Published online Sep 15, 2022. doi: 10.4251/wjgo.v14.i9.1844
Peer-review started: March 14, 2022
First decision: April 17, 2022
Revised: April 29, 2022
Accepted: August 15, 2022
Article in press: August 15, 2022
Published online: September 15, 2022
Processing time: 179 Days and 4.6 Hours
Genetic variants of Helicobacter pylori (H. pylori) are involved in gastric cancer occurrence. Single nucleotide polymorphisms (SNPs) of H. pylori that are assoc
To assess the performance of a polygenic risk score (PRS) based on H. pylori SNPs in predicting the risk of gastric cancer.
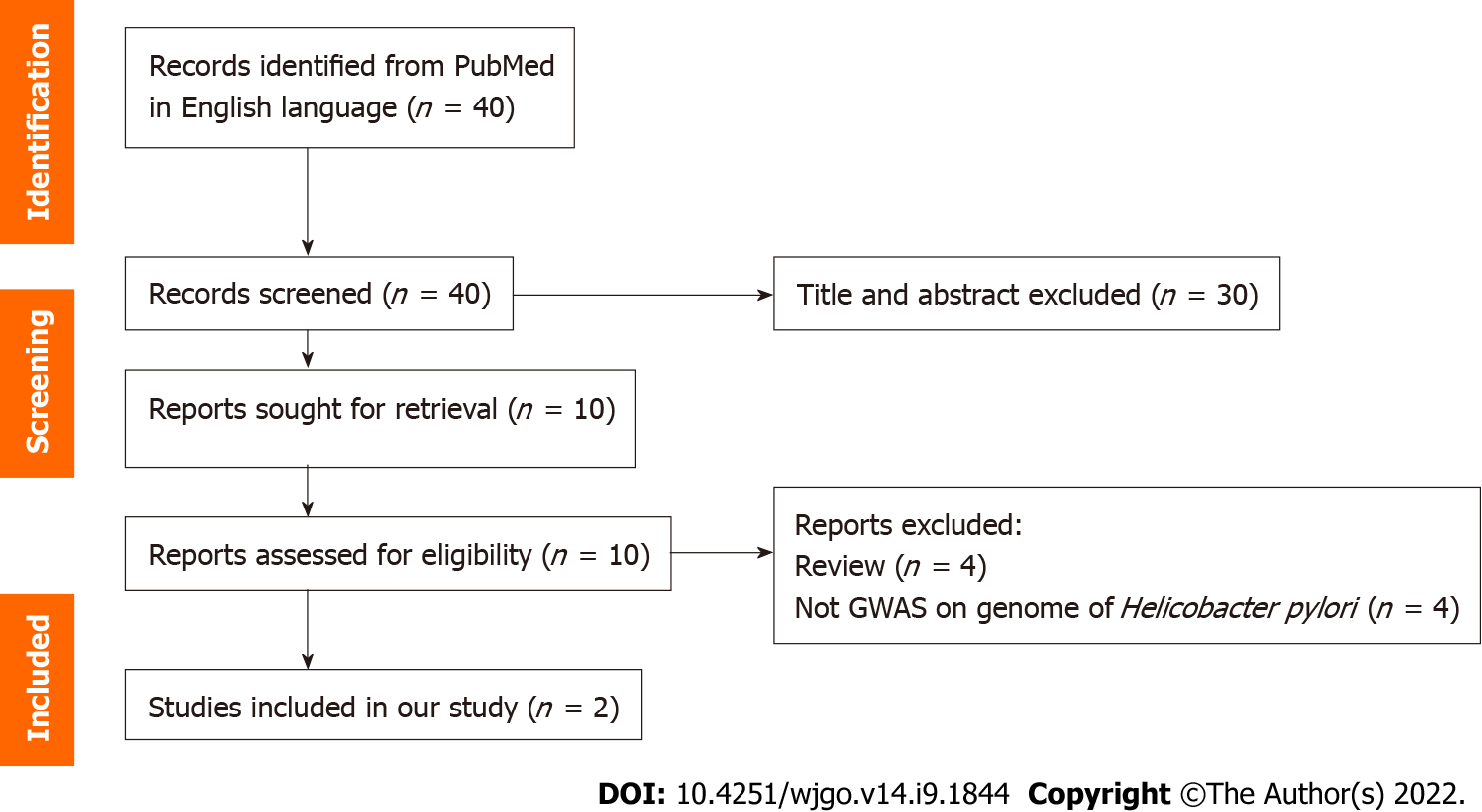
A total of 15 gastric cancer-associated H. pylori SNPs were selected. The associations between these SNPs and gastric cancer were further validated in 1022 global strains with publicly available genome sequences. The PRS model was established based on the validated SNPs. The performance of the PRS for predicting the risk of gastric cancer was assessed in global strains using quintiles and random forest (RF) methods. The variation in the performance of the PRS among different populations of H. pylori was further examined.
Analyses of the association between selected SNPs and gastric cancer in the global dataset revealed that the risk allele frequencies of six SNPs were significantly higher in gastric cancer cases than non-gastric cancer cases. The PRS model constructed subsequently with these validated SNPs produced significantly higher scores in gastric cancer. The odds ratio (OR) value for gastric cancer gradually increased from the first to the fifth quintile of PRS, with the fifth quintile having an OR value as high as 9.76 (95% confidence interval: 5.84-16.29). The results of RF analyses indicated that the area under the curve (AUC) value for classifying gastric cancer and non-gastric cancer was 0.75, suggesting that the PRS based on H. pylori SNPs was capable of predicting the risk of gastric cancer. Assessing the performance of the PRS among different H. pylori populations demonstrated that it had good predictive power for cancer risk for hpEurope strains, with an AUC value of 0.78.
The PRS model based on H. pylori SNPs had a good performance for assessment of gastric cancer risk. It would be useful in the prediction of final consequences of the H. pylori infection and beneficial for the management of the infection in clinical settings.
Core Tip: Prediction of cancer risk is of importance in the clinical management of populations with a high risk of gastric cancer. This study constructed a polygenic risk score (PRS) model based on Helicobacter pylori (H. pylori) single nucleotide polymorphisms (SNPs) to predict the risk of gastric cancer. Associations between previously reported H. pylori SNPs and gastric cancer were validated in global strains. A PRS model constructed with validated SNPs had a high predictive power for gastric cancer at a global level and for individuals infected with hpEurope strains. It has potential for clinical use in the management of the H. pylori infection.
- Citation: Wang XY, Wang LL, Liang SZ, Yang C, Xu L, Yu MC, Wang YX, Dong QJ. Prediction of gastric cancer risk by a polygenic risk score of Helicobacter pylori. World J Gastrointest Oncol 2022; 14(9): 1844-1855
- URL: https://www.wjgnet.com/1948-5204/full/v14/i9/1844.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i9.1844
Helicobacter pylori (H. pylori) infection affects more than half of the world’s population[1,2]. The outcomes of H. pylori infection vary among individuals. The consequences of most infections are benign. However, a minority of infected individuals may eventually develop gastric cancer[3,4]. Predicting the outcomes of H. pylori infection is a major concern in the management of the infection. Substantial genetic variation has been found in the pathogen. Mutations cause increased virulence in certain strains, enhancing their carcinogenic potential[5,6]. It has been demonstrated that typing H. pylori strains based on the genetic variations of virulent genes has the potential to predict the risk of gastric cancer[7,8].
