Published online Aug 15, 2023. doi: 10.4251/wjgo.v15.i8.1366
Peer-review started: February 12, 2023
First decision: March 28, 2023
Revised: April 11, 2023
Accepted: June 19, 2023
Article in press: June 19, 2023
Published online: August 15, 2023
Processing time: 178 Days and 23.8 Hours
Long non-coding RNAs (lncRNAs) with differential expression characteristics have been found to be closely related to the tumorigenesis and development of gastric cancer (GC), but their specific mechanisms and roles still need to be further elucidated.
To investigate the expression of LINC01268 in GC and its mechanism of affecting GC progression.
Real-time quantitative polymerase chain reaction was used to detect the expression of LINC01268 in GC tissues, cell lines and plasma. The Kaplan-Meier method was used to evaluate the value of LINC01268 in the prognostication of GC patients. An receiver operating characteristic curve was constructed to evaluate the value of LINC01268 in the diagnosis of GC. Transwell migration and invasion assays and wound healing assays were used to confirm the effect of LINC01268 on the invasion and migration of GC cells. The regulatory relationship between LINC01268 and myristoylated alanine rich protein kinase C substrate (MARCKS), the PI3K/Akt signaling pathway, and the epithelial-mesenchymal transition (EMT) process in GC was demonstrated by western blot analysis.
The expression of LINC01268 was increased in GC tissues and cell lines. The expression level of LINC01268 was significantly correlated with lymph node metastasis, TNM stage, and tumor differentiation in patients with GC. Over-expression of LINC01268 indicated a poor prognosis for patients with GC, and it had a certain auxiliary diagnostic value for GC. In vitro functional experiments proved that the abnormal expression of LINC01268 further activated the PI3K/Akt signaling pathway and promoted EMT by targeting and regulating MARCKS and ultimately promoted the invasion and metastasis of GC.
This study elucidates that LINC01268 in GC may be an oncogene that further activates the PI3K/Akt signaling pathway and EMT by targeting and regulating MARCKS, and ultimately promotes the invasion and metastasis of GC. LINC01268 may be a potential effective target for the treatment of GC.
Core Tip: Overexpression of LINC01268 was related to the prognosis of patients with gastric cancer (GC) and showed the value of auxiliary diagnosis. Overexpression of LINC01268 promoted the invasion and metastasis of GC cells. LINC01268 activated the PI3K/Akt signaling pathway and promoted epithelial-mesenchymal transition by targeting myristoylated alanine rich protein kinase C substrate and ultimately promoted the invasion and metastasis of GC.
- Citation: Tang LH, Ye PC, Yao L, Luo YJ, Tan W, Xiang WP, Liu ZL, Tan L, Xiao JW. LINC01268 promotes epithelial-mesenchymal transition, invasion and metastasis of gastric cancer via the PI3K/Akt signaling pathway and targeting MARCKS. World J Gastrointest Oncol 2023; 15(8): 1366-1383
- URL: https://www.wjgnet.com/1948-5204/full/v15/i8/1366.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i8.1366
On a global scale, the number of new cancer patients is increasing year by year[1]. Among these malignant tumors, the morbidity and mortality of gastric cancer (GC) cannot be ignored[2]. For patients with clinically advanced GC, we have carried out comprehensive treatment, such as surgery combined with chemotherapy or targeted therapy, and even customized individualized schemes for some patients through multiple disciplinary teams[3]. However, there are still a large number of patients with advanced GC who have difficulty obtaining a better prognosis. Therefore, it is necessary to explore new directions for early screening, early intervention, and prognosis monitoring of GC to continuously improve the prognosis of GC patients as a whole.
Long non-coding RNAs (lncRNAs) are a class of RNA molecules with a length of more than 200 nucleotides and some of them have the ability to encode functional micropeptides[4-6]. An increasing number of studies have shown that lncRNAs are directly or indirectly involved in the molecular mechanism of tumorigenesis and development, regulate different cell signaling pathways, and exhibit carcinogenic or tumor suppressor effects[7]. For example, the highly expressed lncRNA-ZFAS1 in colon cancer attenuates the inhibition of vascular endothelial growth factor A expression by competitively binding to miR-150-5p and ultimately promotes the proliferation and metastasis of colon cancer cells[8]. As an antisense transcript encoding the oxidative stress-activated ion channel protein TRPM2, lncRNA TRPM2-AS is abnormally highly expressed in prostate cancer. It inhibits cancer cell apoptosis in response to oxidative stress, and also has a positive correlation with the targets of some existing therapeutic drugs, and it can be used to identify invasive prostate cancer early and provide valuable prognostic information[9]. LncRNA and GC are also inextricably linked. What different roles lncRNAs play in the process of GC, what signal transduction pathways they affect, and their application value in the clinic remain to be further studied and clarified.
In this study, the differentially expressed lncRNA LINC01268 in GC tissues was screened by preliminary microarray analysis. LINC01268, which is located at 6q21 and encoded by 3 exons, has been mapped to chromosome 6 region 113868013-113873347. We detected the expression level of LINC01268 in GC tissue samples, plasma of GC patients, and GC cell lines and found that the expression of LINC01268 in GC showed an up-regulation trend. Combined with the clinicopathological data and survival data of GC patients, we analyzed the possible role of LINC01268 in the invasion and metastasis of GC and evaluated its value and significance in the auxiliary diagnosis and prognostic evaluation of GC. At the same time, in cytological experiments in vitro, we investigated the role of LINC01268 in the tumorigenesis and development of GC, evaluated its potential in the clinical diagnosis and prognostication of GC, and elucidated the possible molecular mechanisms by which it regulated the biological behavior of GC.
A total of 62 human GC tissues and paired adjacent noncancerous tissues (ANTs) (5 cm away from the tumor margin) were obtained at the time of surgery from March 2015 to May 2018 at the Affiliated Hospital of North Sichuan Medical College (Sichuan, China). Plasma was isolated from 31 GC patients before surgery and 19 healthy volunteers. Following resection of the specimens, the samples were immediately frozen in liquid nitrogen and preserved at -80 °C until use. The detailed clinicopathologic parameters of the GC patients were collected. All GC patients and healthy volunteers provided written informed consent, and the entire study protocol was approved by the Ethics Committee of the Affiliated Hospital of North Sichuan Medical College, Nanchong, China.
The survival information of 62 patients with GC was followed up by telephone. Among them, 33 patients died, 23 patients survived, and 6 patients were lost to follow-up. The date of the operation was taken as the starting point for the follow-up, and the follow-up ended on August 20, 2019. The follow-up periods were 0.5-52 mo.
The human GC cell lines MGC80-3 and AGS were purchased from Shanghai Cell Bank (Shanghai, China). Cells were cultured in RPMI-1640 or Ham’s F-12 (Procell, Wuhan, China) supplemented with 10% fetal bovine serum (FBS) (Gibco BRL, Gaithersburg, MD, United States) and antibiotics (100 IU/mL penicillin and 100 μg/mL streptomycin) at 37 °C in a humidified atmosphere with 5% CO2. Short hairpin RNA for LINC01268 (NR_038863) was ligated into the GV493 plasmid (GeneChem, Shanghai, China). The sh-LINC01268 or nonspecific sh-negative control (sh-NC) plasmid was transfected into MGC80-3 and AGS cells using X-tremeGENE HP DNA Transfection Reagent (Roche, Germany). After infection for 48 h, the infected cells were harvested for the extraction of total RNA and protein.
Total RNA was isolated from GC tissues, ANTs and GC cell lines using TRIzol reagent (Invitrogen, Carlsbad, CA, United States). Reverse transcription reactions were performed using GoScript™ Reverse Transcription System (Promega, Madison, WI, United States) according to the manufacturer’s instructions. Quantitative polymerase chain reaction (PCR) was performed using a standard SYBR Green PCR kit (Roche, United States) and a Roche LightCycler®96 Instrument (Roche, United States) according to the respective manufacturer’s protocol. ACTB, GAPDH and 18S RNA were used as the internal controls for lncRNA and other mRNA expression normalization and quantification. Primers for LINC01268, MARCKS, PIK3CA, Akt, GAPDH, ACTB and 18S RNA were synthesized by Sangon Biotech (Shanghai, China), and their sequences are listed in Supplementary Table 1. All experiments were repeated three times. The relative gene expression level was calculated with the 2-ΔΔCt method.
Total protein was extracted, and the concentration was determined using a BCA protein assay kit (Thermo Fisher Scientific, Waltham, MA, United States). Sample lysates (10 μg of protein) were separated by SDS-PAGE and transferred to a PVDF membrane. The membrane was incubated with specific antibodies against MARCKS (1:2000) (Proteintech Group, China), p-MARCKS (1:1000) (Proteintech Group, China), PI3K (1:2000) (CST, United States), p-PI3K (1:500) (BIOSS, China), Akt (1:2000) (CST, United States), p-Akt (1:1000) (CST, United States), β-Catenin (1:2000) (CST, United States), p-β-Catenin (1:500) (CST, United States), Vimentin (1:2000) (CST, United States), matrix metalloproteinase (MMP)-9 (1:500) (Abcam, United States), N-Cadherin (1:1000) (Abcam, United States) or E-Cadherin (1:2000) (CST, United States) at 4 °C overnight, followed by incubation with the appropriate secondary antibody. Protein levels were normalized to those of total GAPDH, which was detected with a monoclonal anti-GAPDH antibody (1:10000) (Sigma-Aldrich Corporation, St. Louis, MO, United States). Autoradiograms were quantified by densitometry (Quantity One software; Bio-Rad).
