Published online Dec 15, 2023. doi: 10.4251/wjgo.v15.i12.2225
Peer-review started: August 27, 2023
First decision: October 18, 2023
Revised: October 23, 2023
Accepted: November 17, 2023
Article in press: November 17, 2023
Published online: December 15, 2023
Processing time: 108 Days and 21.5 Hours
The MBOAT7 rs641738 single-nucleotide polymorphism (SNP) has been proven to influence various liver diseases, but its association with hepatocellular carcinoma (HCC) susceptibility has been debated. To address this discrepancy, we conducted the current systematic review and meta-analysis.
To perform a systematic review and meta-analysis on association of MBOAT7 SNP and HCC susceptibility.
We performed a systematic review in PubMed, Web of Science, Scopus, and EMBASE; applied specific inclusion and exclusion criteria; and extracted the data. Meta-analysis was conducted with the meta package in R. Sensitivity and subgroup analyses were also performed. This meta-analysis was registered in PROSPERO (CRD42023458046).
Eight studies were included in the systematic review, and 12 cohorts from 6 studies were included in the meta-analysis. Our meta-analysis revealed an association between the MBOAT7 SNP and HCC susceptibility in both the dominant [odds ratio (OR): 1.14, 95% confidence interval (95%CI): 1.02-1.26, P = 0.020] and recessive (OR: 1.21, 95%CI: 1.05-1.39, P = 0.008) models. Subgroup analysis revealed that stratification of the included patients by geographical origin showed a significant association in Asia (OR: 1.20, 95%CI: 1.03-1.39).
This meta-analysis underscores the contribution of the MBOAT7 rs641738 SNP to hepatocarcinogenesis, especially in Asian populations, which warrants further investigation.
Core Tip: We conducted the current systematic review and meta-analysis to address the association between MBOAT7 rs641738 SNP and HCC susceptibility. We demonstrated the correlation between MBOAT7 rs641738 SNP and HCC susceptibility both in the dominant and recessive models, and subgroup analysis revealed the significant association especially in Asia populations, which could guide clinical practice in the identification of at-risk population.
- Citation: Lai M, Qin YL, Jin QY, Chen WJ, Hu J. Association of MBOAT7 rs641738 polymorphism with hepatocellular carcinoma susceptibility: A systematic review and meta-analysis. World J Gastrointest Oncol 2023; 15(12): 2225-2236
- URL: https://www.wjgnet.com/1948-5204/full/v15/i12/2225.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i12.2225
Hepatocellular carcinoma (HCC), which ranks as the fourth leading cause of cancer-related mortality worldwide, presents a substantial challenge in the health care landscape[1]. Over 80% of HCC cases occur in low-resource and middle-resource countries, particularly in eastern Asia, where medical and social care resources are often constrained[2,3]. To address this challenge, early detection, improved management, and careful monitoring of high-risk populations are essential strategies.
Germline DNA single-nucleotide polymorphisms (SNPs) likely represent etiology-specific host factors that determine HCC susceptibility, including SNPs within PNPLA3 (patatin like phospholipase domain containing 3), TM6SF2 (transmembrane 6 superfamily 2), and MBOAT7 (membrane-bound O-acyltransferase domain-containing 7)[4]. The association between the PNPLA3 rs738409 SNP and HCC has already been demonstrated by Singal et al[5], who indicated that PNPLA3 is an independent risk factor for HCC. Furthermore, the association between the TM6SF2 rs58542926 SNP and HCC has been illustrated by Tang et al[6], who also suggested a significant association of the TM6SF2 SNP with HCC risk.
Interestingly, another previously studied SNP is the missense rs641738 C > T variant positioned downstream of the MBOAT7 locus. The MBOAT7 rs641738 SNP has been proven to influence histological liver damage in alcoholic liver disease, nonalcoholic fatty liver disease (NAFLD), hepatitis C, and hepatitis B[7]. However, its association with HCC has been debated. Thabet et al[8] performed a large case-control study in patients with hepatitis C virus (HCV)-related HCC and found that the role of rs641738 is limited to the early stages of liver disease but not to further progression or occurrence of HCC. Conversely, Donati et al[9] found that the MBOAT7 rs641738 T allele may predispose patients without cirrhosis to HCC.
To address this discrepancy, we conducted the current systematic review and meta-analysis on the association of the MBOAT7 SNP and HCC susceptibility, aiming to provide an updated and comprehensive assessment of the evolving evidence in this area.
We performed a systematic review using the following search strategy in four different databases (PubMed, Web of Science, Scopus, and EMBASE, searched in August 2023): (1): Neoplasms, Hepatic OR Neoplasms, Liver OR Liver Neoplasm OR Neoplasm, Liver OR Hepatic Neoplasms OR Hepatic Neoplasm OR Neoplasm, Hepatic OR Cancer of Liver OR Hepatocellular Cancer OR Cancers, Hepatocellular OR Hepatocellular Cancers OR Hepatic Cancer OR Cancer, Hepatic OR Cancers, Hepatic OR Hepatic Cancers OR Liver Cancer OR Cancer, Liver OR Cancers, Liver OR Liver Cancers OR Cancer of the Liver OR Cancer, Hepatocellular; (2): Membrane bound O-acyltransferase domain-containing 7 protein, human OR MBOAT7 protein, human OR MBOAT7 OR membrane bound O-acyltransferase domain-containing 7; and (3): (1) AND (2).
The inclusion criteria were as follows: (1) Case-control or cohort study evaluating the association between MBOAT7 rs641738 and HCC risk; and (2) reported odds ratios (ORs) and 95% confidence intervals (95%CIs) and/or allele frequencies and/or genotypes. If duplicate research reports were retrieved, the most comprehensive report was selected to avoid repeated statistics. Studies were excluded if they met one of the following criteria: (1) Review, comment, or conference abstract; and (2) insufficient data to estimate OR and 95%CI.
The following data were extracted from each included study: Author, publication year, country, cohort characteristics, sample size, genotype distribution, allele distribution, minor allele frequency, genotyping method, genetic models, adjustment, and Hardy-Weinberg equilibrium (HWE) data. Two authors independently assessed the studies, and disagreements were resolved through discussion with a third author.
We further conducted a meta-analysis using the cohorts of the included studies. We used the HardyWeinberg package (1.7.5) to calculate HWE in the control group of each cohort and performed the meta-analysis using the "metabin", "metagen", and "metainf" functions of the meta package (6.5-0) in R (4.3.0). Between-study heterogeneity was examined using the Q test and quantified using I2. If I2 > 50%, heterogeneity was considered significant, and a random-effects model was applied. Otherwise, a common-effects model was used. Sensitivity analysis was carried out by re-estimating pooled ORs and 95%CIs after excluding each eligible study in turn to assess the stability of the pooled results. Publication bias was evaluated using a funnel plot.
This meta-analysis was performed according to the guidelines of the PRISMA[10]. The meta-analysis was registered in PROSPERO (CRD42023458046).