Two studies have been recently conducted to investigate the association between H. pylori genomic variations and gastric cancer within the hpEurope and hpEastAsia populations, respectively[9,10]. The first study contained 173 hpEurope strains and found 11 cancer risk-associated variants, including gene loss variants and single nucleotide polymorphisms (SNPs). Risk scores calculated based on the status of the cag11, cag12 and cag20 genes were increased during the progression from inflammation to gastric cancer. The other study identified 11 SNPs and three DNA motifs associated with gastric cancer through examination of 240 hpEastAsia strains. It is unclear whether the association between these variations and gastric cancer exists for all H. pylori strains. However, the findings from these studies suggest that SNPs from the H. pylori genome have the potential to predict the risk of gastric cancer.
To explore the combined effect of multiple SNPs on disease susceptibility, the polygenic risk score (PRS) model has been developed[11]. A PRS is calculated as a sum of the effects of multiple SNPs on disease. PRS models composed of SNPs from the human genome have been successfully used to predict the risk of cancers such as gastric cancer, colorectal cancer, and breast cancer[12-15]. Few studies, however, have been conducted to explore the capacities of PRS model constructed with SNPs from bacterial genomes in predicting the risk of cancer. Our study aimed to construct a PRS model based on validated risk alleles of H. pylori to predict the risk of gastric cancer.
A total of 2022 H. pylori genome sequences deposited in GenBank at the National Center for Biotechnology Information by December 8, 2021 (https://www.ncbi.nlm.nih.gov/genome/browse#!/prokaryotes/169/), and the figshare website (https://figshare.com/s/2174da1fa20ae71c71e0)[10] were downloaded for further analyses. Of them, 1187 H. pylori strains had relevant clinical information of patients. Subsequently, duplicate strains and strains isolated from peptic ulcer disease, mucosa-associated lymphoma or stromal tumors were excluded from further analyses. This led to a final dataset of 1022 global strains included in the study. They were divided into gastric cancer (n = 253) and non-gastric cancer (n = 769) groups. Patients in the latter group were diagnosed with functional dyspepsia (n = 46), or chronic gastritis with or without intestinal metaplasia (n = 143 and n = 580, respectively). A total of 15 H. pylori SNPs or genetic variants from the two previous genome-wide association studies (GWASs) were selected for further analyses (Figure 1, Table 1)[9,10]. We cited high-quality articles in Reference Citation Analysis (https://www.referencecitationanalysis.com). The selection criteria were as follows: (1) The length of the variants was no longer than five contiguous nucleotides; and (2) The SNP selected was located in a protein-coding region.
| SNP | Corresponding locus in the strain 26695 | Gene name | Description | Position in gene | Position in chromosome | Risk allele | Amino acid change | Prevalence of risk allele | P value | LogOR-value |
| 11 | HP00821 | tlpC1 | Chemotaxis sensor1 | 1631 | 880291 | A1 | K217E,Q1 | 16.2%1 | 2.88E-151 | 1.291 |
| 21 | HP01301 | triH1 | BIR, Dps/NapA, RAD21 similarity1 | 3451 | 1407971 | C1 | Synonymous1 | 16.2%1 | 2.88E-151 | 2.261 |
| 31 | HP02311 | dsbG/K1 | Thiol:disulfide interchange protein1 | 4331 | 2416251 | A1 | T145A1 | 23.2%1 | 5.34E-151 | 1.241 |
| 4 | HP0468 | Unknown | 729 | 489762 | A | Synonymous | 34.6% | 0.110 | ||
| 5 | HP0468 | Unknown | 705-708 | 489783-489786 | CGCC | A236T | 1.2% | 0.313 | ||
| 6 | HP0709 | Adenosyl-chloride synthase | 145 | 762953 | A | N49D | 14.6% | 0.08 | ||
| 7 | HP0709 | Adenosyl-chloride synthase | 159 | 762967 | A | Synonymous | 90.0% | 0.274 | ||
| 81 | HP07471 | trmB1 | tRNA ([guanine-N(7)-]-methyltransferase1 | (934-937)1 | (803467-803470)1 | GGAA1 | G312K,G,R+T313A,T,S1 | 38.7%1 | 1.17E-141 | 1.201 |
| 9 | HP0797 | hpaA | Neuraminyllactose-binding hemagglutinin | 334 | 854406 | T | S112A | 26.8% | 0.567 | |
| 101 | HP07971 | hpaA1 | Neuraminyllactose-binding hemagglutinin1 | 3251 | 8544151 | C1 | L109F1 | 40.1%1 | 1.12E-141 | 2.161 |
| 11 | HP0807 | fecA-2 | Iron importer in outer membrane | 2010 | 861345 | C | Synonymous | 96.7% | 0.158 | |
| 121 | HP10551 | Outer membrane protein1 | 7981 | 11174021 | A1 | Synonymous1 | 34.6%1 | 2.93E-141 | 2.581 | |
| 13 | HP1250 | csd5 | Cell shape determinant | 370 | 1325727 | A | N116H,D | 65.8% | 1 | |
| 14 | HP1440 | isp | Inactive Ser protease | 533 | 1513405 | G | G173E | 92.0% | 0.505 | |
| 15 | HP1467 | ompA101 | Outer membrane protein of OmpA family | 53 | 1538114 | T | I18T | 98.4% | 0.246 |
Based on the 1022 H. pylori genomes, the SNPs in the core genome (present in > 99% isolates) were identified by aligning the assembled genomes against the reference genome (26695-1MET, accession number: CP010436.1) using MUMmer as previously described[16,17]. A neighbour-joining tree was then constructed based on the sequences of concatenated SNPs using TreeBeST software (http://treesoft.sourceforge.net/treebest.shtml) with default parameters.
The chi-square test was used to test the difference in the prevalence of risk alleles in strains isolated from gastric cancer and non-gastric cancer. Student’s t test was used to compare the PRS values between the gastric cancer and non-cancer groups. These tests were performed using SPSS 18.0 software. Odds ratios (ORs) and 95% confidence intervals (CIs) of the selected SNPs were calculated using logistic regression analysis in R (version 3.6.3).
A PRS was created for each strain using the following equation: PRS = β1 + β2 + … βk… + βn. Briefly, in this equation, βk is the value obtained from the regression analysis of the risk allele and disease, and n is the total number of SNPs included in the PRS[18]. Logistic regression analysis was performed to evaluate the association between PRS and gastric cancer risk and by quintiles of the PRS risk distribution, standardized by the controls, and using the 3rd quintile, 40%-60%, as the reference[18].
A random forest (RF) model was built using the AUC-RF algorithm[19]. The input variables were the scores of each of the validated SNPs. A 20-times repeated 10-fold cross-validation of the RF model was performed. The performance of the RF model was demonstrated by receiver operating characteristic curve analysis[20].
Previous studies have identified two sets of H. pylori SNPs that are associated with gastric cancer[9,10]. The association between these SNPs and gastric cancer has been verified only in strains from the hpEurope or hpEastAsia populations, respectively. We selected 15 SNPs to validate the association between selected SNPs and gastric cancer in global strains (Table 1). The risk alleles were defined as those with a higher prevalence in strains from gastric cancer. Statistical analyses revealed that the risk alleles of six SNPs showed a significant increase in prevalence in the gastric cancer group compared with the non-gastric cancer group. These SNPs, validated in the global dataset, were used for subsequent analyses.
