Published online Aug 15, 2023. doi: 10.4251/wjgo.v15.i8.1400
Peer-review started: April 26, 2023
First decision: June 15, 2023
Revised: June 16, 2023
Accepted: July 17, 2023
Article in press: July 17, 2023
Published online: August 15, 2023
Processing time: 106 Days and 6.3 Hours
Resistance to sorafenib has become a challenge in clinical treatment of hepatocellular carcinoma (HCC). Physcion is a common bioactive anthraquinone that has potential as an anticancer agent.
To study the effect of physcion on sensitizing HCC cells to sorafenib.
Sorafenib-resistant HCC cells were established and treated with sorafenib and/or physcion. The cell viability, proliferation and apoptosis were measured by cell counting kit-8, colony formation, flow cytometry, and in vivo xenograft model. Glucose uptake, lactate acid production, extracellular acidification rate (ECAR), and oxygen consumption rate (OCR) were measured to analyze glycolysis. Expression of glycolysis-related regulators was assessed by western blotting.
The addition of physcion significantly enhanced the antitumor effects of sorafenib on sorafenib-resistant HCC cells, manifested by enhanced apoptosis and suppre
Our data indicated that physcion enhanced the sensitivity of HCC cells to sorafenib by enhancing miR-370 to suppress PIM1-promoted glycolysis.
Core tip: This study investigated the effects of physcion on sorafenib resistance of hepatocellular carcinoma (HCC) cells. Through utilizing in vitro and in vivo models, we found that physcion significantly enhanced the antitumor effects of sorafenib on sorafenib-resistant HCC cells, inducing apoptosis and suppressing cell growth. Further exploration on the mechanisms identified that physcion repressed HCC cells glycolysis by targeting the miRNA-370/PIM1 axis, which consequently sensitized HCC cells to sorafenib.
- Citation: Pan XP, Jiya BR, Wang F, Lan Z. Physcion increases the sensitivity of hepatocellular carcinoma to sorafenib through miRNA-370/PIM1 axis-regulated glycolysis. World J Gastrointest Oncol 2023; 15(8): 1400-1411
- URL: https://www.wjgnet.com/1948-5204/full/v15/i8/1400.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i8.1400
Hepatocellular carcinoma (HCC) is one of the top three malignant cancers with high morbidity and mortality[1,2]. The global incidence of HCC has increased in recent years and commonly presents in patients with chronic liver diseases, including liver fibrosis and cirrhosis[3,4]. The epidemiology indicates that liver cancer is closely associated with alcohol overuse, viral infection, and metabolic syndrome[5]. Currently, patients with advanced HCC usually require local syste
The excessive glycolysis of cancer cells compared with normal cells leads to higher glucose consumption and facilitates the enhanced ATP production for metabolic activities, which is termed as Warburg effect[10]. Aerobic glycolysis is a well-recognized hallmark of cancer and plays a critical role in cancer initiation and development[11,12]. The activated glucose uptake and metabolism is regulated by multiple genes and enzymes, such as the glucose transporters (GLUTs), hexo
Physcion is a common bioactive anthraquinone in various plants[16]. Accumulating studies have reported the anti-inflammatory, antimicrobial, hepatoprotective properties, and anticancer functions of physcion with minimal or no adverse effects[16]. Physcion participates in the regulation of multiple intracellular signaling pathways via targeting protein kinases, cell cycle, transcriptional factors, miRNAs, and apoptosis-related proteins[17]. Moreover, physcion acts as a suppressor of metastasis and plays a pivotal role in chemosensitization[16]. Nevertheless, the role of physcion during sorafenib resistance has not been studied.
In this study, we explored the effects of physcion on sorafenib resistance of HCC cells in vitro and in vivo, determined the changes in glycolysis, and explored the potential mechanism of the miR-370/PIM1 regulatory axis. Our results may provide evidence for an effective therapeutic strategy for HCC.
Huh7 and HepG2 cell lines were purchased from Procell (Wuhan, China) and maintained in Dulbecco’s modified Eagle’s medium (DMEM; BI, Israel) supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin (Sigma, St. Louis, MO, United States). All cells were incubated in a 37 °C humidified atmosphere with 5% CO2. The sorafenib-resistant cell lines were obtained by long-term stimulation with increasing drug doses. Sorafenib was purchased from Selleck (Houston, TX, United States) and stocked at 10 mmol/L in dimethyl sulfoxide. Physcion was purchased from MedChemExpress (Monmouth Junction, NJ, United States) and stored at -20 °C. For treatment, cells were incubated in culture medium that contained 10 μM sorafenib and 20 μM physcion for 24 h.
miR-370 inhibitors were purchased from RiboBio (Guangzhou, China) and transfected into cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, United States).
Male BALB/c nude mice aged 4–6 wk were purchased from Huafukang (Beijing, China) and fed in standard pathogen-free conditions. After acclimation for 1 wk, Huh7 or Huh7-SR cells (107 cells in 50 μL saline) were subcutaneously injected into the right fat pad on the back. One week after tumor inoculation, sorafenib (30 mg/kg) and physcion (50 mg/kg) were orally administrated daily. The width and length of tumors were measured, and the tumor size was calculated every 5 d.
The mice were killed after treatment for 25 d and tumors were fixed in 4% formaldehyde, dehydrated and cut into 5-μm paraffin slices. The tissues samples were analyzed with hematoxylin and eosin (SolarBio, China) and immunohistochemical (IHC) staining. For IHC analysis, the tissues were stained with anti-Ki67 antibody overnight at 4 °C after antigen retrieval, incubation with endogenous peroxidase blocking solution, and blocking with goat serum. The slides were incubated with biotin-conjugated secondary antibody and horseradish peroxidase (HRP)–streptavidin. The samples were visualized by incubating with diamonobenzidine (ZhongShanJinQiao, China). The nuclei were counter-stained with hematoxylin.
Cell viability was measured by cell counting kit 8 (CCK-8) (Beyotime, China), and optical density at 450 nm was measured by microplate reader (Thermo, United States).
