Published online Jan 15, 2024. doi: 10.4251/wjgo.v16.i1.197
Peer-review started: September 24, 2023
First decision: October 16, 2023
Revised: November 5, 2023
Accepted: December 7, 2023
Article in press: December 7, 2023
Published online: January 15, 2024
Processing time: 108 Days and 8.3 Hours
Colorectal cancer (CRC) is the third most frequent and the second most fatal cancer. The search for more effective drugs to treat this disease is ongoing. A better understanding of the mechanisms of CRC development and progression may reveal new therapeutic strategies. Ubiquitin-specific peptidases (USPs), the largest group of the deubiquitinase protein family, have long been implicated in various cancers. There have been numerous studies on the role of USPs in CRC; however, a comprehensive view of this role is lacking.
To provide a systematic review of the studies investigating the roles and functions of USPs in CRC.
We systematically queried the MEDLINE (via PubMed), Scopus, and Web of Science databases.
Our study highlights the pivotal role of various USPs in several processes implicated in CRC: Regulation of the cell cycle, apoptosis, cancer stemness, epithelial–mesenchymal transition, metastasis, DNA repair, and drug resistance. The findings of this study suggest that USPs have great potential as drug targets and noninvasive biomarkers in CRC. The dysregulation of USPs in CRC contributes to drug resistance through multiple mechanisms.
Targeting specific USPs involved in drug resistance pathways could provide a novel therapeutic strategy for overcoming resistance to current treatment regimens in CRC.
Core Tip: Here, we provide a systematic review of the role of ubiquitin-specific peptidases (USPs) in colorectal cancer (CRC). Our study highlights the pivotal role of various USPs in several processes implicated in CRC: Regulation of the cell cycle, apoptosis, cancer stemness, epithelial–mesenchymal transition, metastasis, DNA repair, and drug resistance. The findings of this study suggest that USPs have the potential to be drug targets and noninvasive biomarkers for CRC.
- Citation: Al-Balushi E, Al Marzouqi A, Tavoosi S, Baghsheikhi AH, Sadri A, Aliabadi LS, Salarabedi MM, Rahman SA, Al-Yateem N, Jarrahi AM, Halimi A, Ahmadvand M, Abdel-Rahman WM. Comprehensive analysis of the role of ubiquitin-specific peptidases in colorectal cancer: A systematic review. World J Gastrointest Oncol 2024; 16(1): 197-213
- URL: https://www.wjgnet.com/1948-5204/full/v16/i1/197.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i1.197
Cancer is a disease characterized by the uncontrolled proliferation of certain cells throughout the body. In addition to the features of immune evasion, angiogenesis, resistance to growth-inhibitory signals and chemotherapy, and evasion of programmed cell death (apoptosis), cancer cells can also possess the ability to metastasize to distant areas[1]. Colorectal cancer (CRC) is a cancer that occurs in the colon and/or the rectum and is considered the third most frequent and second most fatal cancer in many countries. In 2020, CRC accounted for approximately 1.9 million cases of cancer and 0.9 million deaths worldwide[1-6]. The current management strategies have not successfully reduced these numbers in recent decades. Therefore, there is an ongoing search for more effective therapeutic strategies. Identification of the various factors involved in the emergence of CRC can be of paramount importance in this search[7-10].
Posttranslational protein modifications play a key role in the regulation of various cellular functions, including tumor growth and metastasis. This has motivated researchers to better understand these modifications, as such studies may reveal novel therapeutic strategies. Ubiquitination is a type of such posttranslational modification. It is a reversible process in which ubiquitin, a 76-amino-acid protein, is attached to other proteins. Ubiquitination regulates many cellular processes and maintains homeostasis[11]. In recent years, an increasing number of studies highlighted the pivotal role of aberrant protein ubiquitination in carcinogenesis; it can prompt a variety of processes in tumor cells, such as proliferation, apoptosis, and cell differentiation. Ubiquitin chains are encoded by 4 different genes: UBA52, RPS27A, UBB, and UBC[11]. Ubiquitination is mediated by 3 catalytic enzymes: ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin ligases (E3). Ubiquitin is activated in an ATP-dependent manner by an E1 enzyme and afterward is transferred to an E2 enzyme and finally ligated covalently to the substrate by an E3 ligase[12]. Ubiquitination can be reversed through deubiquitination, a process in which deubiquitinases (DUBs) catalyze the removal of ubiquitin (single or polyubiquitin chains) from the substrate. DUBs act as antagonists for E3 ligases and are thought to stabilize several oncogenes and tumor suppressor genes and protect them from ubiquitin-mediated degradation[13]. DUBs account for approximately 20% of all protease-encoding genes; thus, they are considered the largest family of proteases in humans. Moreover, there are approximately 100 DUBs that belong to the cysteine protease family that can be either cysteine-dependent proteases or metalloproteases. DUBs are categorized into six families (the ZUP1 family and five other families, all of which are cysteine-dependent proteases). The five other families include ubiquitin-specific peptidases (USPs) (the largest family of DUBs), ovarian tumor proteases, ubiquitin carboxy-terminal hydrolases, Machado–Joseph disease proteases, and the newly identified motif interacting with ubiquitin-containing novel DUB family[14,15]. JAB1/MPN/Mov34 metalloenzymes make up the only metalloprotease family[16]. Alterations in the expression levels of these deubiquitinating enzymes are associated with the development of cancer and metastasis in addition to other disorders. A specific DUB might be upregulated in some types of cancers but downregulated in some other types of cancer[17,18]. DUBs are also involved in several processes that enable the spread and metastasis of cancer cells to distant organs, mostly by regulating epithelial-mesenchymal transition (EMT), in which cells shift from the epithelial phenotype to the mesenchymal phenotype and start to migrate through the body[19].
Because of the considerable number of studies carried out in recent years on these topics and the importance of obtaining a comprehensive view for guiding future studies and the search for novel therapeutic approaches, in this study, we aimed to provide a comprehensive review of the role of USPs in CRC development and progression through the reliable methodology of systematic review.
This systematic review was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines[20]. Its protocol was registered on the International Prospective Register for Systematic Reviews as CRD42022348183 and it was published[21]. We included articles published in English from 2008 to Feb 2023. We searched three databases [MEDLINE (via PubMed), Web of Science, and Scopus] for relevant studies. The exact search strategies used are shown in Table 1. EndNote version 20 was used to manage and deduplicate the retrieved articles.
| Database | Search strategy |
| MEDLINE, 124 | (((Colorectal Cancer, Hereditary Nonpolyposis, Type 8, Colorectal Cancer, Hereditary Nonpolyposis, Type 6, Colorectal Cancer, Hereditary Nonpolyposis, Type 7, Colorectal Cancer, Hereditary Nonpolyposis, Type 3, Colorectal Cancer, Hereditary Nonpolyposis, Type 4, Colorectal Cancer, Hereditary Nonpolyposis, Type 5) OR (("Colorectal Neoplasms"[Mesh] OR "Colorectal Neoplasms, Hereditary Nonpolyposis"[Mesh] OR "Colitis-Associated Neoplasms"[Mesh])) OR ((Colorectal Cancer[Title/Abstract])) OR (Colorectal Neoplasms[Title/Abstract]))) AND ((((((((((((((Ubiquitin Specific Proteases[Title/Abstract]) OR (Ubiquitin-Specific Peptidase[Title/Abstract])) OR (Peptidase, Ubiquitin-Specific[Title/Abstract])) OR (Ubiquitin Specific Peptidase[Title/Abstract])) OR (Ubiquitin-Specific Peptidases[Title/Abstract])) OR (Peptidases, Ubiquitin-Specific[Title/Abstract])) OR (Ubiquitin Specific Peptidases[Title/Abstract])) OR (Ubiquitin-Specific Protease[Title/Abstract])) OR (Protease, Ubiquitin-Specific[Title/Abstract])) OR (Ubiquitin Specific Protease[Title/Abstract])) OR (Ubiquitin-Specific Protease Family[Title/Abstract])) OR (Protease Family, Ubiquitin-Specific[Title/Abstract])) OR (Ubiquitin Specific Protease Family[Title/Abstract])) OR ((Ubiquitin-Specific Peptidase[MeSH Terms]) OR (Ubiquitin Specific Proteases[MeSH Terms]))) |
| Scopus, 139 | (TITLE-ABS-KEY ((((colorectal AND neoplasms) OR (colorectal AND neoplasms, AND hereditary AND nonpolyposis) OR (colitis-associated AND neoplasms) OR (colorectal AND cancer, AND hereditary AND nonpolyposis, AND type 8) OR (colorectal AND cancer, AND hereditary AND nonpolyposis, AND type 6) OR (colorectal AND cancer, AND hereditary AND nonpolyposis, AND type 7) OR (colorectal AND cancer, AND hereditary AND nonpolyposis, AND type 3) OR (colorectal AND cancer, AND hereditary AND nonpolyposis, AND type 4) OR (colorectal AND cancer, AND hereditary AND nonpolyposis, AND type 5) OR (colorectal AND cancer) ) ) ) ) AND ((TITLE-ABS-KEY (ubiquitin AND specific AND proteases) OR TITLE-ABS-KEY (ubiquitin-specific AND peptidase) OR TITLE-ABS-KEY (peptidase, AND ubiquitin-specific) OR TITLE-ABS-KEY (ubiquitin AND specific AND peptidase) OR TITLE-ABS-KEY (ubiquitin-specific AND peptidases) OR TITLE-ABS-KEY (peptidases, AND ubiquitin-specific) OR TITLE-ABS-KEY (ubiquitin AND specific AND peptidases) OR TITLE-ABS-KEY (ubiquitin-specific AND protease) OR TITLE-ABS-KEY (protease, AND ubiquitin-specific) OR TITLE-ABS-KEY (ubiquitin AND specific AND protease) OR TITLE-ABS-KEY (ubiquitin-specific AND protease AND family) OR TITLE-ABS-KEY (protease AND family, AND ubiquitin-specific) OR TITLE-ABS-KEY (ubiquitin AND specific AND protease AND family) ) ) AND PUBYEAR > 2007 AND PUBYEAR < 2024 |
| Web of Science, 473 | (((((((((TS=(Colorectal Neoplasms)) OR TS=(Colorectal Neoplasms, Hereditary Nonpolyposis)) OR TS=(Colitis-Associated Neoplasms)) OR TS=(Colorectal Cancer, Hereditary Nonpolyposis, Type 8)) OR TS=(Colorectal Cancer, Hereditary Nonpolyposis, Type 6)) OR TS=(Colorectal Cancer, Hereditary Nonpolyposis, Type 7)) OR TS=(Colorectal Cancer, Hereditary Nonpolyposis, Type 3)) OR TS=(Colorectal Cancer, Hereditary Nonpolyposis, Type 4)) OR TS=(Colorectal Cancer, Hereditary Nonpolyposis, Type 5)) OR TS=(Colorectal Cancer) AND ((((((((((((TS=(Ubiquitin Specific Proteases)) OR TS=(Ubiquitin-Specific Peptidase)) OR TS=(Peptidase, Ubiquitin-Specific)) OR TS=(Ubiquitin Specific Peptidase)) OR TS=(Ubiquitin-Specific Peptidases)) OR TS=(Peptidases, Ubiquitin-Specific)) OR TS=(Ubiquitin Specific Peptidases)) OR TS=(Ubiquitin-Specific Protease)) OR TS=(Protease, Ubiquitin-Specific)) OR TS=(Ubiquitin Specific Protease)) OR TS=(Ubiquitin-Specific Protease Family)) OR TS=(Protease Family, Ubiquitin-Specific)) OR TS=(Ubiquitin Specific Protease Family) |
No limitations based on the setting, country, or study design were imposed on the search. Studies with the following features were excluded: letters to editors, duplicate studies, systematic reviews and meta-analyses, articles published before 2008, and articles not written in English.
