Published online Dec 15, 2023. doi: 10.4251/wjgo.v15.i12.2120
Peer-review started: July 27, 2023
First decision: August 8, 2023
Revised: September 22, 2023
Accepted: October 16, 2023
Article in press: October 16, 2023
Published online: December 15, 2023
Processing time: 140 Days and 1.9 Hours
This study investigate the anti-tumor effect of curcumin and whether its mediated by hsa_circ_0136666 through miR-1301-3p/CXCL1 in colorectal carcinoma (CRC). Through multiple experiments, we have drawn the conclusion that curcumin inhibited CRC development through the hsa_circ_0136666/miR-1301-3p/CXCL1 axis, hinting at a novel treatment option for curcumin to prevent CRC deve
To determine whether hsa_circ_0136666 involvement in curcumin-triggered CRC progression was mediated by sponging miR-1301-3p.
Cell counting kit-8, colony-forming cell, 5-ethynyl-2’-deoxyuridine, and flow cytometry assays were carried out to determine cell proliferation, apoptosis, and cell cycle progression. Real-time quantitative polymerase chain reaction quantified hsa_circ_0136666, miR-1301-3p, and chemokine (C-X-C motif) ligand 1 (CXCL1), and western blot analysis determined CXCL1, B-cell lymphoma-2 (Bcl-2), and Bcl-2 related X protein (Bax) protein levels. CircBank or starbase software was first used for the prediction of miR-1301-3p binding with hsa_circ_0136666 and CXCL1, followed by RNA pull-down, RNA immunoprecipitation, and dual-luciferase reporter assay validation. In vivo experiments were implemented in a murine xenograft model.
Curcumin blocked CRC cell proliferation but boosted apoptosis. Moreover, ele
Curcumin inhibited CRC development through the hsa_circ_0136666/miR-1301-3p/CXCL1 axis, hinting at a novel treatment option for curcumin to prevent CRC development.
Core Tip: This study investigate the anti-tumor effect of curcumin and whether its mediated by hsa_circ_0136666 through miR-1301-3p/CXCL1 in colorectal carcinoma (CRC). Through multiple experiments, we have drawn the conclusion that curcumin inhibited CRC development through the hsa_circ_0136666/miR-1301-3p/CXCL1 axis, hinting at a novel treatment option for curcumin to prevent CRC development.
- Citation: Chen S, Li W, Ning CG, Wang F, Wang LX, Liao C, Sun F. Hsa_circ_0136666 mediates the antitumor effect of curcumin in colorectal carcinoma by regulating CXCL1 via miR-1301-3p. World J Gastrointest Oncol 2023; 15(12): 2120-2137
- URL: https://www.wjgnet.com/1948-5204/full/v15/i12/2120.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i12.2120
Colorectal carcinoma (CRC), afflicting an estimated 104270 new cases and causing 52980 deaths in 2021 in the United States, is still considered the prime reason for cancer-related death in the United States[1,2]. Clinically, despite substantial advances made in surgical resection and adjuvant chemoradiotherapy, the conventional treatment method for CRC, the overall patient prognosis is still unfavorable[3,4]. Hence, it is urgent to find a more effective therapy that has high clinical significance for CRC patients. Currently, the chemo-preventive properties of traditional Chinese medicine have drawn great attention in cancer therapeutics. As a naturally occurring medicine, curcumin is derived from the medicinal plant Curcuma longa L.[5]. The salient features of curcumin include chemical stability, low toxicity, and extensive distribution, so the application of curcumin in the management of various diseases has gradually been understood[6,7]. Of note, curcumin exhibited powerful antitumor activity in diverse cancers[8-10], including CRC[11]. However, further investigation is needed regarding the mechanism through which curcumin affects CRC development.
Over the last decade, circular RNAs (circRNAs) have been identified as a novel noncoding RNA subgroup that are produced by exon or intron back-splicing and have a closed continuous loop[12,13]. CircRNAs are correlated with the initiation and development of various tumors, including CRC. For example, circ-FARSA aggravates malignant behavior by promoting CRC cell growth[14]. Consistently, circRNA_0000392 exerted oncogenic properties in CRC by regulating PIK3R3/AKT[15]. Hsa_circ_0136666 has been identified as a highly circRNA that is a carcinogenic factor in breast cancer and osteosarcoma[16,17]. Moreover, it facilitated CRC progression by enhancing cell growth and metastasis[18,19]. Interestingly, some studies indicated that curcumin could participate in the regulation of tumor progression by interacting with noncoding RNAs, including circRNAs[20,21]. However, the exact role played by hsa_circ_0136666 in curcumin-mediated CRC progression is still unclear.
Currently, the increasing focus on the competing endogenous RNA (ceRNA) hypothesis is that circRNAs serve as microRNA (miRNA) sponges to derepress target mRNA levels[22,23]. Here, some binding sites between hsa_circ_0136666 and miR-1301-3p were identified by bioinformatics analysis. In addition, miR-1301-3p has been suggested to dampen CRC cell growth ability[24]. Hence, this article aims to illuminate whether hsa_circ_0136666 involvement in curcumin-triggered CRC progression is mediated by sponging miR-1301-3p.
Under a consistent temperature (37 °C) and humidified atmosphere containing 5% CO2, a normal human colon mucosal epithelial cell line (NCM460, ATCC, Manassas, VA, United States) and two CRC cell strains (SW480 and SW620, ATCC) were cultivated in DMEM (PAN Biotech, Aidenbach, Germany) that was mixed with FBS (10%; HyClone, Logan, UT, United States) and penicillin/streptomycin (1%; KeyGen, Nanjing, China). In addition, 20 μM curcumin (Sigma-Aldrich, St. Louis, MO, United States) was added to CRC cells for 24 h after dilution with dimethyl sulfoxide (DMSO, Sigma-Aldrich).
Cell viability assessment was performed as per the cell counting kit (CCK)-8 reagent (Dojindo, Osaka, Japan) guidebook. A total of 2000 cells were plated into a 96-well culture plate for curcumin (20 μM) treatment, followed by the addition of CCK-8 solution (0 μL) into each well for another 4 h after 24 h of curcumin treatment. At 450 nm, the absorbance value was determined with the use of a microplate reader.
After incubation for 2 wk, 5 × 102 treated or untreated cells in 6-well plates were mixed with paraformaldehyde (4%) and dyed with crystal violet (0.1%). After an incubation period of 30 min, the colony number was observed using a microscope.
Apoptosis assessment was implemented based on the Annexin V-FITC/PI Apoptosis kit (Bender Med System, Vienna, Austria) manuals. In short, tumor cells were trypsinized, followed by resuspension in binding buffer. After the addition of Annexin V-FITC and PI at volumes of 5 μL and 10 μL, respectively, a FACSCalibur (BD Biosciences, Heidelberg, Germany) was applied for apoptosis detection according to the operation manual.
In this assay, total cellular RNAs prepared by TRIzol reagent (Invitrogen, Carlsbad, CA, United States) were quantified with a NanoDrop 2000 instrument (Thermo Scientific, Waltham, MA, United States) spectrophotometer, followed by synthesis into cDNA with a PrimeScript RT reagent kit (Takara, Tokyo, Japan) and the subsequent determination of the relative gene expression with the use of a SYBR Green PCR Kit (TaKaRa). After normalization against GAPDH (for circRNA and mRNA) and U6 (for miRNA), the relative fold changes were counted with the 2–ΔΔCt formula. See Table 1 for primers.
| Names | Sequences (5’-3’) |
| hsa_circ_0136666: Forward | AGGTGCTCACTGTGCTGAAA |
| hsa_circ_0136666: Reverse | CAGATGTTCATTGGGTCCAT |
| hsa_circ_0000896: Forward | ACTTCATTGAGAGCTCCTTCTGG |
| hsa_circ_0000896: Reverse | CTTCAGAGTCCTCGAAGGAAGA |
| hsa_circ_0000392: Forward | TCAAGTTACTGAGAAGAAAAAGCTG |
| hsa_circ_0000392: Reverse | GTCCTCGAGGCACTCACAAT |
| miR-1301-3p: Forward | TTACAGCTGCCTGAGAGTGACTTA |
| miR-1301-3p: Reverse | CTCTACAGCTATATTGCCAGCCA |
| miR-34a-5p: Forward | TCCGAGTGGCAGTGTCTTAG |
| miR-34a-5p: Reverse | CTCAACTGGTGTCGTGGAG |
| miR-216a-3p: Forward | ATAGTCACAGTGGTCTCTGG |
| miR-216a-3p: Reverse | CTCAACTGGTGTCGTGGAG |
| CXCL1: Forward | AACCGAAGTCATAGCCACAC |
| CXCL1: Reverse | GTTGGATTTGTCACTGTTCAGC |
| U6: Forward | CTCGCTTCGGCAGCACA |
| U6: Reverse | AACGCTTCACGAATTTGCGT |
| GAPDH: Forward | GGTCACCAGGGCTGCTTT |
| GAPDH: Reverse | GGAAGATGGTGATGGGATT |
For hsa_circ_0136666 stable upregulation, CRC cells were transfected with hsa_circ_0136666 overexpression vector (oe-hsa_circ_0136666, Geenseed Biotech, Guangzhou, China). In addition, 40 nM each of miR-1301-3p mimic and inhibitor, as well as chemokine (C-X-C motif) ligand 1 (CXCL1) small interfering RNA (si-CXCL1) and their corresponding controls (mimic NC, inhibitor NC, si-NC), were transfected. These oligonucleotides were provided by RiboBio (Guangzhou, China). Lipofectamine 3000 reagent (Invitrogen) was utilized for transfection, and further analysis was conducted after a 48-hour incubation.
