Published online Sep 15, 2022. doi: 10.4251/wjgo.v14.i9.1675
Peer-review started: January 18, 2022
First decision: March 8, 2022
Revised: March 23, 2022
Accepted: August 6, 2022
Article in press: August 6, 2022
Published online: September 15, 2022
Processing time: 234 Days and 8.3 Hours
Immune cells play a role in the regulation of tumor cell behavior, and accumulating evidence supports their significance in predicting outcomes and therapeutic efficacy in colorectal cancers (CRC). Human six-transmembrane epithelial antigen of the prostate (STEAP) proteins have been recognized and utilized as promising targets for cell- and antibody-based immunotherapy. One STEAP family member, STEAP4, is expected to be an attractive biomarker for the immunotherapy of prostate and breast cancer. However, the immunotherapeutic role of STEAP4 for colorectal carcinomas has not been demonstrated.
To explore the expression pattern of STEAPs in CRC and their relationship with immune infiltration, and investigate the potential utilization of STEAPs as novel prognostic indicators in colorectal carcinomas.
The expression level of STEAPs in CRC was evaluated using various open-resource databases and online tools to explore the expression characteristics and prognostic significance of STEAPs, as well as their correlation with immune-related biomarkers, such as immune infiltration. Immunohistochemical (IHC) experiments were subsequently performed to verify the database conclusions.
The levels of STEAPs in CRC were inconsistent. The expression of STEAPs 1-3 in CRC was not significantly different from that in normal tissues. However, STEAP4 mRNA levels were significantly lower in CRC than in normal tissue and were positively correlated with immune-related biomarkers, such as immune cell infiltration, immune stimulation, major histocompatibility complex levels, and chemokines. Interestingly, the expression of STEAP4 in microsatellite instability-high CRC subtype was higher than that in microsatellite stability subtype. IHC staining was performed on colon cancer tissue samples and showed that high expression of STEAP4 in adjacent tissues positively correlated with immune-related biomarkers, including MLH1, MLH6, and PMS2, but negatively correlated with programmed death ligand 1, to varying degrees.
Our results provide an analysis of the expression of STEAP family members in CRC. Among different STEAP family members, STEAP4 plays a different role in CRC compared to STEAPs 1-3. In CRC, STEAP4 expression is not only lower than that in normal tissues, but it is also positively correlated with immune infiltration and immune-related biomarkers. These findings suggest that STEAP4 may be a potential biomarker for predicting CRC immune infiltration status.
Core Tip: This study analyzed the expression levels of six-transmembrane epithelial antigen of the prostate (STEAP) family members in colorectal cancer (CRC) and explored the potential biological function of STEAP4. It was found that the expression of STEAP4 in CRC tissues had a positive correlation with immune infiltration and immune-related biomarkers, such as MLH1, MLH6, and PMS2, and a negative correlation with programmed death ligand 1. STEAP4 is expected to be a novel and potential prognostic biomarker for CRC.
- Citation: Fang ZX, Li CL, Chen WJ, Wu HT, Liu J. Potential of six-transmembrane epithelial antigen of the prostate 4 as a prognostic marker for colorectal cancer. World J Gastrointest Oncol 2022; 14(9): 1675-1688
- URL: https://www.wjgnet.com/1948-5204/full/v14/i9/1675.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i9.1675
Colorectal cancer (CRC) ranks third in incidence and second in cancer mortality among malignant tumors worldwide[1]. Currently, large changes in lifestyle and dietary habits are thought to have contributed to the increased incidence and mortality of CRC. For example, in China, the annual average increase of new CRC cases was estimated to be 4.2%[2]. Although gender and regional differences are considered as the prognostic factors for patients with CRC[3], the etiology of CRC oncogenesis and development is still complex and unclear.
With the development of standardized treatment for patients with CRC, the prognosis of CRC has greatly improved. However, the control of progression of metastatic disease is still intractable. Immune cells play an important role in the regulation of tumor cell behavior, and accumulating evidence supports their significance in predicting outcomes and therapeutic efficacy in CRC patients[4]. In this regard, attention is currently being paid to the immune microenvironment and immunotherapy of CRC, mainly focusing on T cells and therapeutic response as related to promising treatment strategies[5].
Immunotherapy serves as an alternative treatment for cancer patients, especially for those whose tumors overexpress antigens recognized by immune cells. The human six-transmembrane epithelial antigen of the prostate (STEAP) family of proteins belongs to a class of cellular transmembrane proteins and has been used to derive epitope peptides that stimulate T lymphocytes in patients with renal cell or bladder cancer[6]. Importantly, STEAPs are present at the intercellular junctions of the prostate secretory epithelium, and are overexpressed in prostate cancer, serving as attractive targets for prostate cancer immunotherapy[7].
Although STEAPs have been reported to be overexpressed in CRC[8-11], research on STEAPs in CRC remains limited and the immunotherapeutic role of STEAPs in colorectal carcinomas has not been shown. This research investigated the biological function of STEAPs in CRCs, in addition to the relationship between STEAPs and immune infiltration, and demonstrated the potential of STEAPs to serve as novel and prognostic biomarkers for immunotherapy in colorectal carcinomas.
A tissue microarray with 87 matched primary CRC tissues and their corresponding adjacent normal colorectal tissue samples, and six extra samples of cancer cases without the corresponding paracancerous tissue were purchased from the Shanghai OUTDO Biotech Company, Shanghai, China (XT17-025, HColA180Su18). Pathological type was classified according to the prognostic degree of cancers. This study was approved by the Ethics Committee of Shantou University Medical College.
TCGA datasets were used to evaluate the expression of STEAPs in normal and different cancerous tissues through the Tumor IMmune Estimation Resource (TIMER2.0) online source (http://timer.cistrome.org/)[12]. The UCSC Xena database (https://genome-cancer.ucsc.edu/)[13] was applied to analyze STEAP expression differences in colon adenocarcinomas (COAD) and rectal adenocarcinomas (READ) and related normal tissues. Regarding the different subtypes of CRC, namely, microsatellite instability-high (MSI-H), microsatellite instability-low (MSI-L), and microsatellite stable (MSS)[14], MSI is a biomarker for response to immune checkpoint inhibitors (ICIs); high disease control rates and good progression-free survival were observed in patients with MSI-H CRC[15]. MSI-L tumors are phenotypically indistinguishable from MSS tumors, and the biological significance of MSI-L is unclear[16], so emphasis has been placed on MSI-H and MSS. The expression of STEAPs in MSI-H and MSS was also evaluated in the GEPIA2 database (http://gepia2.cancer-pku.cn/)[17].
