Published online Jul 15, 2023. doi: 10.4251/wjgo.v15.i7.1215
Peer-review started: February 1, 2023
First decision: March 21, 2023
Revised: March 31, 2023
Accepted: May 8, 2023
Article in press: May 8, 2023
Published online: July 15, 2023
Processing time: 161 Days and 2.5 Hours
Single-cell sequencing technology provides the capability to analyze changes in specific cell types during the progression of disease. However, previous single-cell sequencing studies on gastric cancer (GC) have largely focused on immune cells and stromal cells, and further elucidation is required regarding the alterations that occur in gastric epithelial cells during the development of GC.
To create a GC prediction model based on single-cell and bulk RNA sequencing (bulk RNA-seq) data.
In this study, we conducted a comprehensive analysis by integrating three single-cell RNA sequencing (scRNA-seq) datasets and ten bulk RNA-seq datasets. Our analysis mainly focused on determining cell proportions and identifying differentially expressed genes (DEGs). Specifically, we performed differential expression analysis among epithelial cells in GC tissues and normal gastric tissues (NAGs) and utilized both single-cell and bulk RNA-seq data to establish a prediction model for GC. We further validated the accuracy of the GC prediction model in bulk RNA-seq data. We also used Kaplan–Meier plots to verify the correlation between genes in the prediction model and the prognosis of GC.
By analyzing scRNA-seq data from a total of 70707 cells from GC tissue, NAG, and chronic gastric tissue, 10 cell types were identified, and DEGs in GC and normal epithelial cells were screened. After determining the DEGs in GC and normal gastric samples identified by bulk RNA-seq data, a GC predictive classifier was constructed using the Least absolute shrinkage and selection operator (LASSO) and random forest methods. The LASSO classifier showed good performance in both validation and model verification using The Cancer Genome Atlas and Genotype-Tissue Expression (GTEx) datasets [area under the curve (AUC)_min = 0.988, AUC_1se = 0.994], and the random forest model also achieved good results with the validation set (AUC = 0.92). Genes TIMP1, PLOD3, CKS2, TYMP, TNFRSF10B, CPNE1, GDF15, BCAP31, and CLDN7 were identified to have high importance values in multiple GC predictive models, and KM-PLOTTER analysis showed their relevance to GC prognosis, suggesting their potential for use in GC diagnosis and treatment.
A predictive classifier was established based on the analysis of RNA-seq data, and the genes in it are expected to serve as auxiliary markers in the clinical diagnosis of GC.
Core Tip: In this study, we integrated and analyzed three single-cell RNA sequencing datasets and 10 bulk RNA sequencing datasets of gastric cancer (GC) from the Gene Expression Omnibus database. We conducted a differential expression analysis of epithelial cell subpopulations from GC tissue and normal gastric mucosa tissue and constructed GC prediction classifiers using the Least absolute shrinkage and selection operator (LASSO) method and random forest method. The LASSO prediction model was further validated in the Cancer Genome Atlas stomach adenocarcinoma dataset. TIMP1, PLOD3, CKS2, TYMP, TNFRSF10B, CPNE1, GDF15, BCAP31, and CLDN7 were selected as the predictive genes for GC. This study provides a new approach for constructing prediction models based on single-cell sequencing data and offers new reference targets for the clinical diagnosis and treatment of GC.
- Citation: Wen F, Guan X, Qu HX, Jiang XJ. Integrated analysis of single-cell and bulk RNA-seq establishes a novel signature for prediction in gastric cancer. World J Gastrointest Oncol 2023; 15(7): 1215-1226
- URL: https://www.wjgnet.com/1948-5204/full/v15/i7/1215.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i7.1215
Gastric cancer (GC) is the second leading cause of cancer-related mortality globally[1-3]. Endoscopy remains the most prevalent and reliable method for GC diagnosis[3]. Nevertheless, due to the invasiveness of the procedure and the often asymptomatic nature of early-stage GC, patients are frequently diagnosed in advanced stages, resulting in poor survival and prognosis rates. Thus, the development of effective diagnostic methods and specific biomarkers for GC is urgently needed.
Serological markers and liquid biopsies (circulating tumor cells, circulating tumor DNA or RNA, microRNA, exosomes) are used to diagnose GC[2,4,5]. However, due to the small amount of circulating tumor cells and tumor DNA and the uneven distribution in the peripheral circulation, the repeatability of liquid biopsy is greatly limited[2,6]. Serological markers such as carcinoembryonic antigen (CEA), carbohydrate antigen 19-9 and carbohydrate antigen 72-4 are not sensitive enough to diagnose GC and have little importance in the diagnosis of early GC[6,7].
The tumor microenvironment (TME) consists of tumor cells and stromal cells, including fibroblasts, pericytes, mesenchymal stem cells, and various types of immune cells[8,9]. Tumorigenesis and progression result from the collective action of multiple cells[10]. Single-cell RNA sequencing (scRNA-seq) provides a promising avenue for under
Based on the scRNA-seq data, we identified genes that were differentially expressed in epithelial cell populations between normal gastric tissue (NAG) and GC tissue. Subsequently, using bulk RNA-seq data, we developed a predictive classifier. Our findings suggest that developing a prediction model for GC based on epithelial cells is a viable approach and that the results could serve as promising biomarkers for the diagnosis and prognosis of this disease.