For colony formation assay, single cells were placed into six-well plates with 1000 cells per well and left for one night for adhesion. The cells were treated with 10 μM sorafenib and/or 20 μM physcion for 10 d. The colonies were fixed with methanol and stained with 0.2% (w/v) crystal violet for 20 min. Colonies were captured by a digital camera and counted.
Cell apoptosis was measured by TUNEL assay and flow cytometry. The cells were seeded into confocal well at 104 cells/well and treated. Cells were fixed with 4% polyformaldehyde and stained with TUNEL assay kit (Beyotime). Images were captured under a confocal microscope (CarlZeiss, Germany). Apoptosis was examined by Annexin V/PI Apoptosis Detection Kit (Beyotime). Cells were collected after treatment and suspended in binding buffer that contained Annexin V and PI for 30 min. Samples were directly detected by a C6 flow cytometer (BD Biosciences, United States).
To measure glucose uptake, cells were seeded into a six-well plate at a density of 5 × 105 per well and incubated overnight. After treatment with sorafenib or physcion for 24 h, the cells were starved in glucose-free DMEM for 1 h, and 50 μM D-glucose (Sigma) was added for another 30 min. The cells were washed with phosphate-buffered saline and intracellular accumulation of 2-NBDG was checked by flow cytometer (BD Biosciences).
Lactate production was measured using Lactate Assay Kit (Sigma). Cells were placed into a six-well plate at 5 × 105 cells per well. After treatment with sorafenib or physcion for 24 h, cells were suspended in lactate assay buffer and homogenized by sonication.
The extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) were examined by Extracellular Acidification assay kit and OCR assay kit (Bestbio, China). The fluorescence was measured by flow cytometer.
Total RNA was isolated using Trizol reagent (Invitrogen) and a total of 1 μg RNA was reverse transcribed using Superscript IV transcriptase (Thermo). RNA expression was measured by real-time quantitative polymerase chain reaction (qPCR) using SYBR green. Gene expression was normalized to the level of β-actin gene.
HCC cells were homogenized with RIPA lysis buffer to extract total proteins. An equal amount of proteins was loaded and separated with 8%–12% SDS-PAGE and transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA, United States). The blots were blocked with 5% skimmed milk and probed with primary antibody against LDHA, PGK1, PKM2, HK1, HK2, and GLUT (Abcam, Cambridge, MA, United States) overnight at 4 °C. Next day, the blots were incubated with HRP-conjugated secondary anti-mouse or anti-rabbit antibody and visualized using the ECL reagent (Thermo).
All statistical analysis was conducted using GraphPad Prism 7.0 (La Jolla, CA, United States). Comparison between two or more groups was analyzed by two-tailed student’s t test or one-way analysis of variance. P < 0.05 was set as statistically significant.
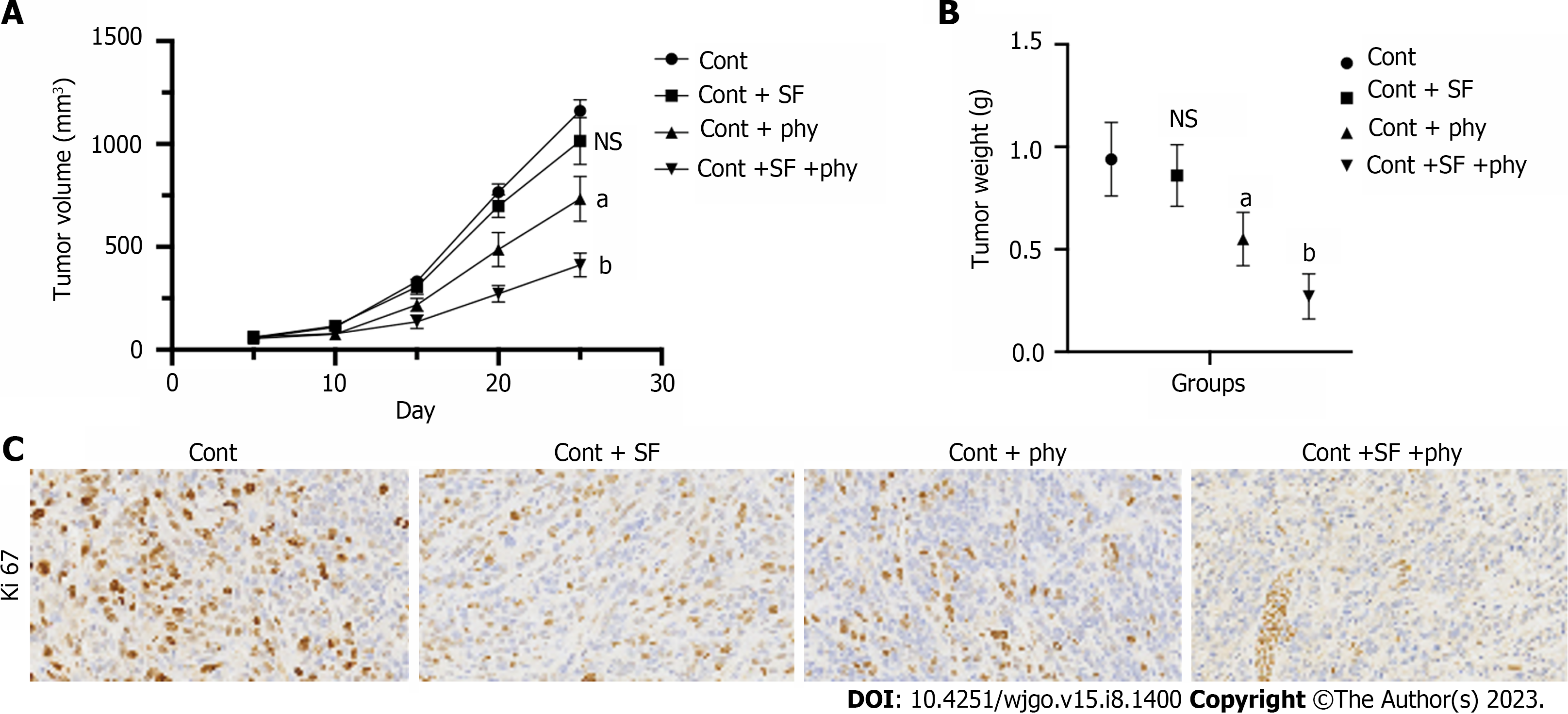
We first established the sorafenib-resistant cell lines to construct an in vivo xenograft model and evaluated the therapeutic effects of physcion. The xenograft model was established using parental Huh7 cell line and sorafenib-resistant Huh7 cell line (Huh7-SR). Sorafenib and physcion both suppressed tumor volume and weight (Supplementary Figure 1A and B) of Huh7 cells and suppressed expression of proliferative biomarker Ki67 (Supplementary Figure 1C). The combined administration of sorafenib and physcion further decreased tumor volume and weight, as well as Ki67 expression in the Huh7 xenograft tumor model (Supplementary Figure 1). Sorafenib treatment did not affect tumor growth (Figure 1A), tumor volume (Supplementary Figure 2), tumor weight (Figure 1B), and level of Ki67 (Figure 1C) in the Huh7-SR xenograft mouse model. Physcion significantly suppressed the in vivo growth of Huh7-SR cells compared with the control and sorafenib groups (Figure 1). Combination of physcion with sorafenib enhanced the tumor suppressive effects of physcion. These data indicated that physcion enhanced the sensitivity of sorafenib-resistant HCC cells to sorafenib.