Immune cells involved in CRC development were evaluated using the TIMER2.0 database to predict the association between the expression of STEAPs and the abundance of immune cells, including B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells, in the tumor microenvironment. In terms of immune characteristics of lymphocytes, immunostimulants, major histocompatibility complex (MHC), and chemokines, TISIDB, an integrated repository portal for tumor-immune system interactions (http://cis.hku.hk/TISIDB/)[18], was applied to explore the potential function of STEAPs in CRC immune infiltration. After being downloaded from the TISIDB, the dataset was analyzed and drawn with matrix2png (https://matrix2png.msl.ubc.ca/bin/matrix2png.cgi), an online mapping program[19].
The immunohistochemical (IHC) staining for STEAP4 in tissue microarrays was conducted as described before[20]. The tissue microarray slide was dewaxed in xylene, hydrated in graded alcohols, and processed with 2% ethylenediamine tetraacetic acid antigen-repair solution (Fuzhou Maixin Biotechnology Development Co. LTD, Fuzhou, China) by microwave heating for epitope retrieval. After blocking endogenous peroxidase with 3% H2O2, the slide was incubated with anti-STEAP4 antibody (dilution: 1:1000, Proteintech 11944-AP) at 4 °C overnight. Stained tissues with DAB reagent were mounted and underwent nuclear counterstaining with hematoxylin for visualization.
Sections were visualized under a bright-field microscope (Axio Imager A2, Zeiss, Germany) and evaluated independently by two investigators with no prior knowledge of the CRC patient information. For tissue expression of STEAP4, staining intensity was scored as 0, 1, 2, and 3 for colorless, light yellow, brown yellow, and dark brown, respectively, while the percentage of positive cells, equaling 0%, 1%-25%, 26%-50%, 51%-75% and 76%-100%, was scored as 0, 1, 2, 3, and 4 points, respectively. The final staining score for STEAP4 expression was calculated as the sum of staining intensity and the percentage of positive cells, and divided into low expression (scores 0-4) and high expression (scores 5-7) groups. The expression levels of MLH1/2/6, PMS2, and programmed death ligand 1 (PDL1) were included in the patient information and the cutoffs were described before[21].
SPSS 25.0 statistical software was used to analyze the results. Enumerated data are recorded as the number of cases (n = 93), and the relationship between STEAP4 and the clinicopathological parameters of CRC patients was analyzed by the χ2 and Fisher’s exact probability tests. The relationship between expression of STEAP4 in CRC and adjacent normal tissues (n = 87 cases) was examined by the χ2 test. Likewise, the correlation between highly expressed STEAP4 in CRC and the immune-related factors MLH1, MLH2, MLH6, PMS2, and PDL1 was determined by the χ2 test. To investigate the prognostic value of STEAP4 in CRC patients, the Kaplan-Meier survival curve and log-rank test were used to evaluate the association of STEAP4 expression with CRC patient prognosis by using SPSS 25.0 software. The difference was considered statistically significant at P < 0.05.
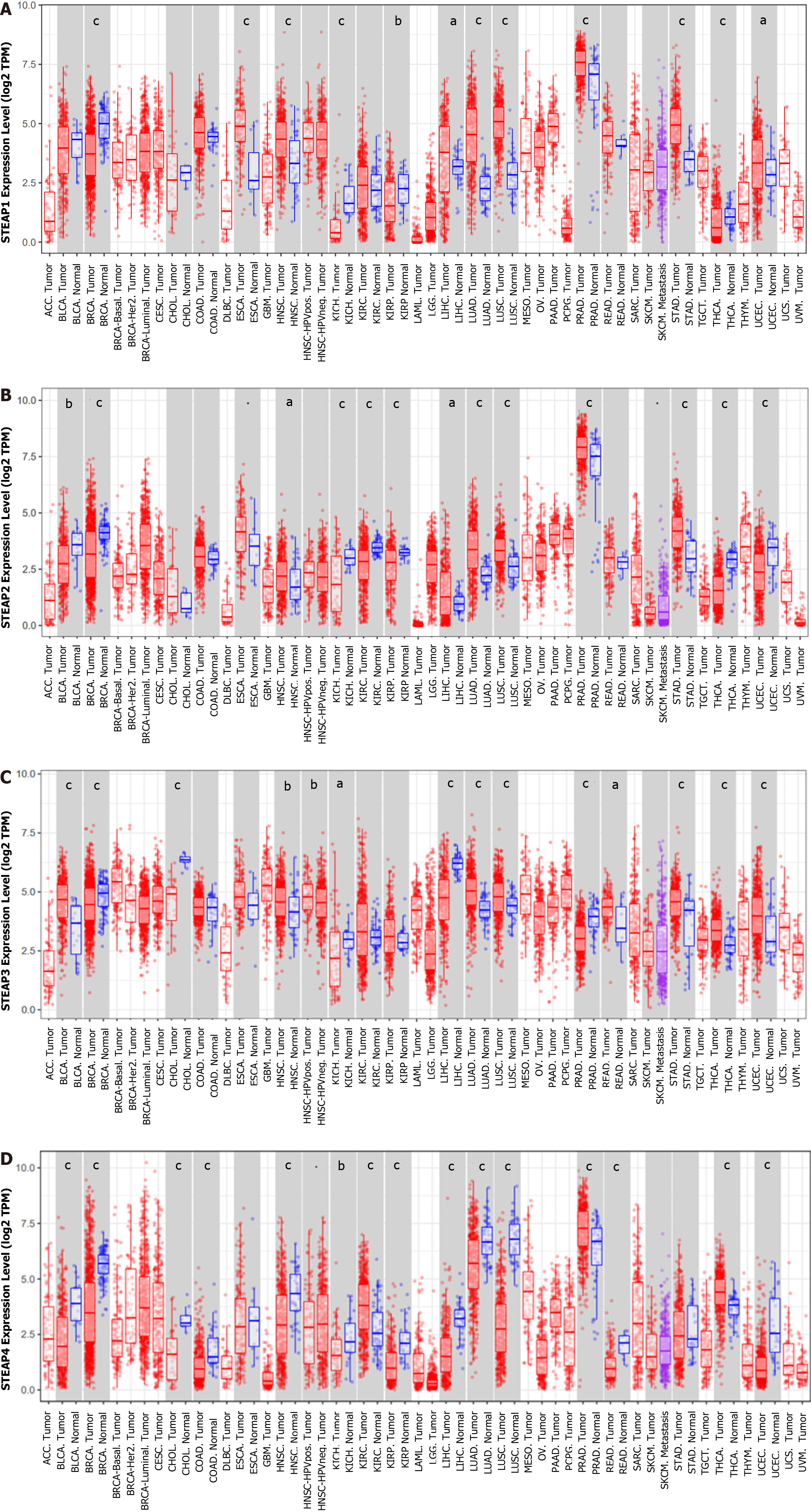
To determine the expression pattern of STEAPs in different types of malignant tumors, TIMER2.0 was used to analyze the difference between normal and cancerous tissues in TCGA. All STEAPs were found to be expressed at low levels in breast invasive carcinoma and kidney chromophobe compared to corresponding normal tissues, while in other types of malignancies, the expression patterns of STEAPs differed (Figure 1). Interestingly, in COAD, head and neck squamous cell carcinoma, lung adenocarcinoma, lung squamous cell carcinoma, and READ, the expression of STEAP4 was lower than that in normal tissues. However, in such cancerous tissues, the levels of STEAPs 1-3 were higher or not significantly different from those in normal tissues, suggesting that STEAP4 may perform a different function from STEAPs 1-3 in patients with such cancers.