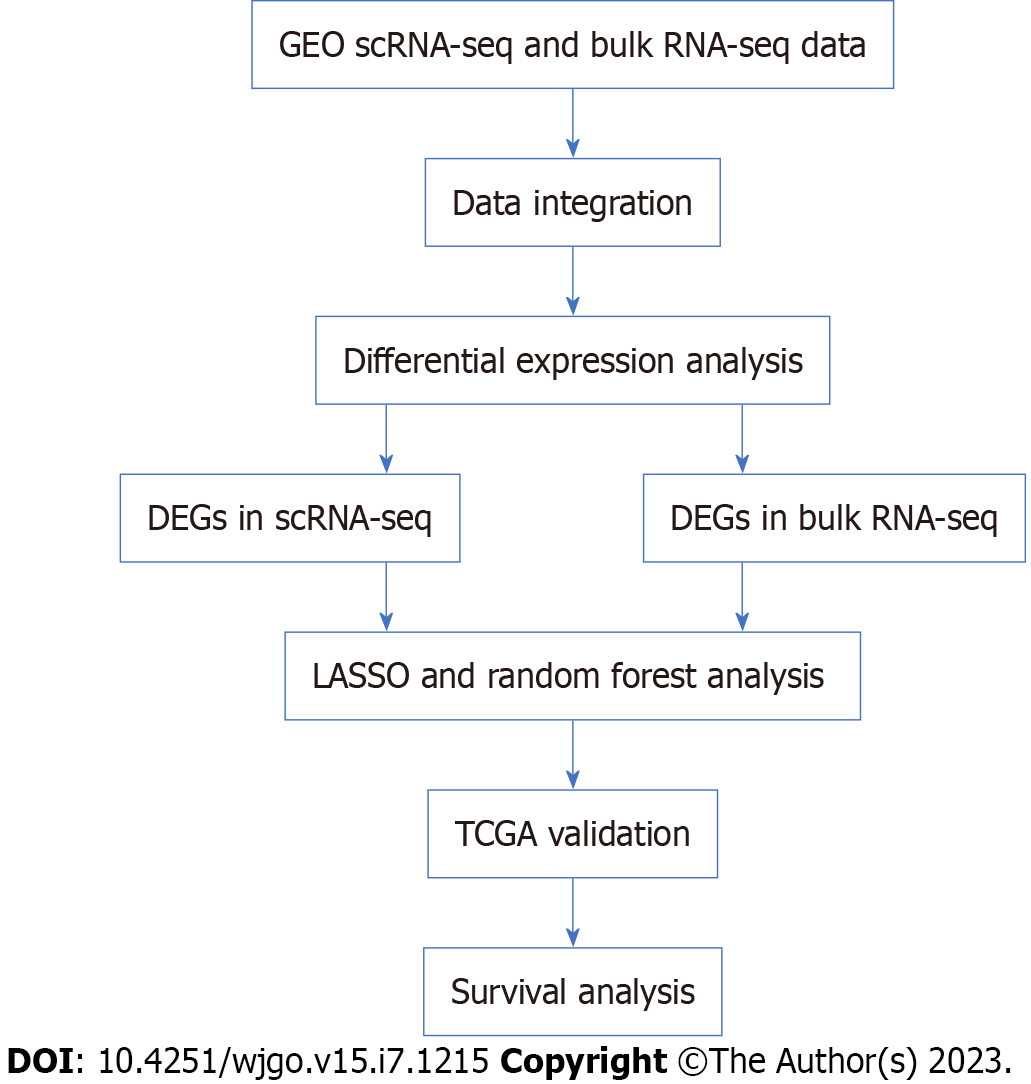
We utilized three scRNA-seq datasets (GSE134520, GSE183904, GSE150290) and ten bulk RNA-seq datasets (GSE79973, GSE66229, GSE64951, GSE57303, GSE38749, GSE35809, GSE34942, GSE19826, GSE13911, GSE15459) that were obtained from the Gene Expression Omnibus (GEO) website (https://www.ncbi.nlm.nih.gov/geo/). We used the stomach adenocarcinoma (STAD) dataset and Genotype-Tissue Expression (GTEx) dataset obtained from the University of California at Santa Cruz website (https://xenabrowser.net/datapages/). The study did not require ethical approval because the data we used came from a publicly accessible database. The workflow of this study is shown in Figure 1.
Cells with fewer than 7000 and more than 400 genes possessing less than 10% mitochondria and less than 20% ribosomes were retained. To ensure adequate data quality, samples with fewer than 800 cells were removed before data integration. Ultimately, a total of 34 samples from three datasets were used for data integration and subsequent analysis, including 2 cases of NAG, 3 cases of chronic atrophic gastritis (CAG), 7 cases of intestinal metaplasia (IM) and 22 cases of GC (13 cases of intestinal GC, 6 cases of diffuse GC and 3 cases of mixed GC) (Supplementary Table 1). For scRNA-seq data analysis, we utilized the Seurat package[13] (https://satijalab.org/seurat/; 4.3.0) and its related functions. We employed the RunUMAP function for dimensionality reduction (using the first 20 PCs), the FindClusters function for cell clustering (resolution = 1.2), and the FindAllMarkers function for differential gene expression analysis. Default parameter values were used for all other functions.
Ten GC chip sequencing datasets based on GPL570 were included in this study, including 834 GC samples and 187 NAG samples. The samples were processed using the robust multichip average algorithm to perform background correction and standardization. To mitigate the effects of batch variation, the COMBAT algorithm was utilized.