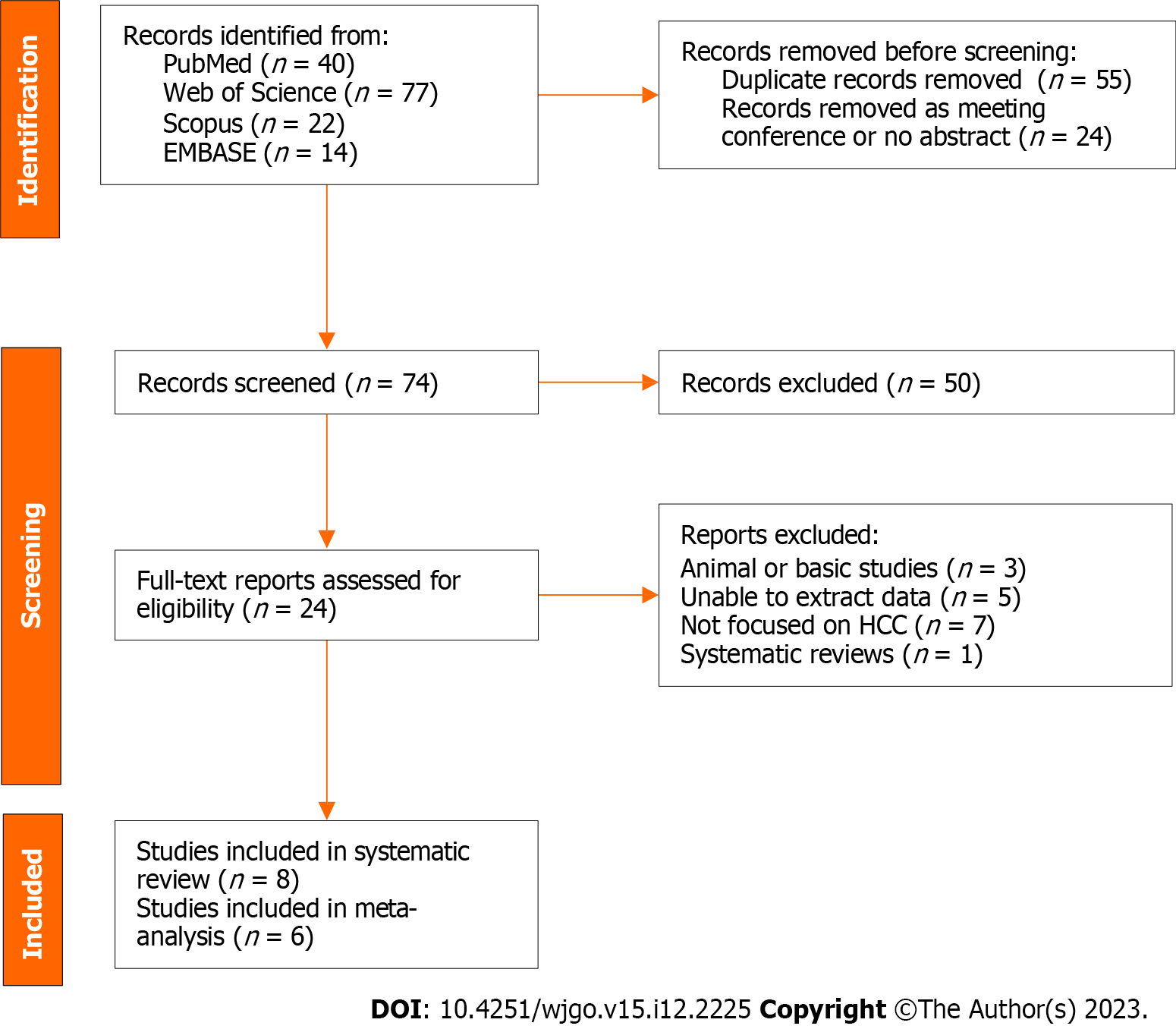
A total of 153 records were found in four different databases; 40 records were found in PubMed, 77 in the Web of Science, 22 in Scopus, and 14 in EMBASE. After removing 55 duplicates and 24 meeting conferences or records without abstracts, we rigorously reviewed the titles and abstracts of the remaining 74 records. We assessed 24 full-text reports for eligibility and included 8 studies in the systematic review (Figure 1).
Detailed information on the included studies is shown in Table 1. Our systematic review encompassed a comprehensive analysis of eight studies, spanning from 2016 to 2022, that explored the connection between the MBOAT7 rs641738 polymorphism and HCC. Notably, the majority of these studies (n = 6) were conducted outside of Asia. Of these, one study[8] concentrated solely on HCV-related HCC, while another[11] exclusively addressed alcohol-related HCC. Furthermore, two studies focused on HCC arising from NAFLD[12] or metabolic dysfunction-associated fatty liver disease[13], while another two[9,14] investigated a mixed cohort of NAFLD, viral, and alcohol-related HCC. One study[15] encompassed HCC occurring in the context of compensated cirrhosis (HCV or alcohol), and one[16] did not specify the particular characteristics of the included patient cohort. The genetic analysis techniques employed were diverse, with the TaqMan genotyping method being utilized in most studies (n = 5), with one study employing the MassARRAY[16] and two[12,13] employing United Kingdom Biobank Axiom array methodologies. A subset of studies (n = 4) explicitly delineated the genetic models employed in their analysis. Specifically, two studies[9,11] employed additive models, one[8] adopted a dominant model, and another[16] performed an investigation across dominant, additive, recessive, and allelic models. Conversely, the remaining four studies did not expound upon the genetic models utilized.
| Ref. | Country | Cohort characteristics | Study design | Genotyping method | Genetic model(s) | Main results | Conclusion |
| Thabet et al[8], 2016 | Multi-countries | HCV-related HCC | 1706 with chronic HCV infection, divided into two cohorts: Discovery cohort (n = 931) and validation cohort (n = 775) | TaqMan | Dominant model | No significant association was observed with HCC (OR: 0.96; 95%CI: 0.58-1.57) | The role of rs641738 is limited to the early stages of liver disease, but not to further progression or occurrence of HCC |