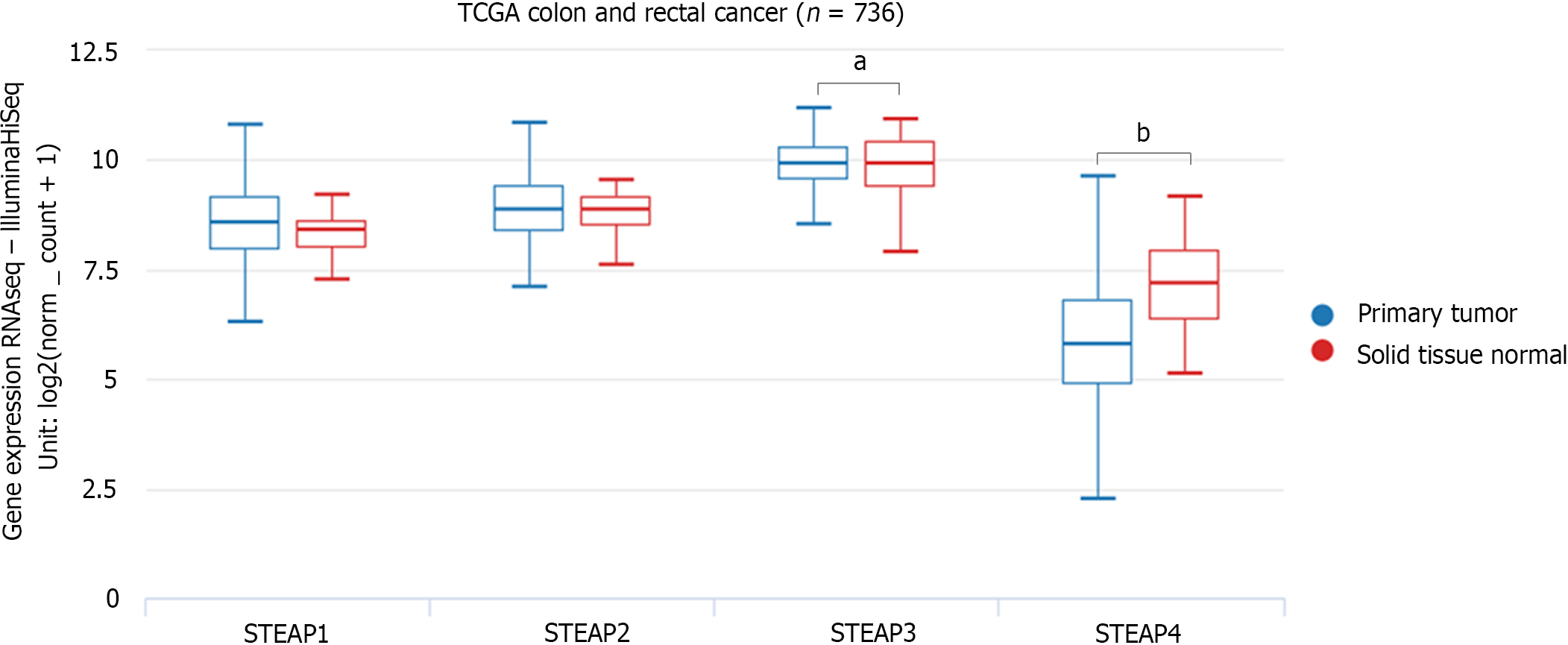
To verify the expression pattern of STEAPs in CRC, another database, UCSC Xena, was analyzed to confirm the findings. Although expression of STEAPs 1 and 2 in COAD and READ were not different compared to their corresponding normal tissues, STEAP4 expression was lower, while STEAP3 expression was higher compared to that in corresponding normal tissues (Figure 2).
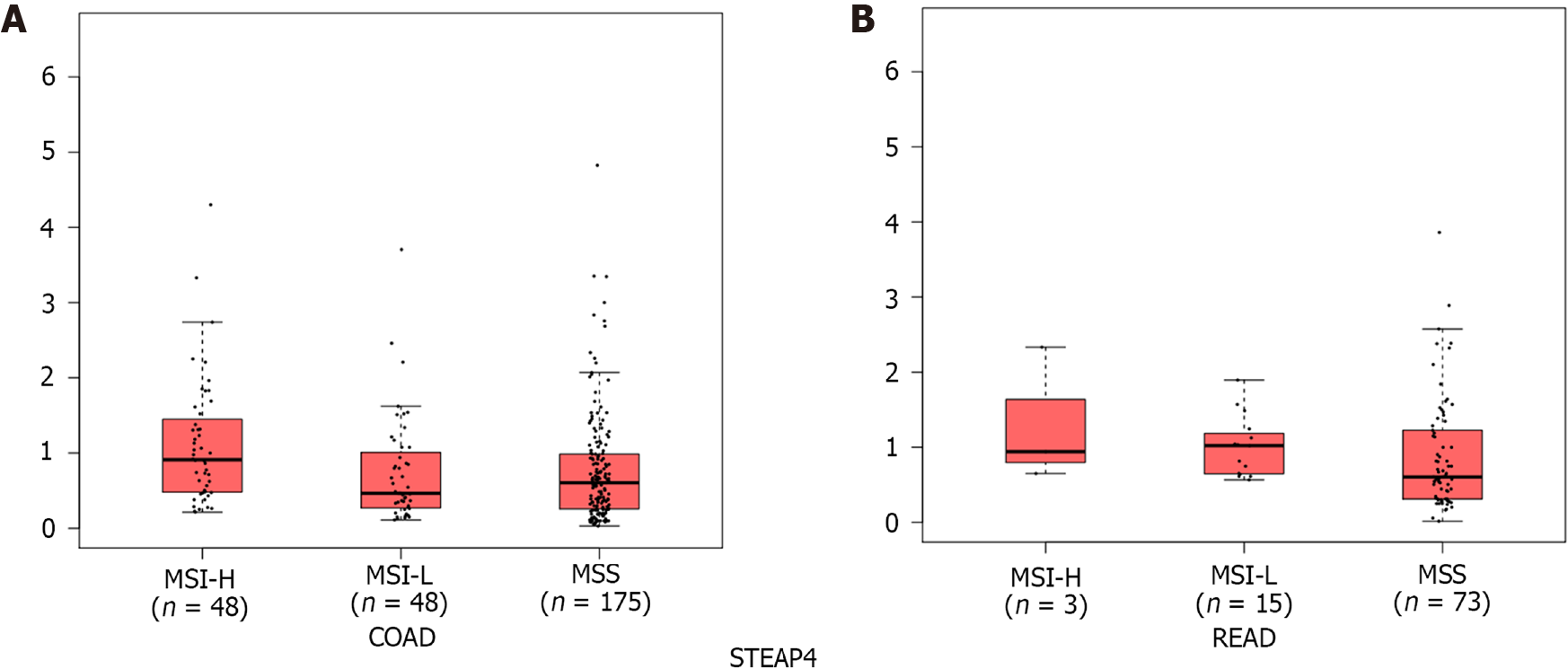
CRC is highly heterogeneous at the genetic and molecular levels, which affects the efficacy of clinical therapy. A subset of CRCs exhibit MSI, indicating defective DNA mismatch repair and high mutational burden, different from the majority of MSS subtypes[22]. CRC patients, especially those with MSI-H tumors, are more sensitive to ICIs than those with MSS tumors, and MSI-H tumors have greater infiltration of immune cells, higher expression of immune-related genes, and higher immunogenicity than MSS tumors[23]. Since the expression of STEAP4 is consistently low in CRC, different from the other three genes, we focused on STEAP4 for the remainder of this study. To explore STEAP4 expression levels in different subtypes of CRCs, the GEPIA2 database was used. Interestingly, the mRNA level of STEAP4 was high in MSI-H CRCs compared to MSI-L and MSS tumors (Figure 3).