Cell invasion was examined with Matrigel (BD Pharmingen, San Jose, CA, United States) using Transwell plates (8 μm pore size, Corning, United States). GC cells (MGC80-3 and AGS) were transfected with sh-LINC01268 and sh-NC plasmids, respectively, for 48 h. A total of 3 × 105 cells were collected and placed in the upper chambers with 200 μL serum-free medium, and 600 μL of medium with 10% FBS was added to the lower chambers. Then, the Transwell plates were placed in a cell incubator (37 °C, 5% CO2) for 24 h. The cells in the upper chamber were gently wiped off with cotton swabs. Cells attached to the lower membrane surface were fixed with 4% paraformaldehyde (Sigma Aldrich, St. Louis, MO, United States) for 15 min and stained with 1% crystal violet (Beyotime, Shanghai, China). The migration assays were also performed in the same way without using Matrigel. Three fields were randomly selected for cell counting under a microscope.
Transfected GC cells were seeded in 6-well plates and grown to a density of 70%. Pipette tips (200 μL) were used to make artificial wounds, and then the cell fragments were gently washed away with sterile phosphate buffered saline. The 6-well plates were returned to the incubator (37 °C, 5% CO2). Wounds were observed and photographed separately at 0 h, 24 h, and 48 h.
The datasets supporting the conclusions of this article are available in the Kaplan-Meier Plotter, circlncRNAnet, GeneCards, starBase and lncLocator. The Kaplan-Meier Plotter[10] (http://kmplot.com/) online database was used for gene-related prognostic survival assessment. The circlncRNAnet online analysis tool[11] (http://app.cgu.edu.tw/cir
SPSS 22.0 software (SPSS, Chicago, IL, United States) and GraphPad Prism 7.0 (GraphPad Software, La Jolla, CA, United States) were used for statistical analysis of all data. Differences between the two groups of measurement data were calculated by student’s t-test, and the comparison of count data was analyzed by the Chi-Square test or Fisher’s exact probability method. Logistic regression analysis was used to identify factors associated with tumor invasion and migration. A receiver operating characteristic (ROC) curve was constructed to evaluate the diagnostic value. The Kaplan-Meier method was used to draw survival curves, and the log-rank test was used to compare the differences between the two groups. All of the tests were two-tailed, and P < 0.05 was considered statistically significant.
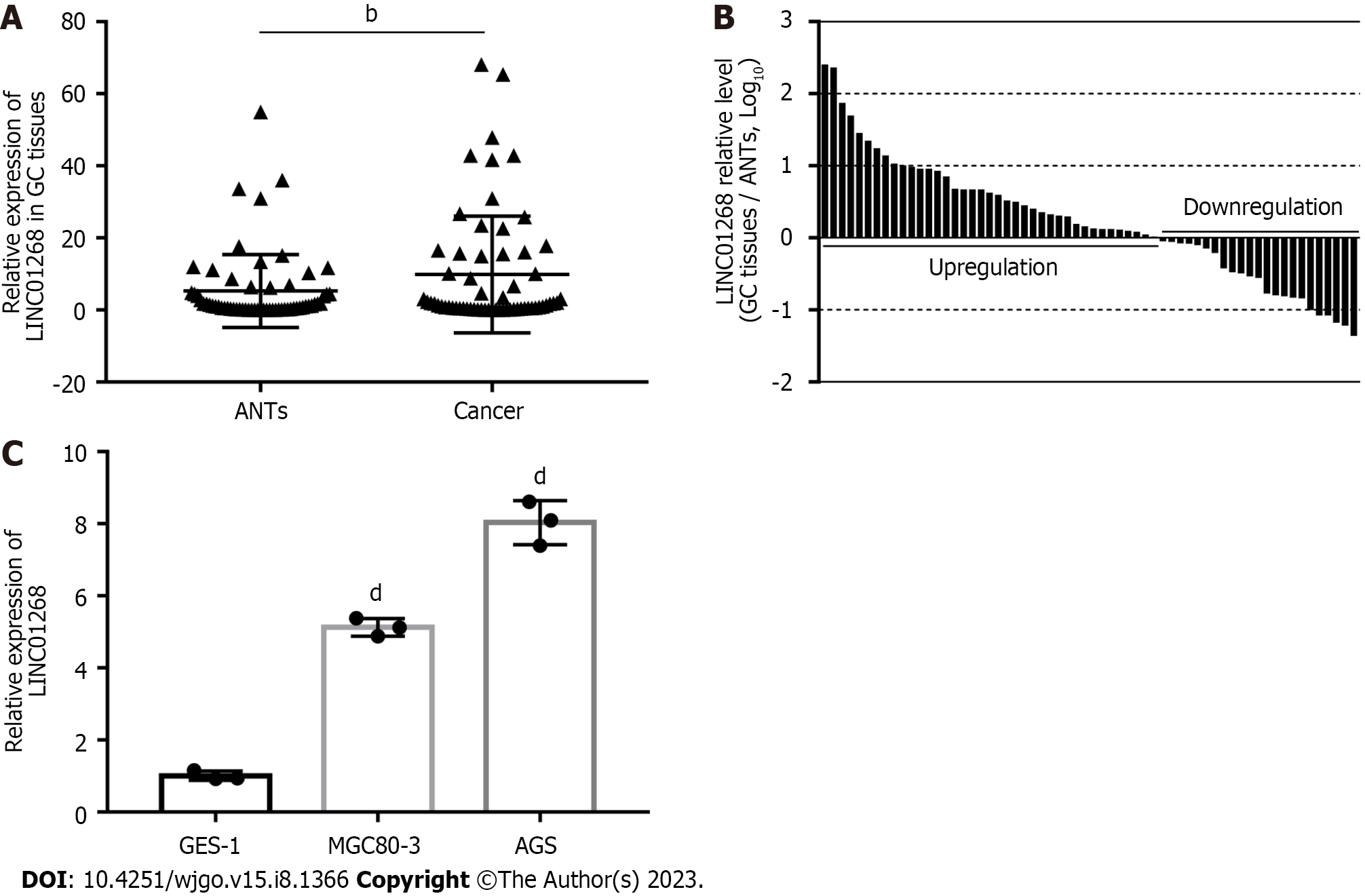
We used real-time quantitative reverse-transcription PCR (RT-qPCR) to detect 62 cases of GC tissues and paired ANTs. As shown in Figure 1A, the expression level of LINC01268 in GC tissues was higher than that in ANTs (P = 0.0065). Among them, the expression of LINC01268 in 39 GC tissues (62.90%) was higher than that in ANTs, and the expression of LINC01268 in 23 GC tissues (37.10%) was lower than that in ANTs, as shown in Figure 1B. At the same time, RT-qPCR was used to compare the relative expression levels of LINC01268 between two GC cell lines and a normal gastric mucosal epithelial cell line, as shown in Figure 1C. The results indicate that the expression levels of LINC01268 in the GC cell lines MGC80-3 and AGS were higher than those in the normal gastric mucosa epithelial cell line GES-1 (P < 0.0001).
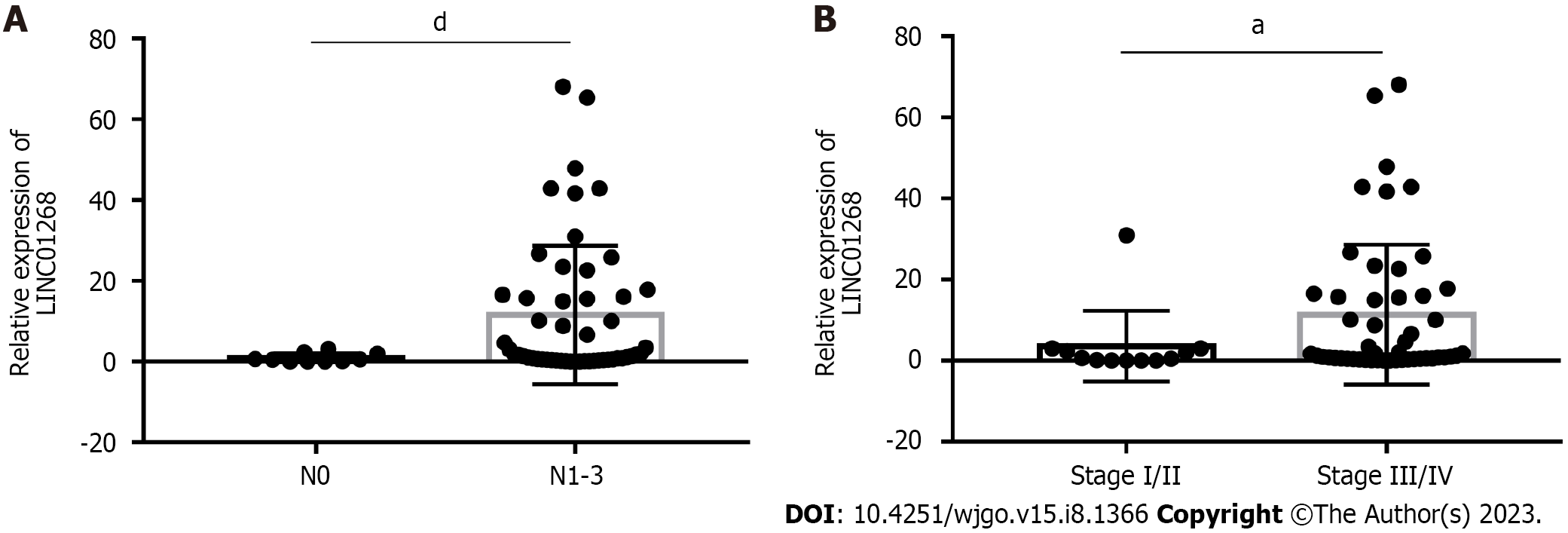
To investigate the clinical significance of changes in the expression level of LINC01268 in GC tissues, according to the expression of LINC01268 in GC tissues and ANTs, we divided them into 39 cases for the high expression group and 23 cases for the low expression group for comparison. As shown in Table 1, high expression of LINC01268 was positively correlated with positive lymph node metastasis, later TNM staging, and poorly differentiated cancer tissue (P < 0.05). However, the difference in the LINC01268 expression level was not significantly correlated with smoking, drinking, gender, age, maximum tumor diameter, depth of invasion, distant metastasis, pathological classification of GC, intraoperative ascites volume, fatty cancer nodules, liver metastasis, venous/lymphatic invasion or nervous invasion. Then, we excluded the ANTs and analyzed the GC tissues separately. As shown in Figure 2, the expression level of LINC01268 in the lymph node metastasis group N1-3 (n = 52 cases) in the GC tissue was significantly higher than that in the N0 group (n = 10 cases) (P < 0.0001), and the expression level of LINC01268 in the III/IV group (n = 50 cases) in the GC tissue was significantly higher than that in the GC tissue in the I/II group (n = 12 cases) (P = 0.0327). These results suggested that LINC01268 may play an important role in the progression of GC, especially in the invasion and metastasis of GC.