This paper includes the STAD data and the GTEx data. For both datasets, 'log2 (fpkm + 1)' data were used for subsequent analysis, and the normalizeBetweenArrays function was used to remove batch effects. The STAD dataset contained 375 GC samples and 32 paracancerous samples. The GTEx contains 174 samples of normal stomach tissue.
The FindMarkers function and the scCODE package[14] (https://github.com/XZouProjects/scCODE; version 1.0.1.0) were used to identify differentially expressed genes (DEGs) in scRNA-seq. The lmfit function was used to identify DEGs in the bulk RNA-seq data. Genes with a P value > 0.05 and an absolute logFC value greater than 0.5 were considered DEGs and subjected to functional enrichment analysis. The clusterProfiler package (version 4.2.0) was used to functionally annotate DEGs to identify significantly enriched Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways.
We conducted molecular interaction analysis utilizing the STRING database (https://cn.string-db.org/). To present the results, we utilized Cytoscape (https://cytoscape.org/).
We screened DEGs in epithelial cells obtained from GC and normal adjacent gastric tissue (NAG), preserving genes with logFC > 0.5 and detected-times = 5. These genes were then compared with DEGs identified in bulk RNA-seq data between GC and NAG (logFC > 0.5 and P value < 0.05) to obtain an overlapping set. Subsequently, we used these genes to build a LASSO regression model and random forest model in the GEO training set and verified them in the GEO test set and The Cancer Genome Atlas (TCGA)-GTEx dataset. The GEO data were randomly divided into a training set and test set in a 6:4 ratio. The LASSO model was established using the glmnet function (version 4.1-6). The randomForest function (version 4.7-1.1) was used to build the random forest model. Finally, we evaluated the relationship between the gene and GC survival rates using Kaplan–Meier plotter (http://kmplot.com/analysis/).
The molecular interactions were illustrated using Cytoscape software, while all other visualizations were created using ggplot2.
After applying quality control criteria, our analysis included 70707 cells that were classified into 43 clusters (Figure 2A). We assigned each cluster to a specific cell type based on cluster-specific genes and DEGs (Figure 2B-D): T cells (CD3D and CD3E), myeloid cells (C1QA, S100A8), mast cells (KIT, TPSAB1), B cells (CD79A), endothelial cells (VWF, PLVAP), epithelial cells (MUC5AC, EPCAM), chief cells (PGC, PGA3), endocrine cells (CHGA, GAST), fibroblasts (ACTA2, DCN), and SMCs (RGS5) (Figure 2B).