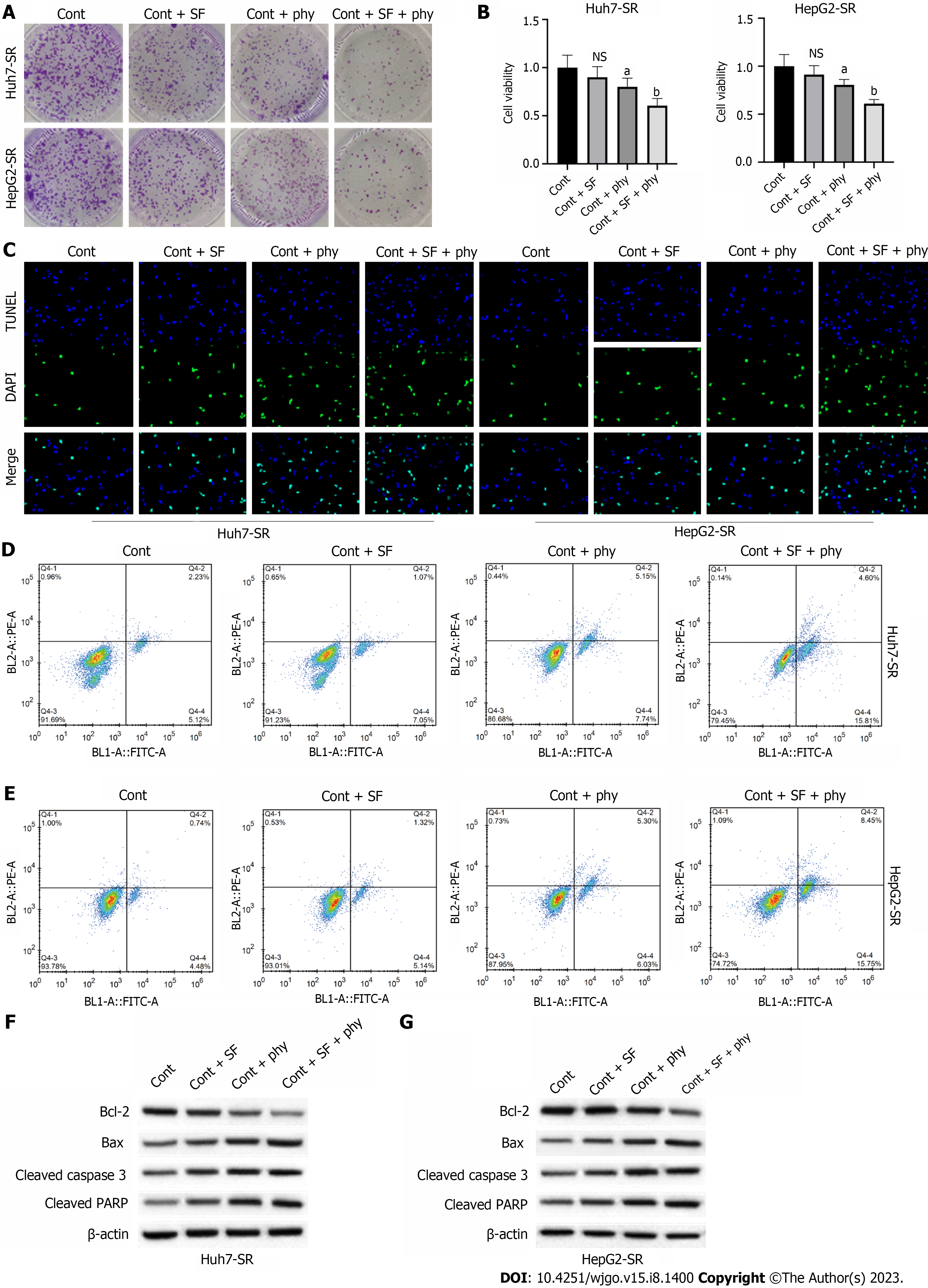
We determined the effects of physcion on elevating sorafenib sensitivity of HCC cells. We observed that the colony number (Figure 2A) and cell viability (Figure 2B) of sorafenib-resistant Huh7-SR and HepG2-SR cells did not change after sorafenib treatment and slightly decreased after physcion treatment, whereas combined physcion and sorafenib treatment enhanced the suppressive effect on cell growth compared with the monotherapy group (Figure 2A and B). Sorafenib did not affect apoptosis (Figure 2C–E). Combination of physcion and sorafenib significantly induced the TUNEL-positive staining (Figure 2C) and the apoptotic proportion (Figure 2D and E) of sorafenib-resistant Huh7-SR and HepG2-SR cells compared with the physcion or sorafenib group, along with decreased expression of Bcl-2 and elevated levels of Bax, cleaved caspase-3 and cleaved poly (ADP-ribose) polymerase (Figure 2F and G).