Screening of the titles and abstracts was performed by two reviewers (Al-Balushi E and Abdel-Rahman WM). In case of any disagreement, the conflict was resolved by a third author (Jarrahi AM).
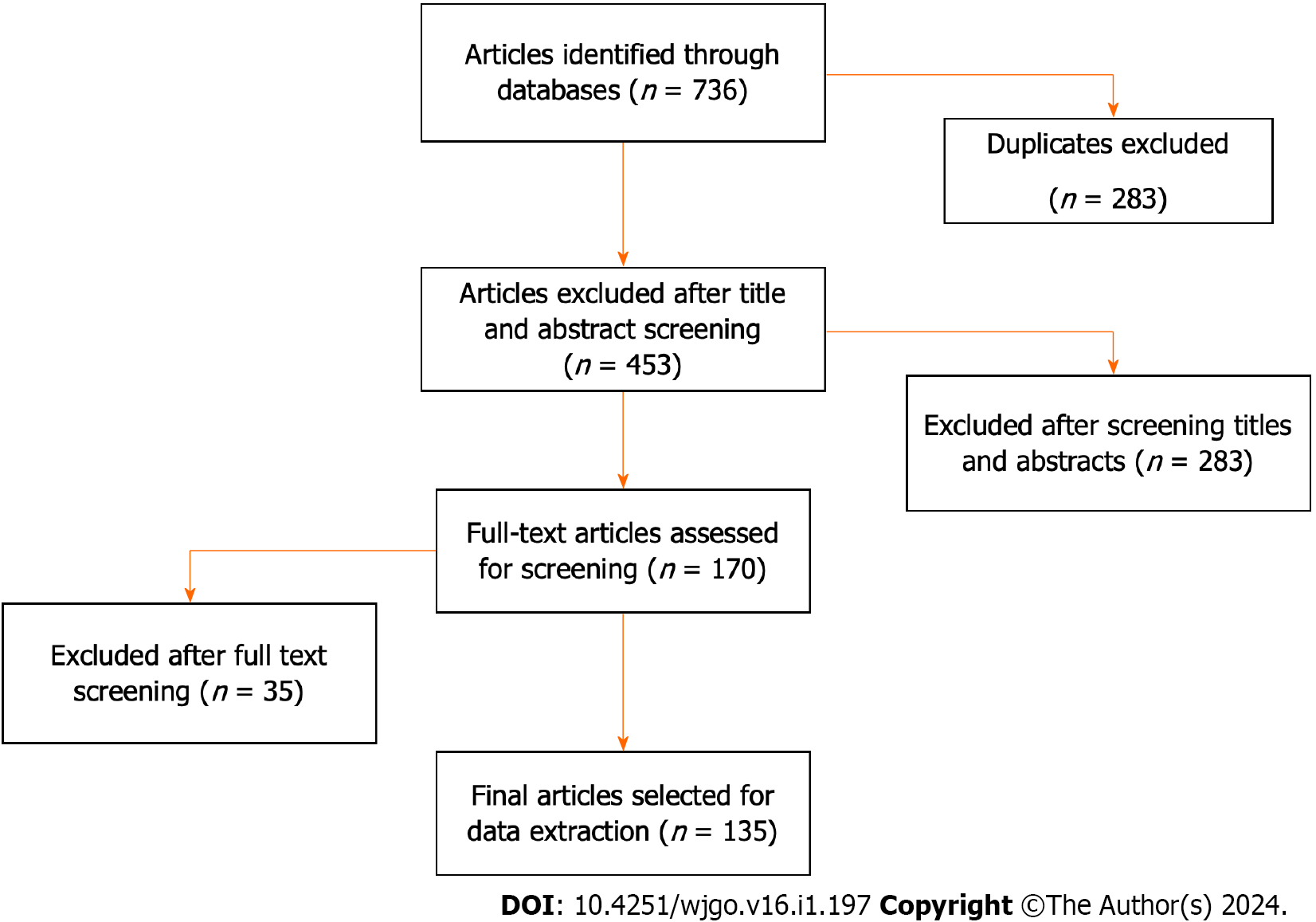
The study selection process is depicted in Figure 1. A total of 736 records were identified from the databases. After excluding duplicate articles, 453 studies were initially included for screening titles and abstracts; of these 283 were excluded and 170 studies were included for full-text screening. Of these, 35 were excluded because they did not investigate the role of USPs in CRC (Figure 1).
The results of the reviewed studies can be categorized into seven sections: USPs regulate cell cycle progression; the role of USPs in apoptosis; USPs are involved in stemness of cancer; USPs are involved in EMT and metastasis; USPs are involved in DNA repair; USPs are involved in drug resistance; and USPs as targets for anticancer drug development. Table 2 provides a summary of the data extracted from the 135 included articles.
| No. | Gene symbol | Role in tumors | Cellular localizatioa | Target(s) | Functions and pathways | Inhibitor(s) | Ref. |
| 1 | USP1 | Oncogene | Nucleus | Cyclin A/D/E, Bcl2, MCL1, FANCD1, ID1 | Cell cycle, DNA Repair, apoptosis | ML323 | Xu et al[24] |
| 2 | USP2 | Oncogene | Cytoplasm, cytoplasm (perinuclear region), isoform 4: nucleus, membrane, cytoplasm | Cyclin D1, PER1, CRY1, HDM2/p53, FASN, LDLR, TRAF6, TBK1, TGFBR1-TGFBR2 complex | Cell survival, circadian rhythm, cell metabolism, inflammatory response, antiviral response, metastasis, cell growth, cell cycle (G0-G1), HR-mediated DNA repair | ML364 | Davis et al[26] and Luo et al[76] |
| USP2a | Oncogene | Cyclin D1 | Cell cycle progression, development | LCAHA | Magiera et al[23] | ||
| 4 | USP3 | Tumor suppressor | Nucleus | SMAD4 | TGF-β pathway, CRC migration, metastasis, invasion | miR-224 | Das et al[33], Wang et al[71], and Zheng et al[87] |
| Oncogene | Androgen receptor, RASGRP3, Cdc25A | MAPK pathway, apoptosis, cell cycle progression, CRC non-invasive biomarker | AC092894.1 | ||||
| 5 | USP4 | Oncogene | Cytoplasm, nucleus | PRL-3, β-catenin, Rheb | AKT, EMT, E-Cadherin, Wnt/β-catenin pathway, colony formation, mTORC1 activation, tumorigenesis, progression | Vialinin A, PR619, NR (neutral red) | Yun et al[56], Xing et al[65], Nguyen et al[90], Deng et al[97], Wang et al[98], and Zhao et al[99] |
| 6 | USP5 | Tumor suppressor | Cytosol, lysosome, nucleus | ORP8 | ER stress, apoptosis, progression/drug resistance | G9, vialinin A, WP1130, EOAI3402143, AM146, RA-9, RA-14, PR619, doxorubicin | Xu et al[81], Zhang et al[100], and Ning et al[101] |
| Oncogene | TUFM | Cell growth | |||||
| 7 | USP6 | Oncogene | Cell membrane, cytoplasm, endosome | Deubiquitination, development, progression | Zeng et al[72] | ||
| 8 | USP6NL | Oncogene | Golgi apparatus, cytoplasmic vesicle | β-catenin (P27, c-MYC, CyclinD1) | Wnt/β-catenin pathway, proliferation, metastasis, progression | Sun et al[64] | |
| 9 | USP7 (HAUSP) | Tumor suppressor | Nucleus, cytoplasm, nucleus (PML body), chromosome | MDM2, p53 | Tumor growth, development, progression, drug resistance | IL-6/STAT3/HDAC-1, P22077, HBX41, P0045204, P005091, N-benzylpiperidinol, HBX19,FT671, GNE6640, GNE6776, C7, C19, parthenolide | Choi et al[36], Novellasdemunt et al[58], Novellasdemunt et al[59], Sarkar[60], Basu et al[61], Hou et al[62], Zhou et al[80], An et al[91], Weinstock et al[92], Li et al[93], Nicholson et al[102], Colland et al[103], Qian et al[104], Bufalieri et al[105], Li et al[106], Becker et al[107], Yang et al[108], Du et al[109], Zhu et al[110] |
| Oncogene | DNMT1, β-catenin, FAM188B, DDX3X, c-MYC, P53, HDM2, DNA damage repair proteins DNA polymerase eta (Polη), PCNA | Tumor growth, WNT/β-catenin pathway, apoptosis, cell growth, stress-tolerance pathway, tumorigenesis, proliferation | |||||
| 10 | USP9X | Tumor suppressor | Cytosol, cell projection (growth cone), cytoplasm (cytoskeleton, cilium axoneme) | FBW7 | C-MYC, C-JUN, cyclin E, development, progression, drug resistance (5-FU) | G9, WP1130, PR619 | Harris et al[27], Peddaboina et al[40], and Tian et al[111] |
| Oncogene | BCL-XL, MCL-1 | Apoptosis | |||||
| 11 | USP10 | Tumor suppressor | Cytoplasm, nucleus, early endosome | SIRT6, P53 | c-MYC, progression | P22077, HBX19818, spautin-1, PR619 | Lin et al[28], Kim et al[29], Li et al[34], Reissland et al[51], Li et al[78], Ouyang et al[112], and Ye et al[113] |
| Oncogene | NLRP7, MSI2, miR-1286, β-catenin | Tumor promotion, NF-kB pathway, cancer proliferation, invasion, metastasis, WNT/β-catenin pathway | |||||
| 12 | USP11 | Tumor suppressor | Nucleus, cytoplasm, chromosome | P21 | Apoptosis | miR-132-3P | Zhang et al[42], Sun et al[66], Liao et al[67], Tian et al[111], Lee et al[114], Huang et al[115], and Sun et al[116] |
| Oncogene | PPP1CA, IGF2BP3, Cyclin D1, c-IAP2, VCP | Tumor growth, metastasis, apoptosis, ERK/MAPK pathway, autophagy AMPK/Akt/mTOR pathway, proliferation, migration, development, progression, drug resistance (5-FU) | |||||
| 13 | USP13 | Tumor suppressor | Cytosol, nucleoplasm, nucleus | PTEN | Cell proliferation, glycolysis, development | miR-135b | Xiang et al[117] |
| 14 | USP14 | Oncogene | Cytoplasm, cell membrane | JNK, β-catenin, Dvl, NLRC5, RIG-I, FASN, CXCR4 | MAPK/JNK signaling, immune regulation, tumorigenesis, proliferation, Wnt/β-catenin pathway, liver/lymph node metastases, progression, drug resistance (IDO1), correlated with pathological stage | b-AP15 | Sarkar[60], Liu et al[118], Mofers et al[119], Du et al[120], and Shi et al[121] |
| 15 | USP15 | Tumor suppressor | Cytoplasm, nucleus, mitochondrion | APC | Wnt/β-catenin pathway, development | Huang et al[122] | |
| 16 | USP17 | Tumor suppressor | Nucleus, endoplasmic reticulum | DNA damage response, chemosensitization | ZEB1 | Wang et al[123] | |
| 17 | USP18 | Oncogene | Cytoplasm, nucleus | SNAIL | EMT, development, progression, drug resistance | Huang et al[68], and Zhang et al[124] | |
| 18 | USP19 | Oncogene | Endoplasmic reticulum membrane | ME1 | Lipogenesis, NADPH production, inhibiting ROS, tumorigenesis, carcinogenesis, development | Zhu et al[125] | |
| 19 | USP20 | Oncogene | Cytoplasm, endoplasmic reticulum, cytoplasm (perinuclear region), cytoplasm (centrosome) | SOX4, ADORA2A, CD160, CD27, TNFRSF25, MRP1, MRP3, MRP5 | E-Cadherin, N-Cadherin, SNAIL, Slug, migration, Invasion, Metastasis, Notch/Hedgehog/β-catenin pathway, progression, drug Resistance | PR619 | Guan et al[53], Jin et al[79], and Ming et al[86] |
| 20 | USP21 | Oncogene | Cytoplasm, nucleus | Fra-1, RIG-I, NANOG, Foxp3 | Metastasis, apoptosis, DNA repair, MMP1, tumor development/growth, NF-κB pathways, progression | Gallic Acid, BAY-805 | Kang et al[52], Yun et al[73], Deng et al[94], and Göricke et al[95] |
| 21 | USP22 | Tumor suppressor | Nucleus | SIRT1, SPARC(H2B) | STAT3-MMP9 pathway, mTOR, inflammation-associated CRC | PR619, miR-30a-5p | Lin et al[25], Jiang et al[48], Yan et al[49], Miao et al[50], Li et al[54], Guo et al[96], Liu et al[126], Jiang et al[127], Xu et al[128], Ao et al[129], Ao et al[130], Kosinsky et al[131], Liu et al[132], Sippl et al[133], Liu et al[134], Kapoor[135], Liu et al[136], Wang et al[137], Gennaro et al[138], Kosinsky et al[139], He et al[140], Feng et al[141], Kosinsky et al[142], Xian et al[143], and Kotelevets et al[144] |
| Oncogene | AP-4, BMI-1, PD-L1, β-catenin, ZRANB1, Sox9, FOXP3, Akt, INK4a, ARF, CCND1, SNHG16, Axin2 miR-132-3p, MVP, cyclinD2 c-MYC, Pakt, CCND1(CyclinD1), H2B(RNF2), HSP90, AB1 | Cell transition (G1-S)/(G2-M), EMT, Wnt/β-catenin pathway, RNF-22, proliferation, invasion, stemness, INK4a/ARF pathway, AKT signaling, apoptosis, immune escape, tumorigenesis, lymph node metastasis, development, progression, drug resistance (5-FU, Ganetespib), therapy resistant phenotype | |||||
| 22 | USP25 | Oncogene | Cytoplasm, nucleus | STAT3, SOCS3, EGFR | WNT pathway, EGF pathway, tumorigenesis, development | Vismodegib | Wang et al[30] and Wang et al[55] |
| 23 | USP28 | Oncogene | Nucleus (nucleoplasm) | FBW7α, FOX1, NICD1, FOXC1, FBP1, Checkpoint | c-MYC, c-JUN, aerobic glycolysis, notch signaling, proliferation, tumorigenesis, progression | Vismodegib, FT206 | Wang et al[30], Diefenbacher et al[31], Bernhard et al[32], Zhou et al[89], Popov et al[145], Liu et al[146], Xu et al[147], and Ren et al[148] |