To construct a PRS model for predicting the risk of gastric cancer, the logOR values of each validated SNP were calculated (Table 1). A PRS model was subsequently constructed with the sum of the logOR values of six validated SNPs. The mean PRS value was 8.64 ± 1.71 and 6.99 ± 1.27 in the gastric cancer and non-gastric cancer groups, respectively. The PRS value in the gastric cancer group was significantly higher (P = 5.6E-36).
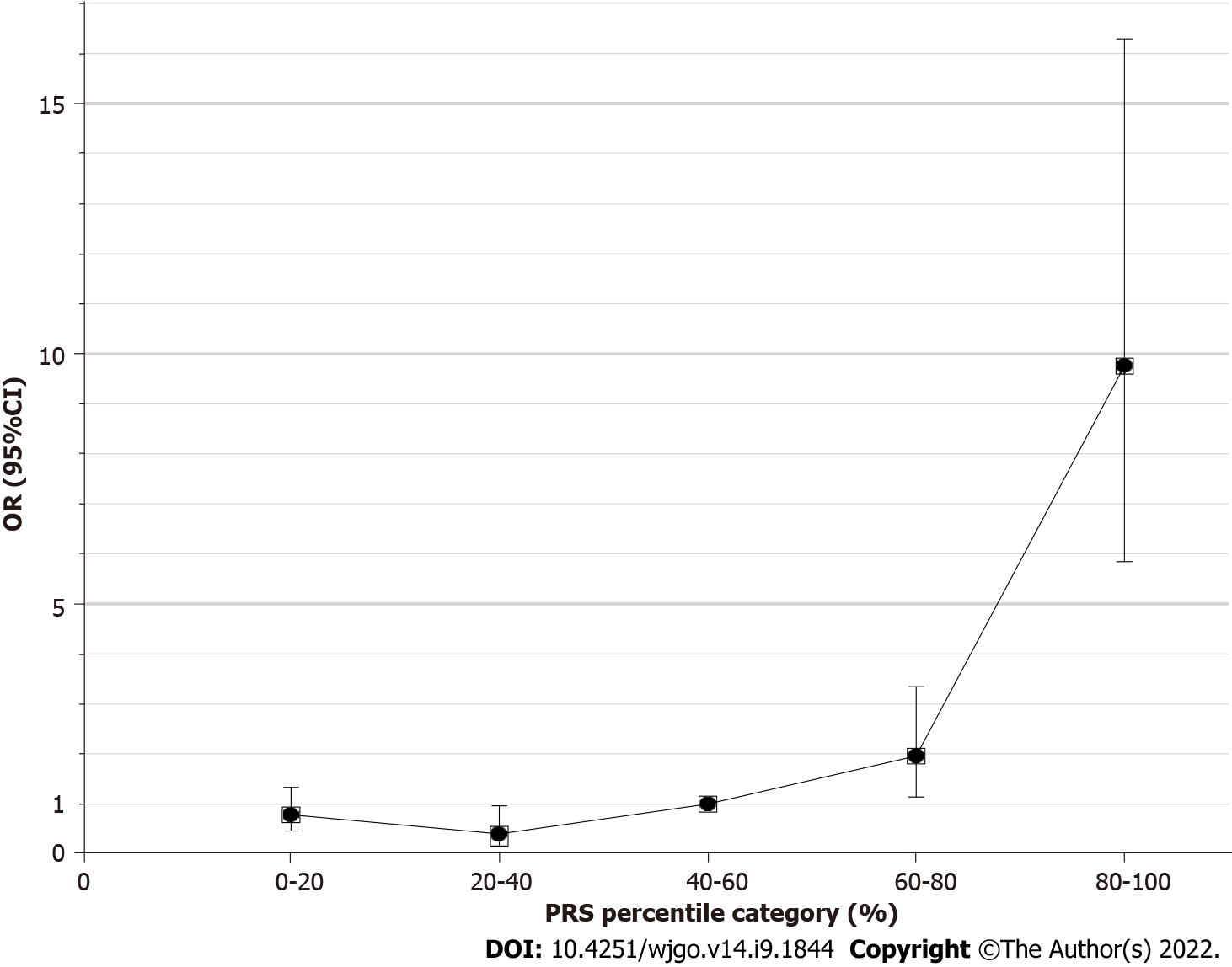
To evaluate the performance of the 6-SNP PRS model for predicting the risk of gastric cancer, the PRS values for each of the selected 1022 strains were grouped according to the quintile method. With the third quintile as the reference, the estimated OR value gradually increased from the first quintile (< 20%) to the fifth quintile (> 80%) (Figure 2, Table 2). The fifth quintile had an OR value as high as 9.76 (95%CI: 5.84-16.29).
| Quintile | Non-GC | GC | OR (95%CI) | P value |
| 1 | 315 (41.0%) | 44 (17.3%) | 0.79 (0.46-1.34) | 0.405 |
| 2 | 98 (12.7%) | 7 (2.8%) | 0.40 (0.17-0.97) | 0.052 |
| 3 | 141 (18.3%) | 25 (9.9%) | 1 | - |