| Donati et al[9], 2017 | Multi-countries | NAFLD, HCV, and alcohol-related HCC | Italian (n = 765) and United Kingdom (n = 358) NAFLD patients; combined cohort of chronic hepatitis C (n = 597) or alcoholic liver disease (n = 524) | TaqMan | Additive models | In Italian NAFLD patients, the T allele was associated with NAFLD-HCC (OR: 1.65, 95%CI: 1.08-2.55; n = 765). In United Kingdom & Italian NAFLD cohort with non-cirrhotic NAFLD, the T allele remained associated with HCC (OR: 2.10, 95%CI: 1.33-3.31; n = 913). In combined cohort of chronic hepatitis C or alcoholic liver disease, the T allele was independently associated with HCC risk (OR: 1.93, 95%CI: 1.07-3.58; n = 1121) | The MBOAT7 rs641738 T allele may predispose to HCC in patients without cirrhosis |
| Stickel et al[11], 2018 | Multi-countries | Alcohol-related cirrhotic HCC | 751 cases with alcohol-related cirrhosis and HCC and 1165 controls with alcohol-related cirrhosis without HCC | TaqMan | Additive models | The risk associated with carriage of MBOAT7 rs641738 was not significant (OR: 1.04, 95%CI: 0.88-1.24) | Neither heterozygous nor homozygous carriage of the MBOAT7 rs641738 T allele was associated with HCC risk |
| Raksayot et al[14], 2019 | Thailand | HBV, HCV, and NBNC-related HCC | Healthy controls (n = 105), HBV-related HCC (n = 270), HCV-related HCC (n = 131), and NBNC-related HCC (n = 129) | TaqMan | NA | The genotype distribution and T allele frequencies of MBOAT7 rs641738 were similar between groups (NBNC-HCC vs NBNC-cirrhosis OR: 0.86, 95%CI: 0.51-1.46) | The data did not reveal any association between MBOAT7 rs641738 and the development of NBNC-HCC |
| Wang et al[16], 2021 | China | Unspecified | 779 HCC cases and 1412 cancer-free controls (controls consist of 678 persistent HBV carriers and 734 spontaneously recovered subjects) | MassARRAY | Dominant, additive, recessive, and allelic models | The results suggested no association between MBOAT7-TMC4 rs641738 and HCC risk in most genetic models | The work highlights that MBOAT7-TMC4 rs641738 is not associated with the risk of HCC |
| Bianco et al[12], 2021 | Multi-countries | NAFLD-related HCC | At-risk individuals (NAFLD cohort, n = 2566 and a replication cohort of 427 German patients with NAFLD). The general population (UKBB cohort, n = 364048). | TaqMan & United Kingdom BiLEVE and UKBB Axiom array | NA | In NAFLD cohort, OR: 1.0, 95%CI: 0.7-1.5; in overall UKBB, OR: 1.3, 95%CI: 0.9-1.8; in non-viral UKBB, OR: 1.3, 95%CI 0.9-1.9 | Variants in PNPLA3-TM6SF2-GCKR-MBOAT7 were combined in a hepatic fat PRS and PRS predicted HCC more robustly than single variants |