| Characteristics | No. of each group | LINC01268 expression | P value | |
| High | Low | |||
| All cases | 62 | 39 | 23 | |
| Age (yr) | ||||
| ≥ 60 | 38 | 23 | 15 | 0.626 |
| < 60 | 24 | 16 | 8 | |
| Gender | ||||
| Male | 53 | 32 | 21 | 0.531 |
| Female | 9 | 7 | 2 | |
| Smoking | ||||
| Yes | 23 | 15 | 8 | 0.772 |
| No | 39 | 24 | 15 | |
| Drinking alcohol | ||||
| Yes | 21 | 14 | 7 | 0.661 |
| No | 41 | 25 | 16 | |
| Maximum tumor diameter | ||||
| ≥ 5 | 25 | 19 | 6 | 0.079 |
| < 5 | 37 | 20 | 17 | |
| Depth of invasion | ||||
| pT1-pT2 | 8 | 5 | 3 | 1.000 |
| pT3-pT4 | 54 | 34 | 20 | |
| Lymph node metastasis | ||||
| pN0 | 10 | 2 | 8 | 0.007 |
| pN1-pN3 | 52 | 37 | 15 | |
| Distant metastasis | ||||
| pM0 | 59 | 36 | 23 | 0.453 |
| pM1 | 3 | 3 | 0 | |
| Tumor TNM stage | ||||
| I-II | 12 | 4 | 8 | 0.043 |
| III-IV | 50 | 35 | 15 | |
| Histology | ||||
| Middle differentiated and well differentiated | 23 | 10 | 13 | 0.015 |
| Poor differentiated | 39 | 29 | 10 | |
| Macroscopic types | ||||
| Mass | 3 | 0 | 3 | 0.378 |
| Ulcerative | 12 | 8 | 4 | |
| Infiltrative ulcerative | 47 | 31 | 16 | |
| Diffuse infiltrative | 0 | 0 | 0 | |
| Venous/lymphatic invasion | ||||
| Positive | 10 | 7 | 3 | 0.881 |
| Negative | 52 | 32 | 20 | |
| Nervous invasion | ||||
| Positive | 7 | 5 | 2 | 0.936 |
| Negative | 55 | 34 | 21 | |
| Fatty nodules | ||||
| Positive | 8 | 7 | 1 | 0.250 |
| Negative | 54 | 32 | 22 | |
| Liver metastasis | ||||
| Present | 3 | 2 | 1 | 1.000 |
| Absent | 59 | 37 | 22 | |
| Ascitic fluid | ||||
| Negative | 48 | 30 | 18 | 0.903 |
| Positive | 14 | 9 | 5 | |
According to the analysis of clinicopathological factors, it was speculated that the highly expressed LINC01268 may be involved in the regulation of the biological behavior of GC invasion and metastasis, so we proceeded to use logistic regression analysis for further analysis. As shown in Table 2, in the univariate analysis, tumor metastasis was related to macroscopic type [odds ratio (OR) = 7.167, 95% confidence interval (CI): 1.673-30.703, P = 0.008), depth of invasion (OR = 8.000, 95%CI: 1.575-40.632, P = 0.012), and the expression level of LINC01268 (OR = 9.867, 95%CI: 1.873-51.974, P = 0.007). Factors such as gender, age, smoking, drinking and ascitic fluid were not related to the invasion and metastasis of GC. Furthermore, multivariate regression models demonstrated that the following factors increased the risk of tumor metastasis: macroscopic type (OR = 9.291, 95%CI: 1.465-58.928, P = 0.018), depth of invasion (OR = 21.490, 95%CI: 1.815-254.398, P = 0.015) and expression level of LINC01268 (OR = 24.726, 95%CI: 2.167-282.097, P = 0.010). These results further indicated that LINC01268 was involved in the regulation of the invasion and metastasis of GC.
| Factors | Univariate analysis | Multivariate analysis | ||
| OR (95%CI) | P value | OR (95%CI) | P value | |
| Age | 1.067 (0.268-4.253) | 0.927 | ||
| Gender | 1.607 (0.281-9.176) | 0.593 | ||
| Smoking | 0.324 (0.081-1.302) | 0.112 | ||
| Drinking alcohol | 0.444 (0.113-1.753) | 0.247 | ||
| Macroscopic types | 7.167 (1.673-30.703) | 0.008 | 9.291 (1.465-58.928) | 0.018 |
| Maximum tumor diameter | 7.714 (0.911-65.352) | 0.061 | ||
| Histology | 1.889 (0.482-7.396) | 0.361 | ||
| Depth of invasion | 8.000 (1.575-40.632) | 0.012 | 21.490 (1.815-254.398) | 0.015 |
| Venous/lymphatic invasion | 1.884 (0.211-16.787) | 0.570 | ||
| Ascitic fluid | 3.000 (0.346-25.993) | 0.319 | ||
| LINC01268 | 9.867 (1.873-51.974) | 0.007 | 24.726 (2.167-282.097) | 0.010 |
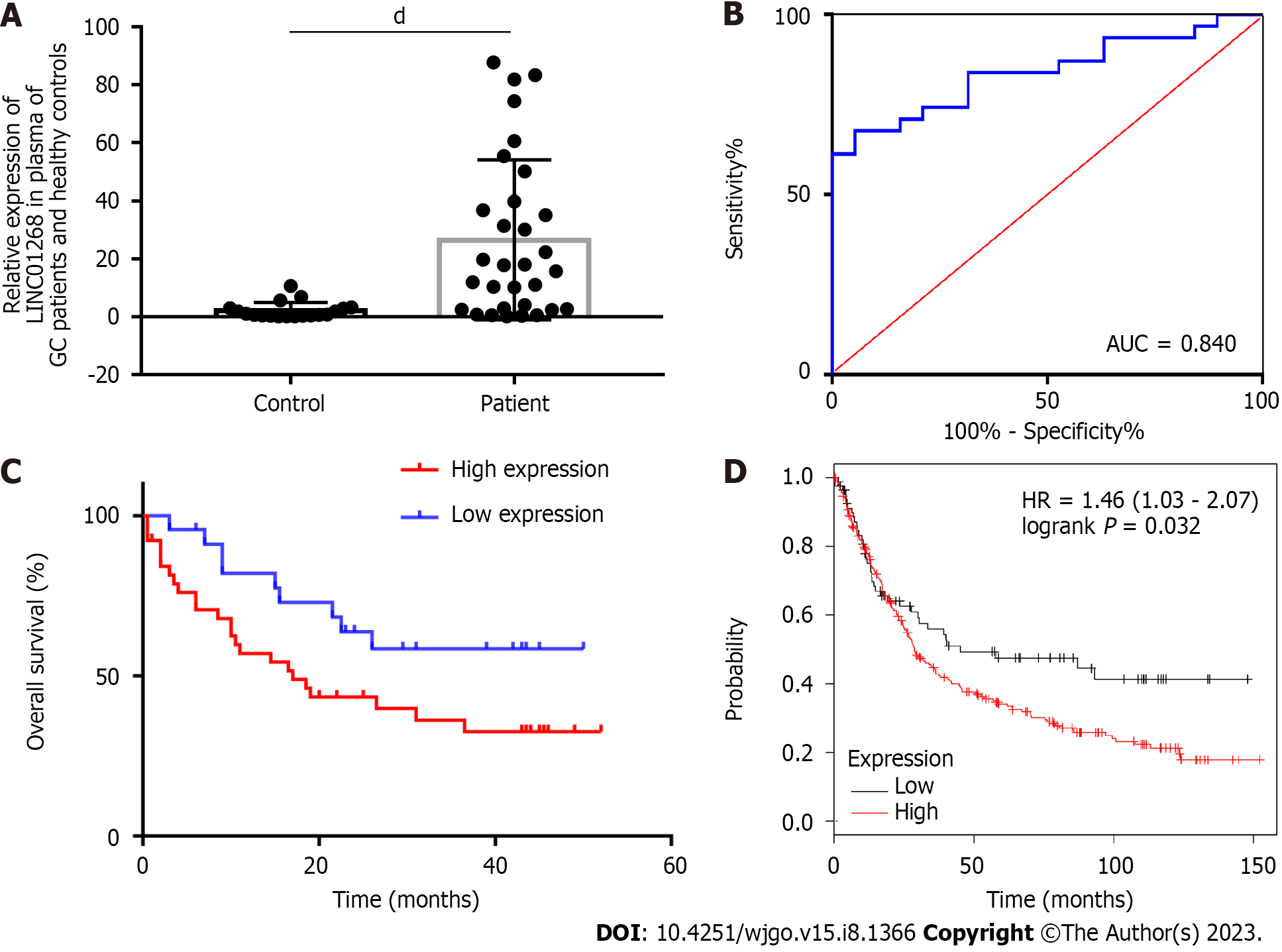
RT-qPCR was used to detect the expression level of LINC01268 in the plasma of 31 patients with GC and 19 healthy volunteers, as shown in Figure 3A. By comparison, the expression level of LINC01268 in the plasma of patients with GC was significantly higher than that of healthy controls (P < 0.0001). To explore whether LINC01268 has a certain value in the auxiliary diagnosis of GC, an ROC curve of the expression level of LINC01268 in plasma was constructed, as shown in Figure 3B. In the plasma of patients with GC, the sensitivity of LINC01268 in diagnosing GC was 67.74%, the specificity was 94.74%, the Youden index was 0.624, and the area under the curve (AUC) was equal to 0.840 (95%CI: 0.733-0.948, P < 0.0001). These results suggested that the expression of LINC01268 in plasma may be related to the progression and prognosis of GC patients, and it may become a new molecular marker for auxiliary diagnosis of GC.