| 4 | 141 (18.3%) | 49 (19.4%) | 1.96 (1.15-3.35) | 0.013 |
| 5 | 74 (9.6%) | 128 (50.6%) | 9.76 (5.84-16.29) | 1.25E-14 |
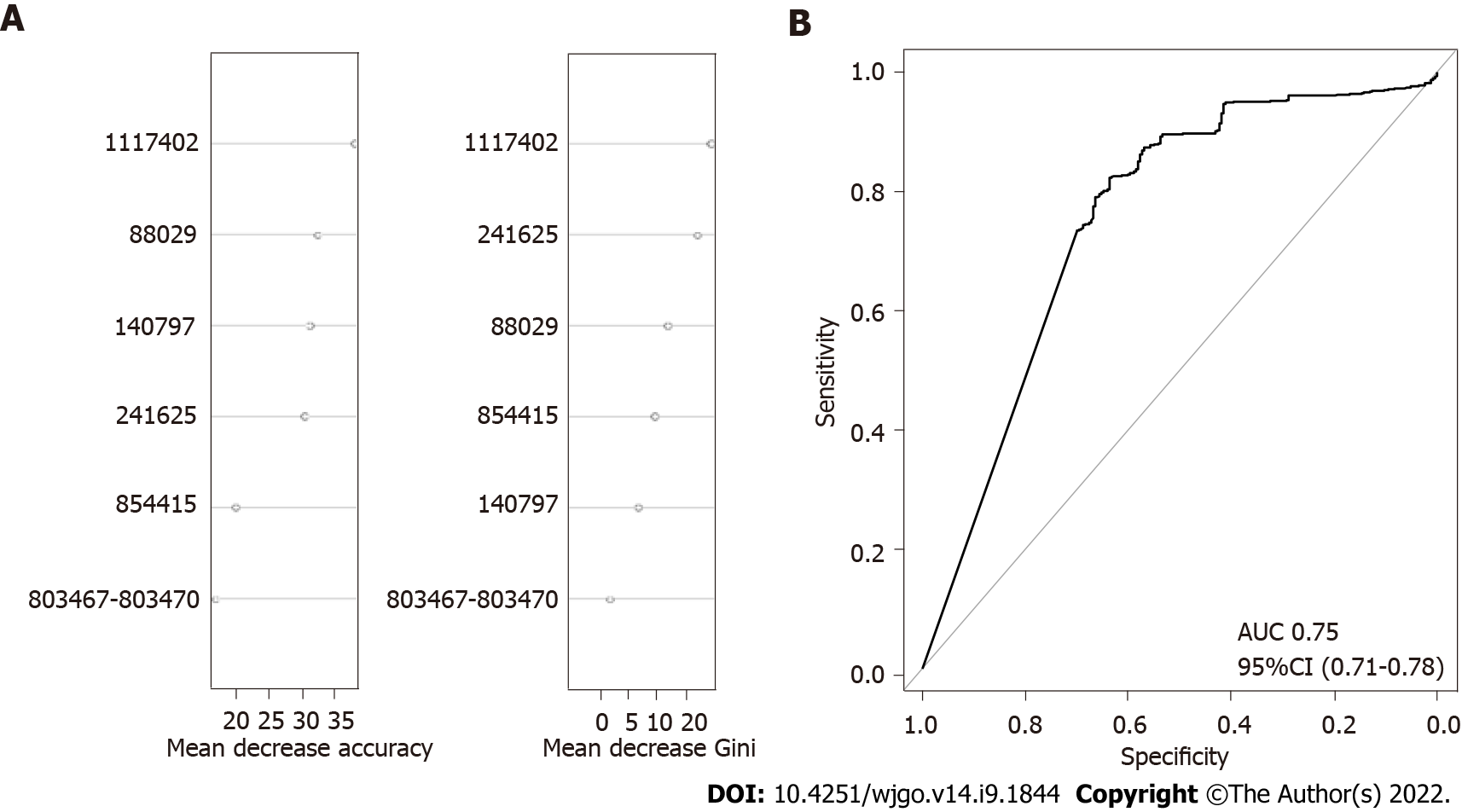
To further confirm the combined effect of the validated SNPs for prediction of gastric cancer risk, an RF model was constructed with logOR values from each SNP as input. The classification potentials of the combined logOR values of validated SNPs were then analysed. The importance of each SNP is shown in Figure 3. The AUC value was 0.75 (DeLong 95%CI: 0.71-0.78), suggesting a good classifying capacity of the combined SNPs.
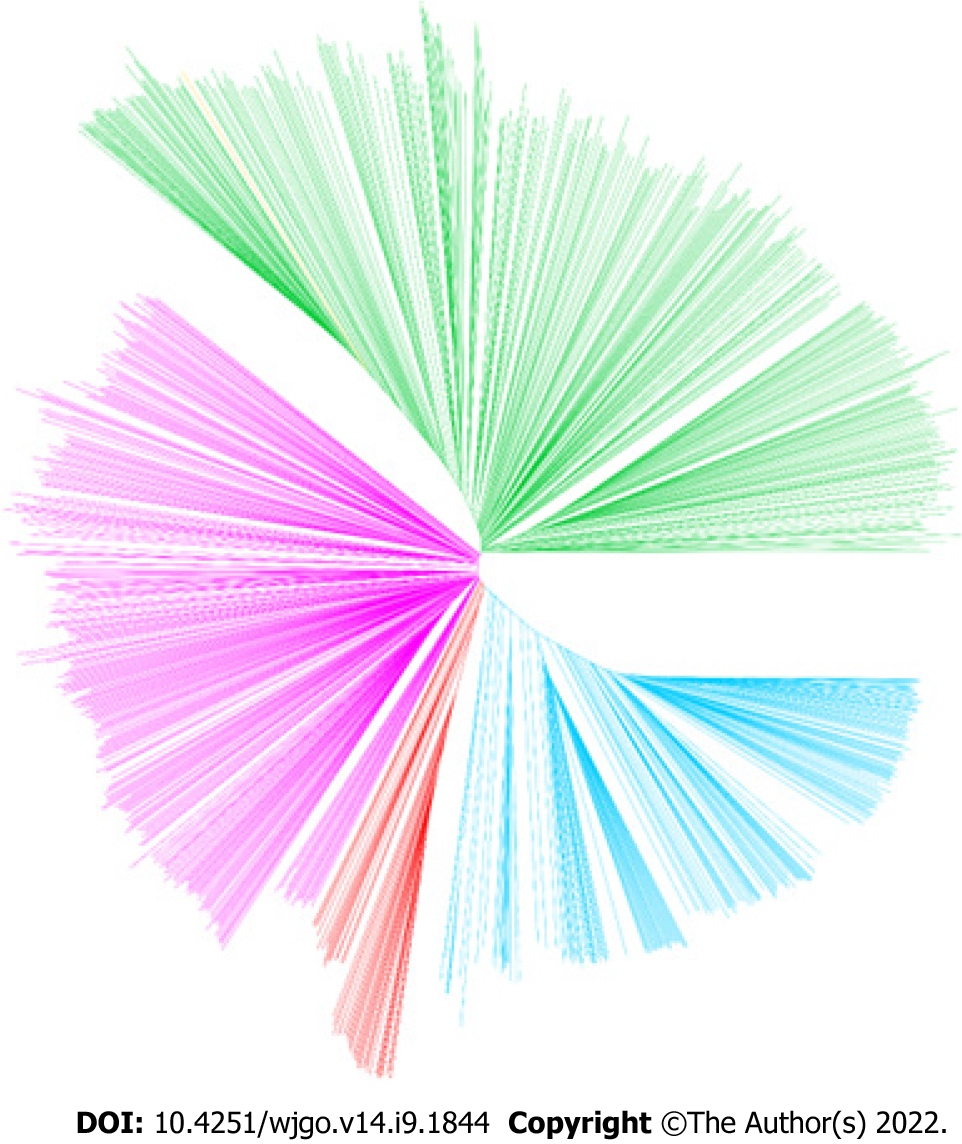
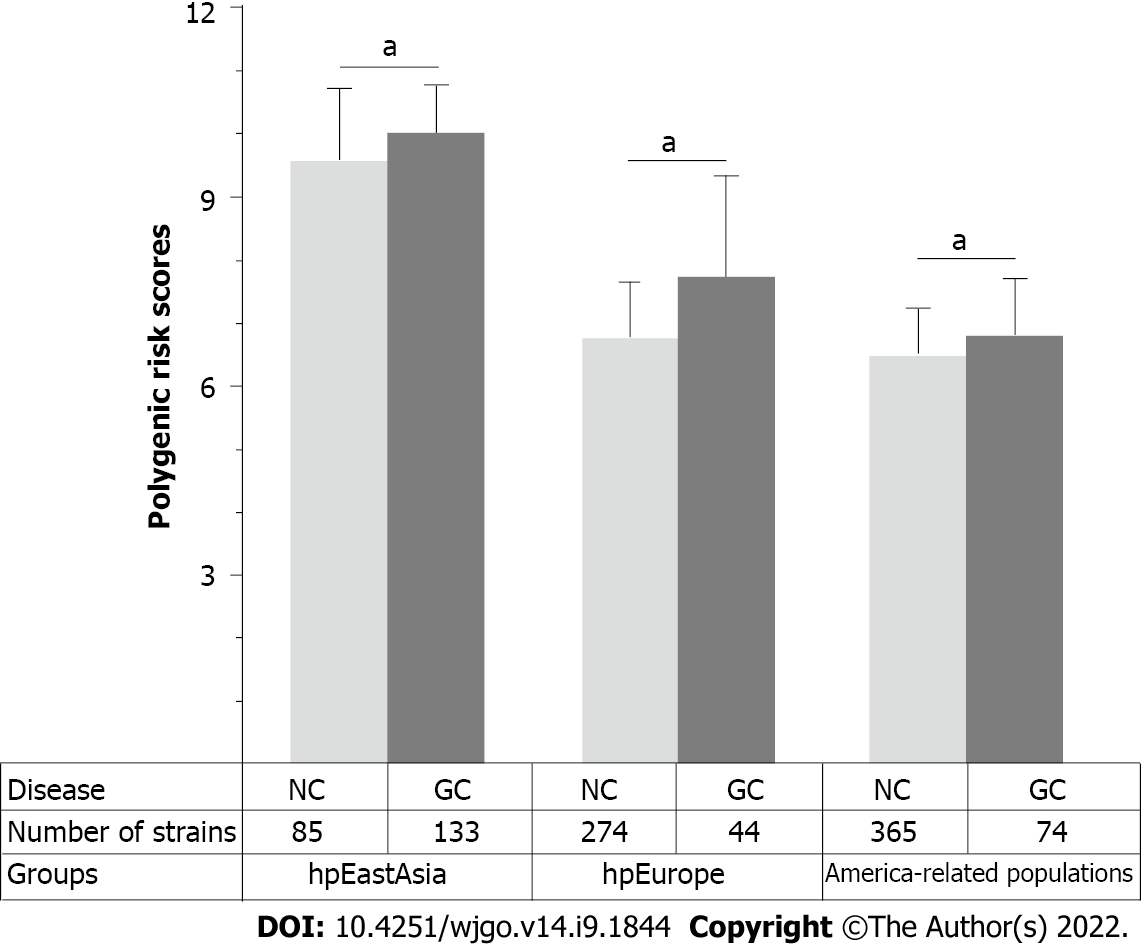
Considering the remarkable genomic variations among strains from different H. pylori populations, the performance of PRS for predicting the risk of gastric cancer was subsequently assessed in different H. pylori populations. The results of the phylogenetic analyses divided the 1022 global strains into five groups, namely, the hpEastAsia, hpAsia2, hpEurope, America-related and Africa-related populations (Figure 