Analysis of cell composition revealed that the proportions of T cells, myeloid cells, fibroblasts, endothelial cells, and SMCs increased during the progression from nonatrophic gastritis to atrophic gastritis, intestinal metaplasia, and GC (Figure 2E and F). Both nonatrophic gastritis and atrophic gastritis without intestinal metaplasia exhibited a high proportion of epithelial cells (Figure 2F). There was no significant difference in cell composition among different Lauren subtypes of GC (Figure 2F).
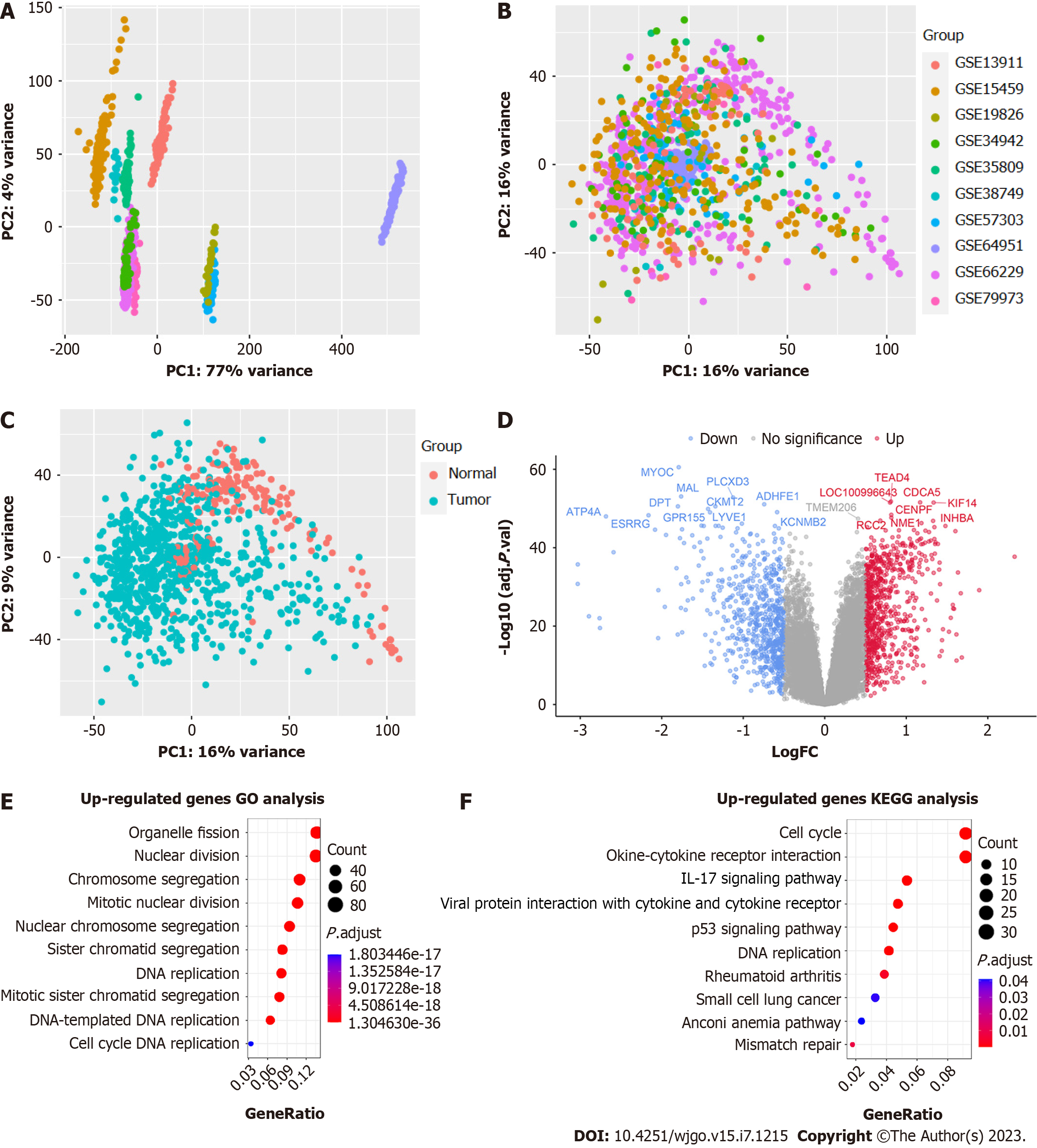
We performed an analysis of bulk RNA-seq data to identify genes that were differentially expressed. Our analysis involved the integration of 10 bulk RNA-seq datasets, and the principal component analysis (PCA) results before and after using COMBAT indicated that the batch effect was successfully eliminated (Figure 3A-C). Our differential expression analysis between GC and NAGs identified 757 genes that were highly expressed in GC tissues (P < 0.05, logFC > 0.5). We sorted the DEGs by ‘-log10 (P value)’ and displayed the top 20 genes in the volcano plot (Figure 3D). Enrichment analyses of highly expressed genes in GC tissues using GO and KEGG pathway databases showed that pathways related to cell proliferation, such as nuclear division and DNA replication, were enriched (Figure 3E). Additionally, we observed enrichment of pathways related to tumorigenesis, such as the p53 signaling pathway and IL17 signaling pathway (Figure 3E).
We screened for DEGs in epithelial cells obtained from GC and NAG, retaining 934 genes with logFC > 0.5 and detected-times = 5. Then, these genes were compared to the 757 DEGs identified between GC and NAG (logFC > 0.5 and P value < 0.05), resulting in an overlapping set of 69 genes. Among the 69 genes, EPCAM, CLDN7, CLDN3, and CLDN4, essential components of gastrointestinal tract, were found (Figure 4, Supplementary Table 2). Additionally, we identified immune-related genes, such as CEACAM6, MIF, C1QBP, EPCAM, TNFRSF10B, CXCL16 (Figure 4, Supplementary Table 2).
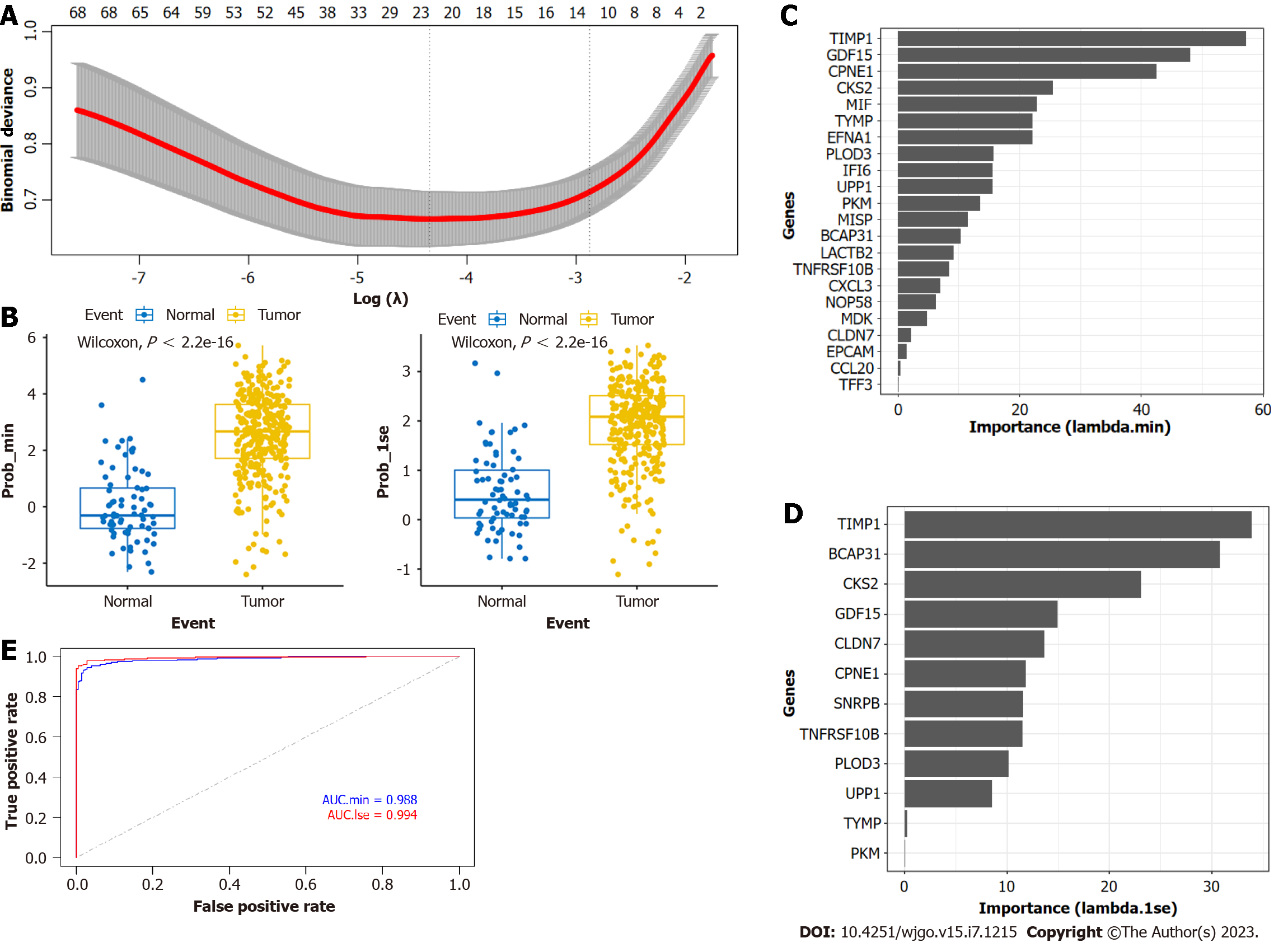
Using LASSO regression analysis, we selected "prob_min" and "prob_1se" to calculate the prediction model (Figure 5A). The "prob_min" model consisted of 22 genes, including CLDN7, TFF3, TYMP, PLOD3, NOP58, CCL20, IFI6, LACTB2, TNFRSF10B, CPNE1, PKM, EFNA1, GDF15, UPP1, MISP, TIMP1, EPCAM, CXCL3, MIF, MDK, CKS2, and BCAP31 (Supplementary Table 3). The "prob_1se" model included 12 genes, such as CLDN7, TYMP, PLOD3, TNFRSF10B, CPNE1, PKM, GDF15, UPP1, TIMP1, CKS2, BCAP31, and SNRPB (Supplementary Table 4). Notably, the