Briefly, 5-Ethynyl-2’-deoxyuridine (EdU) (50 μM, RiboBio) was placed into CRC cells (4 × 104 cells/well), followed by a 2-hour incubation. After immobilization in a formaldehyde solution (4%), the cells were treated with Apollo and 4',6-diamidino-2-phenylindole (DAPI). Finally, EdU-positive cells were counted by imaging five random fields using a fluorescence microscope (Olympus, Tokyo, Japan). Cell proliferation was calculated after normalizing the EdU-positive cell (red) count against the DAPI-stained cell (blue) count.
In brief, CRC cells were trypsinized, followed by fixation in ice-cold ethanol overnight. The PBS-rinsed cells were then resuspended in PI (Bender Med System) for 30 min at 37 °C. Referring to the operation manual of FACSCalibur (BD Biosciences), the cell cycle distribution was assessed in this assay.
Cell lysates were prepared from 5 × 106 treated or untreated CRC cells in 6-well plates following the RIPA buffer (Beyotime, Nantong, China) instructions. After that, the specimens were subjected to SDS-PAGE (10%) and electrotransfer onto nitrocellulose membranes (Millipore, Molsheim, France). After being probed using the primary antibodies (Abcam, Cambridge, MA, United States): B-cell lymphoma-2 (Bcl-2; ab59348, 1:1000), Bcl-2 related X protein (Bax; ab53154, 1:1000), CXCL1 (1:100, ab206411), and β-actin (1:1000, ab8227) overnight, the immune complexes were treated with a 2-hour incubation with a secondary antibody (ab6721, 1:10000), followed by visualization with an ECL reagent (Amersham Biosciences, Pittsburg, PA, Sweden).
In short, probe-coated beads, generated by 2 h of room temperature incubation of the biotinylated hsa_circ_0136666 or NC probe (GenePharma, Shanghai, China) with magnetic beads, were incubated with SW480 and SW620 Lysates that were collected after sonication. Finally, the bead-bound RNA complexes were subjected to real-time quantitative PCR (RT-qPCR) analysis.
Briefly, SW480 and SW620 cells were cultured to 80% confluency through overnight incubation, followed by immersion in complete RNA immunoprecipitation (RIP) lysis buffer (Millipore). Then, the cell lysates were incubated with anti-Argonaute2 (Ago2) or immunoglobulin G (IgG) and 2 h of treatment with magnetic protein A/G beads. At length, the samples were isolated for RT-qPCR quantification of hsa_circ_0136666 and miR-1301-3p.
In brief, hsa_circ_0136666 and CXCL1 3’ untranslated region (3’UTR) segments possessing miR-1301-3p-matched regions or mismatches were introduced into the psiCHECK2 vector (Promega, Madison, WI, United States) to generate hsa_circ_0136666 wild-type (wt)/mutant-type (mut) and CXCL1 3’UTR wt/mut reporter vectors. Subsequently, the indicated reporter vector (50 ng) was transfected into 293T cells (Sigma-Aldrich) with miR-1301-3p mimic or mimic NC (20 nM), followed by luciferase activity detection with the use of a dual-luciferase reporter (DLR) assay system (Promega).
After obtaining the approval of the Animal Ethics Committee of the Second Affiliated Hospital of Kunming Medical University, we used BALB/C nude mice (Vital River Laboratory, Beijing, China) raised under a specific-pathogen-free environment for experiments. Male mice aged 5 wk were arranged in 4 groups (vector + DMSO, oe-hsa_circ_0136666 + DMSO, vector + curcumin, and oe-hsa_circ_0136666 + curcumin) with 6 mice in each group. SW480 cells (1 × 105) were transfected with vector or oe-hsa_circ_0136666, followed by subcutaneous inoculation into nude mice. Seven days later, these groups were treated with intraperitoneal DMSO or curcumin (25 mg/kg) injection twice a week. Additionally, a caliper was used to examine the tumor size once a week. All mice were euthanized on day 35 after inoculation, and their tumors were resected and weighed for further analysis. Additionally, immunohistochemical staining (IHCS) was executed in xenograft tissue sections as per the prior description[25], with Ki67- and proliferating cell nuclear antigen (PCNA)-specific antibodies to assess proliferation. In addition, section observation and image recording were performed using a BX51 system microscope (Olympus) and a digital microscope camera (DP70; Olympus), respectively.
A CXCL1-dedicated enzyme-linked immunosorbent assay (ELISA) kit was used to quantify CXCL1. After curcumin (20 μM, 24 h) treatment, the culture media of SW480 and SW620 cells were collected and measured using a human CXCL1 ELISA kit (ab190805, Abcam) according to the manufacturer’s instructions.
Data were obtained from experiments run independently in triplicate at least with the results analyzed using GraphPad Prism 7 (GraphPad Prism software, San Diego, CA, United States), and statistical significance was indicated by P < 0.05. Meanwhile, the mean ± SD was used for statistical description of the data. Intergroup and multigroup differences were identified by two-tailed Student’s t test and one-way analysis of variance (ANOVA) with Tukey’s tests, respectively.
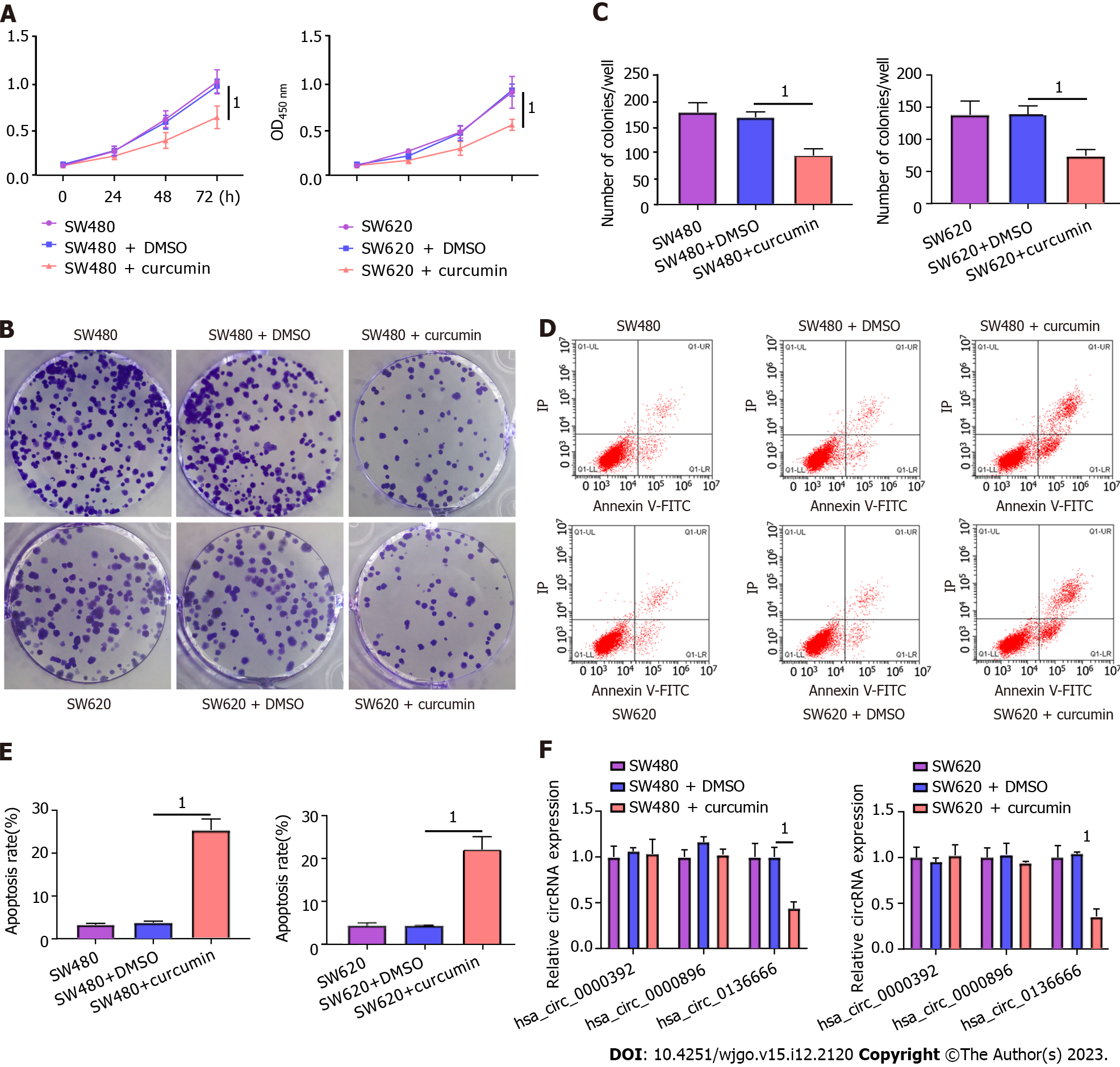
First, CRC SW480 and SW620 cells were treated with 20 μM curcumin to investigate the functional role played by curcumin in CRC. The CCK-8 results showed that 20 μM curcumin intervention was not toxic to NCM460, a normal human colon mucosal epithelial cell line (Supplementary Figure 1). As presented in Figure 1A, decreased cell viability was observed due to curcumin intervention compared to both the control and DMSO-treated groups. Consistently, curcumin treatment distinctly reduced the colony number of SW480 and SW620 cells relative to their control groups (Figure 1B and C). Furthermore, the apoptosis rate in the curcumin group was markedly improved in comparison with that in the other groups (Figure 1D and E). In addition, previous studies indicated the involvement of hsa_circ_0000392, hsa_circ_0000896, and hsa_circ_0136666 in the regulation of CRC progression[26]. Interestingly, curcumin treatment significantly decreased hsa_circ_0136666 expression in SW480 and SW620 cells but had no effect on hsa_circ_0000392 or hsa_circ_0000896 expression (Figure 1F). Therefore, follow-up studies were conducted on hsa_circ_0136666. Together, these data indicated the suppressive role of curcumin in CRC development.
