pancreatic Cancer Development and Prognosis. Pancreas. 2020;49:612-620. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 28. | Yuan S, Chen J, Ruan X, Sun Y, Zhang K, Wang X, Li X, Gill D, Burgess S, Giovannucci E, Larsson SC. Smoking, alcohol consumption, and 24 gastrointestinal diseases: Mendelian randomization analysis. Elife. 2023;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 138] [Article Influence: 46.0] [Reference Citation Analysis (0)] |
| 29. | Li H, Chen X, Hoffmeister M, Brenner H. Associations of smoking with early- and late-onset colorectal cancer. JNCI Cancer Spectr. 2023;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 25] [Reference Citation Analysis (0)] |
| 30. | Uhlenhopp DJ, Then EO, Sunkara T, Gaduputi V. Epidemiology of esophageal cancer: update in global trends, etiology and risk factors. Clin J Gastroenterol. 2020;13:1010-1021. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 497] [Article Influence: 82.8] [Reference Citation Analysis (1)] |
| 31. | McGee EE, Jackson SS, Petrick JL, Van Dyke AL, Adami HO, Albanes D, Andreotti G, Beane-Freeman LE, Berrington de Gonzalez A, Buring JE, Chan AT, Chen Y, Fraser GE, Freedman ND, Gao YT, Gapstur SM, Gaziano JM, Giles GG, Grant EJ, Grodstein F, Hartge P, Jenab M, Kitahara CM, Knutsen SF, Koh WP, Larsson SC, Lee IM, Liao LM, Luo J, Milne RL, Monroe KR, Neuhouser ML, O'Brien KM, Peters U, Poynter JN, Purdue MP, Robien K, Sandler DP, Sawada N, Schairer C, Sesso HD, Simon TG, Sinha R, Stolzenberg-Solomon R, Tsugane S, Wang R, Weiderpass E, Weinstein SJ, White E, Wolk A, Yuan JM, Zeleniuch-Jacquotte A, Zhang X, Zhu B, McGlynn KA, Campbell PT, Koshiol J. Smoking, Alcohol, and Biliary Tract Cancer Risk: A Pooling Project of 26 Prospective Studies. J Natl Cancer Inst. 2019;111:1263-1278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 76] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 32. | Zhang K, Chen J, Song H, Chen LB. SNHG16/miR-140-5p axis promotes esophagus cancer cell proliferation, migration and EMT formation through regulating ZEB1. Oncotarget. 2018;9:1028-1040. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 70] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 33. | Ren L, Fang X, Shrestha SM, Ji Q, Ye H, Liang Y, Liu Y, Feng Y, Dong J, Shi R. LncRNA SNHG16 promotes development of oesophageal squamous cell carcinoma by interacting with EIF4A3 and modulating RhoU mRNA stability. Cell Mol Biol Lett. 2022;27:89. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 34. | Xie X, Xu X, Sun C, Yu Z. Long intergenic noncoding RNA SNHG16 interacts with miR-195 to promote proliferation, invasion and tumorigenesis in hepatocellular carcinoma. Exp Cell Res. 2019;383:111501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 35. | Lin Q, Zheng H, Xu J, Zhang F, Pan H. LncRNA SNHG16 aggravates tumorigenesis and development of hepatocellular carcinoma by sponging miR-4500 and targeting STAT3. J Cell Biochem. 2019;120:11604-11615. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 36. | Ye J, Zhang R, Du X, Chai W, Zhou Q. Long noncoding RNA SNHG16 induces sorafenib resistance in hepatocellular carcinoma cells through sponging miR-140-5p. Onco Targets Ther. 2019;12:415-422. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 37. | Li S, Qi Y, Huang Y, Guo Y, Huang T, Jia L. Exosome-derived SNHG16 sponging miR-4500 activates HUVEC angiogenesis by targeting GALNT1 via PI3K/Akt/mTOR pathway in hepatocellular carcinoma. J Physiol Biochem. 2021;77:667-682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 38. | Zhong JH, Xiang X, Wang YY, Liu X, Qi LN, Luo CP, Wei WE, You XM, Ma L, Xiang BD, Li LQ. The lncRNA SNHG16 affects prognosis in hepatocellular carcinoma by regulating p62 expression. J Cell Physiol. 2020;235:1090-1102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 39. | Chen H, Li M, Huang P. LncRNA SNHG16 Promotes Hepatocellular Carcinoma Proliferation, Migration and Invasion by Regulating miR-186 Expression. J Cancer. 2019;10:3571-3581. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 40. | Li W, Xu W, Song JS, Wu T, Wang WX. LncRNA SNHG16 promotes cell proliferation through miR-302a-3p/FGF19 axis in hepatocellular carcinoma. Neoplasma. 2019;66:397-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 41. | Li S, Peng F, Ning Y, Jiang P, Peng J, Ding X, Zhang J, Jiang T, Xiang S. SNHG16 as the miRNA let-7b-5p sponge facilitates the G2/M and epithelial-mesenchymal transition by regulating CDC25B and HMGA2 expression in hepatocellular carcinoma. J Cell Biochem. 