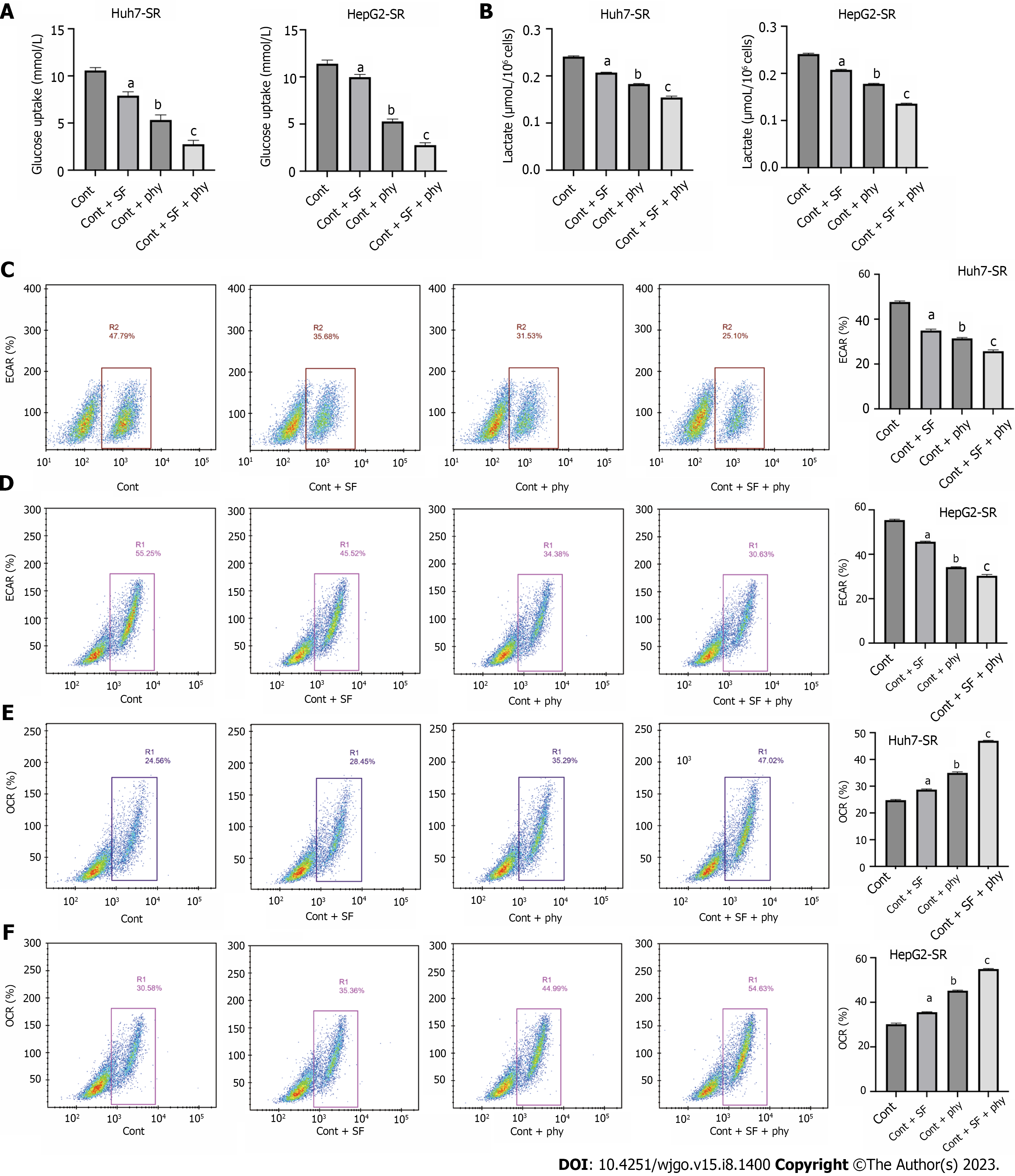
The effects of sorafenib and physcion on glycolysis were determined by glucose uptake test, lactic acid production test, ECAR and OCR. Physcion had little effect on glucose uptake (Figure 3A), lactate production (Figure 3B), ECAR (Figure 3C and D), and OCR (Figure 3E and F) in Huh7-SR and HepG2-SR cells. Physcion inhibited glycolysis, whereas combined administration of sorafenib and physcion suppressed glucose uptake, lactate production and ECAR, and enhanced OCR of sorafenib-resistant HCC cells. These findings indicated that physcion promoted suppression of glycolysis caused by sorafenib.