CLDN7, TYMP, PLOD3, TNFRSF10B, CPNE1, PKM, GDF15, UPP1, TIMP1, CKS2, and BCAP31 genes were present in both models. We validated the performance of the model, and in the validation set, the model could effectively distinguish between tumor tissue and NAG (P < 0.01) (Figure 5B). We ranked the importance values of the feature genes in the model. In the "prob_min" model, TIMP1, GDF15, CPNE1, CKS2, and MIF had the highest importance values (Figure 5C); in the "prob_1se" model, TIMP1, BCAP31, CKS2, GDF15, and CLDN7 had the highest importance values (Figure 5D). Using the TCGA and GTEx datasets for validation, the "prob_min" model had an AUC of 0.988, and the "prob_1se" model had an AUC of 0.994 (Figure 5E). These results suggest that both LASSO models have good predictive performance.
We first applied the Boruta function to further screen the feature genes and sorted them based on their importance values (Importance) (Figure 6A). A total of 57 genes were defined as 'confirmed' and entered the next step of constructing the random forest model as the feature set. We used Caret for hyperparameter tuning and chose mtry = 9 (Figure 6B) to build the final model. The contribution values of each feature in the final model are shown in Figure 6C, where SNRBP, TIMP1, GDF15, PLOD3, and CKS2 had the highest contribution values. Compared with the LASSO model, TIMP1,
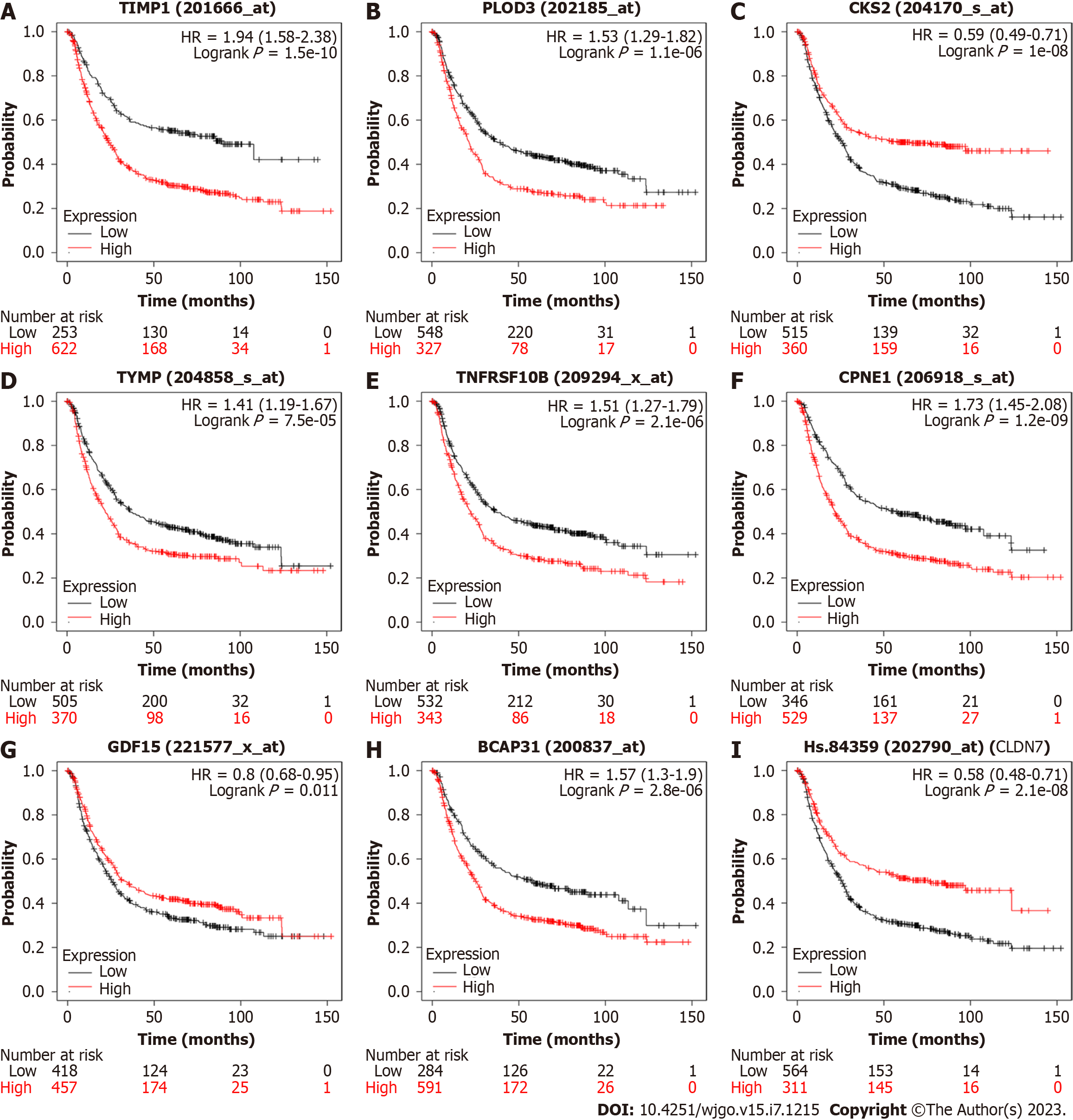
We analyzed the important genes in the model using KM-PLOTTER (Figure 7) and found that several GC-related genes, such as TIMP1, PLOD3, CKS2, TYMP, TNFRSF10B, CPNE1, GDF15, BCAP31, and CLDN7, were associated with the prognosis of GC. Among them, CKS2, CLDN7, and GDF15 were positively correlated with the survival time of GC patients, while the other genes were negatively correlated.
This study combined single-cell sequencing data and bulk RNA-seq data to identify GC-specific genes and construct a GC prediction model.
Our study combined single-cell sequencing data and bulk RNA-seq data to identify GC-specific genes and construct a GC prediction model. Our GC prediction model suggests that genes such as TIMP1, CKS2, and GDF15 have potential for the clinical diagnosis of GC. TIMP1 belongs to the TIMP gene family and encodes a natural inhibitor of matrix metalloproteinases, which can promote tumor cell proliferation and may also have antiapoptotic functions[15,16]. Studies have shown that TIMP6 and TIMP8 can be used as diagnostic markers for colorectal cancer, while the significance of other members of the TIMP family in cancer diagnosis remains unclear[17]. Our study proposes for the first time that TIMP1 may be a