| Liu et al[13], 2022 | United Kingdom | MAFLD-related HCC | 160979 participants were diagnosed as having MAFLD | United Kingdom BiLEVE and UKBB Axiom array | NA | Model 2: Adjusted for gender, age, assessment center, genotyping chip, smoking status, physical activity level, overall health rating, average household income, and alcohol consumption. CT vs CC: OR: 1.16, 95%CI: 0.84-1.62; TT vs CC: OR: 1.36, 95%CI: 0.95-1.95 | MAFLD is independently associated with an increased risk of both intrahepatic and extrahepatic events. The impact of MAFLD on hepatic health events was amplified by variants in fatty liver disease related genes, among which the genetic variations in PNPLA3, TM6SF2, and MBOAT7 play prominent roles |
| Nahon et al[15], 2023 | French | Compensated cirrhosis (HCV or alcohol)-related HCC | Cohort 1 (n = 659): Compensated cirrhosis with HCV sustained virologic response. Cohort 2 (n = 486): Compensated alcohol-related cirrhosis | TaqMan | NA | In HCV-cured cohort, CT/TT vs CC: Subhazard ratio: 1.43, 95%CI: 0.68-3.01; In alcohol cohort, CT/TT vs CC: Subhazard ratio: 1.83, 95%CI: 0.85-3.94 (Fine-Gray regression modelling) | A 7-SNP genetic risk score was established, which contains PNPLA3, TM6SF2, HSD17B13, APOE, MBOAT7, and WNT3A-WNT9A variants |
Overall, the prevailing trend in the majority of studies (n = 7) indicated a lack of significant association between the MBOAT7 rs641738 variant and HCC. Among these, Thabet et al[8] examined a cohort of 1706 patients with chronic HCV infection, suggesting that the role of rs641738 was confined to the early stages of liver disease rather than the progression or emergence of HCC. In contrast, Donati et al[9] conducted an investigation on 765 Italian and 358 United Kingdom NAFLD patients, and they postulated that the MBOAT7 rs641738 T allele might predispose individuals to HCC in the absence of cirrhosis. Notably, they discovered that the T allele was autonomously associated with HCC risk in a combined cohort characterized by chronic HCV infection or alcoholic liver disease. To reconcile these divergent findings, both Stickel et al[11] and Raksayot et al[14] pursued separate inquiries, focusing on alcohol-related and virus-related HCC. Intriguingly, both studies arrived at unanimous negative outcomes. In a similar vein, Wang et al[16] undertook a case-control study with unspecified cohort attributes, and they predominantly identified negative associations across various genetic models. The remaining three[12,13,15] studies adopted the polygenic risk score (PRS) approach, which encom