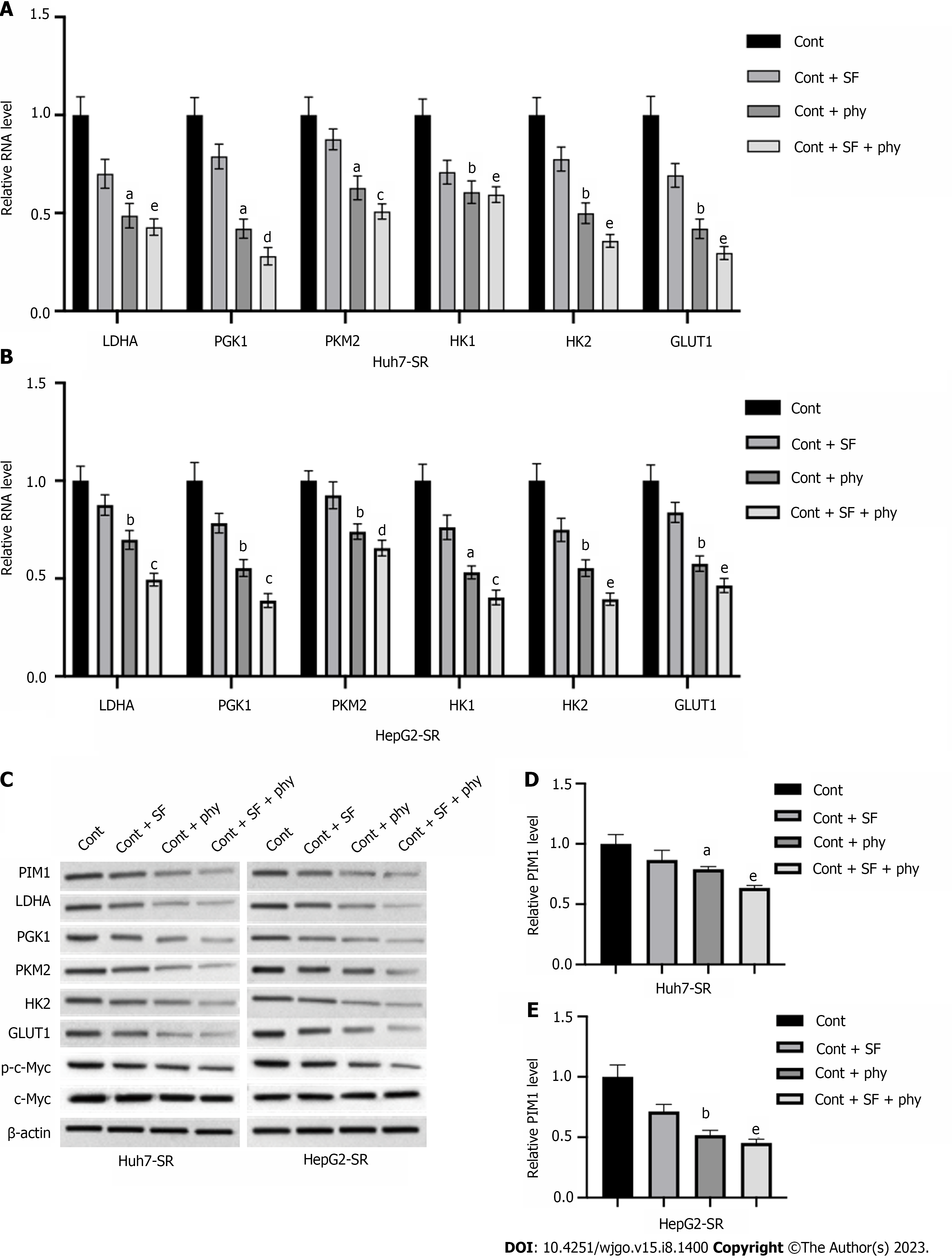
We explored the mechanisms underlying physcion-regulated glycolysis in sorafenib-resistant HCC cells by measuring expression of glycolysis regulators. The RNA (Figure 4A and B) and protein (Figure 4C) levels of LDHA, PGK1, PKM2, HK1, HK2, and GLUT1 in Huh7-SR and HepG2-SR cells were not obviously changed by treatment with sorafenib, whereas addition of physcion significantly suppressed the levels of glycolysis regulators compared with the sorafenib monotherapy group. We observed a significant decrease of PIM1 RNA and protein level in sorafenib-resistant cells upon physcion administration compared with sorafenib (Figure 4C–E). It has been reported that PIM1 regulates glycolysis through regulating phosphorylation of c-Myc. Here, we demonstrated that sorafenib did not induce a notable change in c-Myc phosphorylation, but addition of physcion enhanced suppression of phosphorylated c-Myc level without affecting expression of total c-Myc (Figure 4C). We observed elevation of PIM1 protein level in Huh7-SR and HepG2-SR cells compared with the parental Huh7 and HepG2 cells (Supplementary Figure 3). These findings demonstrated that physcion may affect glycolysis of sorafenib-resistant HCC cells by regulating PIM1 expression.
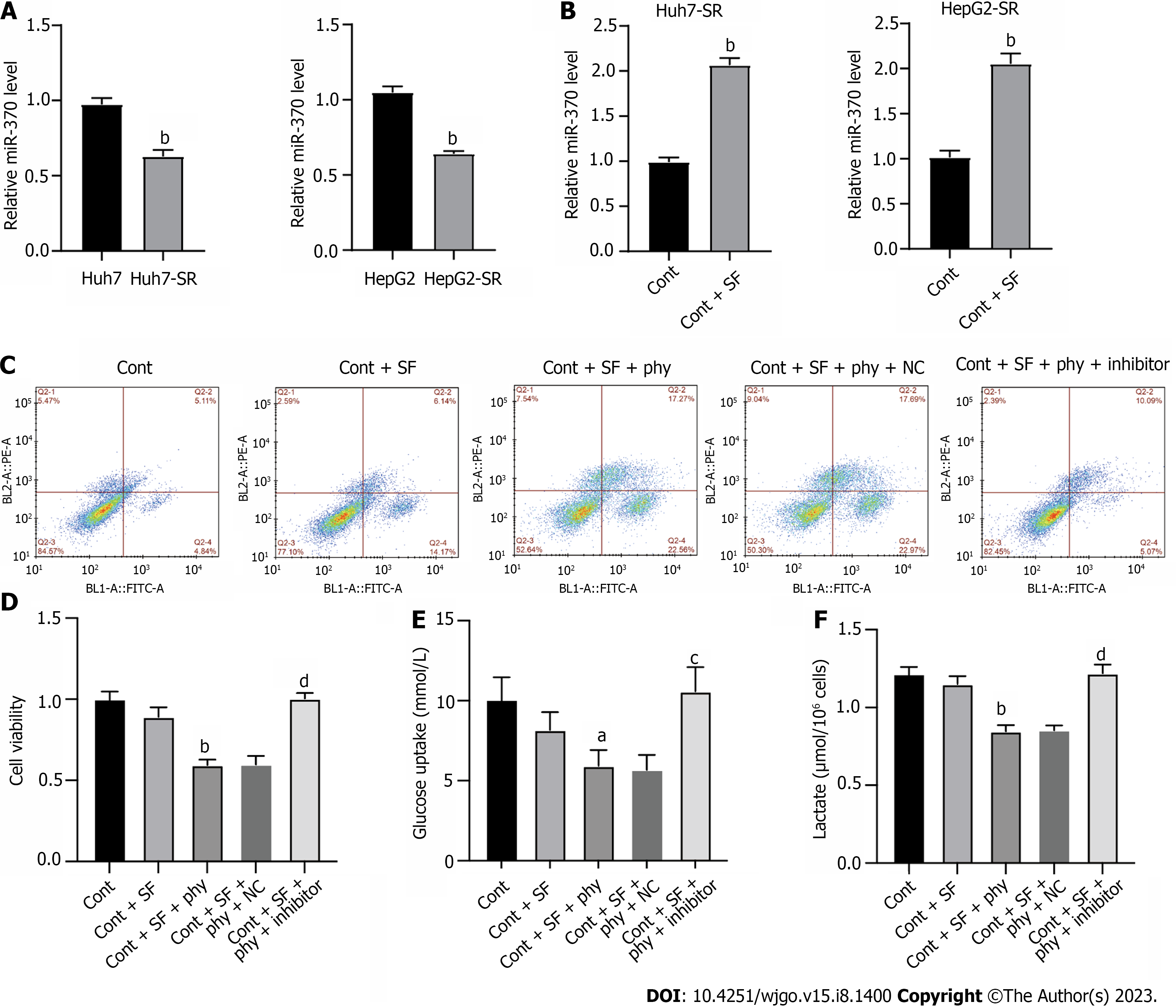
miRNAs play important roles in gene expression regulation. Here, we explored whether miRNA participated in physcion-regulated PIM1 expression. The level of miR-370 was notably lower in Huh7-SR and HepG2-SR cells compared with parental cell lines (Figure 5A), whereas treatment with physcion elevated the level of miR-370 (Figure 5B). To evaluate the function of miR-370 as a mediator of physcion-regulated HCC sorafenib resistance, we added miR-370 inhibitors and evaluated cell phenotypes. Flow cytometry and CCK-8 assay indicated that inhibition of miR-370 abolished the apoptosis and suppressed cell proliferation caused by physcion and sorafenib co-treatment (Figure 5C and D). Physcion- and sorafenib-suppressed glucose uptake (Figure 5E) and lactate production (Figure 5F) was reversed by miR-370 inhibitors. These data suggested that physcion increases sorafenib sensitivity by upregulating miR-370 and negatively modulating PIM1, thereby affecting glycolysis.
Despite the advances in diagnosis and therapy of HCC over the past few years, > 50% of patients are diagnosed at a late stage and 70% of patients relapse within 5 years after initial treatment[18,19]. Sorafenib is a multikinase inhibitor that targets Ras/Raf and downstream signaling cascades and usually induces cell apoptosis, suppresses tumor cell growth, and mitigates angiogenesis[20]. The mechanisms of sorafenib resistance involve epigenetic regulation, drug transportation, tumor microenvironment, and cell death regulation[21]. Currently, strategies to alleviate sorafenib resistance mainly include co-treatment with other drugs in clinical use, such as cytotoxic chemotherapeutic drugs (cisplatin and 5-fluorouracil), immunotherapeutic drugs, and reagents that target specific molecules (anti-EGFR antibodies)[4]. Nevertheless, these therapies are usually accompanied with severe side effects[4]. In this study, we established sorafenib-resistant HCC cell lines and identified that physcion enhanced the antitumor effects of sorafenib in vitro and in vivo. monotherapy with sorafenib did not affect the proliferation and death of sorafenib-resistant HCC cells, whereas co-treatment with physcion induced apoptosis of Huh7-SR and HepG2-SR cells.