| 24 | USP29 | Oncogene | Cytoplasm (perinuclear region) | KIAA1429, SOX8 | Proliferation, DNA damage, apoptosis, migration, invasion, progression | Chandrasekaran et al[149] and Li et al[150] | |
| 25 | USP33 | Tumor suppressor | Cytoplasm (perinuclear region), cytoplasm (centrosome), Golgi apparatus | ROBO1 | SLIT2 pathway, progression | Huang et al[74] | |
| 26 | USP34 | Oncogene | Cytosol, nucleus | AXIN1, AXIN2 | WNT pathway, development | Lui et al[151] | |
| 27 | USP35 | Oncogene | Cytosol, nucleus | FUCA1, NER | Proliferation, drug resistance (5-FU/OXA) | Xiao et al[85] | |
| 28 | USP38 | Tumor suppressor | Cytosol, nucleus | HDAC3, HMX3 | Stemness markers, apoptosis, KRAS pathways, proliferation, migration, invasion, development, progression, drug resistance | Zhan et al[45], Wang et al[77], and Liu et al[82] | |
| Oncogene | LSD1 | Proliferation, cell colony formation | |||||
| 29 | USP39 | Oncogene | Nucleus | β-catenin, TCF-4, MMP2, MMP9, KRAS, P21, CHK2, TCF/LEF, Aurora B | Wnt/β-catenin pathway, cell Cycle progression, c-MYC, cell Growth, cyclin D1, PPARd, invasion, tumorigenesis, proliferation, apoptosis, angiogenesis, spindle checkpoint and cytokinesis, colony formation, development | Yuan et al[41], Yuan et al[63], Fraile et al[152], Remitha et al[153] | |
| 30 | USP42 | Tumor suppressor | Cytosol, nucleoplasm, nucleus | RNF43, ZNRF3 | WNT pathway | Giebel et al[57] | |
| Mesenchymal genes, Endothelial Genes | EMT, development, progression | ||||||
| 31 | USP43 | Oncogene | Nucleoplasm | ZEB1 | EMT pathway, proliferation, migration, invasion, progression | Ye et al[154] | |
| 32 | USP44 | Tumor suppressor | Chromatin, cytoplasm, mitotic spindle, nucleoplasm, nucleus | PCNA, cleaved-caspase3, Axin 1 | Apoptosis, colorectal neoplasia, Wnt/β-catenin pathway, development, progression | Huang et al[38], Lou et al[39], and Sloane et al[155] | |
| 33 | USP46 | Tumor suppressor | Cytosol, nucleus | PHLPP | AKT pathway, development, progression hypoxia-induced drug resistance | Wen et al[83], and Li et al[156] | |
| 34 | USP47 | Oncogene | Cytoplasm | SNAIL, LINC00668, β-catenin, SATB1, SMURF2, YAP, RP52 | EMT, stemness, tumor growth, chromatin remodeling, proliferation, migration, invasion, hippo Pathway, RP52-MDM2 interaction, colony formation, development, progression | Parthenolide, P5091, Cpd14, P22077, PR619 | Cho et al[37], Zhang et al[46], Yu et al[47], Yan et al[75], Choi et al[157], Yu et al[158], Pan et al[159], and Pan et al[160] |
| 35 | USP49 | Tumor suppressor | nucleus | miR-5000-3P | PI3K/AKT pathway, proliferation, migration, progression, drug sensitivity (oxaliplatin) | Tu et al[43], and Zhuang et al[84] | |
| Oncogene | BAG2 | Apoptosis | |||||
| 36 | USP54 | Oncogene | Mitochondria | Proliferation, migration, invasion, tumorigenesis, development, progression | Fraile et al[161] |
The cell cycle is tightly regulated by cell cycle-related factors including cyclin dependent-kinases and cyclins. USPs, as the main members of the DUB family, might disrupt cell cycle progression by stabilizing specific proteins[22]. Cyclins are indispensable elements of the cell cycle, and disruption of their function by USPs causes cell cycle progression. USP2a stabilizes cyclin D1, a proto-oncoprotein overexpressed in numerous cancer types. It has been shown that lithocholic acid can arrest the cyclin D1-dependent growth of cells by inhibiting USP2a[23]. USP1 has also been reported to be highly expressed in CRC and to be able to induce entry into the G2/M phase of the cell cycle. Inhibition of USP1 by ML323 resulted in a reduction in cyclin A/D/E expression and growth arrest in CRC cells[24].
Several cancers, especially CRC, are associated with elevated levels of CCNB1 (cyclin B1) and consequent uncontrolled growth. USP22 can inhibit the ubiquitination of CCNB1 and induce the G2/M transition of CRC cells. Therefore, USP22 is considered as a major oncogenic factor in CRC[25]. USP2 can promote cell cycle progression through its effect on cyclin D1. ML364, a USP2 inhibitor, has been reported to increase cellular cyclin D1 degradation and arrest the cell cycle[26].
C-MYC is another well-known oncogene that plays an important role in cell cycle progression through the regulation of genes related to cell cycle control. USP9X suppresses CRC by interacting with a tumor suppressor called FBXW7 (F-box and WD repeat domain-containing 7). Knockdown of USP9X in colon cancer cells led to a decrease in FBXW7 levels and, consequently, an increase in the stability of several oncoproteins, such as c-MYC, c-JUN, and cyclin E[27].
USP10 deubiquitinates and stabilizes SIRT6 and helps inhibit cancer progression by antagonizing c-MYC activity[28]. Kim et al[29] noted that the absence of USP10 expression was associated with distant metastasis and lymph vascular invasion. In contrast, one study showed that USP10 enhanced CRC cell proliferation by stabilizing MSI2, an oncogenic factor.
Vismodegib, a drug used for treating basal cell carcinoma, has been shown to decrease the activity of two DUBs, USP25 and USP28, as well as the levels of their substrates, such as c-MYC, in CRC cells[30]. Moreover, USP28 participates in CRC progression and oncogenesis by increasing c-MYC levels[31]. Conversely, one study showed that loss of USP28 reduces checkpoint activation, thus facilitating cell proliferation[32]. This indicates a tumor suppressor aspect of USP28.
Cell division cycle 25 A (Cdc25A) is a protein phosphatase that positively regulates the activities of CDKs and promotes cell cycle progression. USP3 has been identified as a DUB of Cdc25A. USP3 depletion has been reported to reduce the Cdc25A protein level, resulting in a significant delay in cell cycle progression[33].
A recent study revealed that USP10 expression is elevated in CRC patients and is positively related to the JNK and MAPK/JNK pathways as well as CRC development and progression[34].
Many factors have been identified to play vital roles in apoptosis. Among these is the tumor suppressor protein p53, that regulates the expression of a wide variety of genes involved in apoptosis and is critical in cancer formation[35]. USPs exert a well characterized role in the apoptotic cell death pathways. However, their role in other forms of cell death is much less characterized. USP7 and USP10 exert their tumor suppressor function by protecting p53 and inducing apoptosis in HCT116 cells. Conversely, USP7 interacts with FAM188B and enhances tumor growth and survival of colon cancer cells[28,36]. In addition, the ubiquitination and subsequent degradation of p53 are largely controlled by MDM2 (mouse double minute 2). USP47 inhibits the interaction between the ribosomal proteins RPS2 and MDM2 by RPS2 deubiquitination, which alleviates RPS2-mediated suppression of MDM2. This results in p53 degradation and promotion of carcinogenesis[37].
USP44 overexpression promotes apoptosis in CRC by inactivating the Wnt/β-catenin pathway and increasing Axin1 protein and cleaved caspase-3 protein levels[38,39]. Most CRC specimens have high levels of Bcl-xL and Mcl-1, whose expression is associated with the stage of the tumor. USP9X deubiquitinates these two antiapoptotic (pro-survival) proteins and enhances the survival of CRC cells[40]. Conversely, it has been observed that when CRC cells are transiently transfected with siUSP1, the expression of Bcl-2 and Mcl-1 is significantly downregulated[24].
p21 is another important tumor suppressor that acts downstream of p53 to mediates the DNA damage-induced checkpoint. It is a cyclin-dependent kinase inhibitor that inhibits cell cycle progression and induces cell cycle arrest. Its level is negatively correlated with that of USP39, and knockdown of USP39 prolongs the half-life of p21 and induces apoptosis in colon cancer cell lines[41].
USP11 is an oncoprotein that, according to a recent study, is inhibited by Circ_DOCK1 and miR-132-3p[42].
USP49 promotes the proliferation and chemoresistance of CRC cells by deubiquitinating and stabilizing Bcl-2-associated athanogene 2 (BAG2). BAG2 is a protein that inhibits apoptosis and promotes the adaptive response of cancer cells[43].