diagnostic marker for GC. TYMP is highly expressed in various solid tumors compared to adjacent noncancerous tissues, and research has found it to be related to tumor angiogenesis and immune regulation[18,19], but its importance in cancer diagnosis is not yet clear. GDF15 controls hematopoietic growth, energy homeostasis, adipose tissue metabolism, organismal growth, bone remodeling, and response to stress signals, and its role in cancer development and progression is complex[20]. Studies have shown that GDF15 can be used as a diagnostic marker for early-stage liver cancer[21,22]. BCAP31 is associated with the proliferation and metastasis of breast cancer, lung cancer and other tumors[23,24]. Based on our study, using the above genes as markers for predicting or diagnosing GC has potential feasibility, but further validation is required through experimental and clinical exploration.
The rapid development of scRNA-seq technology has enabled researchers to explore the molecular characteristics of cells in TME. However, most of this work has focused on immune cells and mesenchymal cells[25,26], and the study of epithelial cells has not received enough attention. Our study analyzed the DEGs of GC from the perspective of epithelial cells for the first time and identified GC-specific genes. However, our study did not carry out cytological and histological verifications, and the clinical guidance significance of the above feature genes needs further exploration.
Our study confirmed that combining single-cell sequencing technology with bulk RNA-seq technology to analyze GC-related marker genes from the perspective of cell subpopulations is feasible. However, during the study, we observed that technical noise and batch effects from single-cell sequencing affected the results (such as a small number of cells from the epithelial cell subpopulation mixing into the T/B-cell group). We also observed that the sequencing results were more enriched in immune cells, while the loss of epithelial cells was significant, especially in tumor tissues. The reasons for these limitations are related to many factors, such as the high sequencing depth of single-cell sequencing introducing technical noise, mechanical damage to cells during sample processing, and differences in cell size and morphology. Studies have shown that single-nucleus RNA sequencing (snRNA-seq) has a significant advantage over single-cell sequencing in identifying epithelial cells[27,28]. On the basis of existing studies, the inclusion of snRNA-seq results may supplement the findings of this study and provide better clinical guidance.