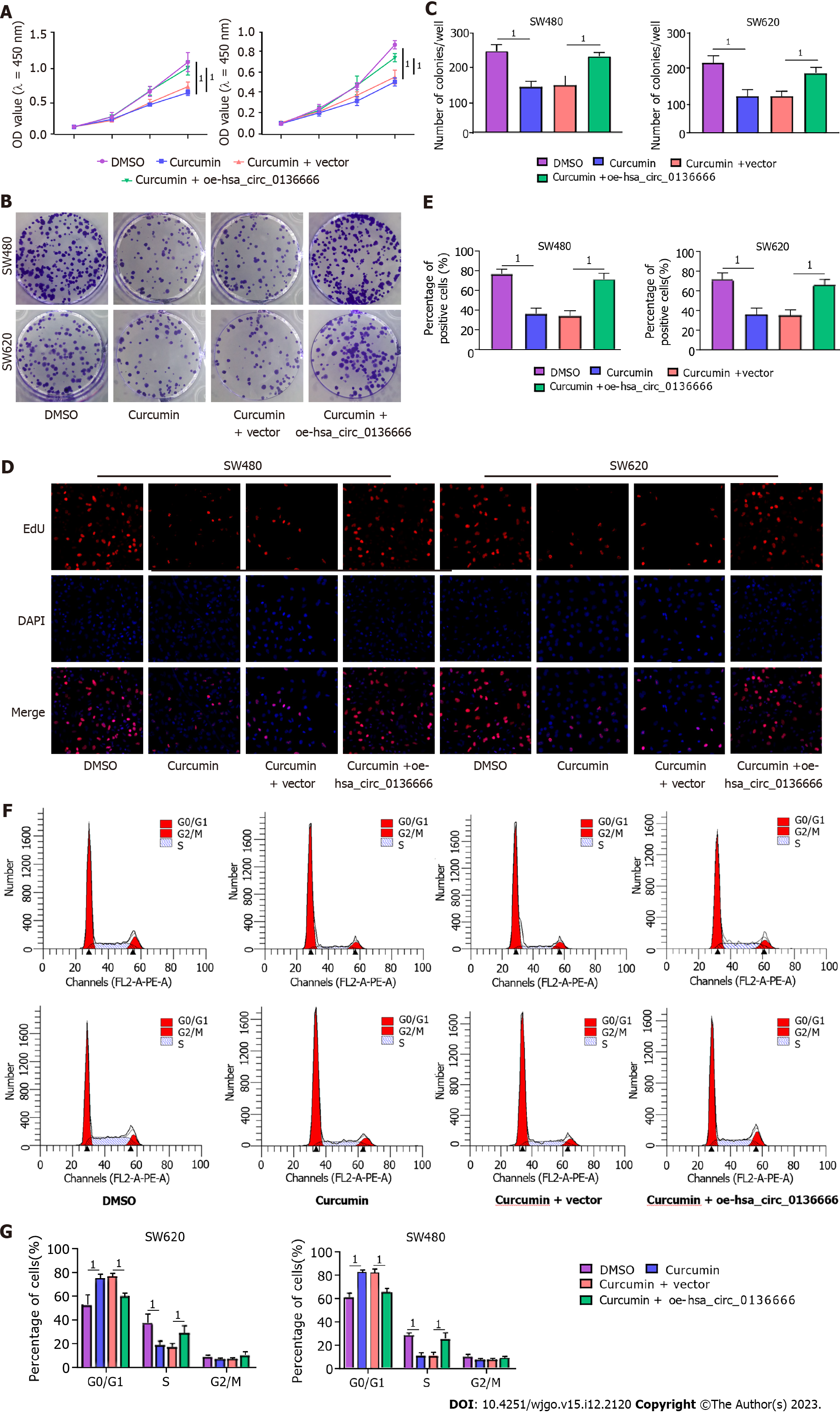
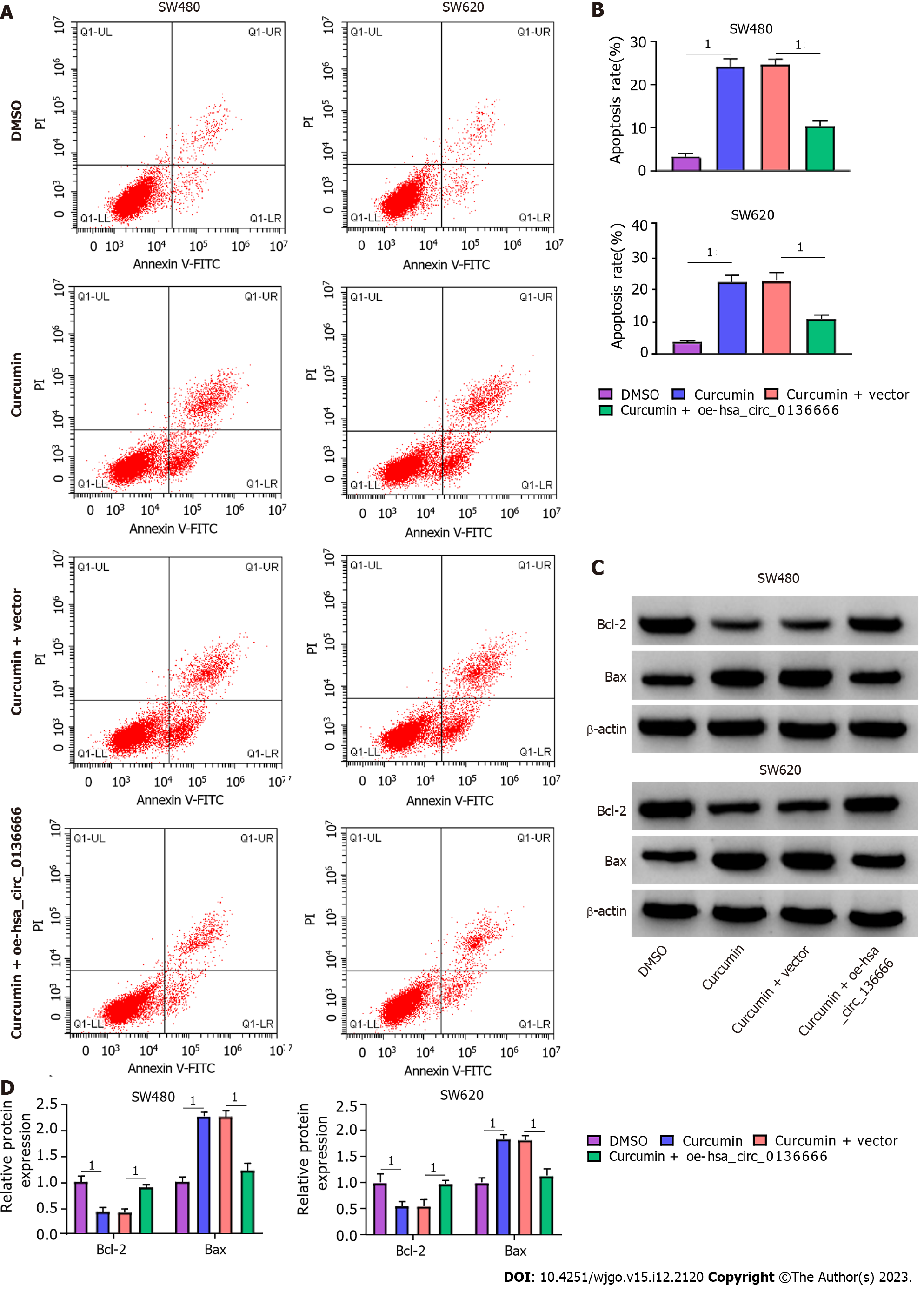
Subsequently, the influence of hsa_circ_013666 and curcumin on CRC cell malignant biological behaviors was determined. First, the overexpression efficiency of hsa_circ_0136666 was successful (Supplementary Figure 2A). The data suggested that hsa_circ_0136666 upregulation could abrogate the inhibition of SW480 and SW620 cell viability and colony counts by curcumin (Figure 2A-C). Similarly, the reduction in EdU-positive cells caused by curcumin was also evidently ameliorated by hsa_circ_0136666 overexpression (Figure 2D and E). In addition, curcumin treatment might block cell cycle progression in SW480 and SW620 cells, which was significantly counteracted by oe-hsa_circ_0136666 transfection (Figure 2F and G). In addition, elevated hsa_circ_0136666 significantly mitigated the positive effect of curcumin on the apoptosis rate in SW480 and SW620 cells (Figure 3A and B). Meanwhile, curcumin treatment induced a marked decrease in Bcl-2 (an anti-apoptosis factor) and a substantial increase in Bax (a pro-apoptosis factor), which was reversed by hsa_circ_0136666 enrichment (Figure 3C and D). Collectively, hsa_circ_0136666 upregulation abolished the impacts of curcumin on CRC cell growth and apoptosis.