Stemness refers to the molecular processes underlying the fundamental stem cell (SC) properties of self-renewal and the ability to generate differentiated daughter cells[44]. USP38 negatively regulates cancer SCs in CRC via HDAC3 (histone deacetylase 3) deubiquitination and is downregulated in clinical CRC samples and cell lines. Ubiquitination of HDAC3 decreases histone acetylation and upregulates cancer SC-related genes or markers such as SOX2 [SRY (sex determining region Y)-Box 2], NANOG, OCT4 (octamer-binding transcription factor 4), and BIM1[45].
USP47 is an oncogene that is essential for regulating the stemness, development, and pathogenesis of CRC and is associated with a poor prognosis[46,47].
USP22 induces cancer stemness, and its knockdown inhibits SC proliferation and CRC tumorigenesis[48]. A study reported that ring finger 220 (RNF220) is upregulated in CRC tissues and cell lines and is associated with increased CRC stemness and tumor growth in vivo. This oncogenic activity of RNF220 is indirectly mediated by USP22[49]. USP22 also stabilizes ZRANB1 (zinc finger RANBP2-type containing 1), a positive regulator of the Wnt/β-catenin pathway, and promotes SC-like features of CRC cells[50].
It has been observed that USP10 acts as an APC (adenomatous polyposis coli)-truncation-specific enhancer of β-catenin stability, modulates Wnt signaling and promotes cancer cell stemness and intestinal tumorigenesis[51].
NANOG is a critical factor for maintaining cancer SC-like properties and is stabilized by USP21[52].
Previous studies have established that different members of the USP family have roles in inducing EMT as essential step in metastasis. USP20 has been shown to regulate the stability of SOX4, which is considered an EMT transcription factor. Knockdown of USP20 suppressed cell proliferation, migration, and invasion and decreased the levels of SOX4, N-cadherin, Snail, and Slug but increased the E-cadherin level[53]. USP22 overexpression enhances colon cancer migration and invasiveness by inducing EMT via activating AP4 (activating enhancer binding protein-4) transcription by binding to its promoter[54].
The aberrant Wnt pathway is a crucial factor in EMT. Overexpression of USP25 activates Wnt signaling and enhances CRC tumorigenesis. In several mouse models of colon cancer, USP25 deletion decreased the expression of Wnt pathway genes, suppressed tumor formation, and decreased tumor size[55]. Moreover, USP4 contributes to the progression of CRC by deubiquitinating and controlling the stability of β-catenin, a key component of the Wnt/β-catenin pathway[56]. USP42 deubiquitinates RNF43 (ring finger protein 43) and ZNRF3 (zinc and ring finger 3), and USP42 knockout blocks Wnt activity[57]. USP7 has two binding sites on the N-terminus through which it interacts with β-catenin and deubiquitinates it in APC-mutated cells. This results in continuous activation of the Wnt signaling pathway[58,59] and affects CRC tumor progression[60,61]. USP7 also stabilizes c-MYC and, subsequently, EMT signaling by binding MYH9 (myosin heavy chain 9); however, ENKUR, a tumor suppressor, prevents the progression of cancer by preventing MYH9/USP7-mediated c-MYC deubiquitylation[62]. Knockdown of USP39 inhibits CRC tumor formation and migration by reducing downstream factors of the Wnt/β-catenin pathway, such as β-catenin, TCF4, MMP2, and MMP9[63]. USP6NL, which is reported to be highly expressed in CRC patients, facilitates CRC carcinogenesis through the Wnt/β-catenin pathway[64].
It has been observed that USP4 regulates tumor formation by stabilizing PRL-3. PRL-3 enhances the invasiveness and metastasis of CRC by activating the Akt pathway and reducing the level of E-cadherin, a hallmark of EMT[65].
USP11 promotes tumor growth and metastasis in CRC via the ERK/MAPK pathway by stabilizing PPP1CA[66,67].
USP18 exerts oncogenic effects on CRC cells by regulating the stability of the Snail1 protein and promoting CRC cell proliferation, invasion, and migration[68].
SMAD4 (SMAD family member 4/Mothers against decapentaplegic homolog 4) is a potent tumor suppressor gene that inhibits CRC metastasis and is often mutated in CRC[69,70]. USP3 modulates SMAD4 expression at the post-transcriptional level. Silencing USP3 reduced SMAD4 expression in LoVo and HCT116 cells[71].
USP6 overexpression in colon cancer cell lines enhanced the invasion and aggregation of cancer cells in vitro and increased liver metastasis in vivo[72].
USP21 has been shown to deubiquitinate and stabilize Fos-related antigen-1 (Fra-1)[73]. Fra-1 is highly expressed in different cancers, including CRC, and is known as a therapeutic target in CRC, especially metastatic CRC.
USP33 has been shown to deubiquitinate and stabilize Robo1 to exert the inhibitory effect of Slit2 on CRC cell migration[74].
USP47 is positively correlated with long intergenic noncoding RNA no. 668 (LINC00668). LINC00668 promotes the tumorigenesis and migration of colon cancer cells and is highly expressed in CRC tissues[75]. It has been demonstrated that the TGF-β pathway has an effective role in metastasis; for instance, USP2 promotes EMT after interacting with TGFBR1 and TGFBR2 after TGF-β stimulation[76].
According to a recent study, USP38 acts as a tumor suppressor by inhibiting CRC migration via HMX3 deubi
USP10 promotes cell migration by deubiquitinating RFC2. USP10 is upregulated by CircCOL1A2, a competing endogenous RNA, via sequestering miR-1286[78].
USP20 also promotes metastasis of CRC cells and is closely correlated with several immune checkpoint genes, including ADORA2A (adenosine A2a receptor), CD160 (cluster of differentiation 160), CD27 (cluster of differentiation 27), and TNFRSF25 (TNF receptor superfamily member 25)[79].
DNA repair pathways are critical to maintaining genetic stability and integrity when mammalian cells are exposed to DNA-damaging agents. Their deregulation is associated with cancer initiation and development. USP1 knockdown has been observed to downregulate the DNA repair-related substrates FANCD2 and ID1; thus, it can be considered a target for regulating DNA repair[24].
USP2 is another key regulator of DNA repair. ML364, a USP2 inhibitor, has been observed to suppress homologous recombination-mediated DNA repair[26].
USP7 also recruits DNA repair proteins in response to DNA damage[80].
The resistance of cancer cells to drugs is an important challenge in treating cancers. It has been shown that increased activity of USP5, which was induced by EBF1 (early B-cell factor 1) transcription factor overexpression, is associated with decreased sensitivity of cancer cells to the anticancer drug doxorubicin. USP5 has been found to be upregulated in primary CRC tissues. It promotes CRC by interacting with, deubiquitinating, and stabilizing the Tu translation elongation factor[81].
USP38 is another oncogene that enhances HCT116 cell proliferation, cell colony formation, and drug tolerance[82].
USP46 interacts with PH domain leucine-rich repeat protein phosphatase (PHLPP) and controls its expression via deubiquitination. PHLPP is a tumor suppressor known to negatively regulate signaling pathways that are activated by kinases such as Akt. Knockdown of USP46 in HCT116 cells has been observed to decrease the levels of PHLPP isoforms and, consequently, contribute to hypoxia-induced drug resistance in CRC[83].
USP9X can also act as an oncogene. Its depletion in colon cancer cells led to increased apoptosis and increased susceptibility to 5-fluorouracil (5-FU), the most commonly used drug in CRC treatment[27].
MiR-5000-3p is an oncogene that plays a critical role in resistance to oxaliplatin, and its knockdown enhances sensitivity to oxaliplatin in resistant CRC cells. It has been observed that the mechanism by which miR-5000-3p confers drug resistance is through regulating the PI3K/AKT pathway via targeting USP49[84].
It has also been observed that USP1 inhibition sensitizes CRC cells to chemotherapeutics that act directly on DNA, such as doxorubicin[24].
In 5-FU-resistant CRC cells, the level of USP22 is increased along with the levels of Wnt target genes; however, after USP22 knockdown, chemoresistant CRC cells showed reduced viability and sphere formation ability but no increase in Wnt target gene levels[48].
USP11 can induce resistance to 5-FU in CRC by activating autophagy via stabilizing VCP[67].
It has been shown that USP35 overexpression enhances CRC cell proliferation and resistance to oxaliplatin and 5-FU[85].
USP20 has been shown to be related to several multidrug resistance genes, such as MRP1, MRP3, and MRP5[79,86].
USP3 is also recruited by a novel lncRNA, AC092894.1, to promote AR (androgen receptor) deubiquitination and, subsequently, chemosensitivity to oxaliplatin[87].
A variety of USP inhibitors have been developed as potential anticancer agents, but to date, no USP inhibitor has been approved for clinical use[88]. Vismodegib is a drug used for treating basal cell carcinoma. Indeed, vismodegib decreases the activity of USP25 and USP28 as well as their substrates, such as c-MYC, in CRC cells[30,89]. Additionally, neutral red has been identified as a noncompetitive inhibitor of USP4. This supports the possibility of developing specific DUB inhibitors as therapeutic agents[90].
USPs, such as USP7, are essential in Wnt signaling regulation, and their inhibitors have great potential in CRC therapy. USP7 inhibition by P5091 has been shown to attenuate the activity of Wnt signaling by promoting the ubiquitination and subsequent degradation of β-catenin[91]. Inhibitors of USP7 and USP47 are considered to have potential as cancer therapeutics because of their ability to stabilize the tumor suppressor p53 and reduce the levels of DNA polymerase β (Polβ)[92]. Some Nbenzylpiperidine derivatives that have shown in vivo antitumor immunity activity act as inhibitors of USP7[93].
USP21 has been reported as a novel therapeutic target for cancer treatment. One study showed that gallic acid causes PD-L1 downregulation by inhibiting USP21-mediated deubiquitination. BAY-805 is another potent and selective USP21 inhibitor[94,95].
USP22 is abnormally expressed in various malignant tumors, such as prostate cancer, lung cancer, liver cancer, and CRC. It can exert oncogenic effects in many of these tumor types via multiple mechanisms including immune evasion, and drug resistance. These data suggest that combining USP22 inhibition with chemotherapeutic, targeted, and immunosuppressive drugs can be effective approach to reduce drug resistance in cancer therapy[96].
While USPs can be utilized as drug targets because of their pivotal role in CRC, they can also be considered potential noninvasive biomarkers in the diagnosis and prognosis of CRC.
As the major group of the DUB family, USPs are crucial regulators of numerous biological processes. Our study highlights their pivotal roles in various processes implicated in CRC; these roles include regulation of the cell cycle, apoptosis, cancer stemness, EMT, metastasis, DNA repair, and drug resistance. This study highlights the great potential of USPs as drug targets and noninvasive biomarkers in CRC.
The dysregulation of USPs in CRC contributes to drug resistance through multiple mechanisms. Targeting specific USPs involved in drug resistance pathways could provide a novel therapeutic strategy for overcoming resistance to current treatment regimens in CRC including immunotherapy and targeted therapy. Further research is warranted to elucidate the precise mechanisms of action and potential inhibitors of these USPs, as well as their effectiveness in preclinical and clinical settings.
Colorectal cancer (CRC) is a major health problem and a leading cause of cancer death. Accumulating evidence shows that ubiquitin-specific peptidases (USPs) play a role in the development and progression of various cancers, including CRCs.
There is a need to identify new therapeutic agents in CRC to tackle the problem of resistance to existing therapy. USPs could be promising targets in this regard.
To provide a systematic review of the available studies on the role of USPs in CRC.
Systematic search in the MEDLINE (via PubMed), Scopus, and Web of Science databases followed by data analysis and discussion.