In summary, we have successfully established a predictive classifier based on the analysis of RNA-seq data, and the genes included in it are expected to serve as auxiliary markers in the clinical diagnosis of GC. This research achievement provides valuable references and guidance for the early diagnosis and treatment of GC.
Improving early diagnosis rates of gastric cancer (GC) is of great importance for reducing GC-related deaths. This study aimed to construct a predictive model for GC by integrating single-cell sequencing data and bulk RNA sequencing (bulk RNA-seq) data to identify potential targets for GC prediction.
Identifying predictive targets for GC is an important approach to reduce GC-related deaths, which is the driving force behind this study.
The objective of this study was to develop a predictive model for GC by combining single-cell sequencing data and bulk RNA-seq data and to identify potential targets for predicting GC.
We downloaded GC single-cell sequencing and bulk RNA-seq datasets from the Gene Expression Omnibus and University of California at Santa Cruz databases. The single-cell sequencing data were analyzed using the Seurat package, and the bulk RNA-seq data were analyzed using the limma package. The construction of the GC prediction model was based on the Least absolute shrinkage and selection operator (LASSO) and random forest methods. Survival analysis was conducted using the KM-PLOTTER online database.
By analyzing single-cell RNA sequencing data from 70707 cells from GC tissue, normal gastric tissue, and chronic gastric tissue, we identified 10 different cell types and screened for genes differentially expressed between GC and normal epithelial cells. After determining differentially expressed genes identified from batch RNA sequencing data of GC and normal gastric samples, we constructed a GC prediction classifier using LASSO and random forest methods. The LASSO classifier performed well when validated and when the model was verified using The Cancer Genome Atlas and Genotype-Tissue Expression datasets [area under the curve (AUC)_min = 0.988, AUC_1se = 0.994], and the random forest model also achieved good results with the validation set (AUC = 0.92). We identified genes such as TIMP1, PLOD3, CKS2, TYMP, TNFRSF10B, CPNE1, GDF15, BCAP31, and CLDN7 with significant importance in multiple GC prediction models, and KM-PLOTTER analysis showed their relevance to GC prognosis, indicating their potential value in GC diagnosis and treatment. However, the limitation of our study is the lack of clinical sample validation for the GC prediction models.
This study demonstrates that the combination of single-cell sequencing data and bulk RNA-seq data is feasible for constructing a GC prediction model.
Using single-nucleus sequencing to assist in constructing GC prediction models may lead to more reliable results, as it has advantages in identifying epithelial cells.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B, B
Grade C (Good): C, C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Cheng YH, United States; El-Arabey AA, Egypt; Emran TB, Bangladesh; Li Q, China S-Editor: Li L L-Editor: Filipodia P-Editor: Ju JL
| 1. | Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4235] [Cited by in RCA: 11903] [Article Influence: 2975.8] [Reference Citation Analysis (9)] |
| 2. | Ma S, Zhou M, Xu Y, Gu X, Zou M, Abudushalamu G, Yao Y, Fan X, Wu G. Clinical application and detection techniques of liquid biopsy in gastric cancer. Mol Cancer. 2023;22:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 125] [Reference Citation Analysis (0)] |
| 3. | Joshi SS, Badgwell BD. Current treatment and recent progress in gastric cancer. CA Cancer J Clin. 2021;71:264-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 382] [Cited by in RCA: 1172] [Article Influence: 234.4] [Reference Citation Analysis (9)] |
| 4. | Izumi D, Zhu Z, Chen Y, Toden S, Huo X, Kanda M, Ishimoto T, Gu D, Tan M, Kodera Y, Baba H, Li W, Chen J, Wang X, Goel A. Assessment of the Diagnostic Efficiency of a Liquid Biopsy Assay for Early Detection of Gastric Cancer. JAMA Netw Open. 2021;4:e2121129. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (1)] |
| 5. | Chen H, Huang C, Wu Y, Sun N, Deng C. Exosome Metabolic Patterns on Aptamer-Coupled Polymorphic Carbon for Precise Detection of Early Gastric Cancer. ACS Nano. 2022;16:12952-12963. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 50] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 6. | Ma X, Ou K, Liu X, Yang L. Application progress of liquid biopsy in gastric cancer. Front Oncol. 2022;12:969866. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 7. | Xu Y, Zhang P, Zhang K, Huang C. The application of CA72-4 in the diagnosis, prognosis, and treatment of gastric cancer. Biochim Biophys Acta Rev Cancer. 2021;1876:188634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 68] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 8. | Nallasamy P, Nimmakayala RK, Parte S, Are AC, Batra SK, Ponnusamy MP. Tumor microenvironment enriches the stemness features: the architectural event of therapy resistance and metastasis. Mol Cancer. 2022;21:225. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 148] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 9. | Sherman MH, Beatty GL. Tumor Microenvironment in Pancreatic Cancer Pathogenesis and Therapeutic Resistance. Annu Rev Pathol. 2023;18:123-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 272] [Article Influence: 90.7] [Reference Citation Analysis (0)] |