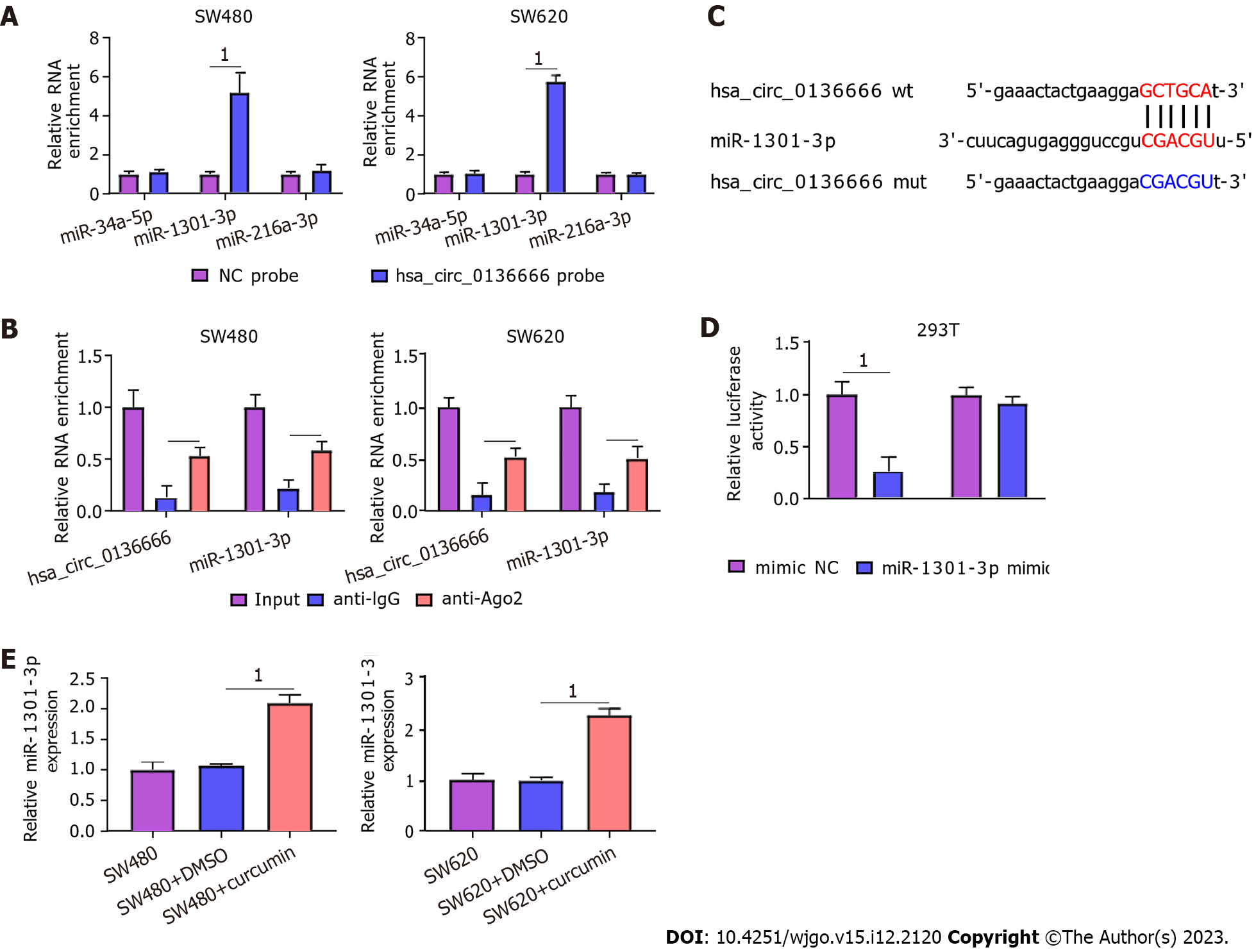
Then, we searched 3 potential target miRNAs of hsa_circ_0136666, namely, miR-34a-5p, miR-1301-3p, and miR-216-3p, from circBank. All these miRNAs were subjected to RNA pull-down analysis. In SW480 and SW620 cells, only miR-1301-3p was substantially pulled down by the biotinylated hsa_circ_0136666 probe (Figure 4A). In addition, the RIP assay showed that hsa_circ_0136666 and miR-1301-3p were both greatly enriched in Ago2 pellets vs the IgG control group (Figure 4B). Thus, miR-1301-3p was chosen for further research. Furthermore, their binding loci are presented in Figure 4C. Furthermore, the miR-1301-3p mimic transfection efficiency is displayed in Supplementary Figure 2B. Then, the DLR assay showed that the miR-1301-3p mimic could decrease hsa_circ_0136666 wt luciferase activity in 293T cells, whereas it had no obvious impact on the mutant group (Figure 4D). Moreover, miR-1301-3p was enhanced by curcumin exposure in SW480 and SW620 cells compared with their control groups (Figure 4E), implying the participation of miR-1301-3p in curcumin-mediated CRC development. Overall, miR-1301-3p acted as a direct target of hsa_circ_0136666.
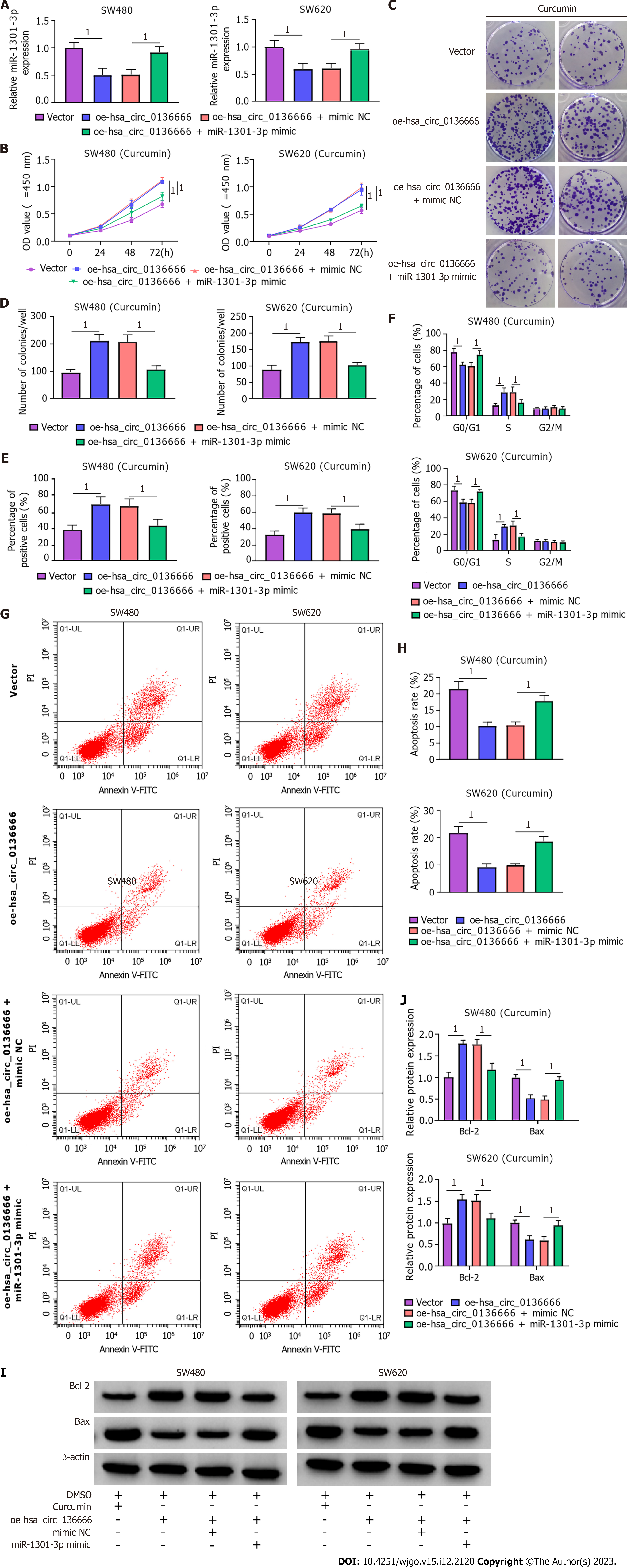
Next, the influence of hsa_circ_0136666/miR-1301-3p on curcumin-medicated proliferation and apoptosis was further explored. According to the RT-qPCR assay, elevated hsa_circ_0136666 repressed miR-1301-3p levels in CRC cells, while miR-1301-3p mimic cotransfection counteracted these effects (Figure 5A). Functionally, miR-1301-3p upregulation weakened the cell growth ability-promoting effect of hsa_circ_0136666 in curcumin-treated SW480 and SW620 cells (Figure 5B-E). Similarly, hsa_circ_0136666 overexpression-induced enhancement of cell cycle progression was partly abated by miR-1301-3p upregulation in curcumin-induced SW480 and SW620 cells (Figure 5F). In addition, miR-1301-3p mimic introduction promoted the suppressive action of hsa_circ_0136666 overexpression against apoptosis in curcumin-stimulated CRC cells (Figure 5G and H), accompanied by lowered Bcl-2 and elevated Bax levels (Figure 5I and J). Overall, hsa_circ_0136666 could partly regulate proliferation and apoptosis by targeting miR-1301-3p in curcumin-treated CRC cells.