We found that USPs play significant roles in several processes and disrupt many pathways in CRC including the cell cycle, apoptosis, cancer stemness, epithelial–mesenchymal transition, metastasis, DNA repair, and drug resistance.
USPs play a major role in CRC development and progression and contribute to drug resistance in CRC via multiple mechanisms.
USPs can be used as biomarkers in CRC and many of them can be therapeutic targets that help overcoming resistance to current CRC therapeutics.
| 1. | Xi Y, Xu P. Global colorectal cancer burden in 2020 and projections to 2040. Transl Oncol. 2021;14:101174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 687] [Cited by in RCA: 1573] [Article Influence: 314.6] [Reference Citation Analysis (7)] |
| 2. | Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19834] [Cited by in RCA: 19783] [Article Influence: 760.9] [Reference Citation Analysis (0)] |
| 3. | Bazarbashi S, Al Eid H, Minguet J. Cancer Incidence in Saudi Arabia: 2012 Data from the Saudi Cancer Registry. Asian Pac J Cancer Prev. 2017;18:2437-2444. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 60] [Reference Citation Analysis (0)] |
| 4. | Shahesmaeili A, Malekpour Afshar R, Sadeghi A, Bazrafshan A. Cancer Incidence in Kerman Province, Southeast of Iran: Report of an ongoing Population-Based Cancer Registry, 2014. Asian Pac J Cancer Prev. 2018;19:1533-1541. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 5. | Elwali NE, Jarrah O, Alzahrani SG, Alharbi MB, Alhejaily AG, Alsharm AA, Elhassan MMA. Colorectal Cancer in Saudi Arabia: The Way Forward. Asian Pac J Cancer Prev. 2023;24:13-19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 22] [Reference Citation Analysis (0)] |
| 6. | Xie Y, Shi L, He X, Luo Y. Gastrointestinal cancers in China, the USA, and Europe. Gastroenterol Rep (Oxf). 2021;9:91-104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 150] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 7. | Chi Z, Lin Y, Huang J, Lv MY, Chen J, Chen X, Zhang B, Chen Y, Hu J, He X, Lan P. Risk factors for recurrence of colorectal conventional adenoma and serrated polyp. Gastroenterol Rep (Oxf). 2022;10:goab038. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 8. | Liang L, Liang Y, Li K, Qin P, Lin G, Li Y, Xu H, Wang S, Jing Q, Liang B, Xu L. A risk-prediction score for colorectal lesions on 12,628 participants at high risk of colorectal cancer. Gastroenterol Rep (Oxf). 2022;10:goac002. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 9. | Parmar S, Easwaran H. Genetic and epigenetic dependencies in colorectal cancer development. Gastroenterol Rep (Oxf). 2022;10:goac035. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 50] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 10. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 56665] [Article Influence: 7083.1] [Reference Citation Analysis (135)] |
| 11. | Kimura Y, Tanaka K. Regulatory mechanisms involved in the control of ubiquitin homeostasis. J Biochem. 2010;147:793-798. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 158] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 12. | Grou CP, Pinto MP, Mendes AV, Domingues P, Azevedo JE. The de novo synthesis of ubiquitin: identification of deubiquitinases acting on ubiquitin precursors. Sci Rep. 2015;5:12836. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 81] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 13. | Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576-1583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1081] [Cited by in RCA: 1154] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 14. | Fraile JM, Quesada V, Rodríguez D, Freije JM, López-Otín C. Deubiquitinases in cancer: new functions and therapeutic options. Oncogene. 2012;31:2373-2388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 302] [Cited by in RCA: 366] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 15. | Abdul Rehman SA, Kristariyanto YA, Choi SY, Nkosi PJ, Weidlich S, Labib K, Hofmann K, Kulathu Y. MINDY-1 Is a Member of an Evolutionarily Conserved and Structurally Distinct New Family of Deubiquitinating Enzymes. Mol Cell. 2016;63:146-155. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 218] [Cited by in RCA: 294] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 16. | Kwasna D, Abdul Rehman SA, Natarajan J, Matthews S, Madden R, De Cesare V, Weidlich S, Virdee S, Ahel I, Gibbs-Seymour I, Kulathu Y. Discovery and Characterization of ZUFSP/ZUP1, a Distinct Deubiquitinase Class Important for Genome Stability. Mol Cell. 2018;70:150-164.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 154] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 17. | Komander D, Clague MJ, Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550-563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1748] [Cited by in RCA: 1682] [Article Influence: 98.9] [Reference Citation Analysis (0)] |
| 18. | Chen YJ, Ma YS, Fang Y, Wang Y, Fu D, Shen XZ. Power and promise of ubiquitin carboxyl-terminal hydrolase 37 as a target of cancer therapy. Asian Pac J Cancer Prev. 2013;14:2173-2179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 19. | He M, Zhou Z, Wu G, Chen Q, Wan Y. Emerging role of DUBs in tumor metastasis and apoptosis: Therapeutic implication. Pharmacol Ther. 2017;177:96-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 83] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 20. | Moher D, Shamseer L, Clarke M, Ghersi D, Liberati A, Petticrew M, Shekelle P, Stewart LA; PRISMA-P Group. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst Rev. 2015;4:1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15040] [Cited by in RCA: 16694] [Article Influence: 1517.6] [Reference Citation Analysis (1)] |
| 21. | Al-Balushi E, Al-Marzouqi1 A, Tavoosi S, Baghsheikhi AH, Sadri A, Aliabadi LS, Salarabedi MM, Rahman SA, Al-Yateem N, Jarrahi AM, Halimi A, Ahmadvand M, Abdel-Rahman WM. The Role of Ubiquitin-specific Peptidases in Colorectal Cancer A Systematic Review Protocol. Asian Pacific Journal of Cancer Biology. 2023;8:417-419. [DOI] [Full Text] |
| 22. | Bonacci T, Emanuele MJ. Dissenting degradation: Deubiquitinases in cell cycle and cancer. Semin Cancer Biol. 2020;67:145-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 93] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 23. | Magiera K, Tomala M, Kubica K, De Cesare V, Trost M, Zieba BJ, Kachamakova-Trojanowska N, Les M, Dubin G, Holak TA, Skalniak L. Lithocholic Acid Hydroxyamide Destabilizes Cyclin D1 and Induces G(0)/G(1) Arrest by Inhibiting Deubiquitinase USP2a. Cell Chem Biol. 2017;24:458-470.e18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 24. | Xu X, Li S, Cui X, Han K, Wang J, Hou X, Cui L, He S, Xiao J, Yang Y. Inhibition of Ubiquitin Specific Protease 1 Sensitizes Colorectal Cancer Cells to DNA-Damaging Chemotherapeutics. Front Oncol. 2019;9:1406. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 25. | Lin Z, Tan C, Qiu Q, Kong S, Yang H, Zhao F, Liu Z, Li J, Kong Q, Gao B, Barrett T, Yang GY, Zhang J, Fang D. Ubiquitin-specific protease 22 is a deubiquitinase of CCNB1. Cell Discov. 2015;1:15028-15028. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 70] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 26. | Davis MI, Pragani R, Fox JT, Shen M, Parmar K, Gaudiano EF, Liu L, Tanega C, McGee L, Hall MD, McKnight C, Shinn P, Nelson H, Chattopadhyay D, D'Andrea AD, Auld DS, DeLucas LJ, Li Z, Boxer MB, Simeonov A. Small Molecule Inhibition of the Ubiquitin-specific Protease USP2 Accelerates cyclin D1 Degradation and Leads to Cell Cycle Arrest in Colorectal Cancer and Mantle Cell Lymphoma Models. J Biol Chem. 2016;291:24628-24640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 128] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 27. | Harris DR, Mims A, Bunz F. Genetic disruption of USP9X sensitizes colorectal cancer cells to 5-fluorouracil. Cancer Biol Ther. 2012;13:1319-1324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 38] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 28. | Lin Z, Yang H, Tan C, Li J, Liu Z, Quan Q, Kong S, Ye J, Gao B, Fang D. USP10 antagonizes c-Myc transcriptional activation through SIRT6 stabilization to suppress tumor formation. Cell Rep. 2013;5:1639-1649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 166] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 29. | Kim K, Huh T, Park Y, Koo DH, Kim H, Hwang I, Choi CH, Yi JM, Chung JY. Prognostic significance of USP10 and p14ARF expression in patients with colorectal cancer. Pathol Res Pract. 2020;216:152988. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (3)] |
| 30. | Wang H, Meng Q, Ding Y, Xiong M, Zhu M, Yang Y, Su H, Gu L, Xu Y, Shi L, Zhou H, Zhang N. USP28 and USP25 are downregulated by Vismodegib in vitro and in colorectal cancer cell lines. FEBS J. 2021;288:1325-1342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 31. | Diefenbacher ME, Popov N, Blake SM, Schülein-Völk C, Nye E, Spencer-Dene B, Jaenicke LA, Eilers M, Behrens A. The deubiquitinase USP28 controls intestinal homeostasis and promotes colorectal cancer. J Clin Invest. 2014;124:3407-3418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 129] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 32. | Bernhard SV, Seget-Trzensiok K, Kuffer C, Krastev DB, Stautmeister LM, Theis M, Keuper K, Boekenkamp JE, Kschischo M, Buchholz F, Storchova Z. Loss of USP28 and SPINT2 expression promotes cancer cell survival after whole genome doubling. Cell Oncol (Dordr). 2022;45:103-119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 33. | Das S, Chandrasekaran AP, Suresh B, Haq S, Kang JH, Lee SJ, Kim J, Lee S, Kim HH, Kim KS, Ramakrishna S. Genome-scale screening of deubiquitinase subfamily identifies USP3 as a stabilizer of Cdc25A regulating cell cycle in cancer. Cell Death Differ. 2020;27:3004-3020. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 34. | Li B, Qi ZP, He DL, Chen ZH, Liu JY, Wong MW, Zhang JW, Xu EP, Shi Q, Cai SL, Sun D, Yao LQ, Zhou PH, Zhong YS. NLRP7 deubiquitination by USP10 promotes tumor progression and tumor-associated macrophage polarization in colorectal cancer. J Exp Clin Cancer Res. 2021;40:126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 80] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 35. | Aubrey BJ, Kelly GL, Janic A, Herold MJ, Strasser A. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ. 2018;25:104-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 509] [Cited by in RCA: 1014] [Article Influence: 112.7] [Reference Citation Analysis (0)] |