In the meta-analysis, 12 cohorts from 6 studies were rigorously assessed to elucidate the connection between the MBOAT7 rs641738 polymorphism and HCC. To ensure data quality, we excluded the Italian NAFLD-related and HCV-related HCC cohorts from the study by Donati et al[9] due to HWE P values < 0.05. Additionally, we excluded the alcohol-related HCC cohort from the same study due to limited sample size (n = 12). Employing the "metabin" function, we performed calculations for ORs and 95%CIs, except for the combined Italian and United Kingdom NAFLD-related HCC cohort and the United Kingdom Biobank (nonviral) cohort in the study by Bianco et al[12], in which genotype counts were unspe
| Ref. | Country | Cohort characteristics | Cases | Controls | Genotype (CC/CT/TT) | Allele (C/T) | Minor allele frequency | Adjustment | HWE of control (P value) | |||
| Case | Control | Case | Control | Case | Control | |||||||
| Thabet et al[8], 2016 | Multi | HCV-related HCC | 75 | 1706 | 24/35/16 | 531/822/353 | 83/67 | 1884/1528 | 0.45 (T) | 0.45 (T) | Age, gender, BMI, and Child-Pugh score | 0.305 |
| Donati et al[9], 2017 | Italian | NAFLD-related HCC | 132 | 633 | 26/69/37 | 213/285/135 | 121/143 | 711/555 | 0.46 (C) | 0.44 (T) | Age, gender, obesity, T2DM, and presence of advanced fibrosis | 0.036 |
| Donati et al[9], 2017 | Italian & United Kingdom | Non-cirrhotic NAFLD-related HCC | 41 | 872 | 5/21/15 | 280/422/170 | 31/51 | 982/762 | 0.38 (C) | 0.44 (T) | NA | 0.664 |
| Donati et al[9], 2017 | Italian | HCV-related HCC | 13 | 584 | 1/7/5 | 177/259/148 | 9/17 | 613/555 | 0.35 (C) | 0.48 (T) | Age, gender, and genetic variants | 0.009 |
| Donati et al[9], 2017 | Italian | Alcohol-related HCC | 12 | 512 | 1/8/3 | 150/251/111 | 10/14 | 551/473 | 0.42 (C) | 0.46 (T) | Age, gender, and genetic variants | 0.805 |
| Stickel et al[11], 2018 | Switzerland, Germany & United Kingdom | Alcohol-related cirrhotic HCC | 751 | 1165 | 203/363/185 | 314/583/268 | 769/733 | 1211/1119 | 0.49 (T) | 0.48 (T) | Age, gender, BMI, and T2DM | 0.967 |
| Raksayot et al[14], 2019 | Thailand | HBV-related HCC | 270 | 105 | 140/114/16 | 66/34/5 | 394/146 | 166/44 | 0.27 (T) | 0.21 (T) | NA | 0.943 |
| Raksayot et al[14], 2019 | Thailand | HCV-related HCC | 131 | 105 | 71/53/7 | 66/34/5 | 195/67 | 166/44 | 0.26 (T) | 0.21 (T) | NA | 0.943 |
| Raksayot et al[14], 2019 | Thailand | NBNC-related HCC (containing NAFLD and alcoholic liver disease) | 129 | 105 | 68/46/15 | 66/34/5 | 182/76 | 166/44 | 0.30 (T) | 0.21 (T) | NA | 0.943 |
| Wang et al[16], 2021 | China | Unspecified | 779 | 1405 | 426/295/58 | 800/528/77 | 1147/411 | 2128/682 | 0.26 (T) | 0.24 (T) | Age, gender, smoking status, and drinking status | 0.437 |
| Bianco et al[12], 2021 | Italian & United Kingdom | NAFLD-related HCC | 226 | 2338 | NA | NA | NA | NA | NA | NA | Age, gender, BMI, and T2DM | NA |