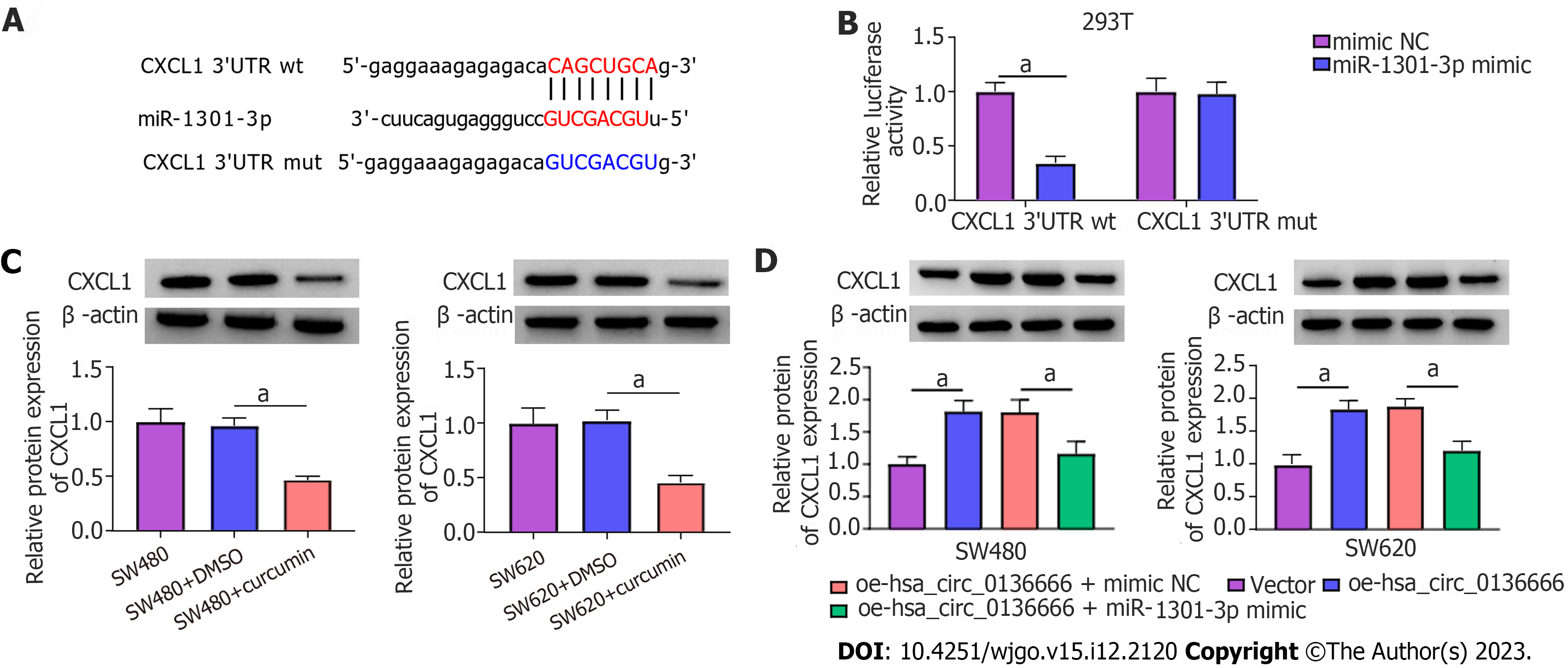
According to starBase analysis, the CXCL1 3’UTR possessed some complementary loci with miR-1301-3p (Figure 6A). The DLR assay suggested that miR-1301-3p upregulation significantly hindered CXCL1 3’UTR reporter luciferase activity but not the mutant group (Figure 6B). Notably, CXCL1 Levels were significantly downregulated by curcumin treatment in CRC cells (Figure 6C). Moreover, ELISA showed that CXCL1 levels were obviously reduced by curcumin treatment in CRC cells (Supplementary Figure 3A). Moreover, elevated hsa_circ_0136666 could facilitate CXCL1 protein levels in CRC cells, and miR-1301-3p upregulation distinctly attenuated this effect (Figure 6D), implying the ability of hsa_circ_0136666 to modulate CXCL1 by sponging miR-1301-3p. Meanwhile, ELISA showed that hsa_circ_0136666 overexpression might increase CXCL1 secretion in CRC cells, which was reversed by miR-1301-3p overexpression (Supplementary Figure 3B). Overall, CXCL1 acted as a direct target of miR-1301-3p.
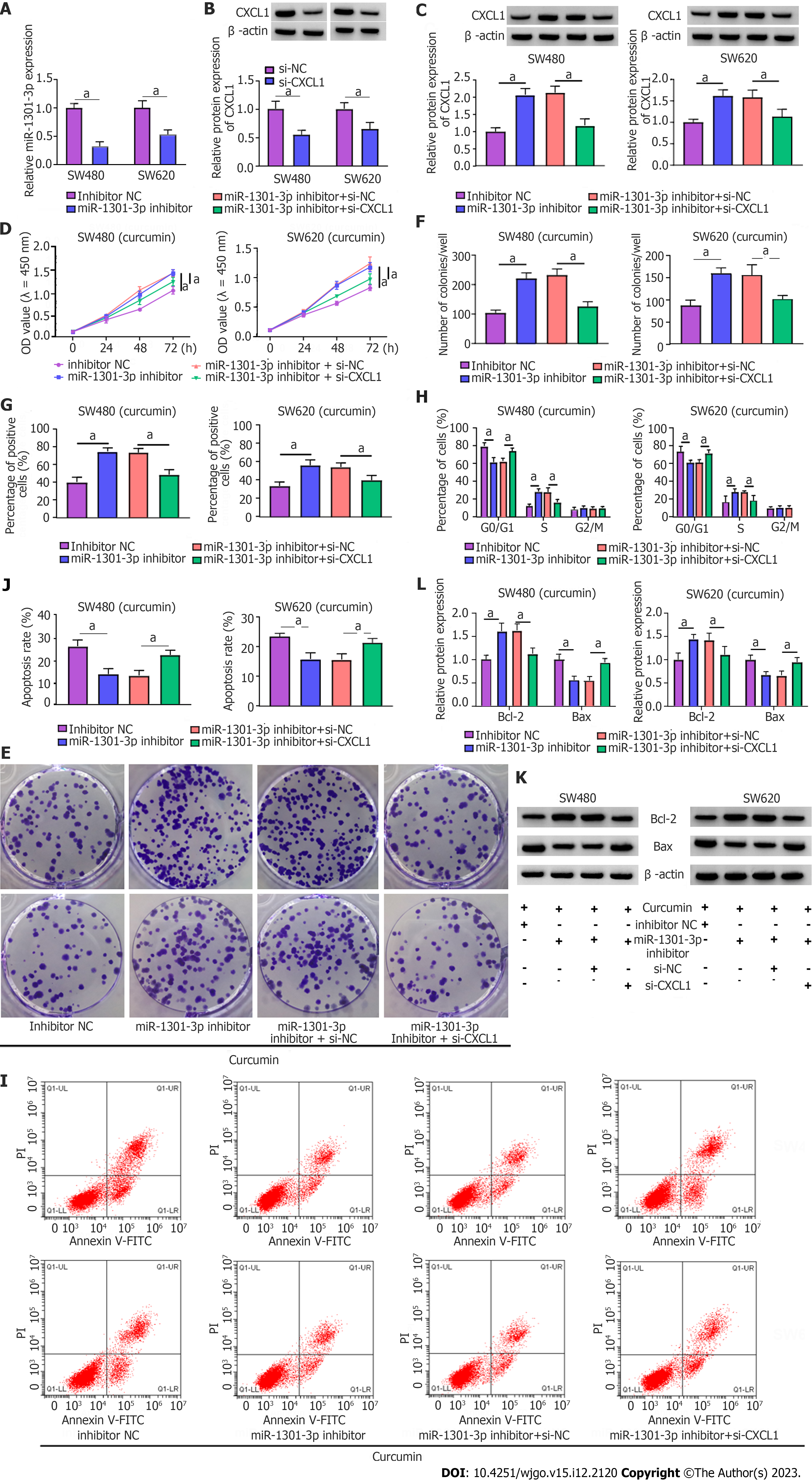
The roles played by miR-1301-3p and CXCL1 in curcumin-mediated proliferation and apoptosis were explored. Simultaneously, the introduction of miR-1301-3p inhibitor and si-CXCL1 into SW480 and SW620 cells was performed, followed by transfection efficiency assessment (Figure 7A and B). Meanwhile, ELISA showed that the introduction of si-CXCL1 might reduce CXCL1 secretion in SW480 and SW620 cells (Supplementary Figure 3C). In addition, reduced miR-1301-3p could reinforce the CXCL1 protein level, while the cotransfection of si-CXCL1 counteracted the effect in SW480 and SW620 cells (Figure 7C). Additionally, ELISA showed that CXCL1 downregulation might significantly abolish the promotion of CXCL1 secretion by the miR-1301-3p inhibitor (Supplementary Figure 3D). Functionally, the augmented cell proliferative ability and cell cycle progression induced by miR-1301-3p downregulation were significantly relieved by CXCL1 knockdown in curcumin-treated SW480 and SW620 cells (Figure 7D-H). In addition, the introduction of si-CXCL1 abolished the negative action of miR-1301-3p knockdown against apoptosis in curcumin-exposed CRC cells (Figure 7I and J), manifested as decreased Bcl-2 and increased Bax levels (Figure 7K and L). In summary, miR-1301-3p regulated proliferation and apoptosis by interacting with CXCL1 in curcumin-treated CRC cells.