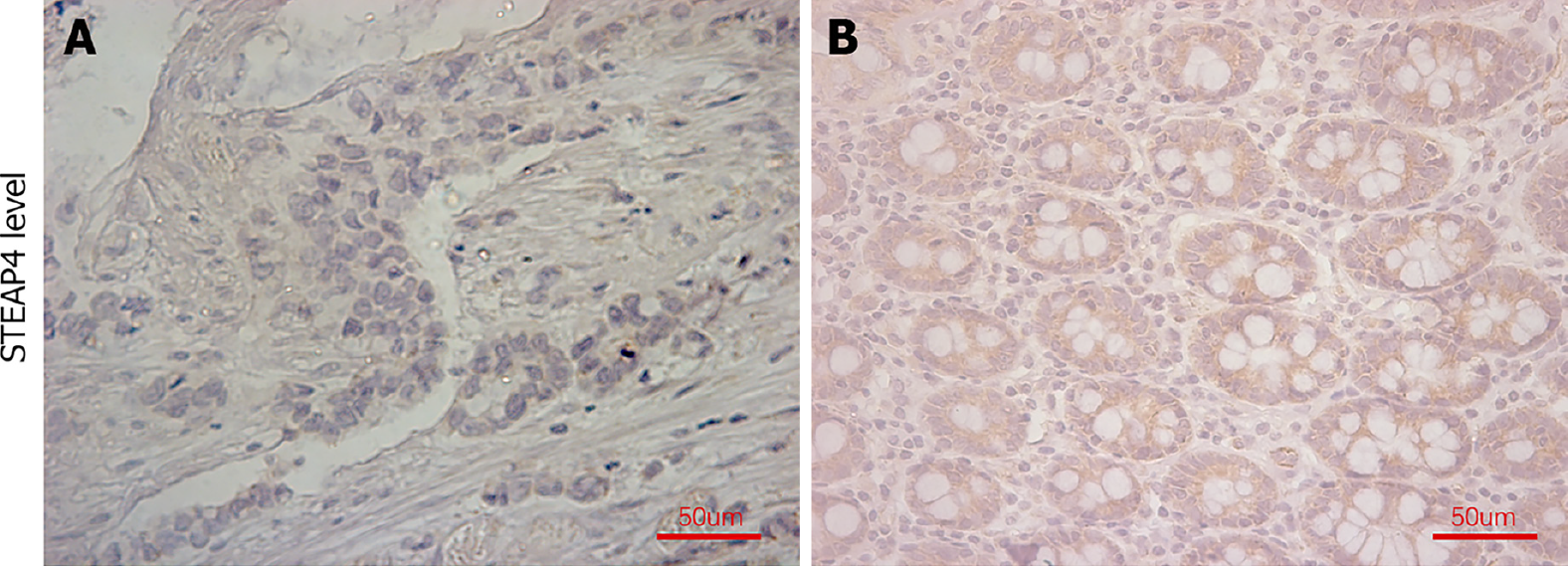
As the expression pattern of STEAP4 seems to be different from those of the other STEAP members, paired normal and CRC tissues were used to evaluate the protein level of STEAP4. Representative images of STEAP4 expression are shown in Figure 4. The percentage of CRC tissues with high levels of STEAP4 was 51.7%, which was still lower than that in normal tissue (73.6%, P = 0.003) (Table 1).
Clinicopathological analysis showed that STEAP4 expression was not associated with gender, primary tumor stage, lymph node status, American Joint Committee on Cancer (AJCC) stage, or pathological type (Table 2). The expression of STEAP4 decreased with the increase of primary tumor stage, lymph node status, and AJCC stage.
| Clinical parameter | STEAP4 | P value | |
| Low (%) | High (%) | ||
| Gender | |||
| Female | 29 (59.2) | 20 (40.8) | 0.123 |
| Male | 19 (43.2) | 25 (56.8) | |
| Primary tumor stage1 | |||
| T1-T3 | 36 (48.0) | 39 (52.0) | 0.213 |
| T4a-T4b | 11 (64.7) | 6 (35.3) | |
| Lymph node status | |||
| N0-N1 | 41 (48.8) | 43 (51.2) | 0.160 |
| N2 | 7 (77.8) | 2 (22.2) | |
| AJCC stage | |||
| Phase 1-2 | 28 (43.1) | 30 (56.9) | 0.407 |
| Phase 3 | 20 (57.1) | 15 (42.9) | |
Based on the IHC results, the expression of STEAP4 and corresponding immune-related biomarkers in CRC was analyzed using χ2 statistical analysis. As shown in Table 3, a low level of STEAP4 was positively related to low levels of MLH1, MLH6, and PMS2, but negatively associated with PDL1 level in CRC patients.