| Bianco et al[12], 2021 | United Kingdom | UKBB (Non-viral) | 202 | 363,846 | NA | NA | NA | NA | NA | NA | Age, gender, BMI, T2DM, ethnicity, array batch, and assessment center | NA |
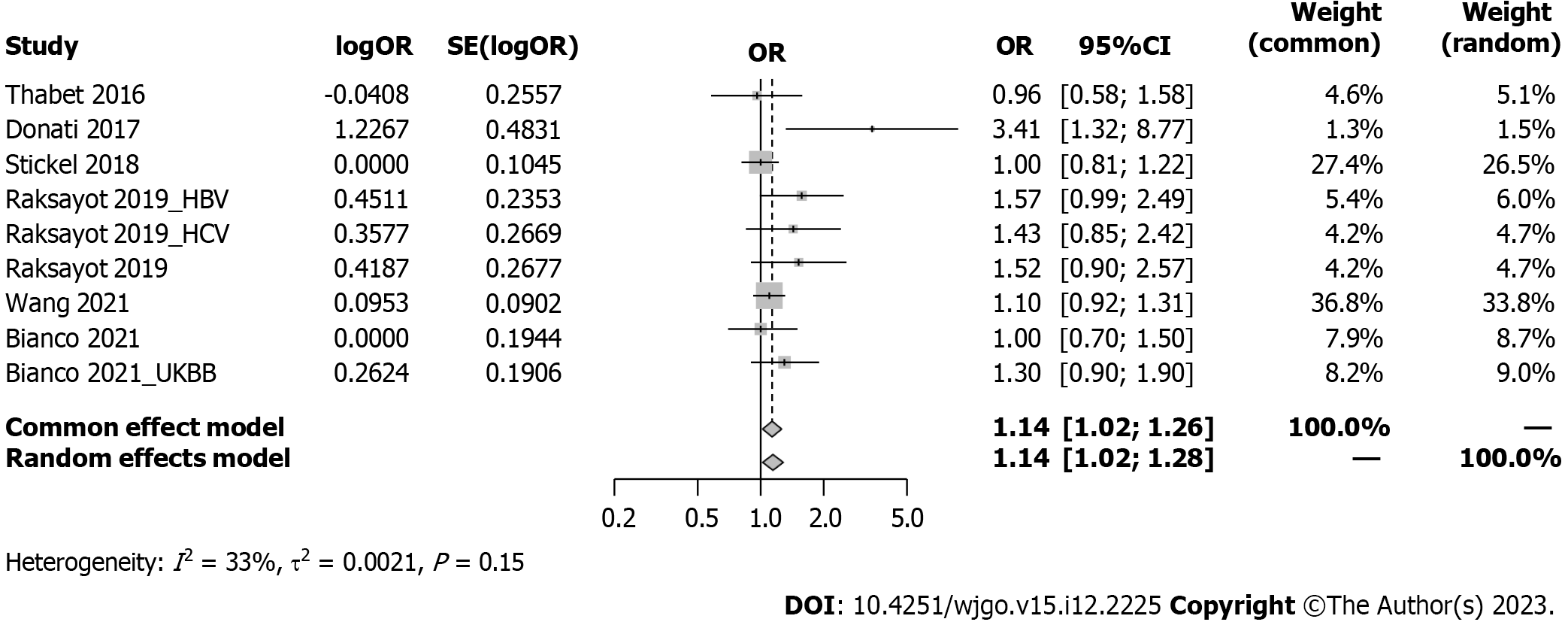
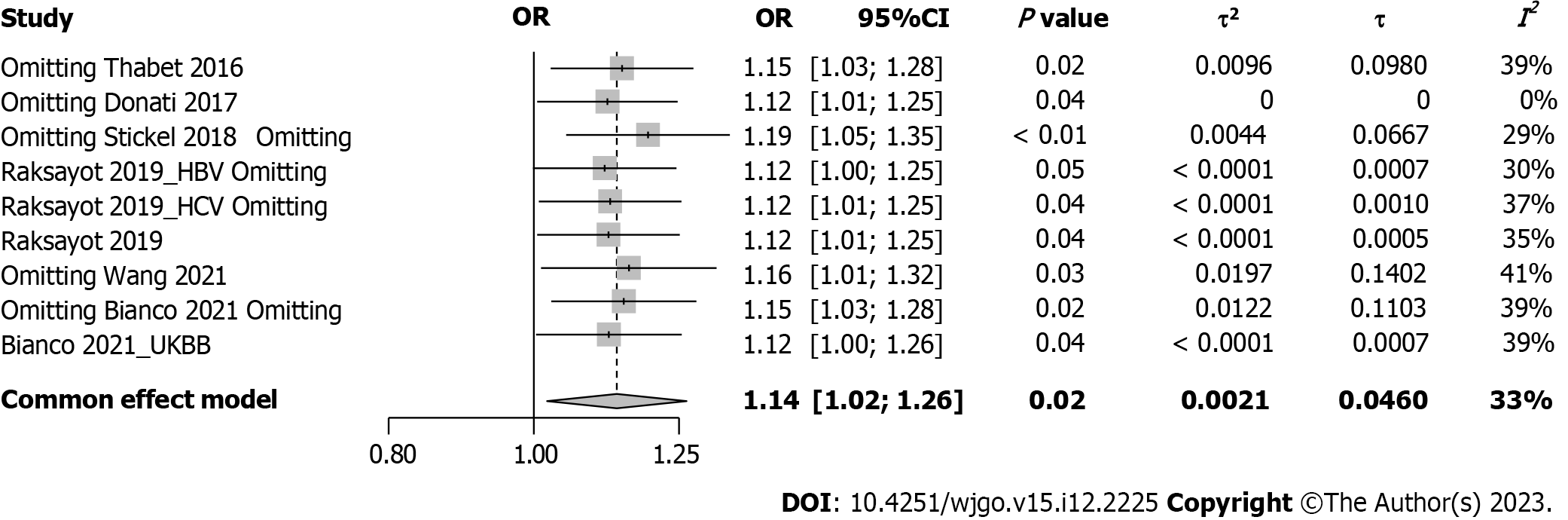
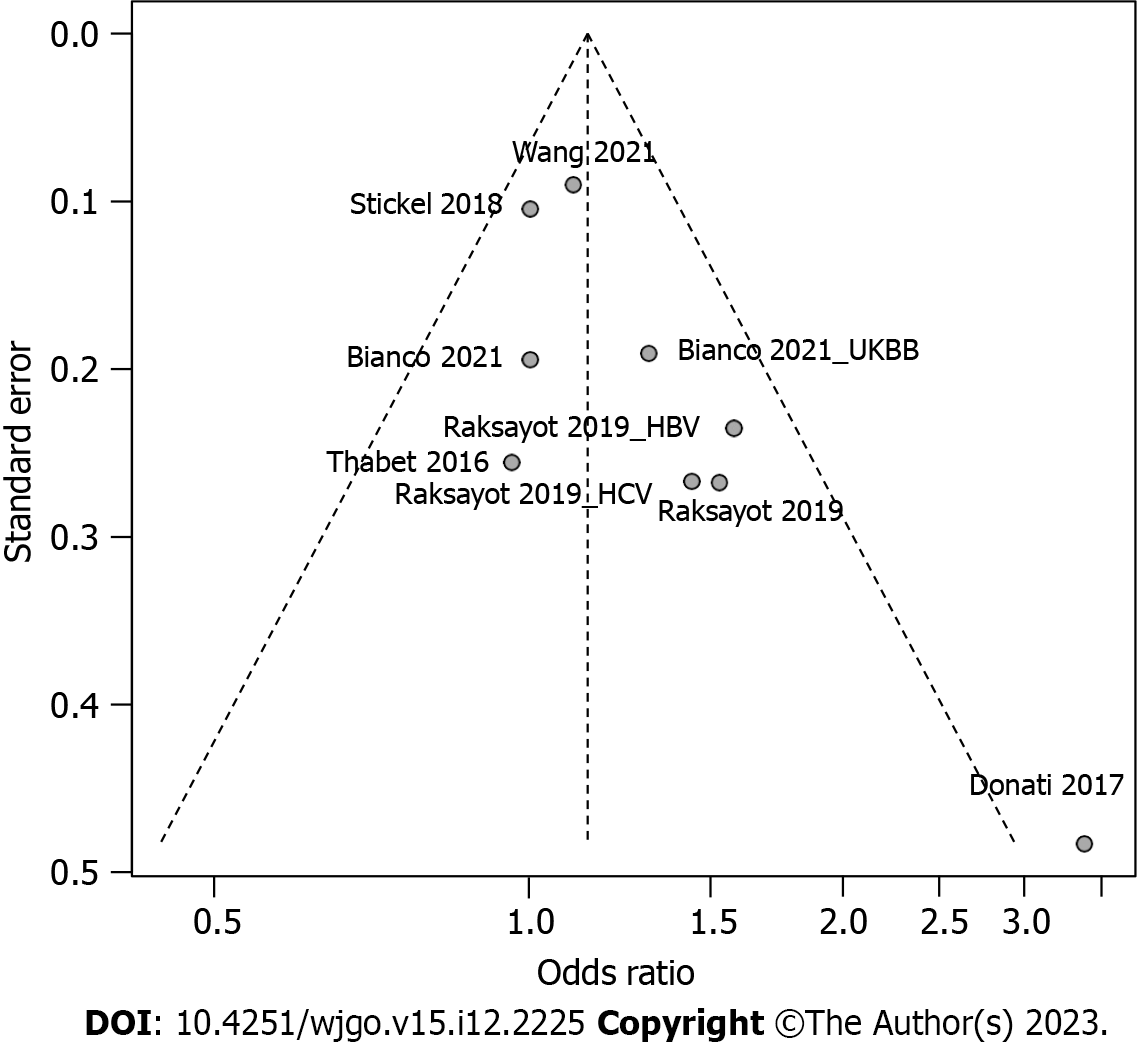
Intriguingly, our meta-analysis revealed an association between the MBOAT7 rs641738 C > T polymorphism and HCC susceptibility in the dominant model (Figure 2). The pooled OR was 1.14 (95%CI: 1.02-1.26, P = 0.020) in the common-effects model used due to low heterogeneity (I2 = 33%, P = 0.15). Robustness was confirmed in the recessive model (Supplementary Figure 1), with a pooled OR of 1.21 (95%CI: 1.05-1.39, P = 0.008). Influential analysis via the leave-one-out method confirmed the stability of our results (Figure 3). Exclusion of the study by Donati et al[9] significantly reduced heterogeneity (I2 = 0%), and the funnel plot indicated this cohort as a source of heterogeneity (Figure 4). Similar outcomes were observed in the recessive model (Supplementary Figures 2 and 3); after omitting Donati et al[9] or Raksayot et al[14], the heterogeneity was significantly reduced (I2 = 0%), and the P value remained < 0.05 in every situation, indicating the robustness of our results.
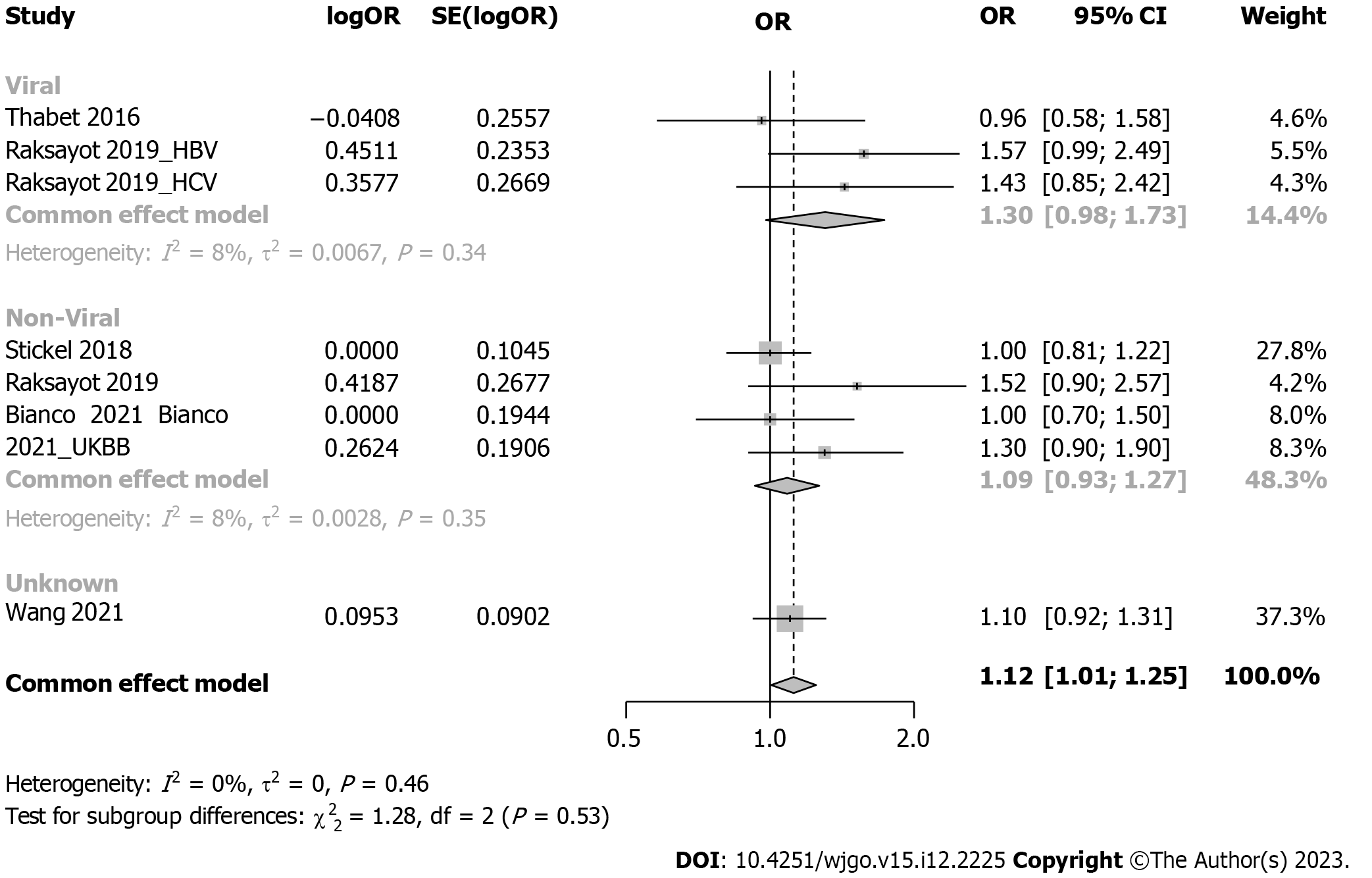
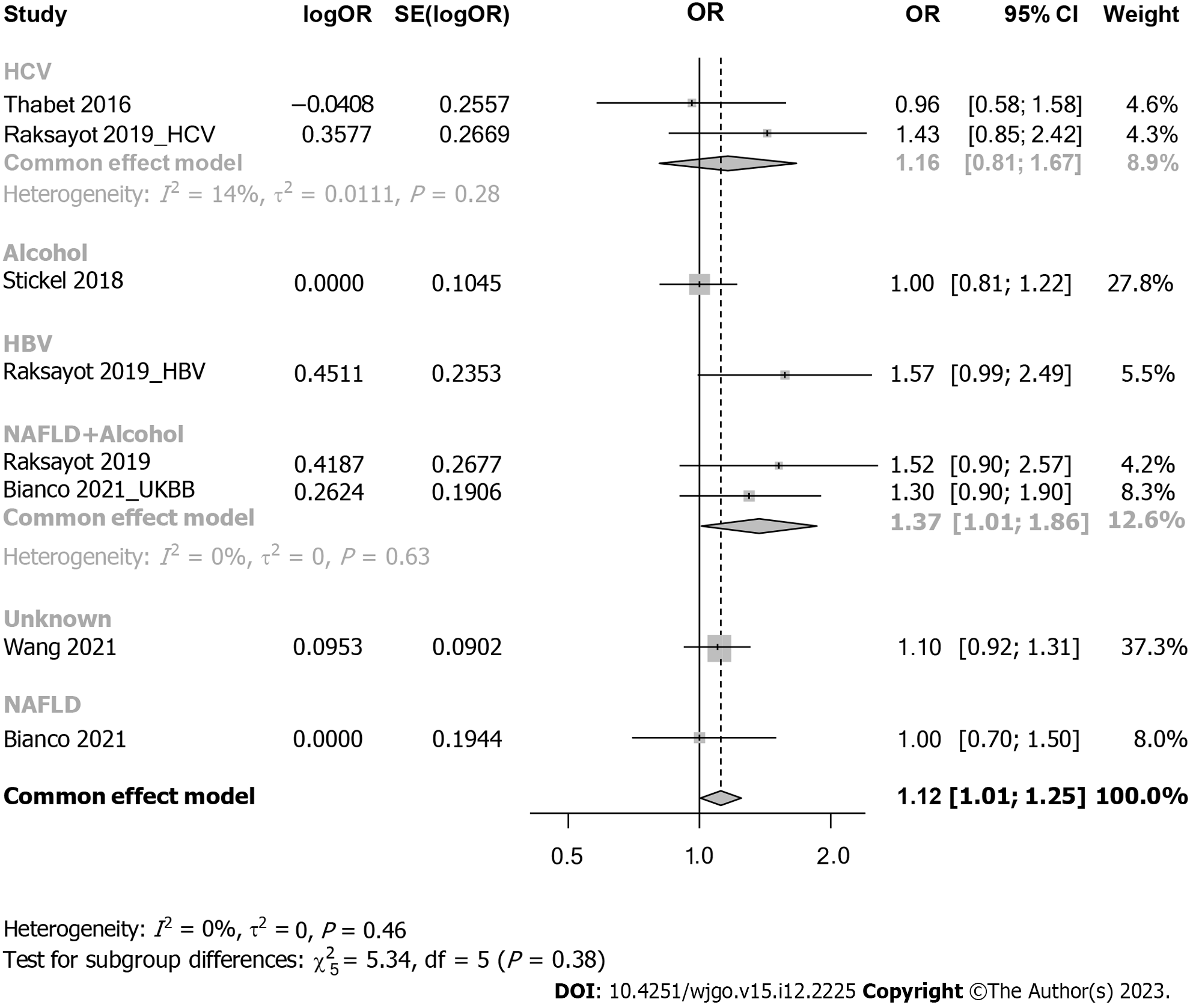
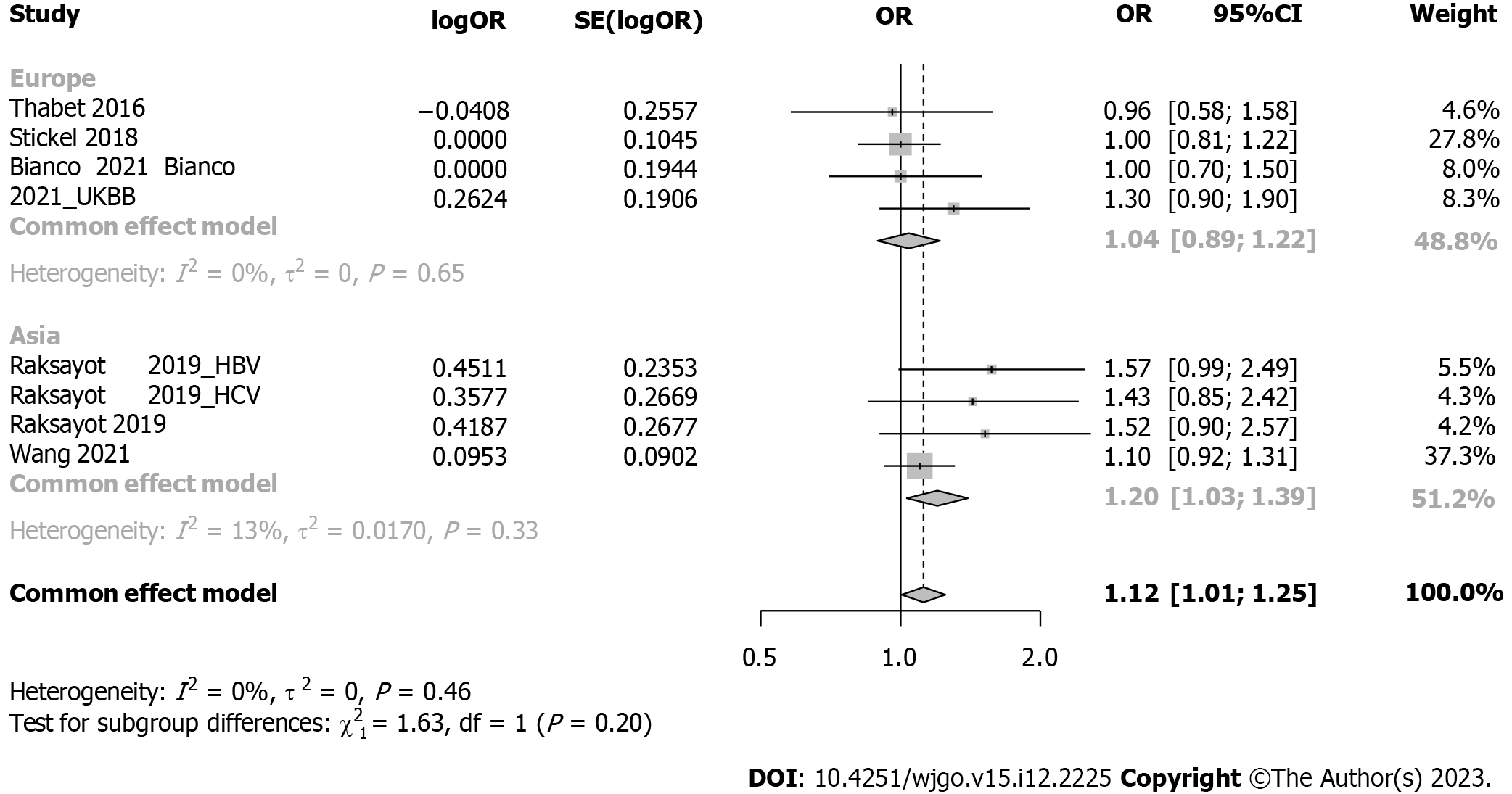
Subgroup analysis considered viral-related and nonviral-related HCC (Figure 5), revealing no significant associations in either the viral (OR: 1.30, 95%CI: 0.98-1.73) or nonviral subgroup (OR: 1.09, 95%CI: 0.93-1.27). Similar trends were observed in the recessive model (Supplementary Figure 4); in the viral subgroup, the OR was 1.09 (95%CI: 0.69-1.73), while in the nonviral subgroup, the OR was 1.14 (95%CI: 0.96-1.34). Studies were categorized into 5 groups by etiology: HCV, alcohol, NAFLD, hepatitis B virus (HBV), and NAFLD plus alcohol (Figure 6). No significant associations were found in the HCV, HBV, alcohol, or NAFLD subgroup. Notably, in the NAFLD plus alcohol subgroup, the OR was 1.37 (95%CI: 1.01-1.86); however, the same phenomenon was not observed with the recessive model (Supplementary Figure 5). Further stratification by the geographical origin of included patients showed a significant association in Asia (OR: 1.20, 95%CI: 1.03-1.39) but not in Europe (Figure 7). In the recessive model, similar results were observed in the Asia subgroup (OR: 1.43, 95%CI: 1.06-1.94, Supplementary Figure 6). This observation underscores the need for more investigation within the Asian population regarding MBOAT7 polymorphisms and HCC susceptibility.