| 36. | Choi ES, Lee H, Sung JY, Lee CH, Jang H, Kim KT, Kim YN, Kim HP, Goh SH. FAM188B enhances cell survival via interaction with USP7. Cell Death Dis. 2018;9:633. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 37. | Cho J, Park J, Shin SC, Jang M, Kim JH, Kim EE, Song EJ. USP47 Promotes Tumorigenesis by Negative Regulation of p53 through Deubiquitinating Ribosomal Protein S2. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 38. | Huang T, Zhang Q, Ren W, Yan B, Yi L, Tang T, Lin H, Zhang Y. USP44 suppresses proliferation and enhances apoptosis in colorectal cancer cells by inactivating the Wnt/β-catenin pathway via Axin1 deubiquitination. Cell Biol Int. 2020;44:1651-1659. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 39. | Lou Y, Ye M, Xu C, Tao F. Insight into the physiological and pathological roles of USP44, a potential tumor target (Review). Oncol Lett. 2022;24:455. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 40. | Peddaboina C, Jupiter D, Fletcher S, Yap JL, Rai A, Tobin RP, Jiang W, Rascoe P, Rogers MK, Smythe WR, Cao X. The downregulation of Mcl-1 via USP9X inhibition sensitizes solid tumors to Bcl-xl inhibition. BMC Cancer. 2012;12:541. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 78] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 41. | Yuan J, Li X, Zhang G, Cheng W, Wang W, Lei Y, Ma Q, Song G. USP39 mediates p21-dependent proliferation and neoplasia of colon cancer cells by regulating the p53/p21/CDC2/cyclin B1 axis. Mol Carcinog. 2021;60:265-278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 42. | Zhang W, Wang Z, Cai G, Huang P. Circ_DOCK1 regulates USP11 through miR-132-3p to control colorectal cancer progression. World J Surg Oncol. 2021;19:67. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 43. | Tu R, Kang W, Kang Y, Chen Z, Zhang P, Xiong X, Ma J, Du RL, Zhang C. c-MYC-USP49-BAG2 axis promotes proliferation and chemoresistance of colorectal cancer cells in vitro. Biochem Biophys Res Commun. 2022;607:117-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 44. | Mushtaq M, Kovalevska L, Darekar S, Abramsson A, Zetterberg H, Kashuba V, Klein G, Arsenian-Henriksson M, Kashuba E. Cell stemness is maintained upon concurrent expression of RB and the mitochondrial ribosomal protein S18-2. Proc Natl Acad Sci U S A. 2020;117:15673-15683. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 45. | Zhan W, Liao X, Liu J, Tian T, Yu L, Li R. USP38 regulates the stemness and chemoresistance of human colorectal cancer via regulation of HDAC3. Oncogenesis. 2020;9:48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 46. | Zhang S, Ju X, Yang Q, Zhu Y, Fan D, Su G, Kong L, Li Y. USP47 maintains the stemness of colorectal cancer cells and is inhibited by parthenolide. Biochem Biophys Res Commun. 2021;562:21-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 47. | Yu L, Fu J, Shen C. Ubiquitin specific peptidase 47 promotes proliferation of lung squamous cell carcinoma. Genes Genomics. 2022;44:721-731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 48. | Jiang S, Song C, Gu X, Wang M, Miao D, Lv J, Liu Y. Ubiquitin-Specific Peptidase 22 Contributes to Colorectal Cancer Stemness and Chemoresistance via Wnt/β-Catenin Pathway. Cell Physiol Biochem. 2018;46:1412-1422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 49. | Yan J, Tan M, Yu L, Jin X, Li Y. Ring finger 220 promotes the stemness and progression of colon cancer cells via Ubiquitin specific peptidase 22-BMI1 axis. Bioengineered. 2021;12:12060-12069. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 50. | Miao D, Wang Y, Jia Y, Tong J, Jiang S, Liu L. ZRANB1 enhances stem-cell-like features and accelerates tumor progression by regulating Sox9-mediated USP22/Wnt/β-catenin pathway in colorectal cancer. Cell Signal. 2022;90:110200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 51. | Reissland M, Hartmann O, Tauch S, Prieto-Garcia C, Schulte C, Solvie D, Loebbert S, Jacomin AC, Pesic M, Bugter JM, Schuelein-Voelk C, Fuss CT, Pahor N, Ade C, Buck V, Potente M, Li V, Beliu G, Wiegering A, Bitman-Lotan E, Grossmann T, Rosenfeldt M, Eilers M, Maric H, Maurice MM, Greten F, Dikič I, Orian A, Gallant P, Diefenbacher ME. Stabilisation of β-Catenin-WNT signalling by USP10 in APC-truncated colorectal cancer drives cancer stemness and enables super-competitor signalling. 2023 Preprint. Available from: bioRxiv:2023.02.10.527983. [DOI] [Full Text] |
| 52. | Kang KT, Shin MJ, Moon HJ, Choi KU, Suh DS, Kim JH. TRRAP Enhances Cancer Stem Cell Characteristics by Regulating NANOG Protein Stability in Colon Cancer Cells. Int J Mol Sci. 2023;24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 53. | Guan Y, Jiang SR, Liu JG, Shi JR, Liu ZB. USP20 regulates the stability of EMT transcription factor SOX4 and influences colorectal cancer metastasis. Pathol Res Pract. 2022;233:153879. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 54. | Li Y, Yang Y, Li J, Liu H, Chen F, Li B, Cui B, Liu Y. USP22 drives colorectal cancer invasion and metastasis via epithelial-mesenchymal transition by activating AP4. Oncotarget. 2017;8:32683-32695. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 55. | Wang XM, Yang C, Zhao Y, Xu ZG, Yang W, Wang P, Lin D, Xiong B, Fang JY, Dong C, Zhong B. The deubiquitinase USP25 supports colonic inflammation and bacterial infection and promotes colorectal cancer. Nat Cancer. 2020;1:811-825. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 61] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 56. | Yun SI, Kim HH, Yoon JH, Park WS, Hahn MJ, Kim HC, Chung CH, Kim KK. Ubiquitin specific protease 4 positively regulates the WNT/β-catenin signaling in colorectal cancer. Mol Oncol. 2015;9:1834-1851. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 107] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 57. | Giebel N, de Jaime-Soguero A, García Del Arco A, Landry JJM, Tietje M, Villacorta L, Benes V, Fernández-Sáiz V, Acebrón SP. USP42 protects ZNRF3/RNF43 from R-spondin-dependent clearance and inhibits Wnt signalling. EMBO Rep. 2021;22:e51415. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 58. | Novellasdemunt L, Foglizzo V, Cuadrado L, Antas P, Kucharska A, Encheva V, Snijders AP, Li VSW. USP7 Is a Tumor-Specific WNT Activator for APC-Mutated Colorectal Cancer by Mediating β-Catenin Deubiquitination. Cell Rep. 2017;21:612-627. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 125] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 59. | Novellasdemunt L, Kucharska A, Baulies A, Hutton C, Vlachogiannis G, Repana D, Rowan A, Suárez-Bonnet A, Ciccarelli F, Valeri N, Li VSW. USP7 inactivation suppresses APC-mutant intestinal hyperproliferation and tumor development. Stem Cell Reports. 2023;18:570-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 60. | Sarkar S. Proteogenomic Approaches to Understand Gene Mutations and Protein Structural Alterations in Colon Cancer. Physiologia. 2023;3:11-29. [DOI] [Full Text] |
| 61. | Basu B, Karmakar S, Basu M, Ghosh MK. A novel role of USP7 in imparting partial EMT state in colorectal cancer through the DDX3X-β-catenin axis. 2018 Preprint. Available from: bioRxiv:2023.01.11.523622. [DOI] [Full Text] |
| 62. | Hou R, Li Y, Luo X, Zhang W, Yang H, Zhang Y, Liu J, Liu S, Han S, Liu C, Huang Y, Liu Z, Li A, Fang W. ENKUR expression induced by chemically synthesized cinobufotalin suppresses malignant activities of hepatocellular carcinoma by modulating β-catenin/c-Jun/MYH9/USP7/c-Myc axis. Int J Biol Sci. 2022;18:2553-2567. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 41] [Reference Citation Analysis (0)] |
| 63. | Yuan X, Sun X, Shi X, Wang H, Wu G, Jiang C, Yu D, Zhang W, Xue B, Ding Y. USP39 promotes colorectal cancer growth and metastasis through the Wnt/β-catenin pathway. Oncol Rep. 2017;37:2398-2404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 64. | Sun K, He SB, Yao YZ, Qu JG, Xie R, Ma YQ, Zong MH, Chen JX. Tre2 (USP6NL) promotes colorectal cancer cell proliferation via Wnt/β-catenin pathway. Cancer Cell Int. 2019;19:102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 65. | Xing C, Lu XX, Guo PD, Shen T, Zhang S, He XS, Gan WJ, Li XM, Wang JR, Zhao YY, Wu H, Li JM. Ubiquitin-Specific Protease 4-Mediated Deubiquitination and Stabilization of PRL-3 Is Required for Potentiating Colorectal Oncogenesis. Cancer Res. 2016;76:83-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 82] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 66. | Sun H, Ou B, Zhao S, Liu X, Song L, Wang R, Peng Z. USP11 promotes growth and metastasis of colorectal cancer via PPP1CA-mediated activation of ERK/MAPK signaling pathway. EBioMedicine. 2019;48:236-247. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 101] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 67. | Liao Y, Zhou D, Wang P, Yang M, Jiang N. Ubiquitin specific peptidase 11 as a novel therapeutic target for cancer management. Cell Death Discov. 2022;8:292. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 24] [Reference Citation Analysis (0)] |
| 68. | Huang F, Zheng C, Huang L, Lin C, Wang J. USP18 directly regulates Snail1 protein through ubiquitination pathway in colorectal cancer. Cancer Cell Int. 2020;20:346. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 69. | Zhang B, Halder SK, Kashikar ND, Cho YJ, Datta A, Gorden DL, Datta PK. Antimetastatic role of Smad4 signaling in colorectal cancer. Gastroenterology. 2010;138:969-80.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 178] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 70. | Inamoto S, Itatani Y, Yamamoto T, Minamiguchi S, Hirai H, Iwamoto M, Hasegawa S, Taketo MM, Sakai Y, Kawada K. Loss of SMAD4 Promotes Colorectal Cancer Progression by Accumulation of Myeloid-Derived Suppressor Cells through the CCL15-CCR1 Chemokine Axis. Clin Cancer Res. 2016;22:492-501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 107] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 71. | Wang Z, Yang J, Di J, Cui M, Xing J, Wu F, Wu W, Yang H, Zhang C, Yao Z, Zhang N, Jiang B, Su X. Downregulated USP3 mRNA functions as a competitive endogenous RNA of SMAD4 by sponging miR-224 and promotes metastasis in colorectal cancer. Sci Rep. 2017;7:4281. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 72. | Zeng H, Yuan F, Mi Y, Xian G, Qin C, Zhang D. As an independent prognostic factor, USP6 promotes the invasion and metastasis of colon cancer. Biochem Biophys Res Commun. 2018;505:816-822. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 73. | Yun SI, Hong HK, Yeo SY, Kim SH, Cho YB, Kim KK. Ubiquitin-Specific Protease 21 Promotes Colorectal Cancer Metastasis by Acting as a Fra-1 Deubiquitinase. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 74. | Huang Z, Wen P, Kong R, Cheng H, Zhang B, Quan C, Bian Z, Chen M, Zhang Z, Chen X, Du X, Liu J, Zhu L, Fushimi K, Hua D, Wu JY. USP33 mediates Slit-Robo signaling in inhibiting colorectal cancer cell migration. Int J Cancer. 2015;136:1792-1802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 55] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 75. | Yan S, Yue Y, Wang J, Li W, Sun M, Gu C, Zeng L. LINC00668 promotes tumorigenesis and progression through sponging miR-188-5p and regulating USP47 in colorectal cancer. Eur J Pharmacol. 2019;858:172464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 76. | Luo H, Ji Y, Gao X, Liu X, Wu Y. Ubiquitin Specific Protease 2: Structure, Isoforms, Cellular Function, Related Diseases and Its Inhibitors. Oncologie. 2022;24:1-15. [RCA] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 77. | Wang J, Gu Y, Yan X, Zhang J, Wang J, Ding Y. USP38 inhibits colorectal cancer cell proliferation and migration via downregulating HMX3 ubiquitylation. Cell Cycle. 2023;22:1169-1181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 78. | Li H, Chai L, Ding Z, He H. CircCOL1A2 Sponges MiR-1286 to Promote Cell Invasion and Migration of Gastric Cancer by Elevating Expression of USP10 to Downregulate RFC2 Ubiquitination Level. J Microbiol Biotechnol. 2022;32:938-948. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 79. | Jin R, Luo Z, Jun-Li, Tao Q, Wang P, Cai X, Jiang L, Zeng C, Chen Y. USP20 is a predictor of poor prognosis in colorectal cancer and associated with lymph node metastasis, immune infiltration and chemotherapy resistance. Front Oncol. 2023;13:1023292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 80. | Zhou L, Ouyang T, Li M, Hong T, Mhs A, Meng W, Zhang N. Ubiquitin-Specific Peptidase 7: A Novel Deubiquitinase That Regulates Protein Homeostasis and Cancers. Front Oncol. 2021;11:784672. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 81. | Xu X, Huang A, Cui X, Han K, Hou X, Wang Q, Cui L, Yang Y. Ubiquitin specific peptidase 5 regulates colorectal cancer cell growth by stabilizing Tu translation elongation factor. Theranostics. 2019;9:4208-4220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 64] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 82. | Liu W, Zhang Q, Fang Y, Wang Y. The deubiquitinase USP38 affects cellular functions through interacting with LSD1. Biol Res. 2018;51:53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 83. | Wen YA, Stevens PD, Gasser ML, Andrei R, Gao T. Downregulation of PHLPP expression contributes to hypoxia-induced resistance to chemotherapy in colon cancer cells. Mol Cell Biol. 2013;33:4594-4605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 84. | Zhuang YY, Zhong W, Xia ZS, Lin SZ, Chan MC, Jiang K, Li WF, Xu XY. miR-5000-3p confers oxaliplatin resistance by targeting ubiquitin-specific peptidase 49 in colorectal cancer. Cell Death Discov. 2021;7:129. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 85. | Xiao Y, Jiang X, Yin K, Miao T, Lu H, Wang W, Ma L, Zhao Y, Liu C, Qiao Y, Zhang P. USP35 promotes cell proliferation and chemotherapeutic resistance through stabilizing FUCA1 in colorectal cancer. Oncogenesis. 2023;12:12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 86. | Ming H, Li B, Jiang J, Qin S, Nice EC, He W, Lang T, Huang C. Protein degradation: expanding the toolbox to restrain cancer drug resistance. J Hematol Oncol. 2023;16:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 35] [Reference Citation Analysis (0)] |