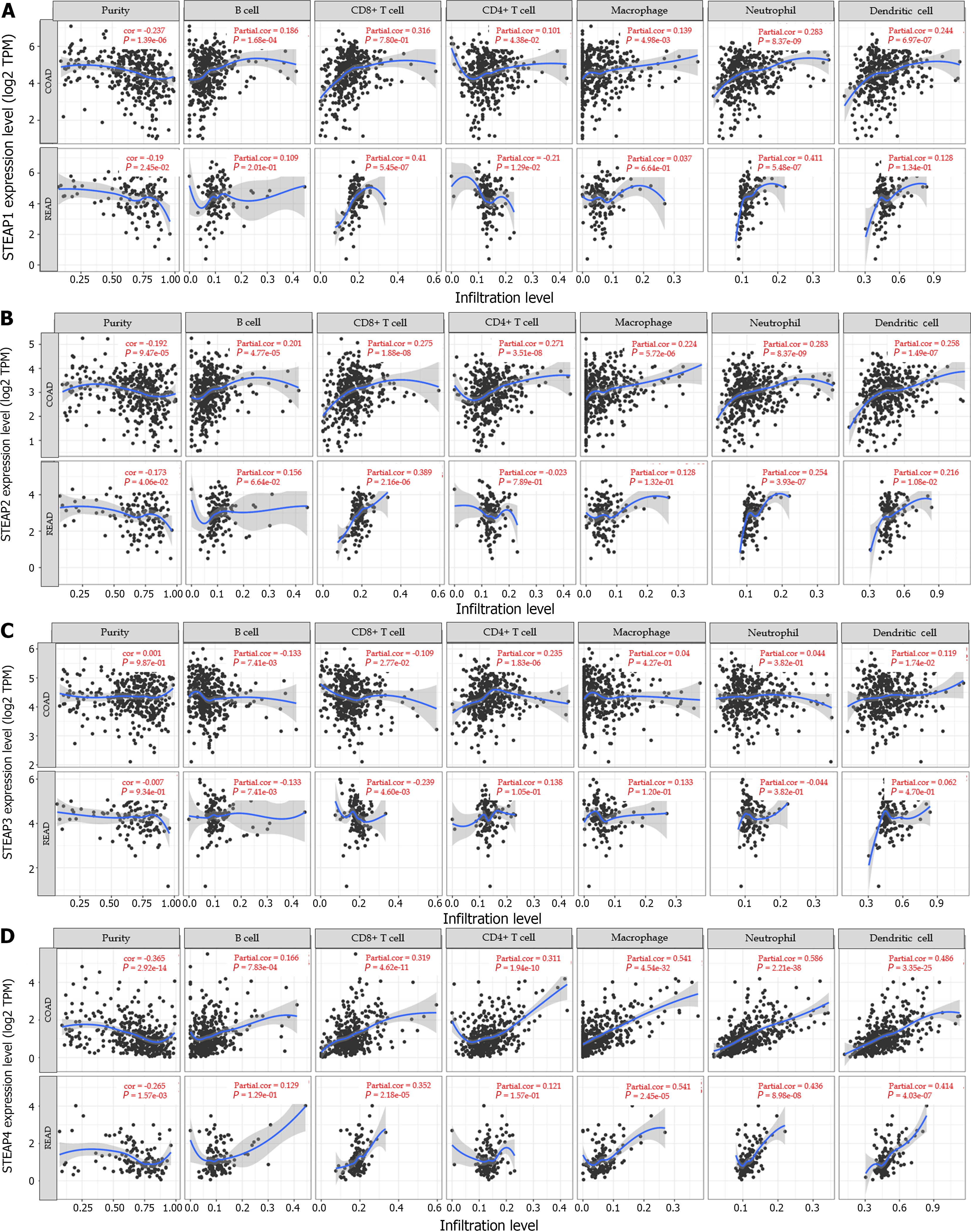
Given the importance of antitumor immunity in the tumor microenvironment, the correlation between expression of STEAP family members and immune cells in CRC was analyzed. Based on the TIMER database, it was found that the expression of STEAP1, STEAP2, and STEAP4 was negatively correlated with tumor purity and positively correlated with six types of immune cells, specifically, B cells, CD8+ T cells, CD4+ T cells, neutrophils, macrophages, and dendritic cells, in COAD and READ (Figures 5A, B, and D). However, the expression of STEAP3 was positively correlated with tumor purity, CD4+ T cells, neutrophils, macrophages, and dendritic cells, and negatively correlated with B cells and CD8+ T cells in COAD and READ (Figure 5C).
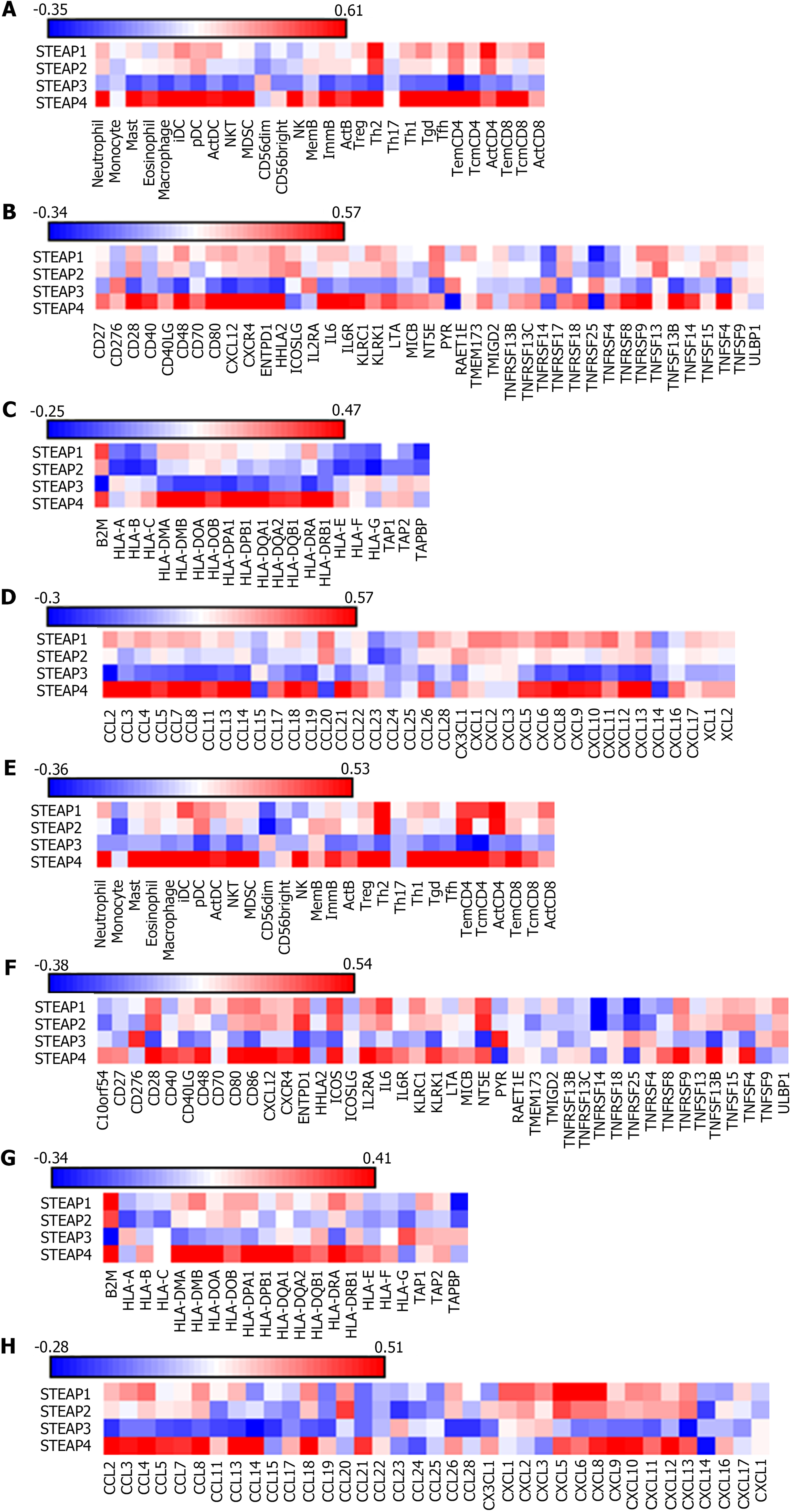
According to the TISIDB database, Spearman correlation analysis showed that the expression of STEAP1, STEAP2, and STEAP4 was positively correlated, but STEAP3 was negatively correlated with lymphocytes, immunostimulants, MHCs, and chemokines (Figures 6A-F), which is consistent with the results obtained based on the TIMER database (Figures 6C and G).