In this systematic review and meta-analysis, a total of 2761 HCC cases and 373376 controls were included. Our results suggested that the MBOAT7 rs641738 polymorphism was positively associated with HCC susceptibility in both dominant and recessive models. Our subgroup analysis indicated that the MBOAT7 rs641738 SNP contributes to hepatocarcinogenesis, especially among Asian populations, warranting further exploration of the underlying mechanisms.
The MBOAT7 rs641738 SNP contributes to hepatocarcinogenesis, which is supported by basic scientific evidence. Longo et al[17] generated a full knockout of MBOAT7 in HepG2 cells (human HCC cell line) and observed an imbalance in mitochondrial dynamics, and the silencing of both MBOAT7 and TM6SF2 impaired mitochondrial activity with a shift toward metabolic reprogramming, which led to hepatocarcinogenesis. Moreover, MBOAT7- and TM6SF2-silenced cells exhibited increased cell survival, proliferation, and invasiveness.
Multiple studies have been conducted to investigate the association between the PRS and HCC susceptibility, and most of them reached a positive conclusion. Most studies established PRS models including genes such as PNPLA3, HSD17B13, TM6SF2, MBOAT7, and GCKR. Thrift et al[18] evaluated the association of PRS with HCC in 1644 patients in the United State and found that HCC risk increased by 134% per unit increase in PRS [hazard ratio (HR): 2.30; 95%CI: 1.35-3.92]. Similarly, Degasperi et al[19] included 509 patients with HCV cirrhosis and found that the PRS was an independent predictor of HCC (HR: 2.30, P = 0.04). In contrast, single genetic risk variants were not useful in stratifying HCC risk. Importantly, the PRS method represents a comprehensive assessment of the multiple aspects involved in cancer development. However, the inclusivity of gene sets within the PRS framework remains inconsistent, necessitating further fundamental investigations to refine the approach.
There remain some limitations in this study. First, the included studies in our meta-analysis exhibited diverse etiologies of HCC, which could influence our outcomes. To address this variability, we conducted subgroup analyses and found that the conspicuously positive outcomes might be attributed to studies focused on Asian patients. Additionally, HCC susceptibility could be influenced by patient characteristics, such as age, sex, and smoking habits. Due to the unavailability of the original data of the included studies, this could generate bias in the interpretation of the results. However, the sensitivity analyses confirmed the robustness of our results.
In summary, this meta-analysis underscores the contribution of the MBOAT7 rs641738 SNP to hepatocarcinogenesis, especially in Asian populations, which warrants further investigation.
The MBOAT7 rs641738 single-nucleotide polymorphism (SNP) has been proven to influence various liver diseases, but its association with hepatocellular carcinoma (HCC) susceptibility has been debated. To address this discrepancy, we conducted the current systematic review and meta-analysis.
Investigating whether MBOAT7 SNP has an association with HCC susceptibility could help identify at-risk population.
We conducted a systematic review and meta-analysis on the association of the MBOAT7 SNP and HCC susceptibility, aiming to provide an updated and comprehensive assessment of the evolving evidence in this area.
We performed a systematic review in PubMed, Web of Science, Scopus, and EMBASE; applied specific inclusion and exclusion criteria; and extracted the data. Meta-analysis was conducted with the meta package in R. Sensitivity and subgroup analyses were also performed.
Eight studies were included in the systematic review, and 12 cohorts from 6 studies were included in the meta-analysis. Our meta-analysis revealed an association between the MBOAT7 SNP and HCC susceptibility in both the dominant [odds ratio (OR): 1.14, 95% confidence interval (95%CI): 1.02-1.26, P = 0.020] and recessive (OR: 1.21, 95%CI: 1.05-1.39, P = 0.008) models. Subgroup analysis revealed that stratification of the included patients by geographical origin showed a significant association in Asia (OR: 1.20, 95%CI: 1.03-1.39).
This meta-analysis underscores the contribution of the MBOAT7 rs641738 SNP to hepatocarcinogenesis, especially in Asian populations, which warrants further investigation.
Future research should focus on what is the specific molecular biological mechanism of MBOAT7 rs641738 SNP leading to HCC and how to prevent it.
| 1. | Global Burden of Disease Cancer Collaboration, Fitzmaurice C, Allen C, Barber RM, Barregard L, Bhutta ZA, Brenner H, Dicker DJ, Chimed-Orchir O, Dandona R, Dandona L, Fleming T, Forouzanfar MH, Hancock J, Hay RJ, Hunter-Merrill R, Huynh C, Hosgood HD, Johnson CO, Jonas JB, Khubchandani J, Kumar GA, Kutz M, Lan Q, Larson HJ, Liang X, Lim SS, Lopez AD, MacIntyre MF, Marczak L, Marquez N, Mokdad AH, Pinho C, Pourmalek F, Salomon JA, Sanabria JR, Sandar L, Sartorius B, Schwartz SM, Shackelford KA, Shibuya K, Stanaway J, Steiner C, Sun J, Takahashi K, Vollset SE, Vos T, Wagner JA, Wang H, Westerman R, Zeeb H, Zoeckler L, Abd-Allah F, Ahmed MB, Alabed S, Alam NK, Aldhahri SF, Alem G, Alemayohu MA, Ali R, Al-Raddadi R, Amare A, Amoako Y, Artaman A, Asayesh H, Atnafu N, Awasthi A, Saleem HB, Barac A, Bedi N, Bensenor I, Berhane A, Bernabé E, Betsu B, Binagwaho A, Boneya D, Campos-Nonato I, Castañeda-Orjuela C, Catalá-López F, Chiang P, Chibueze C, Chitheer A, Choi JY, Cowie B, Damtew S, das Neves J, Dey S, Dharmaratne S, Dhillon P, Ding E, Driscoll T, Ekwueme D, Endries AY, Farvid M, Farzadfar F, Fernandes J, Fischer F, G/Hiwot TT, Gebru A, Gopalani S, Hailu A, Horino M, Horita N, Husseini A, Huybrechts I, Inoue M, Islami F, Jakovljevic M, James S, Javanbakht M, Jee SH, Kasaeian A, Kedir MS, Khader YS, Khang YH, Kim D, Leigh J, Linn S, Lunevicius R, El Razek HMA, Malekzadeh R, Malta DC, Marcenes W, Markos D, Melaku YA, Meles KG, Mendoza W, Mengiste DT, Meretoja TJ, Miller TR, Mohammad KA, Mohammadi A, Mohammed S, Moradi-Lakeh M, Nagel G, Nand D, Le Nguyen Q, Nolte S, Ogbo FA, Oladimeji KE, Oren E, Pa M, Park EK, Pereira DM, Plass D, Qorbani M, Radfar A, Rafay A, Rahman M, Rana SM, Søreide K, Satpathy M, Sawhney M, Sepanlou SG, Shaikh MA, She J, Shiue I, Shore HR, Shrime MG, So S, Soneji S, Stathopoulou V, Stroumpoulis K, Sufiyan MB, Sykes BL, Tabarés-Seisdedos R, Tadese F, Tedla BA, Tessema GA, Thakur JS, Tran BX, Ukwaja KN, Uzochukwu BSC, Vlassov VV, Weiderpass E, Wubshet Terefe M, Yebyo HG, Yimam HH, Yonemoto N, Younis MZ, Yu C, Zaidi Z, Zaki MES, Zenebe ZM, Murray CJL, Naghavi M. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-years for 32 Cancer Groups, 1990 to 2015: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2017;3:524-548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2838] [Cited by in RCA: 3034] [Article Influence: 337.1] [Reference Citation Analysis (0)] |
| 2. | El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264-1273.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2183] [Cited by in RCA: 2564] [Article Influence: 183.1] [Reference Citation Analysis (3)] |
