Published online Sep 15, 2022. doi: 10.4251/wjgo.v14.i9.1654
Peer-review started: March 19, 2022
First decision: May 12, 2022
Revised: May 19, 2022
Accepted: August 9, 2022
Article in press: August 9, 2022
Published online: September 15, 2022
Processing time: 173 Days and 15.9 Hours
Colorectal cancer (CRC) is a major cause of mortality worldwide, associated with a steadily growing prevalence. Notably, the identification of KRAS, NRAS, and BRAF mutations has markedly improved targeted CRC therapy by affording treatments directed against the epidermal growth factor receptor (EGFR) and other anti-angiogenic therapies. However, the survival benefit conferred by these therapies remains variable and difficult to predict, owing to the high level of molecular heterogeneity among patients with CRC. Although classification into consensus molecular subtypes could optimize response prediction to targeted therapies, the acquisition of resistance mutations to targeted therapy is, in part, responsible for the lack of response in some patients. However, the acquisition of such mutations can induce challenges in clinical practice. The utility of liquid biopsy to detect resistance mutations against anti-EGFR therapy has recently been described. This approach may constitute a new standard in the decision algorithm for targeted CRC therapy.
Core Tip: Contemporary management of metastatic colorectal cancer patients with wild type KRAS includes the use of anti-epidermal growth factor receptor (EGFR) agents, such as cetuximab or panitumumab, as first-line treatment. However, a significant number of patients receiving this treatment show disease progression. Some of the relapses could be explained by the presence of acquired resistance mutations in KRAS. Liquid biopsy of circulating tumor cells or circulating cell-free DNA is expected to improve the management of patients undergoing anti-EGFR therapy.
- Citation: Valenzuela G, Burotto M, Marcelain K, González-Montero J. Liquid biopsy to detect resistance mutations against anti-epidermal growth factor receptor therapy in metastatic colorectal cancer. World J Gastrointest Oncol 2022; 14(9): 1654-1664
- URL: https://www.wjgnet.com/1948-5204/full/v14/i9/1654.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i9.1654
Colorectal cancer (CRC) is the third leading cause of cancer-related mortality worldwide[1]. Disseminated disease (stage IV) with metastasis has been associated with poor prognosis, with a mean survival time of 15 mo[2]. The standard treatment for patients with metastatic CRC (mCRC) involves adjuvant chemotherapy with FOLFOX (leucovorin, 5-fluorouracil, and oxaliplatin) or FOLFIRI (leucovorin, 5-fluorouracil, and irinotecan). Furthermore, international guidelines recommend the analysis of KRAS/NRAS and BRAF mutations for targeted therapy[3,4]. Currently, the use of epidermal growth factor receptor (EGFR) inhibitor antibodies (anti-EGFR), such as cetuximab[5] or panitumumab[6], is recommended for patients with KRAS exon 2 wild-type (wt) mCRC. Both monoclonal antibodies exhibit a high affinity for the extracellular domain of EGFR; thus, they can prevent the ligand binding with EGFR[7]. Nevertheless, only 41% of patients with wt KRAS and left-sided colon disease reportedly attained partial or complete response to anti-EGFR treatments[8], as determined by RECIST criteria. The high level of variability in patient responses could be explained by the molecular and genomic va
In real-world clinical settings, given that several patients are not considered suitable candidates for metastatic biopsies, it has been suggested that liquid biopsy could play a role in the early detection of mutations capable of inducing resistance to targeted therapies. Liquid biopsy is a recently described method that involves the analysis of genetic material from various sources, primarily blood (but also from urine, pleural fluid, and ascites). This method affords information on mutations and alterations in the copy number of genes related to the oncogenic process[16]. Several types of liquid biopsies are available, and the most widely used strategies involve the analysis of circulating tumor cells (CTC), circulating tumor DNA (ctDNA), and extracellular vesicles (EVs) or exosomes, exhibiting both advantages and disadvantages[17]. In patients with mCRC, a high correlation has been noted between the primary metastatic tumor sample and ctDNA, approaching approximately 96.15% concordance for the analysis of KRAS, NRAS, and BRAF[18]. The objective of this review was to evaluate the role of liquid biopsy in the early identification of mutations that induce resistance to cetuximab or panitu
Liquid biopsy requires technology capable of extracting tumor genetic material (DNA or RNA) from the blood, along with a technique that can quantify and characterize the molecular sequence. Nucleic acids can be detected by polymerase chain reaction (PCR)-based techniques or next-generation sequencing (NGS)[19]. The advantages of PCR-based techniques include their lower cost, shorter processing time, and easier bioinformatics analysis than NGS techniques[20]. Disadvantages of PCR techniques include the selection of a prior bound study target and the difficulty in examining rare genetic alterations[21].