Based on the expression level of LINC01268 in GC tissues and ANTs, they were divided into 39 cases in the high expression group and 23 cases in the low expression group. The survival curves of the two groups were drawn by the Kaplan-Meier method, and the differences in overall survival between the two groups were compared by the log-rank test. As shown in Figure 3C, the overall survival of GC patients with high expression of LINC01268 was worse than that of patients with low expression of LINC01268 (P = 0.047). In addition, the effect of LINC01268 expression on the prognosis of 348 patients with GC was analyzed through the online database Kaplan-Meier plotter. There were 262 cases in the high expression group and 86 cases in the low expression group, as shown in Figure 3D. The results showed that the prognosis of GC patients with high expression of LINC01268 was worse (P < 0.05). The prediction results of the online database are highly consistent with the real clinical data of our center, which fully indicates that LINC01268 is involved in the regulation of the tumorigenesis and development of GC cells. Its over-expression is correlated with a poor prognosis of patients with GC.
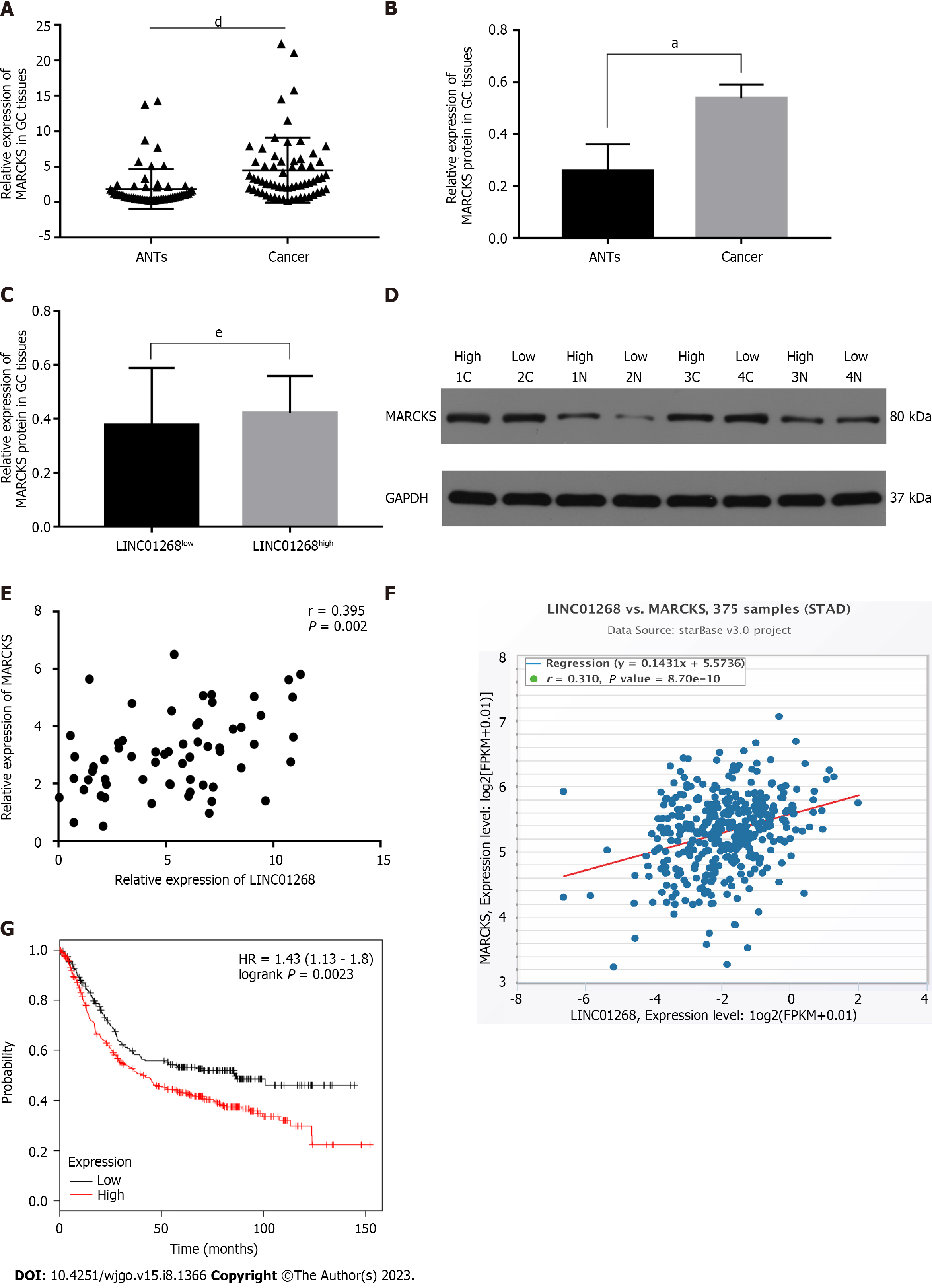
To identify the target genes involved in the regulation of LINC01268, we performed a preliminary analysis by using bioinformatics tools. From the GeneCards database, it was found that LINC01268 was located on the long arm of chromosome 6. The online analysis tool circlncRNA net was used to construct a co-expressed gene network of LINC01268, and the coding gene of MARCKS, also located on chromosome 6 was selected. MARCKS was located at a distance of 4542 bp upstream of the LINC01268 gene locus. Then we compared the expression of the two genes in GC, further verified the correlation between them, and determined whether MARCKS is the target regulatory gene of LINC01268.