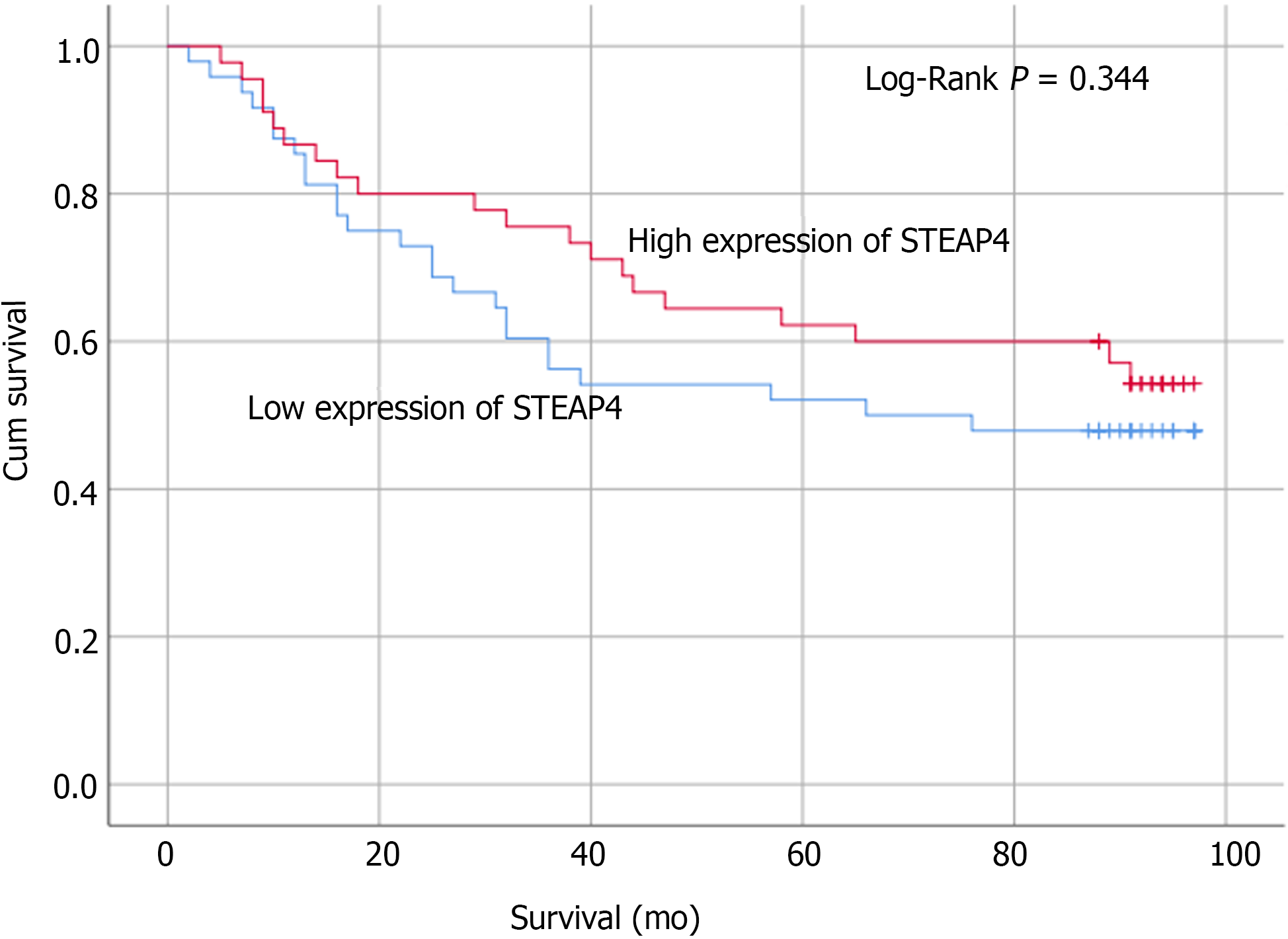
Based on the IHC results, the protein expression of STEAP4 was analyzed by the Kaplan-Meier method with log-rank test, which demonstrated that STEAP4 expression was not significantly associated with the overall survival (OS) (P > 0.05, Figure 7). Although the difference did not meet the statistical criteria, it was found that high expression of STEAP4 tended to predict a longer OS for CRC patients, suggesting that the protein level of STEAP4 could be a predictor of the survival of CRC patients.
Members of the STEAP family were originally identified as metalloreductases in vivo, playing an important role in maintaining iron homeostasis[24]. Abnormal accumulation of iron caused by unbalanced iron metabolism has been reported to lead to the occurrence, progression, and invasion of tumors[25]. Thus, the STEAP family bridges iron homeostasis and cancer[26]. As potential biomarkers and therapeutic targets of tumors, the STEAP family members play an important role in tumor therapy.