Advances in PCR techniques have allowed the development of digital PCR and subsequent evolution toward more advanced technologies such as droplet digital PCR (ddPCR) and Beads, Emulsion, Amplification, Magnetics (BEAMing) digital PCR. Both technologies employ digital PCR principles, which involve sample division or partitioning, where each partition occurs via independent reactions. Subsequently, a digital system allows fluorescence quantification in each partition, and combining the value of each partition affords a final quantification of molecules of interest[22]. In ddPCR, sample reactions occur within water-in-oil droplets, which act as a system of encapsulated molecules, where millions of PCR reactions can be simultaneously quantified[22,23]. The BEAMing technique involves digital PCR in emulsions combined with flow cytometry to quantify DNA molecules. In emulsions, DNA molecules and primers are attached to magnetic beads. Subsequently, amplified fragments are recovered by magnets and recognized by flow cytometry to measure the DNA of interest[24].
NGS techniques are based on massively parallel sequencing of selected or unselected genes; thus, millions of DNA sequences can be read simultaneously[25]. One main advantage of NGS is its ability to detect new mutations or mutations that rarely appear[22]. In addition, NGS offers high sensitivity and specificity for mutation detection; however, it exhibits considerable variability, ranging from 0.1% to 1%, depending on the technique or platform used[26].
Using liquid biopsy, tumor DNA can be obtained from various sources, including ctDNA, CTC, and EV, found in the blood of patients with cancer. Cells normally release nucleotides into patient blood. This genetic material can be isolated and is known as cfDNA. ctDNA is a part of cfDNA derived from tumor cells and can harbor mutations, amplifications, and epigenetic modifications associated with cancer[27]. CTCs are rare tumor cells in the blood that originate from solid tumors or metastases. Enrichment processes allow the elimination of leukocytes from the blood and CTC selection to extract the genetic material to be investigated[28]. Finally, EVs or exosomes are vesicles in the blood and contain DNA, mRNA, or miRNA modulating receptor cells[29].