2020;121:2543-2558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 42. | Assarzadegan N, Montgomery E. What is New in the 2019 World Health Organization (WHO) Classification of Tumors of the Digestive System: Review of Selected Updates on Neuroendocrine Neoplasms, Appendiceal Tumors, and Molecular Testing. Arch Pathol Lab Med. 2021;145:664-677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 129] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 43. | Jing Z, Ye X, Ma X, Hu X, Yang W, Shi J, Chen G, Gong L. SNGH16 regulates cell autophagy to promote Sorafenib Resistance through suppressing miR-23b-3p via sponging EGR1 in hepatocellular carcinoma. Cancer Med. 2020;9:4324-4338. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 44. | Hu YL, Feng Y, Chen YY, Liu JZ, Su Y, Li P, Huang H, Mao QS, Xue WJ. SNHG16/miR-605-3p/TRAF6/NF-κB feedback loop regulates hepatocellular carcinoma metastasis. J Cell Mol Med. 2020;24:7637-7651. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 45. | Xu Y, Luan G, Li Z, Liu Z, Qin G, Chu Y. Correction to: Tumour-derived exosomal lncRNA SNHG16 induces telocytes to promote metastasis of hepatocellular carcinoma via the miR-942-3p/MMP9 axis. Cell Oncol (Dordr). 2023;46:265-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 46. | Zhang J, Lou W. A Key mRNA-miRNA-lncRNA Competing Endogenous RNA Triple Sub-network Linked to Diagnosis and Prognosis of Hepatocellular Carcinoma. Front Oncol. 2020;10:340. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 68] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 47. | Chen S, Zhao Z, Wang X, Zhang Q, Lyu L, Tang B. The Predictive Competing Endogenous RNA Regulatory Networks and Potential Prognostic and Immunological Roles of Cyclin A2 in Pan-Cancer Analysis. Front Mol Biosci. 2022;9:809509. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 48. | Shao X, Zhu J, Shi Y, Fang H, Chen J, Zhang Y, Wang J, Jian H, Lan S, Jiang F, Zhong F, Zhang Y, Cao C. Upregulated UBE4B expression correlates with poor prognosis and tumor immune infiltration in hepatocellular carcinoma. Aging (Albany NY). 2022;14:9632-9646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 49. | Liu Q, Gao P, Li Q, Xu C, Qu K, Zhang J. Long non-coding RNA SNHG16 as a potential biomarker in hepatocellular carcinoma: A meta-analysis. Medicine (Baltimore). 2021;100:e27178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 50. | Lou W, Wang W, Chen J, Wang S, Huang Y. ncRNAs-mediated high expression of SEMA3F correlates with poor prognosis and tumor immune infiltration of hepatocellular carcinoma. Mol Ther Nucleic Acids. 2021;24:845-855. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 51. | Xu F, Zha G, Wu Y, Cai W, Ao J. Overexpressing lncRNA SNHG16 inhibited HCC proliferation and chemoresistance by functionally sponging hsa-miR-93. Onco Targets Ther. 2018;11:8855-8863. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 79] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 52. | Liu S, Zhang W, Liu K, Liu Y. LncRNA SNHG16 promotes tumor growth of pancreatic cancer by targeting miR-218-5p. Biomed Pharmacother. 2019;114:108862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 53. | Yu Y, Dong JT, He B, Zou YF, Li XS, Xi CH, Yu Y. LncRNA SNHG16 induces the SREBP2 to promote lipogenesis and enhance the progression of pancreatic cancer. Future Oncol. 2019;15:3831-3844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 54. | Xu H, Miao X, Li X, Chen H, Zhang B, Zhou W. LncRNA SNHG16 contributes to tumor progression via the miR-302b-3p/SLC2A4 axis in pancreatic adenocarcinoma. Cancer Cell Int. 2021;21:51. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 55. | Yu Y, Zou YF, Hong RQ, Chen WJ, Chen L, Chen WQ, Wang HP, Yu Y. Long non-coding RNA SNHG16 decreased SMAD4 to induce gemcitabine resistance in pancreatic cancer via EZH2-mediated epigenetic modification. Kaohsiung J Med Sci. 2022;38:981-991. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 56. | Ding Y, Gao S, Zheng J, Chen X. Blocking lncRNA-SNHG16 sensitizes gastric cancer cells to 5-Fu through targeting the miR-506-3p-PTBP1-mediated glucose metabolism. Cancer Metab. 2022;10:20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 29] [Reference Citation Analysis (0)] |