The enhanced therapeutic effects of sorafenib upon addition of physcion are accompanied with suppressed glycolysis, as was manifested by decreased levels of LDHA, GLUT1, PGK1, PKM2, and HK2. These decreased gene and enzyme levels lead to suppressed glucose transportation into cells and glucose metabolism[22,23]. Previous studies have reported that the expression of glycolysis-related genes such as HK2 is regulated by c-Myc[24]. Consistent with these studies, our results indicated decreased phosphorylation of c-Myc upon treatment with physcion, indicating the potential therapeutic mechanism of physcion on glycolysis involving c-Myc.
Further study on molecular mechanisms of physcion in HCC cells revealed that PIM1 is overexpressed in sorafenib-resistant HCC cells compared with the parental cells, and administration of physcion significantly decreased its level. PIM1 is a serine/threonine kinase that is constitutively active when expressed in cells, and its activity is directly correlated with the expression level[25,26]. Previous research has found that PIM1 modulates multiple aspects of cellular metabolism in various cell types[27-29]. PIM1 is reported to mediate the phosphorylation of c-Myc[26], which subsequently promotes expression of glycolysis-associated pivotal genes, including LDHA and PGK1, to enhance glycolysis in ovarian cancer cells[30]. Here, we observed that decreased PIM1 expression was accompanied with decreased glucose assumption and lactate production under physcion treatment, suggesting that physcion regulates glycolysis in sorafenib-resistant HCC cells via regulating PIM1.
We also found that miR-370 targets the PIM1 gene expression in HCC cells to mediate the function of physcion. miRNAs have been widely reported as regulators of drug resistance and metabolism in cancer treatment through directly targeting and regulating the expression of various genes[31-33]. Previous studies suggested that miR-370 facilitates the oncogenesis of several cancers, such as breast cancer and liver cancer[34-36]. Wei and Ma[37] reported that enforced miR-370 expression enhanced the glycolysis and proliferation of melanoma cells[37]. Nevertheless, our work indicated decreased level of miR-370 in sorafenib-resistant HCC cells and inhibition of miR-370 suppressed the therapeutic effect of physcion on HCC cells. Since physcion functions through various regulatory mechanisms to modulate cancer cell behavior, PIM1-regulated glycolysis may not be the only mechanism underlying sorafenib sensitivity. It is also possible that physcion enhances chemosensitivity in other therapy resistance. Further studies such as high-throughput sequencing analysis may be conducted to explore the comprehensive and in-depth effects of physcion in cancer.
We identified that physcion enhanced the tumor-suppressive effects of sorafenib on drug-resistant HCC cells and increased the level of miR-370 to directly downregulate PIM1 expression, which suppressed c-Myc-regulated glycolysis in HCC cells. Our study presented physcion as a novel systemic treatment for patients with sorafenib-resistant HCC.