4). Due to the small number of gastric cancer cases (2 cases in hpAsia2 and no cases in Africa-related populations), hpAsia2 and Africa-related populations were excluded from subsequent analyses. In analysing the performance of the established PRS model in different populations, the PRS value was higher in the gastric cancer group for all populations. Statistical analyses revealed a significant difference in PRS between the gastric cancer and non-gastric cancer groups in the hpEastAsia, hpEurope and America-related populations (Figure 5).
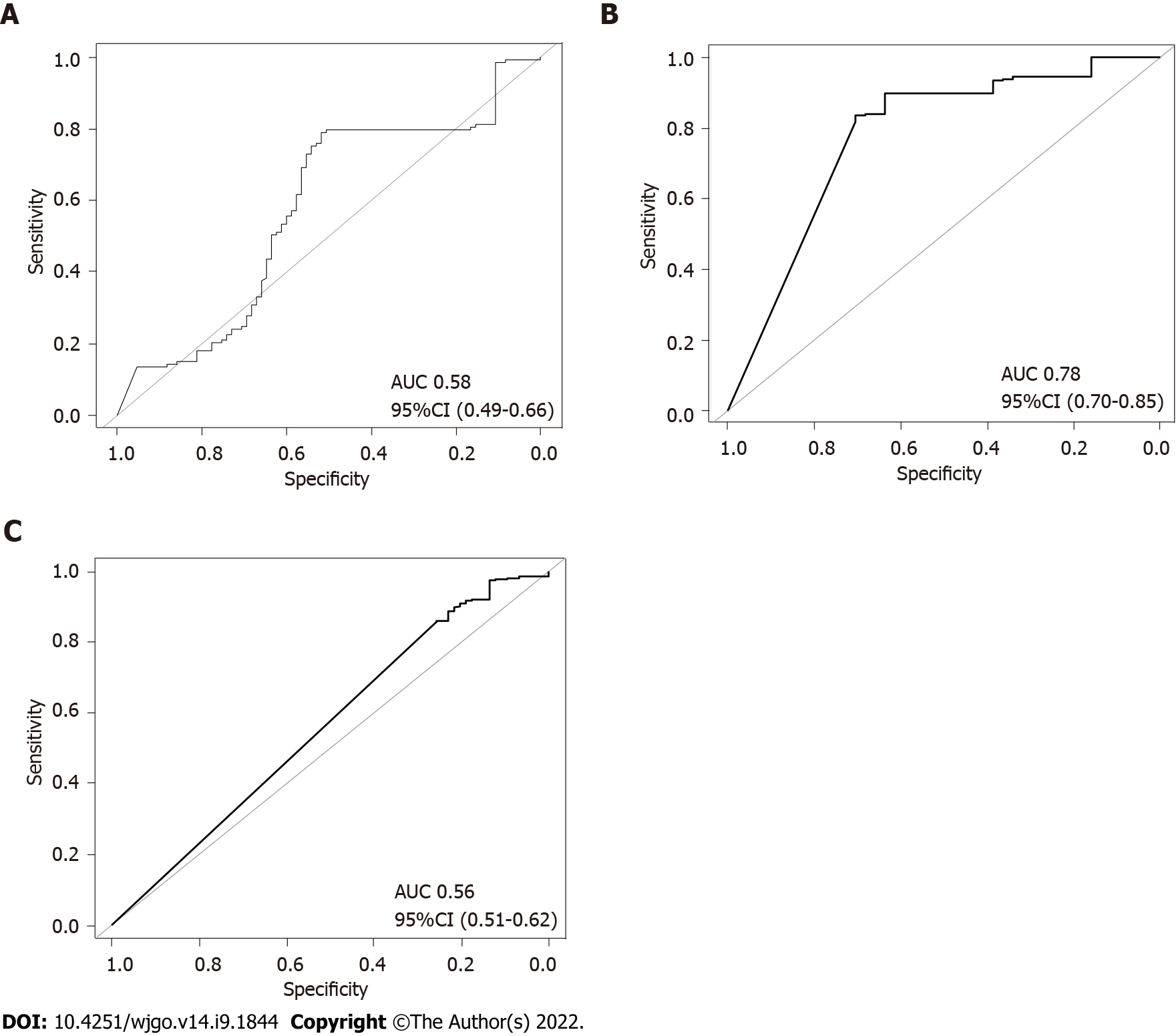
To further verify the combined effects of these SNPs for prediction of gastric cancer risk for different H. pylori populations, a RF classification model was built. The results of RF model analyses demonstrated that the AUC value was highest (0.78, DeLong 95%CI: 0.70-0.85) in the hpEurope population, suggesting a good ability of the combined SNPs to predict the risk of gastric cancer (Figure 6). However, the performance of the combined SNPs for risk prediction in other H. pylori populations was poor (Figure 6).