Advances in methods and technologies for attaining genetic material are expected to complement the limitations of tissue or metastasis biopsies to improve patient prognosis[30].
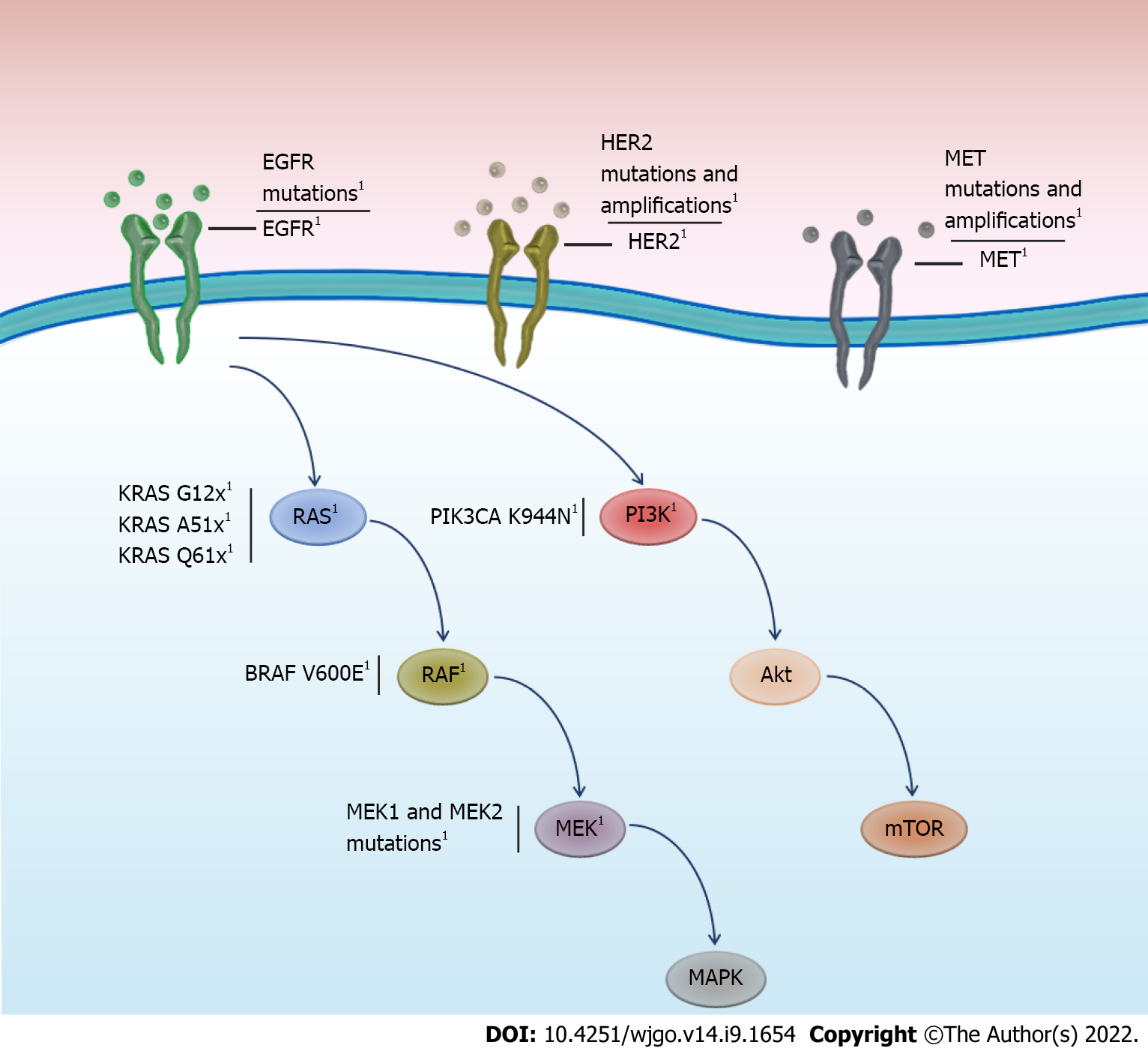
The EGFR receptor is a tyrosine kinase receptor, which, when ligand bound, activates the RAS, RAF, MEK, and ERK pathways[31]. The acquisition of activating mutations in any component of this pathway has been associated with oncogenesis[32]. Initial studies have focused on describing mutations in the KRAS oncogene in patients who relapsed following anti-EGFR therapy. Mutations in KRAS, a member of the small GTP-binding protein family, have been the focus of in-depth study, as the wt KRAS genotype is an indicator for anti-EGFR therapy. In a small number of patients with mCRC presenting disease progression, de novo mutations in KRAS measured by liquid biopsy[33] reached 38% (9/26). Reportedly, 40% of patients with mCRC exhibit KRAS mutations at diagnosis, most frequently in codons 12, 13, 61, and 146[34]. Mutations in codons 12 and 13 alter the position of the KRAS catalytic site at codon 61, reducing GTP hydrolysis and maintaining protein activity, even in the absence of a ligand[35,36]. These activating mutations can induce cellular proliferation and suppress apoptosis[34]. Numerous theories have been proposed to clarify how anti-EGFR antibodies allow the acquisition of resistance mutations. For example, cell culture studies have revealed that prolonged exposure to anti-EGFR treatment allows the survival and selection of clones harboring KRAS mutations[37,38]. In addition, it is postulated that de novo mutations in resistance genes can be generated by genomic instability in cancer[35]. Furthermore, it has been proposed that the same therapeutic drugs can induce mutagenesis[39]. For example, patient studies have revealed that anti-EGFR treatment can induce a distinctive mutational signature, SBS17b, with preferential mutations in KRAS Q61H[15], which is consistent with cell culture studies demonstrating anti-EGFR treatment-induced mutagenesis[40].