| 57. | Lian D, Amin B, Du D, Yan W. Enhanced expression of the long non-coding RNA SNHG16 contributes to gastric cancer progression and metastasis. Cancer Biomark. 2017;21:151-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 57] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 58. | Zhao JJ, Liu JJ, Zhang YY, Xia Y, Du H, Yan ZQ, Zhou CH, Xia WS, Zellmer L, Liao DJ, Wei SX, Huang H. SNHG16 lncRNAs are overexpressed and may be oncogenic in human gastric cancer by regulating cell cycle progression. Neoplasma. 2022;69:49-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 59. | Zhou C, Zhao J, Liu J, Wei S, Xia Y, Xia W, Bi Y, Yan Z, Huang H. LncRNA SNHG16 promotes epithelial- mesenchymal transition via down-regulation of DKK3 in gastric cancer. Cancer Biomark. 2019;26:393-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 60. | Yang Z, Pu M, Dong X, Yang H, Chang W, Liu T, Zhang X. CTCF-activated SNHG16 facilitates gastrointestinal stromal tumor by targeting miR-128-3p/CASC3 axis. Exp Cell Res. 2022;417:113131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 61. | Wang X, Kan J, Han J, Zhang W, Bai L, Wu H. LncRNA SNHG16 Functions as an Oncogene by Sponging MiR-135a and Promotes JAK2/STAT3 Signal Pathway in Gastric Cancer. J Cancer. 2019;10:1013-1022. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 68] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 62. | Li Y, Lu Y, Chen Y. Long non-coding RNA SNHG16 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer via sponging miR-200a-3p. Biosci Rep. 2019;39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 63. | Chen ZY, Wang XY, Yang YM, Wu MH, Yang L, Jiang DT, Cai H, Peng Y. LncRNA SNHG16 promotes colorectal cancer cell proliferation, migration, and epithelial-mesenchymal transition through miR-124-3p/MCP-1. Gene Ther. 2022;29:193-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 64. | Tan P, Xu M, Nie J, Qin J, Liu X, Sun H, Wang S, Pan Y. LncRNA SNHG16 promotes colorectal cancer proliferation by regulating ABCB1 expression through sponging miR-214-3p. J Biomed Res. 2022;36:231-241. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 65. | Zhou W, Zhang S, Li HB, Cai Z, Tang S, Chen LX, Lang JY, Chen Z, Chen XL. Development of Prognostic Indicator Based on Autophagy-Related lncRNA Analysis in Colon Adenocarcinoma. Biomed Res Int. 2020;2020:9807918. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 66. | He X, Ma J, Zhang M, Cui J, Yang H. Long Non-Coding RNA SNHG16 Activates USP22 Expression to Promote Colorectal Cancer Progression by Sponging miR-132-3p. Onco Targets Ther. 2020;13:4283-4294. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 67. | Ke D, Wang Q, Ke S, Zou L, Wang Q. Long-Non Coding RNA SNHG16 Supports Colon Cancer Cell Growth by Modulating miR-302a-3p/AKT Axis. Pathol Oncol Res. 2020;26:1605-1613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 68. | Huang E, Ma T, Zhou J, Ma N, Yang W, Liu C, Hou Z, Chen S, Zong Z, Zeng B, Li Y, Zhou T. A novel senescence-associated LncRNA signature predicts the prognosis and tumor microenvironment of patients with colorectal cancer: a bioinformatics analysis. J Gastrointest Oncol. 2022;13:1842-1863. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 69. | Duan L, Xia Y, Li C, Lan N, Hou X. Identification of Autophagy-Related LncRNA to Predict the Prognosis of Colorectal Cancer. Front Genet. 2022;13:906900. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 70. | Christensen LL, True K, Hamilton MP, Nielsen MM, Damas ND, Damgaard CK, Ongen H, Dermitzakis E, Bramsen JB, Pedersen JS, Lund AH, Vang S, Stribolt K, Madsen MR, Laurberg S, McGuire SE, Ørntoft TF, Andersen CL. SNHG16 is regulated by the Wnt pathway in colorectal cancer and affects genes involved in lipid metabolism. Mol Oncol. 2016;10:1266-1282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 151] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 71. | Xiang Z, Huang G, Wu H, He Q, Yang C, Dou R, Liu Q, Song J, Fang Y, Wang S, Xiong B. SNHG16 upregulation-induced positive feedback loop with YAP1/TEAD1 complex in Colorectal Cancer cell lines facilitates liver metastasis of colorectal cancer by modulating CTCs epithelial-mesenchymal transition. Int J Biol Sci. 2022;18:5291-5308. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 22] [Reference Citation Analysis (0)] |
| 72. | Zhou L, Zhang Y, Jin J, Gu X. Correlation between lncRNA SNHG16 gene polymorphism and its interaction with environmental factors and susceptibility to colorectal cancer. Medicine (Baltimore). 2020;99:e23372. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/