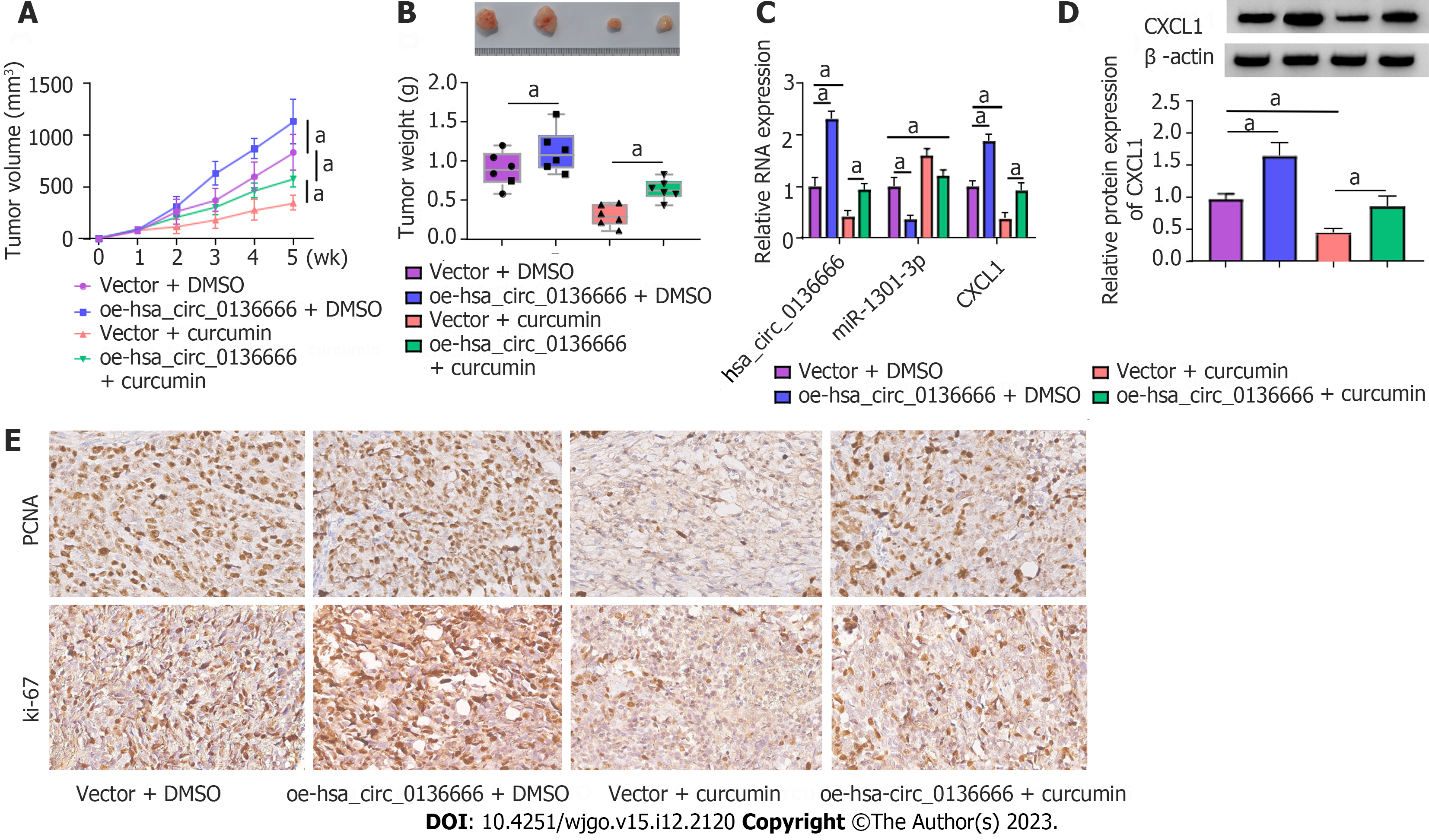
We established mouse xenograft models of CRC to validate the functional effect of hsa_circ_0136666 and curcumin on in vivo tumor growth. As indicated by Figure 8A and B, curcumin could distinctly dampen tumor growth (reduced tumor volume and weight), whereas the overexpression of hsa_circ_0136666 partially attenuated these effects in xenografts (Figure 8A and B). Furthermore, our data showed that curcumin injection could abate hsa_circ_0136666 and CXCL1 Levels in tumor tissues derived from oe-hsa_circ_0136666-transfected SW480 cells (Figure 8C). Synchronously, miR-1301-3p displayed an opposite trend in this xenograft (Figure 8C and D). In addition, IHCS revealed that Ki-67 and PCNA (standard proliferation markers) levels were dampened by hsa_circ_0136666 deficiency in this xenograft (Figure 8E). Together, curcumin suppressed CRC cell growth by regulating hsa_circ_0136666 in vivo.
In this research, the role and mechanism of curcumin in CRC development were explored. Here, we found that curcumin repressed CRC cell proliferation and boosted apoptosis and first verified that it was associated with the hsa_circ_013
Work in several laboratories has revealed that curcumin, a natural polyphenolic compound, presents antioxidant, anti-inflammatory, and anticancer properties[27-29]. Recently, it has become evident that curcumin can repress tumor progression in a variety of cancers[30,31]. In terms of CRC, some studies have indicated that curcumin is a novel agent to prevent cancer development[32-34]. In this regard, the present study suggested the antiproliferative and pro-apoptotic actions of curcumin on CRC, consistent with previous reports[35,36]. Notably, previous documents discovered that circRNAs might be a potential mechanism through which curcumin modulates tumor development[20,21]. In this paper, we found that curcumin could reduce hsa_circ_0136666 levels in CRC cells for the first time. Moreover, recent studies have described that hsa_circ_0136666 acts as a carcinogenic factor by accelerating proliferation and metastasis in CRC[19,37]. Functional analysis suggested that overexpressing hsa_circ_0136666 abolished the curcumin-triggered decline in CRC cell proliferative ability and augmentation of apoptosis in vitro. As expected, hsa_circ_0136666 upregulation also partly reversed the repressive action of curcumin on CRC cell growth in vivo. That is, the regulatory role of curcumin in the development of CRC might be correlated with hsa_circ_0136666.
CircRNAs are mostly stable transcripts with a large number of miRNA-binding loci, enabling them to act as miRNA sponges to repress miRNA modulation of downstream target genes in various human cancers[38,39]. In the current work, we first identified the role of miR-1301-3p as a novel miRNA target of hsa_circ_0136666. Similarly, miR-1301-3p was verified as a tumor-associated miRNA in thyroid papillary, breast, and bladder cancers[40-42]. miR-1301-3p has also been demonstrated to impede CRC cell proliferation and induce apoptosis[24]. Consistent with previous work, miR-1301-3p downregulation was also proven in CRC cells. Interestingly, we observed enhanced miR-1301-3p by curcumin. Furthermore, upregulating miR-1301-3p reversed the effects of hsa_circ_0136666 on curcumin-treated CRC cell growth and apoptosis. These findings imply that curcumin can hinder CRC development by a ceRNA effect of hsa_circ_0136666 and miR-1301-3p.