In this study, we constructed a PRS model based on validated H. pylori SNPs to predict the risk of gastric cancer. To our knowledge, our study is the first to evaluate a PRS model for cancer risk prediction constructed with genomic variants of H. pylori. H. pylori shows substantial genetic variations, resulting in remarkable interstrain differences in carcinogenetic potential[5,21]. The presence/absence or large sequence variation of virulence genes and H. pylori SNPs have been shown to promote gastric carcinogenesis. Few studies have been conducted to assess the predictive power of these cancer-related genetic variations for gastric cancer[9,10]. Moreover, the combined effect of multiple variations on the predictive power for cancer risk has not been explored. Findings from this study demonstrate that a PRS model combining six H. pylori SNPs had a moderate capacity for prediction of gastric cancer risk. This is similar to the findings in studies on PRS model constructed with cancer-associated SNPs from the human genome[14,15].
To assess the combined effects of SNPs on gastric cancer risk prediction, we first selected 15 cancer-associated H. pylori SNPs from two previous GWAS studies. Their association has been validated in strains from specific geographical regions but not in a global strain collection. Our results demonstrated that only six of the SNPs showed a close association with gastric cancer in the global dataset. The SNPs at 88029, 241625, 803467 and 854415 in the reference strain 26695 caused nonsynonymous changes in the corresponding amino acid sequence, whereas the SNPs at 140797 and 1117402 in the reference strain 26695 produced synonymous variations. The hpaA gene, harbouring the SNP at 854415, encodes an adhesion gene of H. pylori[22]. This gene is essential for colonization and is associated with the occurrence of gastric cancer[23-25]. The SNP at 88029 was located on the tlpC gene. TlpC encodes a chemoreceptor that affects the chemotaxis of strains in the mouse gastric environment. It is associated with the induction of mucosal inflammation of the stomach[26,27]. The SNP at 241625 was located in dsbG/K, which has protein disulfide isomerase activity. DsbG/K interacts with a virulence-related factor in vitro[28,29]. In vitro studies have shown that a lack of dsbG/K may cause the loss of T4SS function and inhibit VacA secretion, which are considered the main pathogenic factors in H. pylori[30].
In this study, we constructed a PRS model with six SNPs validated in a global dataset. Assessments of the performance of the PRS model demonstrated that the PRS value was significantly higher in the gastric cancer group than in the non-gastric cancer group. A significant increase in the risk of gastric cancer was found across the quintiles of the PRS. These findings demonstrate that the six-SNPs PRS model is capable of predicting the risk of gastric cancer. In support of this finding, RF analyses demonstrated that the combination of the six SNPs has a high predictive power for gastric cancer, with an AUC value of 0.75. In a recent report, a PRS model constructed with SNPs from the human genome showed unsatisfactory power in classifying gastric cancer from healthy controls, with an AUC value of 0.56[31]. It has been shown that a PRS model derived from 112 SNPs in the human genome and lifestyle factors possesses good predictive capacity for gastric cancer risk[32]. For individuals infected with H. pylori, assessment of their gastric cancer risk is of great concern in the clinical settings. Previous reports have demonstrated that certain genetic variants are associated with increased gastric cancer risk[9,10]. Our study, for the first time, demonstrated the combined effect of H. pylori genomic variations in the assessment of cancer risk. The PRS model derived from H. pylori SNPs would have a high capacity in predicting gastric cancer risk for patients infected with the pathogen. This will benefit the clinical management of the prognosis of the H. pylori infection. It is well known that age, gender and lifestyle factors, including alcohol consuming, smoking, diet habits and economic status, are closely associated with gastric cancer[33-35]. In the future, a PRS model constructed with H. pylori SNPs and those gastric cancer associated risk factors in this study would have substantially increased power in predicting the risk of gastric cancer. The H. pylori genome shows great variations between strains[36,37]. Genetic information differs greatly among H. pylori populations, and their carcinogenic potential is also different[5,21]. We thus evaluated the performance of the PRS model across H. pylori populations. Our results demonstrated a good predictive power of PRS for hpEurope strains.
A limitation of this study is that the performance of the PRS model was not assessed in hpAsia2 and Africa-related H. pylori populations because the number of strains with clinical information available was insufficient. Moreover, we could not consider age, gender, nutrition and other risk factors in the construction of the PRS model, as information on all of these risk factors was not consistently available across databases. A comprehensive risk model enclosing other risk factors of gastric cancer is indicated in future studies. Further in vitro and in vivo exploration of the roles of the combination of H. pylori SNPs identified in this study in gastric cancer would be much helpful in supporting our findings.
In summary, we constructed a PRS model based on H. pylori SNPs, which showed great potential in the prediction of gastric cancer risk globally, especially for individuals infected with hpEurope strains. Findings from this study demonstrated that the PRS model constructed from bacteria genomic variations, in addition to the PRS model established with human SNPs, can be of great value for disease risk prediction. In clinical practice, it is usually difficult to assess gastric cancer risk in patients infected with H. pylori. The model constructed in this study would be beneficial for solving this issue.
Multiple single nucleotide polymorphisms (SNPs) of Helicobacter pylori (H. pylori) associated with gastric cancer have been identified through bacterial genome-wide association studies. Polygenic risk score (PRS) calculated as a sum of effect of SNPs provides a tool for assessing genetic impact on diseases.
Predicting risk of gastric cancer is a major concern in the management of the H. pylori infection.
This study constructed a PRS model based on H. pylori SNPs to predict the risk of gastric cancer.
Associations between previously reported H. pylori SNPs and gastric cancer were validated in global strains. The PRS model based on the validated SNPs was evaluated by quintiles and random forest (RF) methods.
A PRS model was constructed with six validated SNPs. Quintiles and RF methods demonstrated the combination of six SNPs has a high predictive power for gastric cancer.
PRS model constructed from bacterial genomic variations can be of great value for gastric cancer risk prediction.
Comprehensive risk models including personal and genomic information need to be established in future studies.