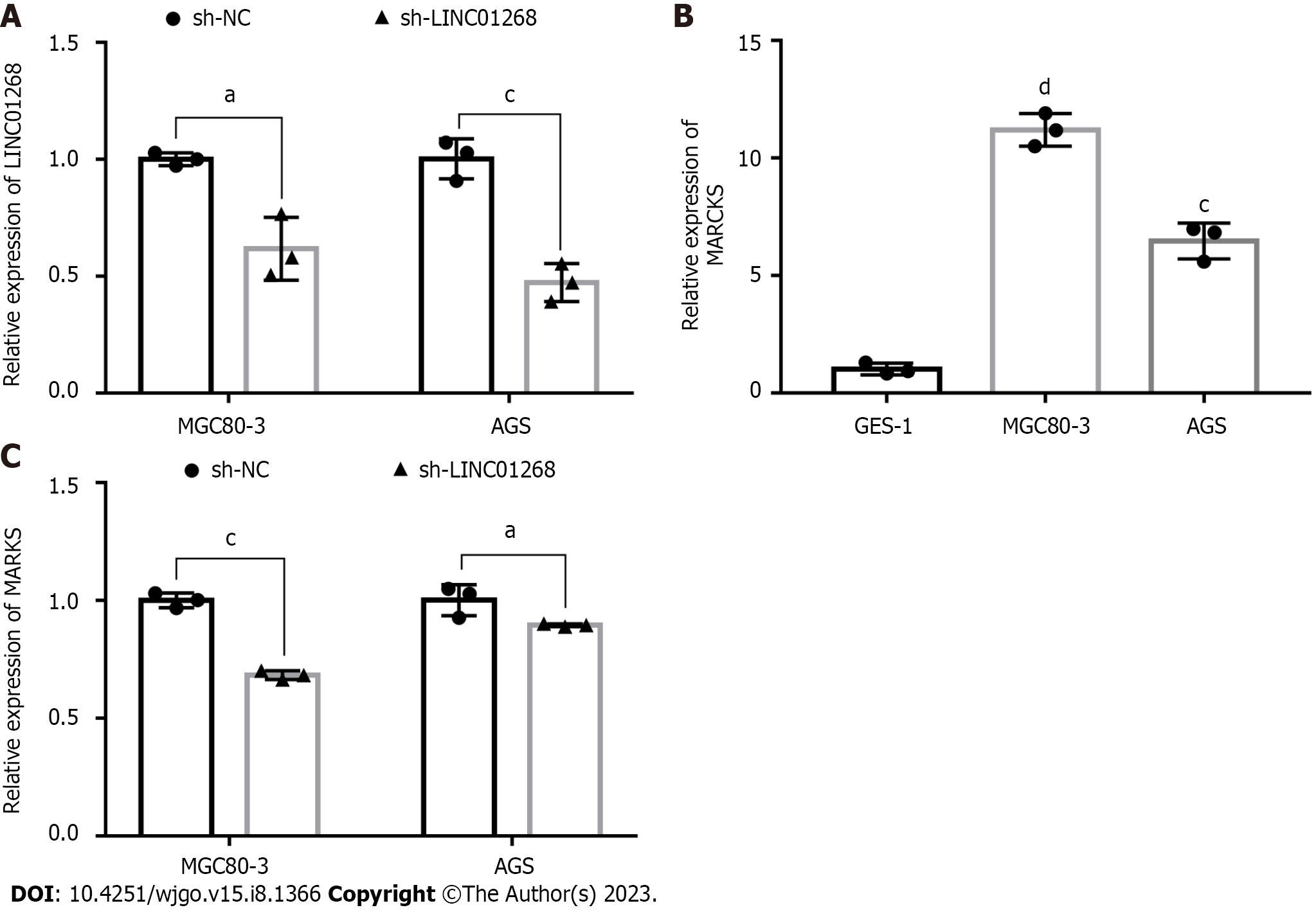
The expression of MARCKS in 62 GC tissues and ANTs was detected by RT-qPCR. As shown in Figure 4A, the relative expression level of MARCKS mRNA in 62 GC tissues was significantly higher than that of matched ANTs (P < 0.0001), which was consistent with the change trend of LINC01268 expression. In the analysis of the protein expression level, we also found that the level of MARCKS protein in GC tissues was also significantly higher than that in ANTs (P = 0.01), but the protein levels of MARCKS in LINC01268 high expression groups were not higher than those in the LINC01268 Low expression groups (P > 0.05) (we divided tested tissue samples into a LINC01268 high expression group and a low expression group according to the expression level of LINC01268 and compared the protein expression level of MARCKS between the two groups), as shown in Figures 4B-D. To further verify the relationship between LINC01268 and MARCKS, we performed a bivariate correlation analysis of the expression of MARCKS and LINC01268 in 62 cases of GC tissues and found that there was a significant positive correlation between them (r = 0.395, P = 0.002), as shown in Figure 4E. In addition, through the analysis of 375 GC samples in the online starBase database, we also found that there was a positive correlation between LINC01268 and MARCKS expression (r = 0.310, P < 0.0001), as shown in Figure 4F, which was consistent with our experimental data. We further analyzed the effect of the difference in the expression level of MARCKS on the prognosis of 631 patients with GC by using the online database Kaplan-Meier plotter. Among them, there were 392 cases in the relatively high expression group and 239 cases in the relatively low expression group. As shown in Figure 4G, the survival of GC patients with high expression of MARCKS was shorter than that of patients with low expression of MARCKS (log-rank P = 0.0023), and the change trend was the same as that of LINC01268. The above results suggested that MARCKS and LINC01268 in GC tissues showed a significant positive correlation.
To verify the interaction between MARCKS and LINC01268, we observed whether the expression level of MARCKS was changed accordingly by knocking down the expression of LINC01268 in GC cells. First, the recombinant plasmids sh-LINC01268 and sh-NC were successfully transfected into two GC cell lines. The RT-qPCR results showed that the expression levels of LINC01268 in MGC80-3 (P = 0.025) and AGS (P < 0.001) cells in the sh-LINC01268 group were significantly lower than those in the control sh-NC group, especially in AGS, as shown in Figure 5A. In addition, the expression of MARCKS in two GC cell lines, MGC80-3 and AGS, and the gastric normal mucosal cell line GES-1 were also detected, as shown in Figure 5B. The expression levels of MARCKS in MGC80-3 (P < 0.0001) and AGS (P < 0.001) GC cell lines were significantly higher than those in GES-1. After down-regulating the expression level of LINC01268 in GC cells by plasmid transfection, we detected the changes in the expression of MARCKS. The results showed that compared with the sh-NC group, the MARCKS expression levels of the sh-LINC01268 group in MGC80-3 (P < 0.001) and AGS (P < 0.05) cells were significantly reduced, as shown in Figure 5C. The above results further verified that the expression level of MARCKS changed with the expression level of LINC01268, and there was a significant positive correlation between them.
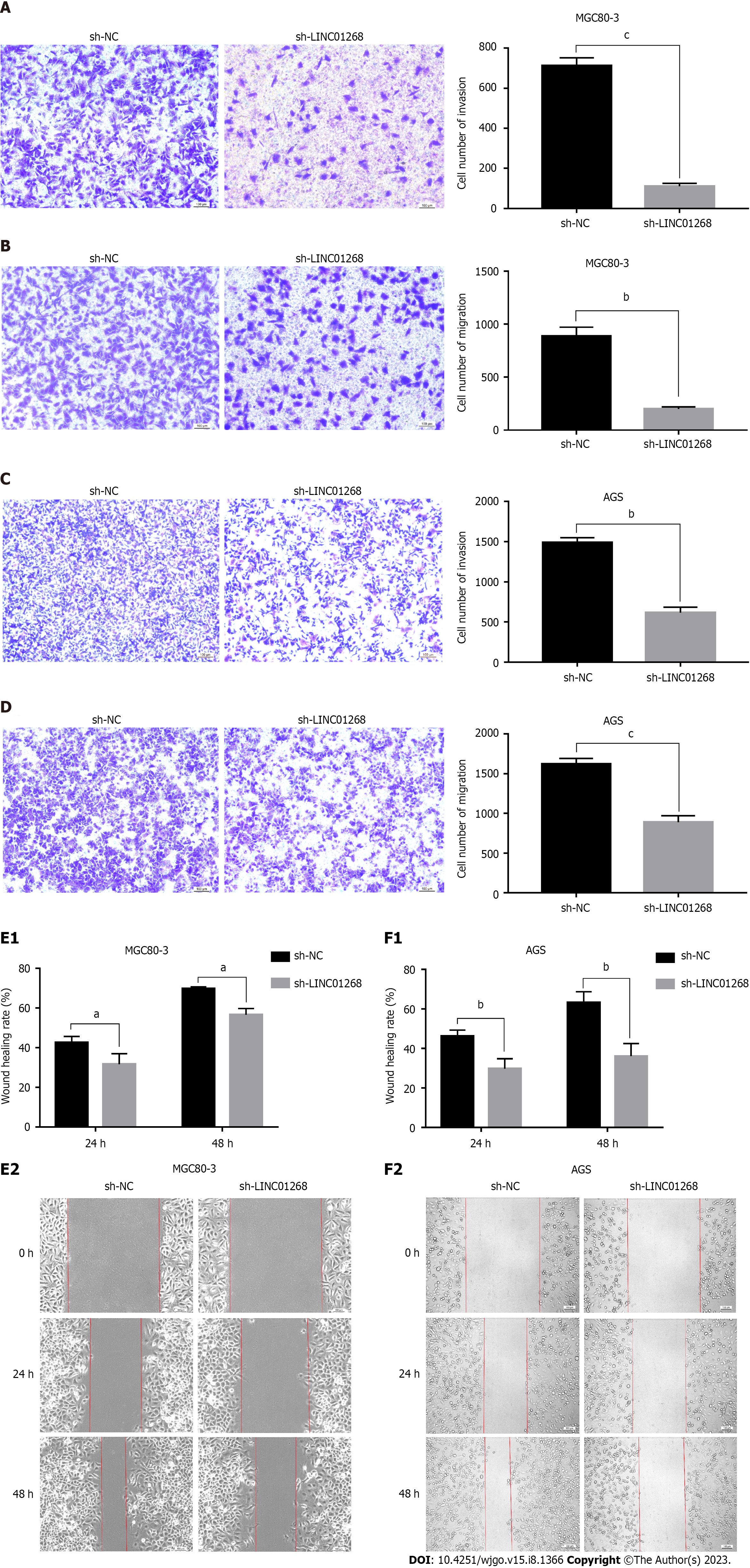
To explore the regulatory effect of LINC01268 on the biological behavior of GC cells, Transwell experiments and wound healing assays were used to compare the changes in the invasion and migration ability of GC cells before and after sh-LINC01268 transfection. As shown in Figures 6A-D, in the invasion and migration experiments of MGC80-3 and AGS cells, the number of GC cells in the sh-LINC01268 group that passed through the chamber to the subventricular surface of the microporous membrane was significantly less than that of the sh-NC group. As shown in Figures 6E and F, the migration ability of MGC80-3 and AGS cells decreased after the down-regulation of LINC01268 expression. Based on the above experimental results, down-regulating the expression of LINC01268 will weaken the invasion and metastasis ability of GC cells, indicating that LINC01268 participates in and promotes the process of invasion and metastasis of GC.