Analogously, the possible miR-1301-3p-interacting target gene was searched based on bioinformatics, and CXCL1 was validated. CXCL1 is a chemokine of epithelial origin in rodents and humans that elevates tumor epithelia-stromal interactions and boosts tumor growth and invasion[43]. Further studies have shown that CXCL1 plays a tumorigenic role in multiple cancers[44,45], including CRC[46]. Meanwhile, the participation of CXCL1 in the modulation of malignant tumor progression by curcumin has been demonstrated[47,48]. In concordance with these findings, CXCL1 was identified to be downregulated by curcumin treatment. Synchronously, the miR-1301-3p inhibitor enhanced cell growth and repressed apoptosis in curcumin-stimulated CRC cells by targeting CXCL1. Additionally, hsa_circ_0136666 modulated CXCL1 by sponging miR-1301-3p, further verifying that hsa_circ_0136666 regulation of tumor development can be mediated through miR-1301-3p/CXCL1 signaling.
Hsa_circ_0136666 relieves curcumin-induced CRC cell growth and apoptosis partly via miR-1301-3p/CXCL1 signaling. This study elucidates a new mechanism of curcumin and sheds light on developing a new therapeutic for CRC treatment.
Colorectal carcinoma (CRC) is the most frequent-occurring malignant tumour in the United States. Curcumin exerts anti-tumor activity in CRC, but the underlying mechanism needs further elucidation. Meanwhile, over the last decade, circular RNAs (circRNAs) is correlated with the initiation and development of various tumors, containing CRC. Also, the increasing focus on the competing endogenous RNAs hypothesis is that circRNAs serve as microRNA sponges to derepress target mRNAs’ levels.
Clinically, it is urgent to find a more effective therapy, which has high clinical significance for CRC patients. Currently, the chemo-preventive properties of traditional Chinese medicine have drawn great attention in cancer therapeutics. As a naturally occurring medicine, curcumin is derived from the medical plant Curcuma longa L. However, it needs further investigation regarding the mechanism through which curcumin affects CRC development.
This study aims at illuminating whether hsa_circ_0136666 involvement on curcumin-triggered CRC progression was mediated via sponging miR-1301-3p.
CRC cells were adopted and treated with 20 μM curcumin for 24 h. Then, for hsa_circ_0136666 stably upregulation, CRC cells were treated with hsa_circ_0136666 overexpression vector transfection or miR-1301-3p mimic and inhibitor. Real-time quantitative PCR was used to quantified the expression of hsa_circ_0136666, miR-1301-3p and CXCL1. Cell counting kit-8, colony-forming cell, 5-ethynyl-2’-deoxyuridine, and flow cytometry assays were carried out to determine cell proliferation, apoptosis, and cell cycle progression. The relationship between hsa_circ_0136666, miR-1301-3p and CXCL1 were validated by RNA pull-down, RIP, and dual-luciferase reporter assay. In vivo experiments were implemented by the murine xenograft model.
Curcumin treatment could distinctly reduce the colony number of SW480 and SW620 cells relative to their control groups. Furthermore, the apoptosis rate in the curcumin group was markedly improved in comparison with other groups Interestingly, curcumin treatment significantly decreased hsa_circ_0136666 in SW480 and SW620 but had no effect on hsa_circ_0000392 and hsa_circ_0000896 expression. These data indicated the suppressive role of curcumin on CRC development. hsa_circ_0136666 upregulation could abolish the impacts of curcumin on CRC cell growth and apoptosis. Meanwhile, miR-1301-3p acted as a direct target of hsa_circ_0136666 and miR-1301-3p directly targeted CXCL1. Hsa_circ_0136666 could reverse curcumin-triggered CRC cell proliferation and apoptosis by interacting with miR-1301-3p while miR-1301-3p knockdown overturned curcumin-induced increase in CRC cell growth and decrease in apoptosis by targeting CXCL1. Curcumin repressed CRC cell growth via regulating hsa_circ_0136666 in vivo.
Curcumin repressed CRC cell proliferation and boosted apoptosis, and first verified it was associated with the regulatory network of the hsa_circ_0136666/miR-1301-3p/CXCL1. Hsa_circ_0136666 relieves curcumin-induced CRC cell growth and apoptosis partly via the miR-1301-3p/CXCL1 signaling.
This study elucidates a new mechanism of curcumin and sheds light on developing a new therapeutic for CRC treatment.
| 1. | Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4235] [Cited by in RCA: 11977] [Article Influence: 2994.3] [Reference Citation Analysis (9)] |
| 2. | Siegel RL, Jakubowski CD, Fedewa SA, Davis A, Azad NS. Colorectal Cancer in the Young: Epidemiology, Prevention, Management. Am Soc Clin Oncol Educ Book. 2020;40:1-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 113] [Article Influence: 18.8] [Reference Citation Analysis (1)] |
| 3. | Baidoun F, Elshiwy K, Elkeraie Y, Merjaneh Z, Khoudari G, Sarmini MT, Gad M, Al-Husseini M, Saad A. Colorectal Cancer Epidemiology: Recent Trends and Impact on Outcomes. Curr Drug Targets. 2021;22:998-1009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 195] [Article Influence: 39.0] [Reference Citation Analysis (3)] |
| 4. | Cardoso R, Guo F, Heisser T, De Schutter H, Van Damme N, Nilbert MC, Christensen J, Bouvier AM, Bouvier V, Launoy G, Woronoff AS, Cariou M, Robaszkiewicz M, Delafosse P, Poncet F, Walsh PM, Senore C, Rosso S, Lemmens VEPP, Elferink MAG, Tomšič S, Žagar T, Marques ALM, Marcos-Gragera R, Puigdemont M, Galceran J, Carulla M, Sánchez-Gil A, Chirlaque MD, Hoffmeister M, Brenner H. Overall and stage-specific survival of patients with screen-detected colorectal cancer in European countries: A population-based study in 9 countries. Lancet Reg Health Eur. 2022;21:100458. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 53] [Reference Citation Analysis (0)] |
| 5. | Ming T, Tao Q, Tang S, Zhao H, Yang H, Liu M, Ren S, Xu H. Curcumin: An epigenetic regulator and its application in cancer. Biomed Pharmacother. 2022;156:113956. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 71] [Reference Citation Analysis (1)] |
| 6. | Mari M, Carrozza D, Ferrari E, Asti M. Applications of Radiolabelled Curcumin and Its Derivatives in Medicinal Chemistry. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 7. | Abd El-Hack ME, El-Saadony MT, Swelum AA, Arif M, Abo Ghanima MM, Shukry M, Noreldin A, Taha AE, El-Tarabily KA. Curcumin, the active substance of turmeric: its effects on health and ways to improve its bioavailability. J Sci Food Agric. 2021;101:5747-5762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 218] [Article Influence: 43.6] [Reference Citation Analysis (0)] |
| 8. | Termini D, Den Hartogh DJ, Jaglanian A, Tsiani E. Curcumin against Prostate Cancer: Current Evidence. Biomolecules. 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 80] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 9. | Ashrafizadeh M, Najafi M, Makvandi P, Zarrabi A, Farkhondeh T, Samarghandian S. Versatile role of curcumin and its derivatives in lung cancer therapy. J Cell Physiol. 2020;235:9241-9268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 89] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 10. | Rutz J, Janicova A, Woidacki K, Chun FK, Blaheta RA, Relja B. Curcumin-A Viable Agent for Better Bladder Cancer Treatment. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 11. | Pricci M, Girardi B, Giorgio F, Losurdo G, Ierardi E, Di Leo A. Curcumin and Colorectal Cancer: From Basic to Clinical Evidences. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 164] [Cited by in RCA: 151] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 12. | Liu CX, Chen LL. Circular RNAs: Characterization, cellular roles, and applications. Cell. 2022;185:2016-2034. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 682] [Article Influence: 170.5] [Reference Citation Analysis (0)] |
| 13. | Lei M, Zheng G, Ning Q, Zheng J, Dong D. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020;19:30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 202] [Cited by in RCA: 549] [Article Influence: 91.5] [Reference Citation Analysis (0)] |