| 87. | Zheng Z, Wu M, Li H, Xu W, Yang M, Pan K, Ni Y, Jiang T, Zheng H, Jin X, Zhang Y, Ding L, Fu J. Downregulation of AC092894.1 promotes oxaliplatin resistance in colorectal cancer via the USP3/AR/RASGRP3 axis. BMC Med. 2023;21:132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 25] [Reference Citation Analysis (0)] |
| 88. | Chen S, Liu Y, Zhou H. Advances in the Development Ubiquitin-Specific Peptidase (USP) Inhibitors. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 104] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 89. | Zhou D, Xu Z, Huang Y, Wang H, Zhu X, Zhang W, Song W, Gao T, Liu T, Wang M, Shi L, Zhang N, Xiong B. Structure-based discovery of potent USP28 inhibitors derived from Vismodegib. Eur J Med Chem. 2023;254:115369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 90. | Nguyen HH, Kim T, Nguyen T, Hahn MJ, Yun SI, Kim KK. A Selective Inhibitor of Ubiquitin-Specific Protease 4 Suppresses Colorectal Cancer Progression by Regulating β-Catenin Signaling. Cell Physiol Biochem. 2019;53:157-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 91. | An T, Gong Y, Li X, Kong L, Ma P, Gong L, Zhu H, Yu C, Liu J, Zhou H, Mao B, Li Y. USP7 inhibitor P5091 inhibits Wnt signaling and colorectal tumor growth. Biochem Pharmacol. 2017;131:29-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 111] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 92. | Weinstock J, Wu J, Cao P, Kingsbury WD, McDermott JL, Kodrasov MP, McKelvey DM, Suresh Kumar KG, Goldenberg SJ, Mattern MR, Nicholson B. Selective Dual Inhibitors of the Cancer-Related Deubiquitylating Proteases USP7 and USP47. ACS Med Chem Lett. 2012;3:789-792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 157] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 93. | Li X, Yang S, Zhang H, Liu X, Gao Y, Chen Y, Liu L, Wang D, Liang Z, Liu S, Dai L, Xu Q, Yuan H, Chen C, Sun H, Wen X. Discovery of Orally Bioavailable N-Benzylpiperidinol Derivatives as Potent and Selective USP7 Inhibitors with In Vivo Antitumor Immunity Activity against Colon Cancer. J Med Chem. 2022;65:16622-16639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 23] [Reference Citation Analysis (0)] |
| 94. | Deng B, Yang B, Chen J, Wang S, Zhang W, Guo Y, Han Y, Li H, Dang Y, Yuan Y, Dai X, Zang Y, Li Y, Li B. Gallic acid induces T-helper-1-like T(reg) cells and strengthens immune checkpoint blockade efficacy. J Immunother Cancer. 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 54] [Reference Citation Analysis (0)] |
| 95. | Göricke F, Vu V, Smith L, Scheib U, Böhm R, Akkilic N, Wohlfahrt G, Weiske J, Bömer U, Brzezinka K, Lindner N, Lienau P, Gradl S, Beck H, Brown PJ, Santhakumar V, Vedadi M, Barsyte-Lovejoy D, Arrowsmith CH, Schmees N, Petersen K. Discovery and Characterization of BAY-805, a Potent and Selective Inhibitor of Ubiquitin-Specific Protease USP21. J Med Chem. 2023;66:3431-3447. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 20] [Reference Citation Analysis (0)] |
| 96. | Guo J, Zhao J, Fu W, Xu Q, Huang D. Immune Evasion and Drug Resistance Mediated by USP22 in Cancer: Novel Targets and Mechanisms. Front Immunol. 2022;13:918314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 97. | Deng L, Chen L, Zhao L, Xu Y, Peng X, Wang X, Ding L, Jin J, Teng H, Wang Y, Pan W, Yu F, Liao L, Li L, Ge X, Wang P. Ubiquitination of Rheb governs growth factor-induced mTORC1 activation. Cell Res. 2019;29:136-150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 104] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 98. | Wang Y, Zhou L, Lu J, Jiang B, Liu C, Guo J. USP4 function and multifaceted roles in cancer: a possible and potential therapeutic target. Cancer Cell Int. 2020;20:298. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 99. | Zhao B, Schlesiger C, Masucci MG, Lindsten K. The ubiquitin specific protease 4 (USP4) is a new player in the Wnt signalling pathway. J Cell Mol Med. 2009;13:1886-1895. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 46] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 100. | Zhang Z, Gao W, Zhou L, Chen Y, Qin S, Zhang L, Liu J, He Y, Lei Y, Chen HN, Han J, Zhou ZG, Nice EC, Li C, Huang C, Wei X. Repurposing Brigatinib for the Treatment of Colorectal Cancer Based on Inhibition of ER-phagy. Theranostics. 2019;9:4878-4892. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 101. | Ning F, Xin H, Liu J, Lv C, Xu X, Wang M, Wang Y, Zhang W, Zhang X. Structure and function of USP5: Insight into physiological and pathophysiological roles. Pharmacol Res. 2020;157:104557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 75] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 102. | Nicholson B, Suresh Kumar KG. The multifaceted roles of USP7: new therapeutic opportunities. Cell Biochem Biophys. 2011;60:61-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 213] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 103. | Colland F, Formstecher E, Jacq X, Reverdy C, Planquette C, Conrath S, Trouplin V, Bianchi J, Aushev VN, Camonis J, Calabrese A, Borg-Capra C, Sippl W, Collura V, Boissy G, Rain JC, Guedat P, Delansorne R, Daviet L. Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Mol Cancer Ther. 2009;8:2286-2295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 273] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 104. | Qian J, Pentz K, Zhu Q, Wang Q, He J, Srivastava AK, Wani AA. USP7 modulates UV-induced PCNA monoubiquitination by regulating DNA polymerase eta stability. Oncogene. 2015;34:4791-4796. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 52] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 105. | Bufalieri F, Lospinoso Severini L, Caimano M, Infante P, Di Marcotullio L. DUBs Activating the Hedgehog Signaling Pathway: A Promising Therapeutic Target in Cancer. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 106. | Li X, Kong L, Yang Q, Duan A, Ju X, Cai B, Chen L, An T, Li Y. Parthenolide inhibits ubiquitin-specific peptidase 7 (USP7), Wnt signaling, and colorectal cancer cell growth. J Biol Chem. 2020;295:3576-3589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 61] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 107. | Becker K, Marchenko ND, Palacios G, Moll UM. A role of HAUSP in tumor suppression in a human colon carcinoma xenograft model. Cell Cycle. 2008;7:1205-1213. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 53] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 108. | Yang Z, Huo S, Shan Y, Liu H, Xu Y, Yao K, Li X, Zhang X. STAT3 repressed USP7 expression is crucial for colon cancer development. FEBS Lett. 2012;586:3013-3017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 109. | Du Z, Song J, Wang Y, Zhao Y, Guda K, Yang S, Kao HY, Xu Y, Willis J, Markowitz SD, Sedwick D, Ewing RM, Wang Z. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci Signal. 2010;3:ra80. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 249] [Cited by in RCA: 269] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 110. | Zhu Y, Gu L, Lin X, Cui K, Liu C, Lu B, Zhou F, Zhao Q, Shen H, Li Y. LINC00265 promotes colorectal tumorigenesis via ZMIZ2 and USP7-mediated stabilization of β-catenin. Cell Death Differ. 2020;27:1316-1327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 72] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 111. | Tian T, Bi H, Liu Y, Li G, Zhang Y, Cao L, Hu F, Zhao Y, Yuan H. Copy number variation of ubiquitin- specific proteases genes in blood leukocytes and colorectal cancer. Cancer Biol Ther. 2020;21:637-646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 112. | Ouyang SW, Liu TT, Liu XS, Zhu FX, Zhu FM, Liu XN, Peng ZH. USP10 regulates Musashi-2 stability via deubiquitination and promotes tumour proliferation in colon cancer. FEBS Lett. 2019;593:406-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 113. | Ye Z, Chen J, Huang P, Xuan Z, Zheng S. Ubiquitin-specific peptidase 10, a deubiquitinating enzyme: Assessing its role in tumor prognosis and immune response. Front Oncol. 2022;12:990195. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 25] [Reference Citation Analysis (0)] |
| 114. | Lee EW, Seong D, Seo J, Jeong M, Lee HK, Song J. USP11-dependent selective cIAP2 deubiquitylation and stabilization determine sensitivity to Smac mimetics. Cell Death Differ. 2015;22:1463-1476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 115. | Huang YY, Zhang CM, Dai YB, Lin JG, Lin N, Huang ZX, Xu TW. USP11 facilitates colorectal cancer proliferation and metastasis by regulating IGF2BP3 stability. Am J Transl Res. 2021;13:480-496. [PubMed] |
| 116. | Sun H, Wang R, Liu Y, Mei H, Liu X, Peng Z. USP11 induce resistance to 5-Fluorouracil in Colorectal Cancer through activating autophagy by stabilizing VCP. J Cancer. 2021;12:2317-2325. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 117. | Xiang S, Fang J, Wang S, Deng B, Zhu L. MicroRNA135b regulates the stability of PTEN and promotes glycolysis by targeting USP13 in human colorectal cancers. Oncol Rep. 2015;33:1342-1348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 118. | Liu B, Chen J, Zhang S. Emerging role of ubiquitin-specific protease 14 in oncogenesis and development of tumor: Therapeutic implication. Life Sci. 2019;239:116875. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 119. | Mofers A, Perego P, Selvaraju K, Gatti L, Gullbo J, Linder S, D'Arcy P. Analysis of determinants for in vitro resistance to the small molecule deubiquitinase inhibitor b-AP15. PLoS One. 2019;14:e0223807. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 120. | Du XH, Ke SB, Liang XY, Gao J, Xie XX, Qi LZ, Liu XY, Xu GY, Zhang XD, Du RL, Li SZ. USP14 promotes colorectal cancer progression by targeting JNK for stabilization. Cell Death Dis. 2023;14:56. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 32] [Reference Citation Analysis (0)] |