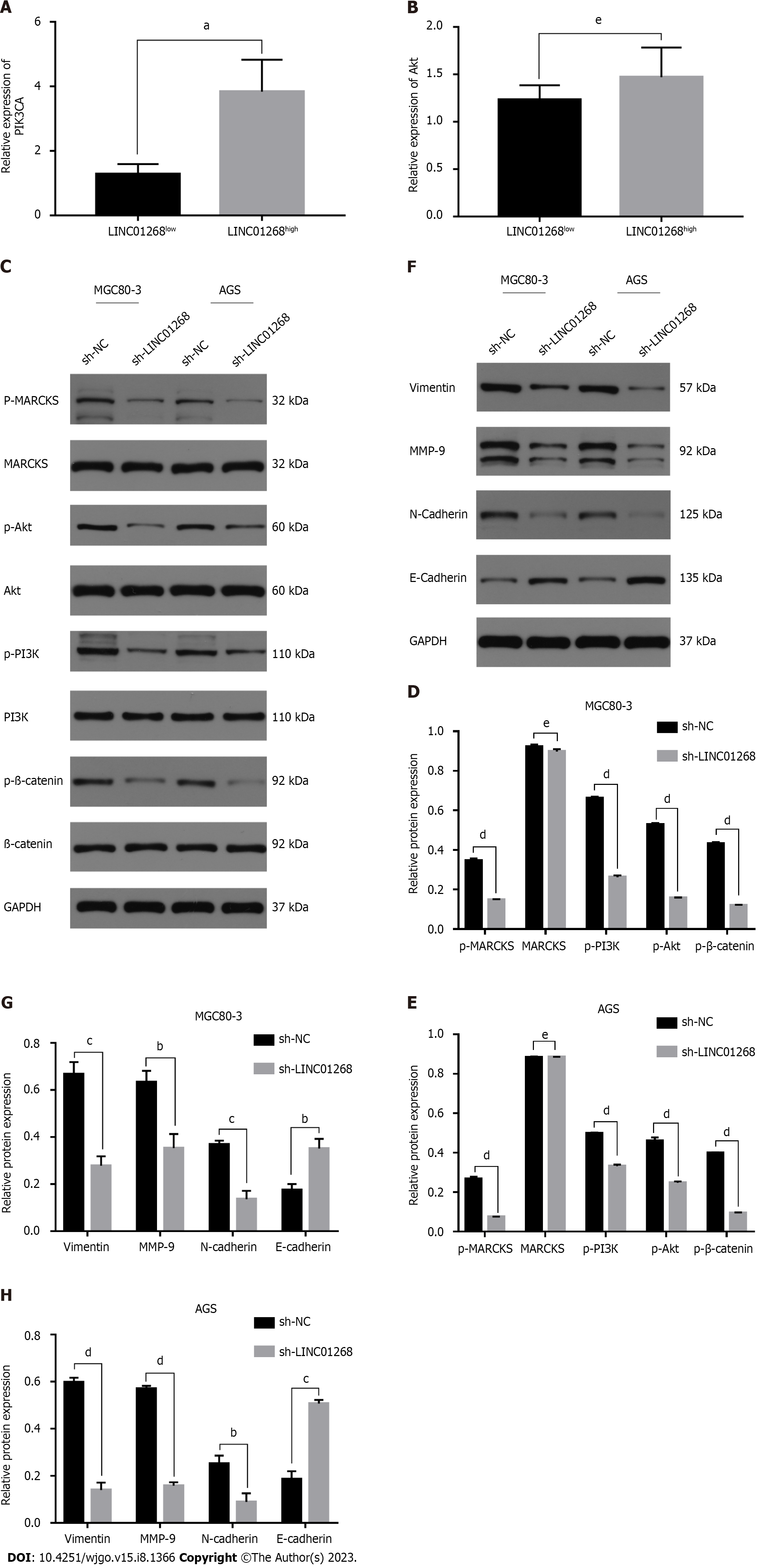
Through reviewing the literature, it was found that MARCKS was closely related to the PI3K/Akt signaling pathway. It was speculated that LINC01268 may also be involved in the regulation of the PI3K/Akt signaling pathway. To understand the relationship between the expression level of LINC01268 and the activation of the PI3K/Akt signaling pathway, we analyzed the results of RT-qPCR detection of the classical signaling pathway in 50 GC tissues and divided them into the LINC01268high group and LINC01268low group according to the relative expression of LINC01268. The mRNA expression levels of PIK3CA and Akt in the two groups were compared (PIK3CA is an important catalytic subunit of PI3K). As shown in Figures 7A and B, the expression level of PIK3CA in LINC01268high cells was significantly higher than that in LINC01268low cells (P = 0.019). The expression level of Akt in LINC01268high cells was higher than that in LINC01268low cells, but there was no significant difference (P > 0.05). These results suggested that the up-regulation of LINC01268 may activate the PI3K/Akt signaling pathway. In addition, when using the online analysis tool circlncRNA net to conduct pathway enrichment analysis (MSigDB database) for LINC01268 and MARCKS, it was found that they were likely to be related to the epithelial-mesenchymal transition (EMT) process (P < 0.0001).
To further explore whether LINC01268 is involved in the activation of the PI3K/Akt signaling pathway and the regulation of the EMT process in GC cells, we knocked down the expression of LINC01268 in GC cells and detected whether the expression level of the PI3K/Akt signaling pathway and key EMT-related proteins changed accordingly. This can indirectly show whether LINC01268 is involved in the activation of the PI3K/Akt signaling pathway and the regulation of the EMT process in GC cells. The results showed that after down-regulating the expression of LINC01268 in MGC80-3 and AGS cells, the expression levels of p-MARCKS, p-PI3K, p-Akt and p-β-catenin in the PI3K/Akt signaling pathway were all down-regulated, as shown in Figures 7C-E. At the same time, after knocking down LINC01268, the levels of vimentin, MMP-9 and N-cadherin proteins related to EMT in MGC80-3 and AGS cells decreased, while the level of E-cadherin protein increased, as shown in Figures 7F-H. These results strongly suggested that the activation of the PI3K/Akt signaling pathway and the EMT process of GC cells were inhibited to varying degrees after down-regulation of LINC01268. Therefore, we speculate that LINC01268 may activate the PI3K/Akt signaling pathway through targeted regulation of MARCKS, participate in the EMT process of GC cells, and ultimately promote tumor invasion and metastasis.
With the deepening of non-coding RNA research, an increasing number of lncRNAs have been found to be specifically expressed in a variety of tumor tissues and may even exist stably in body fluids. This shows good research and application prospects for lncRNAs to become new tumor biomarker molecules and therapeutic targets[15]. At the same time, an increasing number of studies on lncRNAs have shown that they are involved in the regulation of a variety of biological behaviors of tumor cells, such as tumor proliferation, tumor invasion and metastasis, and tumor drug resistance. Among them, many lncRNAs are closely related to the occurrence and development of GC, and it is hoped that they will open up valuable new directions for early clinical screening, targeted therapy, and monitoring of GC. Some studies have established an overall model of differential expression of lncRNAs by analyzing and integrating multiple gene data sets in the GEO database to help predict the outcomes of patients with GC[16]. Another study reported that the highly expressed ZEB1-AS1 promoted the invasion, metastasis and EMT process of GC cells by positively regulating the ZEB1 encoding transcription factor in its adjacent position and found that it was closely related to the malignancy of GC and the prognosis of patients[17]. In addition, some studies showed that overexpression of lncRNA-GAS5 activated miR-34c expression by directly inhibiting E2F1, and ultimately inhibited tumor growth and uncontrolled proliferation in GC[18]. At present, most important lncRNAs in GC have not been identified. Although some lncRNAs with suggestive expression characteristics have been discovered, it is currently difficult to explain the specific mechanism in detail due to its wide range of functions. Therefore, we need to continue to explore the role of lncRNAs related to GC in the occurrence and development of GC to lay a foundation for its clinical application and transformation.