| 1. | Everhart JE. Recent developments in the epidemiology of Helicobacter pylori. Gastroenterol Clin North Am. 2000;29:559-578. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 198] [Article Influence: 7.6] [Reference Citation Analysis (1)] |
| 2. | Schulz C, Schütte K, Mayerle J, Malfertheiner P. The role of the gastric bacterial microbiome in gastric cancer: Helicobacter pylori and beyond. Therap Adv Gastroenterol. 2019;12:1756284819894062. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 3. | González CA, Megraud F, Buissonniere A, Lujan Barroso L, Agudo A, Duell EJ, Boutron-Ruault MC, Clavel-Chapelon F, Palli D, Krogh V, Mattiello A, Tumino R, Sacerdote C, Quirós JR, Sanchez-Cantalejo E, Navarro C, Barricarte A, Dorronsoro M, Khaw KT, Wareham N, Allen NE, Tsilidis KK, Bas Bueno-de-Mesquita H, Jeurnink SM, Numans ME, Peeters PHM, Lagiou P, Valanou E, Trichopoulou A, Kaaks R, Lukanova-McGregor A, Bergman MM, Boeing H, Manjer J, Lindkvist B, Stenling R, Hallmans G, Mortensen LM, Overvad K, Olsen A, Tjonneland A, Bakken K, Dumeaux V, Lund E, Jenab M, Romieu I, Michaud D, Mouw T, Carneiro F, Fenge C, Riboli E. Helicobacter pylori infection assessed by ELISA and by immunoblot and noncardia gastric cancer risk in a prospective study: the Eurgast-EPIC project. Ann Oncol. 2012;23:1320-1324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 102] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 4. | Kuipers EJ, Thijs JC, Festen HP. The prevalence of Helicobacter pylori in peptic ulcer disease. Aliment Pharmacol Ther. 1995;9 Suppl 2:59-69. [PubMed] |
| 5. | Dong QJ, Zhan SH, Wang LL, Xin YN, Jiang M, Xuan SY. Relatedness of Helicobacter pylori populations to gastric carcinogenesis. World J Gastroenterol. 2012;18:6571-6576. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 10] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 6. | Shiota S, Matsunari O, Watada M, Yamaoka Y. Virulence factors or ancestral origin of Helicobacter pylori: which is a better predictor of gastric cancer risk? Gut. 2012;61:469-470. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 7. | Kodaman N, Pazos A, Schneider BG, Piazuelo MB, Mera R, Sobota RS, Sicinschi LA, Shaffer CL, Romero-Gallo J, de Sablet T, Harder RH, Bravo LE, Peek RM Jr, Wilson KT, Cover TL, Williams SM, Correa P. Human and Helicobacter pylori coevolution shapes the risk of gastric disease. Proc Natl Acad Sci U S A. 2014;111:1455-1460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 188] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 8. | Bakhti SZ, Latifi-Navid S, Safaralizadeh R. Helicobacter pylori-related risk predictors of gastric cancer: The latest models, challenges, and future prospects. Cancer Med. 2020;9:4808-4822. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 9. | Berthenet E, Yahara K, Thorell K, Pascoe B, Meric G, Mikhail JM, Engstrand L, Enroth H, Burette A, Megraud F, Varon C, Atherton JC, Smith S, Wilkinson TS, Hitchings MD, Falush D, Sheppard SK. A GWAS on Helicobacter pylori strains points to genetic variants associated with gastric cancer risk. BMC Biol. 2018;16:84. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 10. | Tuan VP, Yahara K, Dung HDQ, Binh TT, Huu Tung P, Tri TD, Thuan NPM, Khien VV, Trang TTH, Phuc BH, Tshibangu-Kabamba E, Matsumoto T, Akada J, Suzuki R, Okimoto T, Kodama M, Murakami K, Yano H, Fukuyo M, Takahashi N, Kato M, Nishiumi S, Azuma T, Ogura Y, Hayashi T, Toyoda A, Kobayashi I, Yamaoka Y. Genome-wide association study of gastric cancer- and duodenal ulcer-derived Helicobacter pylori strains reveals discriminatory genetic variations and novel oncoprotein candidates. Microb Genom. 2021;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 11. | Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 2013;9:e1003348. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 902] [Cited by in RCA: 1034] [Article Influence: 79.5] [Reference Citation Analysis (0)] |
| 12. | Seibert TM, Fan CC, Wang Y, Zuber V, Karunamuni R, Parsons JK, Eeles RA, Easton DF, Kote-Jarai Z, Al Olama AA, Garcia SB, Muir K, Grönberg H, Wiklund F, Aly M, Schleutker J, Sipeky C, Tammela TL, Nordestgaard BG, Nielsen SF, Weischer M, Bisbjerg R, Røder MA, Iversen P, Key TJ, Travis RC, Neal DE, Donovan JL, Hamdy FC, Pharoah P, Pashayan N, Khaw KT, Maier C, Vogel W, Luedeke M, Herkommer K, Kibel AS, Cybulski C, Wokolorczyk D, Kluzniak W, Cannon-Albright L, Brenner H, Cuk K, Saum KU, Park JY, Sellers TA, Slavov C, Kaneva R, Mitev V, Batra J, Clements JA, Spurdle A, Teixeira MR, Paulo P, Maia S, Pandha H, Michael A, Kierzek A, Karow DS, Mills IG, Andreassen OA, Dale AM; PRACTICAL Consortium*. Polygenic hazard score to guide screening for aggressive prostate cancer: development and validation in large scale cohorts. BMJ. 2018;360:j5757. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 142] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 13. | Lecarpentier J, Silvestri V, Kuchenbaecker KB, Barrowdale D, Dennis J, McGuffog L, Soucy P, Leslie G, Rizzolo P, Navazio AS, Valentini V, Zelli V, Lee A, Amin Al Olama A, Tyrer JP, Southey M, John EM, Conner TA, Goldgar DE, Buys SS, Janavicius R, Steele L, Ding YC, Neuhausen SL, Hansen TVO, Osorio A, Weitzel JN, Toss A, Medici V, Cortesi L, Zanna I, Palli D, Radice P, Manoukian S, Peissel B, Azzollini J, Viel A, Cini G, Damante G, Tommasi S, Peterlongo P, Fostira F, Hamann U, Evans DG, Henderson A, Brewer C, Eccles D, Cook J, Ong KR, Walker L, Side LE, Porteous ME, Davidson R, Hodgson S, Frost D, Adlard J, Izatt L, Eeles R, Ellis S, Tischkowitz M; EMBRACE, Godwin AK, Meindl A, Gehrig A, Dworniczak B, Sutter C, Engel C, Niederacher D, Steinemann D, Hahnen E, Hauke J, Rhiem K, Kast K, Arnold N, Ditsch N, Wang-Gohrke S, Wappenschmidt B, Wand D, Lasset C, Stoppa-Lyonnet D, Belotti M, Damiola F, Barjhoux L, Mazoyer S; GEMO Study Collaborators, Van Heetvelde M, Poppe B, De Leeneer K, Claes KBM, de la Hoya M, Garcia-Barberan V, Caldes T, Perez Segura P, Kiiski JI, Aittomäki K, Khan S, Nevanlinna H, van Asperen CJ; HEBON, Vaszko T, Kasler M, Olah E, Balmaña J, Gutiérrez-Enríquez S, Diez O, Teulé A, Izquierdo A, Darder E, Brunet J, Del Valle J, Feliubadalo L, Pujana MA, Lazaro C, Arason A, Agnarsson BA, Johannsson OT, Barkardottir RB, Alducci E, Tognazzo S, Montagna M, Teixeira MR, Pinto P, Spurdle AB, Holland H; KConFab Investigators, Lee JW, Lee MH, Lee J, Kim SW, Kang E, Kim Z, Sharma P, Rebbeck TR, Vijai J, Robson M, Lincoln A, Musinsky J, Gaddam P, Tan YY, Berger A, Singer CF, Loud JT, Greene MH, Mulligan AM, Glendon G, Andrulis IL, Toland AE, Senter L, Bojesen A, Nielsen HR, Skytte AB, Sunde L, Jensen UB, Pedersen IS, Krogh L, Kruse TA, Caligo MA, Yoon SY, Teo SH, von Wachenfeldt A, Huo D, Nielsen SM, Olopade OI, Nathanson KL, Domchek SM, Lorenchick C, Jankowitz RC, Campbell I, James P, Mitchell G, Orr N, Park SK, Thomassen M, Offit K, Couch FJ, Simard J, Easton DF, Chenevix-Trench G, Schmutzler RK, Antoniou AC, Ottini L. Prediction of Breast and Prostate Cancer Risks in Male BRCA1 and BRCA2 Mutation Carriers Using Polygenic Risk Scores. J Clin Oncol. 2017;35:2240-2250. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 135] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 14. | Park B, Yang S, Lee J, Choi IJ, Kim YI, Kim J. Gastric Cancer Risk Prediction Using an Epidemiological Risk Assessment Model and Polygenic Risk Score. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 15. | Duan F, Song C, Wang P, Ye H, Dai L, Zhang J, Wang K. Polygenic Risk Scores for Prediction of Gastric Cancer Based on Bioinformatics Screening and Validation of Functional lncRNA SNPs. Clin Transl Gastroenterol. 2021;12:e00430. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 16. | Delcher AL, Salzberg SL, Phillippy AM. Using MUMmer to identify similar regions in large sequence sets. Curr Protoc Bioinformatics. 2003;Chapter 10:Unit 10.3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 358] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 17. | Yang C, Pei X, Wu Y, Yan L, Yan Y, Song Y, Coyle NM, Martinez-Urtaza J, Quince C, Hu Q, Jiang M, Feil E, Yang D, Zhou D, Yang R, Falush D, Cui Y. Recent mixing of Vibrio parahaemolyticus populations. ISME J. 2019;13:2578-2588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 45] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 18. | Yiangou K, Kyriacou K, Kakouri E, Marcou Y, Panayiotidis MI, Loizidou MA, Hadjisavvas A, Michailidou K. Combination of a 15-SNP Polygenic Risk Score and Classical Risk Factors for the Prediction of Breast Cancer Risk in Cypriot Women. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 19. | Calle ML, Urrea V, Boulesteix AL, Malats N. AUC-RF: a new strategy for genomic profiling with random forest. Hum Hered. 2011;72:121-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 80] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 20. | Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13773] [Cited by in RCA: 12595] [Article Influence: 286.3] [Reference Citation Analysis (1)] |
| 21. | Wen S, Moss SF. Helicobacter pylori virulence factors in gastric carcinogenesis. Cancer Lett. 2009;282:1-8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 121] [Cited by in RCA: 127] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 22. | Evans DG, Karjalainen TK, Evans DJ Jr, Graham DY, Lee CH. Cloning, nucleotide sequence, and expression of a gene encoding an adhesin subunit protein of Helicobacter pylori. J Bacteriol. 1993;175:674-683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 156] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 23. | Carlsohn E, Nyström J, Bölin I, Nilsson CL, Svennerholm AM. HpaA is essential for Helicobacter pylori colonization in mice. Infect Immun. 2006;74:920-926. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 63] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 24. | Cai H, Ye F, Michel A, Murphy G, Sasazuki S, Taylor PR, Qiao YL, Park SK, Yoo KY, Jee SH, Cho ER, Kim J, Chen SC, Abnet CC, Tsugane S, Cai Q, Shu XO, Zheng W, Pawlita M, Epplein M. Helicobacter pylori blood biomarker for gastric cancer risk in East Asia. Int J Epidemiol. 2016;45:774-781. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 64] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 25. | Epplein M, Zheng W, Xiang YB, Peek RM Jr, Li H, Correa P, Gao J, Michel A, Pawlita M, Cai Q, Shu XO. Prospective study of Helicobacter pylori biomarkers for gastric cancer risk among Chinese men. Cancer Epidemiol Biomarkers Prev. 2012;21:2185-2192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 57] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 26. | Andermann TM, Chen YT, Ottemann KM. Two predicted chemoreceptors of Helicobacter pylori promote stomach infection. Infect Immun. 2002;70:5877-5881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 55] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 27. | Williams SM, Chen YT, Andermann TM, Carter JE, McGee DJ, Ottemann KM. Helicobacter pylori chemotaxis modulates inflammation and bacterium-gastric epithelium interactions in infected mice. Infect Immun. 2007;75:3747-3757. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 94] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 28. | Yoon JY, Kim J, Lee SJ, Kim HS, Im HN, Yoon HJ, Kim KH, Kim SJ, Han BW, Suh SW. Structural and functional characterization of Helicobacter pylori DsbG. FEBS Lett. 2011;585:3862-3867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 29. | Lester J, Kichler S, Oickle B, Fairweather S, Oberc A, Chahal J, Ratnayake D, Creuzenet C. Characterization of Helicobacter pylori HP0231 (DsbK): role in disulfide bond formation, redox homeostasis and production of Helicobacter cystein-rich protein HcpE. Mol Microbiol. 2015;96:110-133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 30. | Zhong Y, Anderl F, Kruse T, Schindele F, Jagusztyn-Krynicka EK, Fischer W, Gerhard M, Mejías-Luque R. Helicobacter pylori HP0231 Influences Bacterial Virulence and Is Essential for Gastric Colonization. PLoS One. 2016;11:e0154643. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 31. | Choi J, Jia G, Wen W, Long J, Zheng W. Evaluating polygenic risk scores in assessing risk of nine solid and hematologic cancers in European descendants. Int J Cancer. 2020;147:3416-3423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 32. | Jin G, Lv J, Yang M, Wang M, Zhu M, Wang T, Yan C, Yu C, Ding Y, Li G, Ren C, Ni J, Zhang R, Guo Y, Bian Z, Zheng Y, Zhang N, Jiang Y, Chen J, Wang Y, Xu D, Zheng H, Yang L, Chen Y, Walters R, Millwood IY, Dai J, Ma H, Chen K, Chen Z, Hu Z, Wei Q, Shen H, Li L. Genetic risk, incident gastric cancer, and healthy lifestyle: a meta-analysis of genome-wide association studies and prospective cohort study. Lancet Oncol. 2020;21:1378-1386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 185] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 33. | Mihor A, Tomsic S, Zagar T, Lokar K, Zadnik V. Socioeconomic inequalities in cancer incidence in Europe: a comprehensive review of population-based epidemiological studies. Radiol Oncol. 2020;54:1-13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 81] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 34. | Quach DT, Hiyama T, Gotoda T. Identifying high-risk individuals for gastric cancer surveillance from western and eastern perspectives: Lessons to learn and possibility to develop an integrated approach for daily practice. World J Gastroenterol. 2019;25:3546-3562. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 24] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 35. | Poorolajal J, Moradi L, Mohammadi Y, Cheraghi Z, Gohari-Ensaf F. Risk factors for stomach cancer: a systematic review and meta-analysis. Epidemiol Health. 2020;42:e2020004. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 180] [Cited by in RCA: 186] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 36. | Ge Z, Taylor DE. Contributions of genome sequencing to understanding the biology of Helicobacter pylori. Annu Rev Microbiol. 1999;53:353-387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 41] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 37. | Suerbaum S, Achtman M. Evolution of Helicobacter pylori: the role of recombination. Trends Microbiol. 1999;7:182-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 49] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Invited article; Externally peer reviewed
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Keikha M, Iran; Wang W, China S-Editor: Wang JJ L-Editor: A P-Editor: Wang JJ