| 3. | Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16:589-604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2184] [Cited by in RCA: 3123] [Article Influence: 446.1] [Reference Citation Analysis (17)] |
| 4. | Fujiwara N, Hoshida Y. Hepatocellular Carcinoma Risk Stratification by Genetic Profiling in Patients with Cirrhosis. Semin Liver Dis. 2019;39:153-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 5. | Singal AG, Manjunath H, Yopp AC, Beg MS, Marrero JA, Gopal P, Waljee AK. The effect of PNPLA3 on fibrosis progression and development of hepatocellular carcinoma: a meta-analysis. Am J Gastroenterol. 2014;109:325-334. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 297] [Cited by in RCA: 289] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 6. | Tang S, Zhang J, Mei TT, Guo HQ, Wei XH, Zhang WY, Liu YL, Liang S, Fan ZP, Ma LX, Lin W, Liu YR, Qiu LX, Yu HB. Association of TM6SF2 rs58542926 T/C gene polymorphism with hepatocellular carcinoma: a meta-analysis. BMC Cancer. 2019;19:1128. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 7. | Thabet K, Chan HLY, Petta S, Mangia A, Berg T, Boonstra A, Brouwer WP, Abate ML, Wong VW, Nazmy M, Fischer J, Liddle C, George J, Eslam M. The membrane-bound O-acyltransferase domain-containing 7 variant rs641738 increases inflammation and fibrosis in chronic hepatitis B. Hepatology. 2017;65:1840-1850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 74] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 8. | Thabet K, Asimakopoulos A, Shojaei M, Romero-Gomez M, Mangia A, Irving WL, Berg T, Dore GJ, Grønbæk H, Sheridan D, Abate ML, Bugianesi E, Weltman M, Mollison L, Cheng W, Riordan S, Fischer J, Spengler U, Nattermann J, Wahid A, Rojas A, White R, Douglas MW, McLeod D, Powell E, Liddle C, van der Poorten D, George J, Eslam M; International Liver Disease Genetics Consortium. MBOAT7 rs641738 increases risk of liver inflammation and transition to fibrosis in chronic hepatitis C. Nat Commun. 2016;7:12757. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 109] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 9. | Donati B, Dongiovanni P, Romeo S, Meroni M, McCain M, Miele L, Petta S, Maier S, Rosso C, De Luca L, Vanni E, Grimaudo S, Romagnoli R, Colli F, Ferri F, Mancina RM, Iruzubieta P, Craxi A, Fracanzani AL, Grieco A, Corradini SG, Aghemo A, Colombo M, Soardo G, Bugianesi E, Reeves H, Anstee QM, Fargion S, Valenti L. MBOAT7 rs641738 variant and hepatocellular carcinoma in non-cirrhotic individuals. Sci Rep. 2017;7:4492. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 203] [Cited by in RCA: 204] [Article Influence: 22.7] [Reference Citation Analysis (1)] |
| 10. | Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, Shamseer L, Tetzlaff JM, Akl EA, Brennan SE, Chou R, Glanville J, Grimshaw JM, Hróbjartsson A, Lalu MM, Li T, Loder EW, Mayo-Wilson E, McDonald S, McGuinness LA, Stewart LA, Thomas J, Tricco AC, Welch VA, Whiting P, Moher D. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372:n71. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44932] [Cited by in RCA: 51767] [Article Influence: 10353.4] [Reference Citation Analysis (2)] |
| 11. | Stickel F, Buch S, Nischalke HD, Weiss KH, Gotthardt D, Fischer J, Rosendahl J, Marot A, Elamly M, Casper M, Lammert F, McQuillin A, Zopf S, Spengler U, Marhenke S, Kirstein MM, Vogel A, Eyer F, von Felden J, Wege H, Buch T, Schafmayer C, Braun F, Deltenre P, Berg T, Morgan MY, Hampe J. Genetic variants in PNPLA3 and TM6SF2 predispose to the development of hepatocellular carcinoma in individuals with alcohol-related cirrhosis. Am J Gastroenterol. 2018;113:1475-1483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 87] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 12. | Bianco C, Jamialahmadi O, Pelusi S, Baselli G, Dongiovanni P, Zanoni I, Santoro L, Maier S, Liguori A, Meroni M, Borroni V, D'Ambrosio R, Spagnuolo R, Alisi A, Federico A, Bugianesi E, Petta S, Miele L, Vespasiani-Gentilucci U, Anstee QM, Stickel F, Hampe J, Fischer J, Berg T, Fracanzani AL, Soardo G, Reeves H, Prati D, Romeo S, Valenti L. Non-invasive stratification of hepatocellular carcinoma risk in non-alcoholic fatty liver using polygenic risk scores. J Hepatol. 2021;74:775-782. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 254] [Cited by in RCA: 293] [Article Influence: 58.6] [Reference Citation Analysis (5)] |
| 13. | Liu Z, Suo C, Shi O, Lin C, Zhao R, Yuan H, Jin L, Zhang T, Chen X. The Health Impact of MAFLD, a Novel Disease Cluster of NAFLD, Is Amplified by the Integrated Effect of Fatty Liver Disease-Related Genetic Variants. Clin Gastroenterol Hepatol. 2022;20:e855-e875. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 60] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 14. | Raksayot M, Chuaypen N, Khlaiphuengsin A, Pinjaroen N, Treeprasertsuk S, Poovorawan Y, Tanaka Y, Tangkijvanich P. Independent and additive effects of PNPLA3 and TM6SF2 polymorphisms on the development of non-B, non-C hepatocellular carcinoma. J Gastroenterol. 2019;54:427-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 15. | Nahon P, Bamba-Funck J, Layese R, Trépo E, Zucman-Rossi J, Cagnot C, Ganne-Carrié N, Chaffaut C, Guyot E, Ziol M, Sutton A, Audureau E; ANRS CO12 CirVir and CIRRAL groups. Integrating genetic variants into clinical models for hepatocellular carcinoma risk stratification in cirrhosis. J Hepatol. 2023;78:584-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 52] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 16. | Wang P, Li Y, Li L, Zhong R, Shen N. MBOAT7-TMC4 rs641738 Is Not Associated With the Risk of Hepatocellular Carcinoma or Persistent Hepatitis B Infection. Front Oncol. 2021;11:639438. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 17. | Longo M, Meroni M, Paolini E, Erconi V, Carli F, Fortunato F, Ronchi D, Piciotti R, Sabatini S, Macchi C, Alisi A, Miele L, Soardo G, Comi GP, Valenti L, Ruscica M, Fracanzani AL, Gastaldelli A, Dongiovanni P. TM6SF2/PNPLA3/MBOAT7 Loss-of-Function Genetic Variants Impact on NAFLD Development and Progression Both in Patients and in In Vitro Models. Cell Mol Gastroenterol Hepatol. 2022;13:759-788. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 84] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 18. | Thrift AP, Kanwal F, Liu Y, Khaderi S, Singal AG, Marrero JA, Loo N, Asrani SK, Luster M, Al-Sarraj A, Ning J, Tsavachidis S, Gu X, Amos CI, El-Serag HB. Risk stratification for hepatocellular cancer among patients with cirrhosis using a hepatic fat polygenic risk score. PLoS One. 2023;18:e0282309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 18] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 19. | Degasperi E, Galmozzi E, Pelusi S, D'Ambrosio R, Soffredini R, Borghi M, Perbellini R, Facchetti F, Iavarone M, Sangiovanni A, Valenti L, Lampertico P. Hepatic Fat-Genetic Risk Score Predicts Hepatocellular Carcinoma in Patients With Cirrhotic HCV Treated With DAAs. Hepatology. 2020;72:1912-1923. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Tsoulfas G, Greece; Yoshioka K, Japan S-Editor: Lin C L-Editor: Wang TQ P-Editor: Zhao S