In our previous study, we compared the lncRNA profiles of GC tissues and paired ANTs using a lncRNA expression microarray[19]. LINC01268 displayed a trend of significant differential expression in GC tissues and paired ANTs. In addition, compared with normal gastric mucosal cells, the expression level of LINC01268 in GC cell lines was also up-regulated. To further confirm the authenticity of this difference, we verified that the expression level of LINC01268 in GC tissues was significantly higher than that in ANTs by examining a large number of clinical tissue samples. We further analyzed the clinical data and found that the high expression of LINC01268 was positively correlated with positive GC lymph node metastasis, later TNM staging, and poorly differentiated cancer tissues. Therefore, we speculate that LINC01268 may be involved in the biological behavior regulation of invasion and metastasis of GC. In addition, logistic regression analysis further confirmed that LINC01268 may mainly affect the invasion and metastasis of GC. Similar lncRNAs have been reported in an increasing number of studies. For example, lncRNA XIST over-expression in GC tissue was closely related to tumor size, lymph node invasion, distant metastasis, and TNM staging[20]. Up-regulation of mir4435-2HG expression promoted the invasion and progression of GC[21]. The over-expression of SNHG11 in GC aggravated oncogenic autophagy to facilitate cell proliferation, stemness, migration, invasion, and EMT in GC, and it was correlated with dismal patient outcomes[22]. At the same time, lncRNAs can be used as tumor markers to predict the outcomes of patients and the progression of the disease. Studies have found that some lncRNAs can stably exist in plasma and can be used for independent or combined diagnosis of GC based on their differential expression characteristics for GC[23-25]. Just as the highly expressed lncRNAs PANDAR, FOXD2-AS1 and SMARCC2 in the plasma of patients with GC performed well in the diagnosis of GC, they have the potential to be used as biomarkers for the auxiliary diagnosis of GC[26]. In this study, we found that LINC01268 is highly expressed in the plasma of patients with GC. At the same time, through the evaluation of ROC curves, it was found that the expression of LINC01268 in the plasma had good value in the clinical diagnosis of GC. In addition, through clinical big data analysis, we found that the prognosis of patients with high expression of LINC01268 was significantly worse than that of patients with low expression of LINC01268 in both Kaplan-Meier plotter online database analysis and our center’s own survival data of patients with GC, indicating that the over-expression of LINC01268 is related to a poor prognosis of patients with GC. It is expected that it can be used as one of the indexes for evaluating the disease progression of patients with GC. Based on the above results, we speculate that LINC01268 is involved in the regulation of the invasion and metastasis of GC cells.
To explore the specific molecular mechanism of LINC01268 in the regulation of invasion and metastasis of GC, we first used the online tool circlncRNAnet to initially screen for MARCKS, which was one of the co-expressed genes of LINC01268. Then, we verified that the expression level of MARCKS in GC tissues and GC cells was significantly increased and showed a positive correlation with LINC01268 in terms of expression level and prognostic evaluation of GC, suggesting that MARCKS may be a target regulatory gene of LINC01268. At the same time, the correlation analysis in the starBase online database also confirmed the results of our research. Existing studies have shown that MARCKS is involved in promoting the invasion and metastasis of GC[27], so we speculated that LINC01268 may target and regulate MARCKS to promote the invasion and metastasis of GC. To further confirm our speculation, we knocked down the expression of LINC01268 in GC cells in vitro and observed that the expression and activation of MARCKS and the invasion and migration ability of GC cells were significantly inhibited. This shows that MARCKS is a target regulatory gene of LINC01268 and that LINC01268 affects invasion and metastasis of GC by targeting and regulating MARCKS.
MARCKS is a substrate of protein kinase C, which regulates the cytoskeleton and cell chemotaxis and mediates processes such as inflammation, cell secretion, and exocytosis. The abnormal expression of MARCKS in most cases promotes tumor occurrence and metastasis[28,29]. When MARCKS is not phosphorylated, it uses its phosphorylation site domain (PSD) to combine with PIP2 through electrostatic forces at the cell membrane level to protect PIP2 from being hydrolyzed. When MARCKS is phosphorylated by upstream activated protein kinase C or CaM-Ca2+ (a complex formed by calmodulin CaM and calcium ions) competitively binding to PSD, the newly isolated PIP2 recruits PI3K to transform itself into PIP3, which activates the PI3K/Akt signaling pathway[30-32]. MARCKS has been found to be associated with invasion and metastasis in a variety of tumors, most of which are reflected in its ability to regulate the cytoskeleton. MARCKS affects cytoskeletal reorganization, improves the mobility and migration of colorectal cancer cells, and induces mesenchymal phenotypes[33]. Our enrichment analysis of the co-expression of LINC01268 and MARCKS also found that they were involved in the EMT process of tumor cells. A large amount of existing research data indicates that the function of MARCKS is closely related to the regulation of PI3K/Akt signaling pathway activation. In addition, many studies have found that the PI3K/Akt signaling pathway is also closely related to the invasion and metastasis of GC. Among them, PI3K/Akt can promote the degradation of GSK-3β after activation, thereby activating the Wnt/β-catenin signaling pathway to help promote the invasion and metastasis of adriamycin-resistant GC cells[34]. Therefore, we speculated that LINC01268 may also be involved in the regulation of the PI3K/Akt signaling pathway in GC and promote EMT and invasion and metastasis of GC cells. This speculation was proven by in vitro cell experiments in this study. These research results indicate that LINC01268 promotes the invasion and migration of GC cells by targeting and regulating MARCKS, thereby activating the PI3K/Akt signaling pathway. At the same time, LINC01268 also promotes the expression of mesenchymal phenotype proteins and matrix metalloproteinases in GC cells, enhancing their ability to dissolve extracellular matrix and ultimately promoting the occurrence of EMT.
According to the online tool lncLocator, most LINC01268 exists in the cytoplasm, and the GeneCards database indicates that the MARCKS protein is mainly distributed in the cell membrane and cytoskeleton. Based on these findings, we speculate that LINC01268 may bind to a specific protein and exit the nucleus to affect the phosphorylation state of MARCKS in a manner similar to CaM-Ca2+ or protein kinase C in the cytoplasm, thereby inducing a series of functional changes in downstream signaling pathways. LINC01268 and MARCKS are also both on chromosome 6 and are adjacent to each other. The distance between the two loci is less than 5 kb, which indicates that LINC01268 may also directly affect the expression of MARCKS mRNA. This hypothesis is supported by a study of the inflammatory response. LINC01268 is mainly concentrated in the nucleus of macrophages. Through cis-regulation, it recruits APEX1 to form a complex to bind to the MARCKS promoter sequence, promoting HDAC1-mediated H3K27ac deacetylation, and subsequently inhibited the expression of MARCKS[35]. In this study, we also indirectly decreased the mRNA and phosphorylated protein expression of MARCKS by knocking down LINC01268. At the same time, the expression of several key proteins of the PI3K/Akt signaling pathway were also inhibited, and the invasion and metastasis abilities of GC cells were also decreased. Thus, it is indirectly proven that LINC01268 has a positive regulatory effect on the adjacent gene MARCKS.
In summary, we found that LINC01268 may be an oncogene in GC that activates the PI3K/Akt signaling pathway and EMT by targeting and regulating MARCKS and ultimately promotes the invasion and metastasis of GC cells. These findings indicate that LINC01268 may play an important role in the occurrence and development of GC, and it may become a new molecular marker for assisting in the diagnosis of GC and is a potential target for the treatment of GC.
Long non-coding RNAs (lncRNAs) are a class of RNA molecules with a length of more than 200 nucleotides. It plays an important role in many life activities such as epigenetic regulation, cell cycle regulation, and cell differentiation regulation. More and more studies have found that lncRNAs also affect the occurrence and development of tumors. LncRNAs have also been found to be involved in the regulation of gastric cancer (GC) progression, which may open up new directions for the diagnosis and treatment of GC.
GC is still a malignant tumor of digestive system with high incidence rate and mortality worldwide. Although many lncRNAs related to GC have been discovered in recent years, our understanding of their mechanisms of action is not yet deep enough. Exploring more GC associated lncRNAs and delving into the underlying mechanisms will help us learn more about the development and progression of GC, and hopefully improve the early screening rate and cure rate of GC.
The main objective of this study is to investigate the impact of high expression of LINC01268 in GC on the diagnosis and prognosis evaluation of GC patients, and to explore the impact and mechanism of LINC01268 on the biological behavior of GC cells in vitro experiments. Our study found that GC patients with high expression of LINC01268 have poor prognosis. High expression of LINC01268 can activate the MARCKS and PI3K/Akt signaling pathway, promoting the invasion, metastasis, and epithelial-mesenchymal transition (EMT) processes of GC cells. These may provide potential directions for targeted therapeutic drugs for GC.
Real-time quantitative polymerase chain reaction was used to detect the expression of LINC01268 in GC. The receiver operating characteristic curve and Kaplan-Meier method were used for the analysis of the diagnostic value and prognostic evaluation of LINC01268 in patients with GC. Transwell assays and wound healing assays were used to confirm the effect of LINC01268 on the invasion and migration of GC cells. The regulatory relationship between LINC01268 and MARCKS, the PI3K/Akt signaling pathway, and the EMT process in GC was demonstrated by western blot analysis.
Our study found that high expression of LINC01268 was associated with some pathological features of GC patients and was predictive of poor prognosis. In vitro experiments, after knocking down LINC01268, the invasion and metastasis ability of MGC80-3 and AGS decreased. Meanwhile, molecular mechanism studies have found that LINC01268 may regulate the MARCKS, PI3K/Akt signaling pathway, and EMT process in GC cells.
LINC01268 may promote GC invasion and metastasis by activating MARCKS, PI3K/Akt signaling pathway, and promoting EMT process. High expression of LINC01268 contributed to the poor prognosis of GC patients.