| 121. | Shi D, Wu X, Jian Y, Wang J, Huang C, Mo S, Li Y, Li F, Zhang C, Zhang D, Zhang H, Huang H, Chen X, Wang YA, Lin C, Liu G, Song L, Liao W. USP14 promotes tryptophan metabolism and immune suppression by stabilizing IDO1 in colorectal cancer. Nat Commun. 2022;13:5644. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 140] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 122. | Huang X, Langelotz C, Hetfeld-Pechoc BK, Schwenk W, Dubiel W. The COP9 signalosome mediates beta-catenin degradation by deneddylation and blocks adenomatous polyposis coli destruction via USP15. J Mol Biol. 2009;391:691-702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 90] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 123. | Wang M, He SF, Liu LL, Sun XX, Yang F, Ge Q, Wong WK, Meng JY. Potential role of ZEB1 as a DNA repair regulator in colorectal cancer cells revealed by cancer-associated promoter profiling. Oncol Rep. 2017;38:1941-1948. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 124. | Zhang L, Zhang N, Li X, Wu W, Zhang Y, Wang J. High expression of USP18 is associated with the growth of colorectal carcinoma. Histol Histopathol. 2021;36:697-704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 125. | Zhu Y, Gu L, Lin X, Zhou X, Lu B, Liu C, Lei C, Zhou F, Zhao Q, Prochownik EV, Li Y. USP19 exacerbates lipogenesis and colorectal carcinogenesis by stabilizing ME1. Cell Rep. 2021;37:110174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 126. | Liu YL, Yang YM, Xu H, Dong XS. Aberrant expression of USP22 is associated with liver metastasis and poor prognosis of colorectal cancer. J Surg Oncol. 2011;103:283-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 83] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 127. | Jiang S, Miao D, Wang M, Lv J, Wang Y, Tong J. MiR-30-5p suppresses cell chemoresistance and stemness in colorectal cancer through USP22/Wnt/β-catenin signaling axis. J Cell Mol Med. 2019;23:630-640. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 66] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 128. | Xu H, Liu YL, Yang YM, Dong XS. Knock-down of ubiquitin-specific protease 22 by micro-RNA interference inhibits colorectal cancer growth. Int J Colorectal Dis. 2012;27:21-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 129. | Ao N, Liu Y, Feng H, Bian X, Li Z, Gu B, Zhao X. Ubiquitin-specific peptidase USP22 negatively regulates the STAT signaling pathway by deubiquitinating SIRT1. Cell Physiol Biochem. 2014;33:1863-1875. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 130. | Ao N, Liu Y, Bian X, Feng H. Ubiquitin-specific peptidase 22 inhibits colon cancer cell invasion by suppressing the signal transducer and activator of transcription 3/matrix metalloproteinase 9 pathway. Mol Med Rep. 2015;12:2107-2113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 131. | Kosinsky RL, Zerche M, Saul D, Wang X, Wohn L, Wegwitz F, Begus-Nahrmann Y, Johnsen SA. USP22 exerts tumor-suppressive functions in colorectal cancer by decreasing mTOR activity. Cell Death Differ. 2020;27:1328-1340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 50] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 132. | Liu YL, Yang YM, Xu H, Dong XS. Increased expression of ubiquitin-specific protease 22 can promote cancer progression and predict therapy failure in human colorectal cancer. J Gastroenterol Hepatol. 2010;25:1800-1805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 56] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 133. | Sippl W, Collura V, Colland F. Ubiquitin-specific proteases as cancer drug targets. Future Oncol. 2011;7:619-632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 44] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 134. | Liu YL, Jiang SX, Yang YM, Xu H, Liu JL, Wang XS. USP22 acts as an oncogene by the activation of BMI-1-mediated INK4a/ARF pathway and Akt pathway. Cell Biochem Biophys. 2012;62:229-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 66] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 135. | Kapoor S. Usp22 and its evolving role in systemic carcinogenesis. Lung Cancer. 2013;79:191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 136. | Liu Y, Yang Y, Xu H, Dong X. Implication of USP22 in the regulation of BMI-1, c-Myc, p16INK4a, p14ARF, and cyclin D2 expression in primary colorectal carcinomas. Diagn Mol Pathol. 2010;19:194-200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 137. | Wang Z, Zhu L, Guo T, Wang Y, Yang J. Decreased H2B monoubiquitination and overexpression of ubiquitin-specific protease enzyme 22 in malignant colon carcinoma. Hum Pathol. 2015;46:1006-1014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 138. | Gennaro VJ, Stanek TJ, Peck AR, Sun Y, Wang F, Qie S, Knudsen KE, Rui H, Butt T, Diehl JA, McMahon SB. Control of CCND1 ubiquitylation by the catalytic SAGA subunit USP22 is essential for cell cycle progression through G1 in cancer cells. Proc Natl Acad Sci U S A. 2018;115:E9298-E9307. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 101] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 139. | Kosinsky RL, Helms M, Zerche M, Wohn L, Dyas A, Prokakis E, Kazerouni ZB, Bedi U, Wegwitz F, Johnsen SA. USP22-dependent HSP90AB1 expression promotes resistance to HSP90 inhibition in mammary and colorectal cancer. Cell Death Dis. 2019;10:911. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 140. | He X, Ma J, Zhang M, Cui J, Yang H. Long Non-Coding RNA SNHG16 Activates USP22 Expression to Promote Colorectal Cancer Progression by Sponging miR-132-3p. Onco Targets Ther. 2020;13:4283-4294. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 141. | Feng T, Ling S, Xu C, Ying L, Su D, Xu X. Ubiquitin-specific peptidase 22 in cancer. Cancer Lett. 2021;514:30-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 142. | Kosinsky RL, Saul D, Ammer-Herrmenau C, Faubion WA, Neesse A, Johnsen SA. USP22 Suppresses SPARC Expression in Acute Colitis and Inflammation-Associated Colorectal Cancer. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 143. | Xian D, Niu L, Zeng J, Wang L. LncRNA KCNQ1OT1 Secreted by Tumor Cell-Derived Exosomes Mediates Immune Escape in Colorectal Cancer by Regulating PD-L1 Ubiquitination via MiR-30a-5p/USP22. Front Cell Dev Biol. 2021;9:653808. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 73] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 144. | Kotelevets L, Chastre E. Extracellular Vesicles in Colorectal Cancer: From Tumor Growth and Metastasis to Biomarkers and Nanomedications. Cancers (Basel). 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 28] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 145. | Popov N, Wanzel M, Madiredjo M, Zhang D, Beijersbergen R, Bernards R, Moll R, Elledge SJ, Eilers M. The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol. 2007;9:765-774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 316] [Cited by in RCA: 382] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 146. | Liu Z, Chen M, Xu X, Zhang L, Pan Y, Chen D. USP28 promotes aerobic glycolysis of colorectal cancer by increasing stability of FOXC1. Acta Biochim Pol. 2021;68:633-639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 147. | Xu X, Li J, Sun X, Guo Y, Chu D, Wei L, Li X, Yang G, Liu X, Yao L, Zhang J, Shen L. Tumor suppressor NDRG2 inhibits glycolysis and glutaminolysis in colorectal cancer cells by repressing c-Myc expression. Oncotarget. 2015;6:26161-26176. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 79] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 148. | Ren X, Jiang M, Ding P, Zhang X, Zhou X, Shen J, Liu D, Yan X, Ma Z. Ubiquitin-specific protease 28: the decipherment of its dual roles in cancer development. Exp Hematol Oncol. 2023;12:27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 149. | Chandrasekaran AP, Suresh B, Sarodaya N, Ko NR, Oh SJ, Kim KS, Ramakrishna S. Ubiquitin Specific Protease 29 Functions as an Oncogene Promoting Tumorigenesis in Colorectal Carcinoma. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 150. | Li J, Yang J, Chen Z, Liu L, Wang H, Deng Q, Chen Y, Que Y, Fu Z. Promotive role of USP29-mediated deubiquitination in malignant proliferation of colorectal cancer cells via the KIAA1429/SOX8 axis. Biomol Biomed. 2023;23:483-495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 151. | Lui TT, Lacroix C, Ahmed SM, Goldenberg SJ, Leach CA, Daulat AM, Angers S. The ubiquitin-specific protease USP34 regulates axin stability and Wnt/β-catenin signaling. Mol Cell Biol. 2011;31:2053-2065. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 111] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 152. | Fraile JM, Manchado E, Lujambio A, Quesada V, Campos-Iglesias D, Webb TR, Lowe SW, López-Otín C, Freije JM. USP39 Deubiquitinase Is Essential for KRAS Oncogene-driven Cancer. J Biol Chem. 2017;292:4164-4175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 153. | Remitha NPSI, Wiguna IGWW, Sadvika IGAS, Sadeva IGKA, Supadmanaba IGP, Wihandani DM. Clinicopathological and Prognostic Significance of Ubiquitin-Specific Protease 39 Overexpression in solid Cancers: A Meta-Analysis. Asian Pac J Cancer Prev. 2023;24:1015-1025. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 154. | Ye DX, Wang SS, Huang Y, Wang XJ, Chi P. USP43 directly regulates ZEB1 protein, mediating proliferation and metastasis of colorectal cancer. J Cancer. 2021;12:404-416. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 155. | Sloane MA, Wong JW, Perera D, Nunez AC, Pimanda JE, Hawkins NJ, Sieber OM, Bourke MJ, Hesson LB, Ward RL. Epigenetic inactivation of the candidate tumor suppressor USP44 is a frequent and early event in colorectal neoplasia. Epigenetics. 2014;9:1092-1100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 39] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 156. | Li X, Stevens PD, Yang H, Gulhati P, Wang W, Evers BM, Gao T. The deubiquitination enzyme USP46 functions as a tumor suppressor by controlling PHLPP-dependent attenuation of Akt signaling in colon cancer. Oncogene. 2013;32:471-478. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 99] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 157. | Choi BJ, Park SA, Lee SY, Cha YN, Surh YJ. Hypoxia induces epithelial-mesenchymal transition in colorectal cancer cells through ubiquitin-specific protease 47-mediated stabilization of Snail: A potential role of Sox9. Sci Rep. 2017;7:15918. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 94] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 158. | Yu L, Dong L, Wang Y, Liu L, Long H, Li H, Li J, Yang X, Liu Z, Duan G, Dai X, Lin Z. Reversible regulation of SATB1 ubiquitination by USP47 and SMURF2 mediates colon cancer cell proliferation and tumor progression. Cancer Lett. 2019;448:40-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 65] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 159. | Pan B, Yang Y, Li J, Wang Y, Fang C, Yu FX, Xu Y. USP47-mediated deubiquitination and stabilization of YAP contributes to the progression of colorectal cancer. Protein Cell. 2020;11:138-143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 160. | Pan K, Fu J, Xu W. Role of Ubiquitin-Specific Peptidase 47 in Cancers and Other Diseases. Front Cell Dev Biol. 2021;9:726632. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 161. | Fraile JM, Campos-Iglesias D, Rodríguez F, Español Y, Freije JM. The deubiquitinase USP54 is overexpressed in colorectal cancer stem cells and promotes intestinal tumorigenesis. Oncotarget. 2016;7:74427-74434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 32] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: United Arab Emirates
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Sun X, China; Zhu L, China S-Editor: Lin C L-Editor: A P-Editor: Zhang XD