Hepatocellular carcinoma (HCC) is one of the most malignant cancer types with high morbidity and mortality. Currently, patients with advanced HCC usually receive systemic local treatment with ablation or external irradiation along with sorafenib. Resistance to sorafenib has become a challenge in clinical treatment of HCC.
Physcion is a common bioactive anthraquinone that is a potential anticancer agent that has been reported to regulate multiple intracellular signaling pathways via targeting protein kinases, cell cycle, transcriptional factors, miRNAs, and apoptosis-related proteins. The excessive glycolysis of cancer cells compared with normal cells, named as Warburg effect, is a major characteristic of cancer and play a critical role in cancer initiation and development.
This work aimed to study the effect of physcion on glycolysis metabolism and sensitizing the HCC cells to sorafenib.
Sorafenib-resistant HCC cells were established and treated with sorafenib and/or physcion. The cell viability, proliferation, and apoptosis were measured by cell counting kit-8, colony formation, flow cytometry, and in vivo xenograft model. Glucose uptake, lactate acid production, extracellular acidification rate (ECAR), and oxygen consumption rate (OCR) were measured to analyze glycolysis. Expression of glycolysis-related regulators was assessed by western blotting assay.
The addition of physcion significantly enhanced the antitumor effects of sorafenib on sorafenib-resistant HCC cells, manifested by enhanced apoptosis and suppressed cell growth. The glucose uptake, lactate acid production, and ECAR were elevated, and OCR was suppressed under physcion treatment. The level of PIM1 was elevated and miR-370 was suppressed in sorafenib-resistant HCC cells compared with the parental cells, which was suppressed by physcion treatment. Inhibition of miR-370 notably reversed the effects of physcion on sorafenib-resistant HCC cells.
Physcion enhanced the sensitivity of HCC cells to sorafenib via enhancing miR-370 to suppress PIM1-promoted glycolysis.
As a potential anticancer agent, physcion possibly enhances sensitivity in other therapy resistance. Based on the multiple cellular processes that are regulated by physcion, it is possible that PIM1-regulated glycolysis may not be the only mechanism underlying sorafenib sensitivity. Further studies such as high-throughput sequencing analysis may be conducted to explore the comprehensive and in-depth effects of physcion in cancer.
| 1. | Zhu YJ, Zheng B, Wang HY, Chen L. New knowledge of the mechanisms of sorafenib resistance in liver cancer. Acta Pharmacol Sin. 2017;38:614-622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 403] [Cited by in RCA: 543] [Article Influence: 60.3] [Reference Citation Analysis (0)] |
| 2. | Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, Lencioni R, Koike K, Zucman-Rossi J, Finn RS. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7:6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4432] [Cited by in RCA: 4410] [Article Influence: 882.0] [Reference Citation Analysis (4)] |
| 3. | Wu H, Wang T, Liu Y, Li X, Xu S, Wu C, Zou H, Cao M, Jin G, Lang J, Wang B, Liu B, Luo X, Xu C. Mitophagy promotes sorafenib resistance through hypoxia-inducible ATAD3A dependent Axis. J Exp Clin Cancer Res. 2020;39:274. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 72] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 4. | Anwanwan D, Singh SK, Singh S, Saikam V, Singh R. Challenges in liver cancer and possible treatment approaches. Biochim Biophys Acta Rev Cancer. 2020;1873:188314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 762] [Cited by in RCA: 974] [Article Influence: 162.3] [Reference Citation Analysis (2)] |
| 5. | Sia D, Villanueva A, Friedman SL, Llovet JM. Liver Cancer Cell of Origin, Molecular Class, and Effects on Patient Prognosis. Gastroenterology. 2017;152:745-761. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 619] [Cited by in RCA: 867] [Article Influence: 96.3] [Reference Citation Analysis (2)] |
| 6. | Koshariya M, Agarwal B, Rathod R. Overview of Advancement and Management of Gastrointestinal Stromal Tumors. J Mod Med Oncol. 2021;2:3. [DOI] [Full Text] |
| 7. | Lin Z, Niu Y, Wan A, Chen D, Liang H, Chen X, Sun L, Zhan S, Chen L, Cheng C, Zhang X, Bu X, He W, Wan G. RNA m(6) A methylation regulates sorafenib resistance in liver cancer through FOXO3-mediated autophagy. EMBO J. 2020;39:e103181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 358] [Article Influence: 59.7] [Reference Citation Analysis (0)] |
| 8. | Cheng Z, Wei-Qi J, Jin D. New insights on sorafenib resistance in liver cancer with correlation of individualized therapy. Biochim Biophys Acta Rev Cancer. 2020;1874:188382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 85] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 9. | Bao MH, Wong CC. Hypoxia, Metabolic Reprogramming, and Drug Resistance in Liver Cancer. Cells. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 238] [Article Influence: 47.6] [Reference Citation Analysis (0)] |
| 10. | Chandel NS. Glycolysis. Cold Spring Harb Perspect Biol. 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 256] [Article Influence: 51.2] [Reference Citation Analysis (0)] |
| 11. | Paul S, Ghosh S, Kumar S. Tumor glycolysis, an essential sweet tooth of tumor cells. Semin Cancer Biol. 2022;86:1216-1230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 289] [Reference Citation Analysis (0)] |
| 12. | Brandes RP, Rezende F. Glycolysis and Inflammation: Partners in Crime! Circ Res. 2021;129:30-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 13. | Yang J, Ren B, Yang G, Wang H, Chen G, You L, Zhang T, Zhao Y. The enhancement of glycolysis regulates pancreatic cancer metastasis. Cell Mol Life Sci. 2020;77:305-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 266] [Article Influence: 44.3] [Reference Citation Analysis (0)] |
| 14. | Ganapathy-Kanniappan S, Geschwind JF. Tumor glycolysis as a target for cancer therapy: progress and prospects. Mol Cancer. 2013;12:152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 519] [Cited by in RCA: 1020] [Article Influence: 78.5] [Reference Citation Analysis (0)] |