| 14. | Lu C, Fu L, Qian X, Dou L, Cang S. Knockdown of circular RNA circ-FARSA restricts colorectal cancer cell growth through regulation of miR-330-5p/LASP1 axis. Arch Biochem Biophys. 2020;689:108434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 15. | Xu H, Liu Y, Cheng P, Wang C, Zhou W, Xu Y, Ji G. CircRNA_0000392 promotes colorectal cancer progression through the miR-193a-5p/PIK3R3/AKT axis. J Exp Clin Cancer Res. 2020;39:283. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 106] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 16. | Liu LH, Tian QQ, Liu J, Zhou Y, Yong H. Upregulation of hsa_circ_0136666 contributes to breast cancer progression by sponging miR-1299 and targeting CDK6. J Cell Biochem. 2019;120:12684-12693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 17. | Zhang C, Zhou H, Yuan K, Xie R, Chen C. Overexpression of hsa_circ_0136666 predicts poor prognosis and initiates osteosarcoma tumorigenesis through miR-593-3p/ZEB2 pathway. Aging (Albany NY). 2020;12:10488-10496. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 18. | Li Y, Zang H, Zhang X, Huang G. circ_0136666 Facilitates the Progression of Colorectal Cancer via miR-383/CREB1 Axis. Cancer Manag Res. 2020;12:6795-6806. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 19. | Jin C, Wang A, Liu L, Wang G, Li G. Hsa_circ_0136666 promotes the proliferation and invasion of colorectal cancer through miR-136/SH2B1 axis. J Cell Physiol. 2019;234:7247-7256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 79] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 20. | Xie W, Chu M, Song G, Zuo Z, Han Z, Chen C, Li Y, Wang ZW. Emerging roles of long noncoding RNAs in chemoresistance of pancreatic cancer. Semin Cancer Biol. 2022;83:303-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 86] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 21. | Yang J, Zhu D, Liu S, Shao M, Liu Y, Li A, Lv Y, Huang M, Lou D, Fan Q. Curcumin enhances radiosensitization of nasopharyngeal carcinoma by regulating circRNA network. Mol Carcinog. 2020;59:202-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 22. | Panda AC. Circular RNAs Act as miRNA Sponges. Adv Exp Med Biol. 2018;1087:67-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 494] [Cited by in RCA: 841] [Article Influence: 105.1] [Reference Citation Analysis (0)] |
| 23. | Verduci L, Strano S, Yarden Y, Blandino G. The circRNA-microRNA code: emerging implications for cancer diagnosis and treatment. Mol Oncol. 2019;13:669-680. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 211] [Cited by in RCA: 340] [Article Influence: 48.6] [Reference Citation Analysis (0)] |
| 24. | Xu G, Wang H, Yuan D, Yao J, Meng L, Li K, Zhang Y, Dang C, Zhu K. RUNX1-activated upregulation of lncRNA RNCR3 promotes cell proliferation, invasion, and suppresses apoptosis in colorectal cancer via miR-1301-3p/AKT1 axis in vitro and in vivo. Clin Transl Oncol. 2020;22:1762-1777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 25. | Jiang G, Wu AD, Huang C, Gu J, Zhang L, Huang H, Liao X, Li J, Zhang D, Zeng X, Jin H. Isorhapontigenin (ISO) Inhibits Invasive Bladder Cancer Formation In Vivo and Human Bladder Cancer Invasion In Vitro by Targeting STAT1/FOXO1 Axis. Cancer Prev Res (Phila). 2016;9:567-580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 62] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 26. | Zhang S, Sun J, Gu M, Wang G, Wang X. Circular RNA: A promising new star for the diagnosis and treatment of colorectal cancer. Cancer Med. 2021;10:8725-8740. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 27. | Hasanzadeh S, Read MI, Bland AR, Majeed M, Jamialahmadi T, Sahebkar A. Curcumin: an inflammasome silencer. Pharmacol Res. 2020;159:104921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 222] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 28. | Fernández-Lázaro D, Mielgo-Ayuso J, Seco Calvo J, Córdova Martínez A, Caballero García A, Fernandez-Lazaro CI. Modulation of Exercise-Induced Muscle Damage, Inflammation, and Oxidative Markers by Curcumin Supplementation in a Physically Active Population: A Systematic Review. Nutrients. 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 106] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 29. | Koroth J, Nirgude S, Tiwari S, Gopalakrishnan V, Mahadeva R, Kumar S, Karki SS, Choudhary B. Investigation of anti-cancer and migrastatic properties of novel curcumin derivatives on breast and ovarian cancer cell lines. BMC Complement Altern Med. 2019;19:273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 30. | Zhang X, Zhang C, Ren Z, Zhang F, Xu J, Zhang X, Zheng H. Curcumin Affects Gastric Cancer Cell Migration, Invasion and Cytoskeletal Remodeling Through Gli1-β-Catenin. Cancer Manag Res. 2020;12:3795-3806. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 31. | Ohnishi Y, Sakamoto T, Zhengguang L, Yasui H, Hamada H, Kubo H, Nakajima M. Curcumin inhibits epithelial-mesenchymal transition in oral cancer cells via c-Met blockade. Oncol Lett. 2020;19:4177-4182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 32. | Weng W, Goel A. Curcumin and colorectal cancer: An update and current perspective on this natural medicine. Semin Cancer Biol. 2022;80:73-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 213] [Article Influence: 35.5] [Reference Citation Analysis (0)] |
| 33. | Ruiz de Porras V, Layos L, Martínez-Balibrea E. Curcumin: A therapeutic strategy for colorectal cancer? Semin Cancer Biol. 2021;73:321-330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 34. | Gupta MK, Vadde R, Sarojamma V. Curcumin - A Novel Therapeutic Agent in the Prevention of Colorectal Cancer. Curr Drug Metab. 2019;20:977-987. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 35. | Calibasi-Kocal G, Pakdemirli A, Bayrak S, Ozupek NM, Sever T, Basbinar Y, Ellidokuz H, Yigitbasi T. Curcumin effects on cell proliferation, angiogenesis and metastasis in colorectal cancer. J BUON. 2019;24:1482-1487. [PubMed] |
| 36. | Ismail NI, Othman I, Abas F, H Lajis N, Naidu R. Mechanism of Apoptosis Induced by Curcumin in Colorectal Cancer. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 107] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 37. | Wang G, Li Y, Zhu H, Huo G, Bai J, Gao Z. Circ-PRKDC Facilitates the Progression of Colorectal Cancer Through miR-198/DDR1 Regulatory Axis. Cancer Manag Res. 2020;12:12853-12865. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 38. | Misir S, Hepokur C, Aliyazicioglu Y, Enguita FJ. Circular RNAs serve as miRNA sponges in breast cancer. Breast Cancer. 2020;27:1048-1057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 39. | Zhou J, Wang L, Sun Q, Chen R, Zhang C, Yang P, Tan Y, Peng C, Wang T, Jin C, Ji J, Jin K, Sun Y. Hsa_circ_0001666 suppresses the progression of colorectal cancer through the miR-576-5p/PCDH10 axis. Clin Transl Med. 2021;11:e565. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 80] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 40. | Peng X, Yan B, Shen Y. MiR-1301-3p inhibits human breast cancer cell proliferation by regulating cell cycle progression and apoptosis through directly targeting ICT1. Breast Cancer. 2018;25:742-752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 41. | Qiao DH, He XM, Yang H, Zhou Y, Deng X, Cheng L, Zhou XY. miR-1301-3p suppresses tumor growth by downregulating PCNA in thyroid papillary cancer. Am J Otolaryngol. 2021;42:102920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 42. | Liu Y, Wu G. NNT-AS1 enhances bladder cancer cell growth by targeting miR-1301-3p/PODXL axis and activating Wnt pathway. Neurourol Urodyn. 2020;39:547-557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 43. | Miyake M, Lawton A, Goodison S, Urquidi V, Rosser CJ. Chemokine (C-X-C motif) ligand 1 (CXCL1) protein expression is increased in high-grade prostate cancer. Pathol Res Pract. 2014;210:74-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 44. | Wang N, Liu W, Zheng Y, Wang S, Yang B, Li M, Song J, Zhang F, Zhang X, Wang Q, Wang Z. CXCL1 derived from tumor-associated macrophages promotes breast cancer metastasis via activating NF-κB/SOX4 signaling. Cell Death Dis. 2018;9:880. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 244] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 45. | Yu S, Yi M, Xu L, Qin S, Li A, Wu K. CXCL1 as an Unfavorable Prognosis Factor Negatively Regulated by DACH1 in Non-small Cell Lung Cancer. Front Oncol. 2019;9:1515. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 46. | Wang D, Sun H, Wei J, Cen B, DuBois RN. CXCL1 Is Critical for Premetastatic Niche Formation and Metastasis in Colorectal Cancer. Cancer Res. 2017;77:3655-3665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 282] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 47. | Howells LM, Iwuji COO, Irving GRB, Barber S, Walter H, Sidat Z, Griffin-Teall N, Singh R, Foreman N, Patel SR, Morgan B, Steward WP, Gescher A, Thomas AL, Brown K. Curcumin Combined with FOLFOX Chemotherapy Is Safe and Tolerable in Patients with Metastatic Colorectal Cancer in a Randomized Phase IIa Trial. J Nutr. 2019;149:1133-1139. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 184] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 48. | Killian PH, Kronski E, Michalik KM, Barbieri O, Astigiano S, Sommerhoff CP, Pfeffer U, Nerlich AG, Bachmeier BE. Curcumin inhibits prostate cancer metastasis in vivo by targeting the inflammatory cytokines CXCL1 and -2. Carcinogenesis. 2012;33:2507-2519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 120] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Cardiac and cardiovascular systems
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Garcia K, Spain; Schijven MP, Netherlands S-Editor: Lin C L-Editor: A P-Editor: Wu RR