All four STEAP proteins are increased in prostate cancer and play important roles in the development and progression of prostate cancer[27]. Interestingly, although the structure of STEAP4 is similar to that of the three other STEAP family members, the function of STEAP4 may diverge in different types of cancers[24]. STEAP1 antibody can effectively activate CD8+ T cells, natural killer cells, and other immune-related factors against a broad spectrum of tumors[28]. In this study, there was a great difference between STEAP4 and STEAP1-3; STEAP4 promoted androgen receptor (AR)-positive prostate cancer (PC) and inhibited AR-negative PC, while it was very different from STEAP1-3 in CRC, so there might be different mechanisms. Interestingly, we found dual anti-STEAP1 antibody targeting T cells for cancer immunotherapy[29]. Combined with our study, it is suggested that STEAP4 can be developed as a new therapeutic strategy. Therefore, this study explored the role of STEAP4 at the mRNA and protein levels. STEAP4 was rarely studied in CRC. The current study used clinical tissues from CRC patients and characterized the mRNA and protein levels of STEAP4 to determine the expression of STEAP4 in CRC. Not surprisingly, low expression of STEAP4 was found in CRC tissues compared with adjacent tissues, which is consistent with the low mRNA expression of STEAP4 in CRC and a potential tumor suppressor role for STEAP4 in CRC patients.
To uncover the potential function of STEAP4 in CRC, clinicopathological parameters were analyzed. However, no statistical significance was found between the protein level of STEAP4 and primary tumor stage, lymph node metastasis, or AJCC stage. Reduced STEAP4 expression tended to be associated with advanced CRC stage and increased lymph node metastases, suggesting that the suppression of STEAP4 could play a role in promoting the progression and metastasis of CRC.
Recently, Xue et al[11] investigated the molecular mechanism of STEAP4 involvement in the hypoxic metabolism of inflammatory bowel disease and the linkage with mitochondrial dysfunction in colon cancer. Increasingly, high levels of STEAP4 result from increased levels of hypoxia and are associated with colitis in mouse models and inflammatory bowel disease patients. Inflammatory factors were not examined in the current study, which may influence STEAP4 levels in different types of CRC. Based on the inflammatory environment, hypoxic conditions can be associated with mitochondrial iron dysfunction caused by increased STEAP4[30]. However, in the absence of inflammatory infiltration, the inhibition of STEAP4 was reversed to be a tumor suppressor through interactions with protein kinases[31].
Further analyses were performed to uncover the potential relationship of STEAP4 with immune infiltration. As expected, both the protein and mRNA levels of STEAP4 are associated with immune-related factors, predicting a potential role of STEAP4 in stimulating immune infiltration. MLH1 deficiency has been shown to be associated with cetuximab resistance in CRC[21]. The positive relationship between STEAP4 and MLH1 in CRC suggests an involvement of STEAP4 in the immune response in the tumor microenvironment. Recently, Ijsselsteijn et al[32] reported that, for a cell-based model for Lynch syndrome, DNA mismatch repair deficiency was related to the core DNA mismatch repair genes MSH2, MSH6, MLH1, and PMS2[32]. The protein level of STEAP4 was found to be positively correlated with such DNA mismatch repair genes in the current study. The above results support a role for STEAP4 in the immune response to CRC to prevent further development and metastasis.
In the current study, STEAP4 was found to be a protective factor in the intestinal tract and could be used as a prognostic indicator for patients with CRC. CRC patients with high STEAP4 expression tend to have a longer survival.
Six-transmembrane epithelial antigen of the prostate 4 (STEAP4) is an attractive biomarker for the immunotherapy of prostate and breast cancers. However, the immunotherapeutic role of STEAP4 for colorectal carcinomas has not been demonstrated.
Immunotherapy emerges with predicting outcomes and therapeutic efficacy in colorectal cancers (CRC).
To explore the expression pattern of STEAPs in CRCs and their relationship with immune infiltration, and investigate the potential utilization of STEAPs as novel prognostic indicators in colorectal carcinomas.
CRC patients’ tissues and online datasets were used to analyze the expression level of STEAP4 in different types of CRC and their relationship with immune characteristics.
The expression of STEAP4 was significantly decreased in CRC tissues compared with adjacent normal ones, and was related to immune-related biomarkers. Low STEAP4 level predicted a poor overall survival of CRC patients.
STEAP4 was found to be a protective factor in the intestinal tract and could be used as a prognostic indicator for patients with CRC.
STEAP4 may be a potential biomarker for predicting CRC immune infiltration status.
| 1. | Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71:7-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8287] [Cited by in RCA: 12204] [Article Influence: 2440.8] [Reference Citation Analysis (8)] |
| 2. | Xing XL, Yao ZY, Zhang T, Zhu N, Liu YW, Peng J. MicroRNA-Related Prognosis Biomarkers from High-Throughput Sequencing Data of Colorectal Cancer. Biomed Res Int. 2020;2020:7905380. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 3. | Zahnd WE, Josey MJ, Schootman M, Eberth JM. Spatial accessibility to colonoscopy and its role in predicting late-stage colorectal cancer. Health Serv Res. 2021;56:73-83. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 4. | Zhong F, Lin Y, Jing X, Ye Y, Wang S, Shen Z. Innate tumor killers in colorectal cancer. Cancer Lett. 2022;527:115-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 5. | Chandra R, Karalis JD, Liu C, Murimwa GZ, Voth Park J, Heid CA, Reznik SI, Huang E, Minna JD, Brekken RA. The Colorectal Cancer Tumor Microenvironment and Its Impact on Liver and Lung Metastasis. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 113] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 6. | Azumi M, Kobayashi H, Aoki N, Sato K, Kimura S, Kakizaki H, Tateno M. Six-transmembrane epithelial antigen of the prostate as an immunotherapeutic target for renal cell and bladder cancer. J Urol. 2010;183:2036-2044. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 7. | Esmaeili SA, Nejatollahi F, Sahebkar A. Inhibition of Intercellular Communication between Prostate Cancer Cells by A Specific Anti-STEAP-1 Single Chain Antibody. Anticancer Agents Med Chem. 2018;18:1674-1679. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 8. | Moreaux J, Kassambara A, Hose D, Klein B. STEAP1 is overexpressed in cancers: a promising therapeutic target. Biochem Biophys Res Commun. 2012;429:148-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 67] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 9. | Bhatlekar S, Addya S, Salunek M, Orr CR, Surrey S, McKenzie S, Fields JZ, Boman BM. Identification of a developmental gene expression signature, including HOX genes, for the normal human colonic crypt stem cell niche: overexpression of the signature parallels stem cell overpopulation during colon tumorigenesis. Stem Cells Dev. 2014;23:167-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 42] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 10. | Isobe T, Baba E, Arita S, Komoda M, Tamura S, Shirakawa T, Ariyama H, Takaishi S, Kusaba H, Ueki T, Akashi K. Human STEAP3 maintains tumor growth under hypoferric condition. Exp Cell Res. 2011;317:2582-2591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 37] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 11. | Xue X, Bredell BX, Anderson ER, Martin A, Mays C, Nagao-Kitamoto H, Huang S, Győrffy B, Greenson JK, Hardiman K, Spence JR, Kamada N, Shah YM. Quantitative proteomics identifies STEAP4 as a critical regulator of mitochondrial dysfunction linking inflammation and colon cancer. Proc Natl Acad Sci U S A. 2017;114:E9608-E9617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 91] [Article Influence: 10.1] [Reference Citation Analysis (3)] |