The present study provides a promising direction for targeted therapy of GC. Animal experiments and more comprehensive molecular biology experiments would be better to confirm our conclusions, which is also the direction of our future research.
| 1. | Wild CP. The global cancer burden: necessity is the mother of prevention. Nat Rev Cancer. 2019;19:123-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 79] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 2. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 56640] [Article Influence: 7080.0] [Reference Citation Analysis (134)] |
| 3. | Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654-2664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1282] [Cited by in RCA: 1504] [Article Influence: 150.4] [Reference Citation Analysis (1)] |
| 4. | Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3924] [Cited by in RCA: 4502] [Article Influence: 264.8] [Reference Citation Analysis (0)] |
| 5. | Nelson BR, Makarewich CA, Anderson DM, Winders BR, Troupes CD, Wu F, Reese AL, McAnally JR, Chen X, Kavalali ET, Cannon SC, Houser SR, Bassel-Duby R, Olson EN. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science. 2016;351:271-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 485] [Cited by in RCA: 631] [Article Influence: 63.1] [Reference Citation Analysis (0)] |
| 6. | Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, Kasaragod P, Shelton JM, Liou J, Bassel-Duby R, Olson EN. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595-606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 764] [Cited by in RCA: 968] [Article Influence: 88.0] [Reference Citation Analysis (0)] |
| 7. | Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661-5667. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 757] [Cited by in RCA: 1299] [Article Influence: 144.3] [Reference Citation Analysis (0)] |
| 8. | Chen X, Zeng K, Xu M, Hu X, Liu X, Xu T, He B, Pan Y, Sun H, Wang S. SP1-induced lncRNA-ZFAS1 contributes to colorectal cancer progression via the miR-150-5p/VEGFA axis. Cell Death Dis. 2018;9:982. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 152] [Cited by in RCA: 183] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 9. | Orfanelli U, Jachetti E, Chiacchiera F, Grioni M, Brambilla P, Briganti A, Freschi M, Martinelli-Boneschi F, Doglioni C, Montorsi F, Bellone M, Casari G, Pasini D, Lavorgna G. Antisense transcription at the TRPM2 locus as a novel prognostic marker and therapeutic target in prostate cancer. Oncogene. 2015;34:2094-2102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 71] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 10. | Szász AM, Lánczky A, Nagy Á, Förster S, Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A, Győrffy B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322-49333. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 582] [Cited by in RCA: 765] [Article Influence: 95.6] [Reference Citation Analysis (0)] |
| 11. | Wu SM, Liu H, Huang PJ, Chang IY, Lee CC, Yang CY, Tsai WS, Tan BC. circlncRNAnet: an integrated web-based resource for mapping functional networks of long or circular forms of noncoding RNAs. Gigascience. 2018;7:1-10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (1)] |
| 12. | Safran M, Dalah I, Alexander J, Rosen N, Iny Stein T, Shmoish M, Nativ N, Bahir I, Doniger T, Krug H, Sirota-Madi A, Olender T, Golan Y, Stelzer G, Harel A, Lancet D. GeneCards Version 3: the human gene integrator. Database (Oxford). 2010;2010:baq020. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 718] [Cited by in RCA: 1396] [Article Influence: 87.3] [Reference Citation Analysis (0)] |
| 13. | Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92-D97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2607] [Cited by in RCA: 4294] [Article Influence: 330.3] [Reference Citation Analysis (0)] |
| 14. | Cao Z, Pan X, Yang Y, Huang Y, Shen HB. The lncLocator: a subcellular localization predictor for long non-coding RNAs based on a stacked ensemble classifier. Bioinformatics. 2018;34:2185-2194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 305] [Article Influence: 43.6] [Reference Citation Analysis (0)] |
| 15. | Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017;77:3965-3981. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2088] [Cited by in RCA: 2229] [Article Influence: 247.7] [Reference Citation Analysis (0)] |
| 16. | Song P, Jiang B, Liu Z, Ding J, Liu S, Guan W. A three-lncRNA expression signature associated with the prognosis of gastric cancer patients. Cancer Med. 2017;6:1154-1164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 62] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 17. | Li Y, Wen X, Wang L, Sun X, Ma H, Fu Z, Li L. LncRNA ZEB1-AS1 predicts unfavorable prognosis in gastric cancer. Surg Oncol. 2017;26:527-534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 42] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 18. | Gupta S, Panda PK, Luo W, Hashimoto RF, Ahuja R. Network analysis reveals that the tumor suppressor lncRNA GAS5 acts as a double-edged sword in response to DNA damage in gastric cancer. Sci Rep. 2022;12:18312. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 19. | Luo Y, Wang C, Yong P, Ye P, Liu Z, Fu Z, Lu F, Xiang W, Tan W, Xiao J. Decreased expression of the long non-coding RNA SLC7A11-AS1 predicts poor prognosis and promotes tumor growth in gastric cancer. Oncotarget. 2017;8:112530-112549. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 20. | Chen DL, Ju HQ, Lu YX, Chen LZ, Zeng ZL, Zhang DS, Luo HY, Wang F, Qiu MZ, Wang DS, Xu DZ, Zhou ZW, Pelicano H, Huang P, Xie D, Wang FH, Li YH, Xu RH. Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin Cancer Res. 2016;35:142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 183] [Cited by in RCA: 227] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 21. | Wang H, Wu M, Lu Y, He K, Cai X, Yu X, Lu J, Teng L. LncRNA MIR4435-2HG targets desmoplakin and promotes growth and metastasis of gastric cancer by activating Wnt/β-catenin signaling. Aging (Albany NY). 2019;11:6657-6673. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 68] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 22. | Wu Q, Ma J, Wei J, Meng W, Wang Y, Shi M. lncRNA SNHG11 Promotes Gastric Cancer Progression by Activating the Wnt/β-Catenin Pathway and Oncogenic Autophagy. Mol Ther. 2021;29:1258-1278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 160] [Article Influence: 32.0] [Reference Citation Analysis (1)] |
| 23. | Tsui NB, Ng EK, Lo YM. Stability of endogenous and added RNA in blood specimens, serum, and plasma. Clin Chem. 2002;48:1647-1653. [PubMed] |
| 24. | Zhang K, Shi H, Xi H, Wu X, Cui J, Gao Y, Liang W, Hu C, Liu Y, Li J, Wang N, Wei B, Chen L. Genome-Wide lncRNA Microarray Profiling Identifies Novel Circulating lncRNAs for Detection of Gastric Cancer. Theranostics. 2017;7:213-227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 165] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 25. | Liu J, Wang J, Song Y, Ma B, Luo J, Ni Z, Gao P, Sun J, Zhao J, Chen X, Wang Z. A panel consisting of three novel circulating lncRNAs, is it a predictive tool for gastric cancer? J Cell Mol Med. 2018;22:3605-3613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 26. | Yang Z, Sun Y, Liu R, Shi Y, Ding S. Plasma long noncoding RNAs PANDAR, FOXD2-AS1, and SMARCC2 as potential novel diagnostic biomarkers for gastric cancer. Cancer Manag Res. 2019;11:6175-6184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (1)] |
| 27. | Quan R, Ning Z, Wang Y, Yu W, Zhu H. Prognostic Value of Upregulation of Myristoylated Alanine-Rich C-Kinase Substrate in Gastric Cancer. Med Sci Monit. 2019;25:279-287. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 28. | Blackshear PJ. The MARCKS family of cellular protein kinase C substrates. J Biol Chem. 1993;268:1501-1504. [PubMed] |
| 29. | Fong LWR, Yang DC, Chen CH. Myristoylated alanine-rich C kinase substrate (MARCKS): a multirole signaling protein in cancers. Cancer Metastasis Rev. 2017;36:737-747. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 51] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 30. | Wang J, Arbuzova A, Hangyás-Mihályné G, McLaughlin S. The effector domain of myristoylated alanine-rich C kinase substrate binds strongly to phosphatidylinositol 4,5-bisphosphate. J Biol Chem. 2001;276:5012-5019. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 153] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 31. | Ziemba BP, Burke JE, Masson G, Williams RL, Falke JJ. Regulation of PI3K by PKC and MARCKS: Single-Molecule Analysis of a Reconstituted Signaling Pathway. Biophys J. 2016;110:1811-1825. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 63] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 32. | Ziemba BP, Swisher GH, Masson G, Burke JE, Williams RL, Falke JJ. Regulation of a Coupled MARCKS-PI3K Lipid Kinase Circuit by Calmodulin: Single-Molecule Analysis of a Membrane-Bound Signaling Module. Biochemistry. 2016;55:6395-6405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 33. | Rombouts K, Carloni V, Mello T, Omenetti S, Galastri S, Madiai S, Galli A, Pinzani M. Myristoylated Alanine-Rich protein Kinase C Substrate (MARCKS) expression modulates the metastatic phenotype in human and murine colon carcinoma in vitro and in vivo. Cancer Lett. 2013;333:244-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 36] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 34. | Zhang B, Yang Y, Shi X, Liao W, Chen M, Cheng AS, Yan H, Fang C, Zhang S, Xu G, Shen S, Huang S, Chen G, Lv Y, Ling T, Zhang X, Wang L, Zhuge Y, Zou X. Proton pump inhibitor pantoprazole abrogates adriamycin-resistant gastric cancer cell invasiveness via suppression of Akt/GSK-β/β-catenin signaling and epithelial-mesenchymal transition. Cancer Lett. 2015;356:704-712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 68] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 35. | Zhang Q, Chao TC, Patil VS, Qin Y, Tiwari SK, Chiou J, Dobin A, Tsai CM, Li Z, Dang J, Gupta S, Urdahl K, Nizet V, Gingeras TR, Gaulton KJ, Rana TM. The long noncoding RNA ROCKI regulates inflammatory gene expression. EMBO J. 2019;38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 77] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Gupta S, Brazil; Tanabe S, Japan S-Editor: Wang JJ L-Editor: A P-Editor: Zhang XD