| 15. | Ren Y, Gu YK, Li Z, Xu GZ, Zhang YM, Dong MX, Wang Y, Zhou XB. CXCR3 confers sorafenib resistance of HCC cells through regulating metabolic alteration and AMPK pathway. Am J Transl Res. 2020;12:825-836. [PubMed] |
| 16. | XunLi, Liu Y, Chu S, Yang S, Peng Y, Ren S, Wen B, Chen N. Physcion and physcion 8-O-β-glucopyranoside: A review of their pharmacology, toxicities and pharmacokinetics. Chem Biol Interact. 2019;310:108722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 49] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 17. | Trybus W, Król T, Trybus E, Stachurska A. Physcion Induces Potential Anticancer Effects in Cervical Cancer Cells. Cells. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 18. | Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. 2018;15:599-616. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1487] [Cited by in RCA: 1458] [Article Influence: 182.3] [Reference Citation Analysis (0)] |
| 19. | Sharma BK. Innovative Translation Strategies in the Emergence of Next-Generation Drugs in Precision Cancer Medicine. J Mod Med Oncol. 2022;2:4. [DOI] [Full Text] |
| 20. | Tang W, Chen Z, Zhang W, Cheng Y, Zhang B, Wu F, Wang Q, Wang S, Rong D, Reiter FP, De Toni EN, Wang X. The mechanisms of sorafenib resistance in hepatocellular carcinoma: theoretical basis and therapeutic aspects. Signal Transduct Target Ther. 2020;5:87. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 786] [Article Influence: 131.0] [Reference Citation Analysis (1)] |
| 21. | Xia S, Pan Y, Liang Y, Xu J, Cai X. The microenvironmental and metabolic aspects of sorafenib resistance in hepatocellular carcinoma. EBioMedicine. 2020;51:102610. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 233] [Article Influence: 38.8] [Reference Citation Analysis (11)] |
| 22. | Bose S, Le A. Glucose Metabolism in Cancer. Adv Exp Med Biol. 2018;1063:3-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 165] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 23. | Koppenol WH, Bounds PL, Dang CV. Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325-337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2023] [Cited by in RCA: 2587] [Article Influence: 172.5] [Reference Citation Analysis (0)] |
| 24. | Feng J, Li J, Wu L, Yu Q, Ji J, Wu J, Dai W, Guo C. Emerging roles and the regulation of aerobic glycolysis in hepatocellular carcinoma. J Exp Clin Cancer Res. 2020;39:126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 416] [Cited by in RCA: 485] [Article Influence: 80.8] [Reference Citation Analysis (0)] |
| 25. | Santio NM, Vainio V, Hoikkala T, Mung KL, Lång M, Vahakoski R, Zdrojewska J, Coffey ET, Kremneva E, Rainio EM, Koskinen PJ. PIM1 accelerates prostate cancer cell motility by phosphorylating actin capping proteins. Cell Commun Signal. 2020;18:121. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 26. | Mahata S, Sahoo PK, Pal R, Sarkar S, Mistry T, Ghosh S, Nasare VD. PIM1/STAT3 axis: a potential co-targeted therapeutic approach in triple-negative breast cancer. Med Oncol. 2022;39:74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 27. | Merkel AL, Meggers E, Ocker M. PIM1 kinase as a target for cancer therapy. Expert Opin Investig Drugs. 2012;21:425-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 104] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 28. | Wang BW, Huang CH, Liu LC, Cheng FJ, Wei YL, Lin YM, Wang YF, Wei CT, Chen Y, Chen YJ, Huang WC. Pim1 Kinase Inhibitors Exert Anti-Cancer Activity Against HER2-Positive Breast Cancer Cells Through Downregulation of HER2. Front Pharmacol. 2021;12:614673. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 29. | Brasó-Maristany F, Filosto S, Catchpole S, Marlow R, Quist J, Francesch-Domenech E, Plumb DA, Zakka L, Gazinska P, Liccardi G, Meier P, Gris-Oliver A, Cheang MC, Perdrix-Rosell A, Shafat M, Noël E, Patel N, McEachern K, Scaltriti M, Castel P, Noor F, Buus R, Mathew S, Watkins J, Serra V, Marra P, Grigoriadis A, Tutt AN. PIM1 kinase regulates cell death, tumor growth and chemotherapy response in triple-negative breast cancer. Nat Med. 2016;22:1303-1313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 196] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 30. | Wu Y, Deng Y, Zhu J, Duan Y, Weng W, Wu X. Pim1 promotes cell proliferation and regulates glycolysis via interaction with MYC in ovarian cancer. Onco Targets Ther. 2018;11:6647-6656. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 31. | Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2492] [Cited by in RCA: 3627] [Article Influence: 403.0] [Reference Citation Analysis (0)] |
| 32. | Hill M, Tran N. miRNA interplay: mechanisms and consequences in cancer. Dis Model Mech. 2021;14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 474] [Article Influence: 94.8] [Reference Citation Analysis (0)] |
| 33. | He B, Zhao Z, Cai Q, Zhang Y, Zhang P, Shi S, Xie H, Peng X, Yin W, Tao Y, Wang X. miRNA-based biomarkers, therapies, and resistance in Cancer. Int J Biol Sci. 2020;16:2628-2647. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 310] [Cited by in RCA: 434] [Article Influence: 72.3] [Reference Citation Analysis (0)] |
| 34. | Huang L, Liu X. microRNA-370 Promotes Cell Growth by Targeting WNK2 in Breast Cancer. DNA Cell Biol. 2019;38:501-509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 35. | Lv Q, Xia Q, Li A, Wang Z. circRNA_101277 Influences Cisplatin Resistance of Colorectal Cancer Cells by Modulating the miR-370/IL-6 Axis. Genet Res (Camb). 2022;2022:4237327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 36. | Zhang W, Liu H, Jiang J, Yang Y, Wang W, Jia Z. CircRNA circFOXK2 facilitates oncogenesis in breast cancer via IGF2BP3/miR-370 axis. Aging (Albany NY). 2021;13:18978-18992. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 37. | Wei S, Ma W. MiR-370 functions as oncogene in melanoma by direct targeting pyruvate dehydrogenase B. Biomed Pharmacother. 2017;90:278-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 36] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Asic K, Croatia; Iannone A, Italy; Reiberger T, Austria S-Editor: Fan JR L-Editor: Kerr C P-Editor: Cai YX