| 12. | Chen D, Sun Q, Zhang L, Zhou X, Cheng X, Zhou D, Ye F, Lin J, Wang W. The lncRNA HOXA11-AS functions as a competing endogenous RNA to regulate PADI2 expression by sponging miR-125a-5p in liver metastasis of colorectal cancer. Oncotarget. 2017;8:70642-70652. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 48] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 13. | Sanborn JZ, Benz SC, Craft B, Szeto C, Kober KM, Meyer L, Vaske CJ, Goldman M, Smith KE, Kuhn RM, Karolchik D, Kent WJ, Stuart JM, Haussler D, Zhu J. The UCSC Cancer Genomics Browser: update 2011. Nucleic Acids Res. 2011;39:D951-D959. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 49] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 14. | Picard E, Verschoor CP, Ma GW, Pawelec G. Relationships Between Immune Landscapes, Genetic Subtypes and Responses to Immunotherapy in Colorectal Cancer. Front Immunol. 2020;11:369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 382] [Article Influence: 63.7] [Reference Citation Analysis (1)] |
| 15. | Schrock AB, Ouyang C, Sandhu J, Sokol E, Jin D, Ross JS, Miller VA, Lim D, Amanam I, Chao J, Catenacci D, Cho M, Braiteh F, Klempner SJ, Ali SM, Fakih M. Tumor mutational burden is predictive of response to immune checkpoint inhibitors in MSI-high metastatic colorectal cancer. Ann Oncol. 2019;30:1096-1103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 418] [Cited by in RCA: 475] [Article Influence: 67.9] [Reference Citation Analysis (0)] |
| 16. | Torshizi Esfahani A, Seyedna SY, Nazemalhosseini Mojarad E, Majd A, Asadzadeh Aghdaei H. MSI-L/EMAST is a predictive biomarker for metastasis in colorectal cancer patients. J Cell Physiol. 2019;234:13128-13136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 17. | Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556-W560. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1991] [Cited by in RCA: 3529] [Article Influence: 504.1] [Reference Citation Analysis (0)] |
| 18. | Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC, Chu KC, Wong CY, Lau CY, Chen I, Chan NW, Zhang J. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35:4200-4202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 561] [Cited by in RCA: 1572] [Article Influence: 262.0] [Reference Citation Analysis (0)] |
| 19. | Pavlidis P, Noble WS. Matrix2png: a utility for visualizing matrix data. Bioinformatics. 2003;19:295-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 289] [Cited by in RCA: 292] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 20. | Liu J, Wei XL, Huang WH, Chen CF, Bai JW, Zhang GJ. Cytoplasmic Skp2 expression is associated with p-Akt1 and predicts poor prognosis in human breast carcinomas. PLoS One. 2012;7:e52675. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 47] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 21. | Han Y, Peng Y, Fu Y, Cai C, Guo C, Liu S, Li Y, Chen Y, Shen E, Long K, Wang X, Yu J, Shen H, Zeng S. MLH1 Deficiency Induces Cetuximab Resistance in Colon Cancer via Her-2/PI3K/AKT Signaling. Adv Sci (Weinh). 2020;7:2000112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (1)] |
| 22. | Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073-2087.e3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1677] [Cited by in RCA: 1616] [Article Influence: 101.0] [Reference Citation Analysis (2)] |
| 23. | Lin A, Zhang J, Luo P. Crosstalk Between the MSI Status and Tumor Microenvironment in Colorectal Cancer. Front Immunol. 2020;11:2039. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 273] [Article Influence: 45.5] [Reference Citation Analysis (0)] |
| 24. | Ohgami RS, Campagna DR, McDonald A, Fleming MD. The Steap proteins are metalloreductases. Blood. 2006;108:1388-1394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 403] [Cited by in RCA: 526] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 25. | Legendre C, Garcion E. Iron metabolism: a double-edged sword in the resistance of glioblastoma to therapies. Trends Endocrinol Metab. 2015;26:322-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 61] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 26. | Torti SV, Torti FM. Ironing out cancer. Cancer Res. 2011;71:1511-1514. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 131] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 27. | Hubert RS, Vivanco I, Chen E, Rastegar S, Leong K, Mitchell SC, Madraswala R, Zhou Y, Kuo J, Raitano AB, Jakobovits A, Saffran DC, Afar DE. STEAP: a prostate-specific cell-surface antigen highly expressed in human prostate tumors. Proc Natl Acad Sci U S A. 1999;96:14523-14528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 273] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 28. | Alves PM, Faure O, Graff-Dubois S, Cornet S, Bolonakis I, Gross DA, Miconnet I, Chouaib S, Fizazi K, Soria JC, Lemonnier FA, Kosmatopoulos K. STEAP, a prostate tumor antigen, is a target of human CD8+ T cells. Cancer Immunol Immunother. 2006;55:1515-1523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 56] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 29. | Lin TY, Park JA, Long A, Guo HF, Cheung NV. Novel potent anti-STEAP1 bispecific antibody to redirect T cells for cancer immunotherapy. J Immunother Cancer. 2021;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (3)] |
| 30. | Liao Y, Zhao J, Bulek K, Tang F, Chen X, Cai G, Jia S, Fox PL, Huang E, Pizarro TT, Kalady MF, Jackson MW, Bao S, Sen GC, Stark GR, Chang CJ, Li X. Inflammation mobilizes copper metabolism to promote colon tumorigenesis via an IL-17-STEAP4-XIAP axis. Nat Commun. 2020;11:900. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 164] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 31. | Yan D, Dong W, He Q, Yang M, Huang L, Kong J, Qin H, Lin T, Huang J. Circular RNA circPICALM sponges miR-1265 to inhibit bladder cancer metastasis and influence FAK phosphorylation. EBioMedicine. 2019;48:316-331. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 67] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 32. | Ijsselsteijn R, van Hees S, Drost M, Jansen JG, de Wind N. Induction of mismatch repair deficiency, compromised DNA damage signaling and compound hypermutagenesis by a dietary mutagen in a cell-based model for Lynch syndrome. Carcinogenesis. 2022;43:160-169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kołat D, Poland; Mohamed SY, Egypt S-Editor: Wang JJ L-Editor: Wang TQ P-Editor: Cai YX