| 10. | El-Arabey AA, Abdalla M, Abd-Allah AR. SnapShot: TP53 status and macrophages infiltration in TCGA-analyzed tumors. Int Immunopharmacol. 2020;86:106758. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 57] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 11. | Bridges K, Miller-Jensen K. Mapping and Validation of scRNA-Seq-Derived Cell-Cell Communication Networks in the Tumor Microenvironment. Front Immunol. 2022;13:885267. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 12. | Liu Z, Li H, Dang Q, Weng S, Duo M, Lv J, Han X. Integrative insights and clinical applications of single-cell sequencing in cancer immunotherapy. Cell Mol Life Sci. 2022;79:577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 13. | Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, Hoffman P, Stoeckius M, Papalexi E, Mimitou EP, Jain J, Srivastava A, Stuart T, Fleming LM, Yeung B, Rogers AJ, McElrath JM, Blish CA, Gottardo R, Smibert P, Satija R. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573-3587.e29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7339] [Cited by in RCA: 9859] [Article Influence: 1971.8] [Reference Citation Analysis (0)] |
| 14. | Zou J, Deng F, Wang M, Zhang Z, Liu Z, Zhang X, Hua R, Chen K, Zou X, Hao J. scCODE: an R package for data-specific differentially expressed gene detection on single-cell RNA-sequencing data. Brief Bioinform. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 15. | Jackson HW, Defamie V, Waterhouse P, Khokha R. TIMPs: versatile extracellular regulators in cancer. Nat Rev Cancer. 2017;17:38-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 241] [Cited by in RCA: 358] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 16. | Grünwald B, Schoeps B, Krüger A. Recognizing the Molecular Multifunctionality and Interactome of TIMP-1. Trends Cell Biol. 2019;29:6-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 126] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 17. | Łukaszewicz-Zając M, Mroczko B. Circulating Biomarkers of Colorectal Cancer (CRC)-Their Utility in Diagnosis and Prognosis. J Clin Med. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 18. | Paladhi A, Daripa S, Mondal I, Hira SK. Targeting thymidine phosphorylase alleviates resistance to dendritic cell immunotherapy in colorectal cancer and promotes antitumor immunity. Front Immunol. 2022;13:988071. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 18] [Reference Citation Analysis (0)] |
| 19. | Gu Y, Guo Y, Gao N, Fang Y, Xu C, Hu G, Guo M, Ma Y, Zhang Y, Zhou J, Luo Y, Zhang H, Wen Q, Qiao H. The proteomic characterization of the peritumor microenvironment in human hepatocellular carcinoma. Oncogene. 2022;41:2480-2491. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 20. | Siddiqui JA, Pothuraju R, Khan P, Sharma G, Muniyan S, Seshacharyulu P, Jain M, Nasser MW, Batra SK. Pathophysiological role of growth differentiation factor 15 (GDF15) in obesity, cancer, and cachexia. Cytokine Growth Factor Rev. 2022;64:71-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 116] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 21. | Zhang S, Gao C, Zhou Q, Chen L, Huang GF, Tang H, Song X, Zhang Z, Whittaker K, Chen X, Huang RP. Identification and validation of circulating biomarkers for detection of liver cancer with antibody array. Neoplasma. 2023;70:36-45. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 22. | Guo H, Liu Q. Clinical Value of Growth Differentiation Factor 15 Detection in the Diagnosis of Early Liver Cancer Based on Data Mining. Biomed Res Int. 2022;2022:4448075. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 23. | Fu W, Sun H, Zhao Y, Chen M, Yang X, Liu Y, Jin W. BCAP31 drives TNBC development by modulating ligand-independent EGFR trafficking and spontaneous EGFR phosphorylation. Theranostics. 2019;9:6468-6484. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 24. | Wang J, Jiang D, Li Z, Yang S, Zhou J, Zhang G, Zhang Z, Sun Y, Li X, Tao L, Shi J, Lu Y, Zheng L, Song C, Yang K. BCAP31, a cancer/testis antigen-like protein, can act as a probe for non-small-cell lung cancer metastasis. Sci Rep. 2020;10:4025. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 25. | Yu L, Shen N, Shi Y, Shi X, Fu X, Li S, Zhu B, Yu W, Zhang Y. Characterization of cancer-related fibroblasts (CAF) in hepatocellular carcinoma and construction of CAF-based risk signature based on single-cell RNA-seq and bulk RNA-seq data. Front Immunol. 2022;13:1009789. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 98] [Article Influence: 24.5] [Reference Citation Analysis (1)] |
| 26. | Yang J, Zhang J, Na S, Wang Z, Li H, Su Y, Ji L, Tang X, Yang J, Xu L. Integration of single-cell RNA sequencing and bulk RNA sequencing to reveal an immunogenic cell death-related 5-gene panel as a prognostic model for osteosarcoma. Front Immunol. 2022;13:994034. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 27. | Cuevas-Diaz Duran R, González-Orozco JC, Velasco I, Wu JQ. Single-cell and single-nuclei RNA sequencing as powerful tools to decipher cellular heterogeneity and dysregulation in neurodegenerative diseases. Front Cell Dev Biol. 2022;10:884748. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 45] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 28. | Kim N, Kang H, Jo A, Yoo SA, Lee HO. Perspectives on single-nucleus RNA sequencing in different cell types and tissues. J Pathol Transl Med. 2023;57:52-59. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 46] [Reference Citation Analysis (0)] |