The acquisition of resistance mutations in KRAS is one of the most frequent mechanisms reported in liquid biopsy studies. In a small study, 4 of 11 patients with wt KRAS treated with anti-EGFR antibodies acquired KRAS mutations, as determined by ddPCR of ctDNA. In addition, mutations in other components of the EGFR pathway, such as BRAF, MET, and ERBB2, were detected in three patients[41]. These results were replicated in a study by Vitiello et al[18] (2019), in which 10 new KRAS mutations were identified by automated quantitative reverse-transcription PCR in the ctDNA of 30 mCRC patients with wt KRAS receiving anti-EGFR therapy. In a further study using the BEAMing method, analysis of ctDNA revealed that 7 of 34 patients with wt KRAS, who were treated with anti-EGFR, developed resistance mutations, mainly in KRAS codons 12, 13, and 61[42]. Similarly, a follow-up program using the same methodology showed that, among 31 patients with wt KRAS tumor tissue receiving anti-EGFR treatment, 5 presented mutations in KRAS and 3 in NRAS[43]. Furthermore, an analysis of 62 patients with mCRC treated with cetuximab or panitumumab revealed 27 resistance mutations in KRAS and 5 mutations in EGFR (detected in plasma); mutations in codons 12 and 61 of KRAS were the most common. Interestingly, the authors reported that the longer EGFR inhibitors were discontinued, the more the allelic frequency of these mutations detected in plasma tended to decrease[44]. Finally, an NGS study of ctDNA demonstrated that 69% of 42 patients treated with anti-EGFR had mutations or amplifications in KRAS, with the KRAS Q61H mutation (exon 2) detected in 52% of patients. Extending the analysis to other elements of the EGFR pathway, 91% of patients showed alterations in several pathway components, such as NRAS, BRAF, MAP2K1, ERBB2, MET, and KIT mutations or extensions, with an average of five alterations per patient for these genes[45]. Mutations conferring resistance to anti-EGFR are frequent, specifically in KRAS/NRAS, estimated to account for approximately 30%–89% of patients with mCRC (Table 1).
| Ref. | n wt KRAS patients at baseline | Analysis technique | Mutations or amplifications in KRAS/NRAS | Most frequent mutations |
| Vitiello et al[18], 2019 | 30 | ctDNA/RT-qPCR | 10 (30%) | KRAS Q61x (4) |
| KRAS G12x (3) | ||||
| Diaz et al[33], 2012 | 24 | ctDNA/BEAMing | 9 (36%) | KRAS G12x (9) |
| Pietrantonio et al[41], 2017 | 11 | ctDNA/ddPCR | 4 (36%) | KRAS Q61H (2) |
| Vidal et al[42], 2017 | 18 | ctDNA/BEAMing | 7 (39%) | KRAS G12x (5) |
| NRAS Q61x (3) | ||||
| Morelli et al[44], 2015 | 62 | ctDNA/BEAMing | 27 (43%) | KRAS G12x (10) |
| KRAS Q61x (9) | ||||
| Strickler et al[45], 2018 | 42 | ctDNA/NGS DNAseq | 26 (62%) | KRAS Q61H (22) |
| KRAS G12A (5) | ||||
| Yamada et al[46], 2020 | 19 | ctDNA/ddPCR | 16 (84%) | KRAS Q61H (10) |
| KRAS G12V (9) | ||||
| Kim et al[48], 2018 | 164 | ctDNA/NGS DNA seq | 53 (32.3%) | KRAS exon 3 (A59x o Q61x) (20) |
| Takayama et al[75], 2018 | 25 | ctDNA/ddPCR | 9 (36%) | KRAS Q12S (5) |
| KRAS Q12D (4) |
In addition, the prognostic utility of detecting resistance-acquired mutations during anti-EGFR therapy has been examined. Yamada et al[46] (2020) detected 20 acquired mutations in RAS, BRAF, or EGFR genes in ctDNA of 30 patients with mCRC treated with FOLFOX or FOLFIRI plus anti-EGFR. The authors reported that patients who developed measurable mutations in ctDNA had a worse prognosis for progression-free disease (PFS) than those with wt RAS. Follow-up analysis of patients with chemotherapy-refractory mCRC from the ASPECCT clinical trial[47] treated with panitumumab alone (conducted by liquid biopsy) revealed that 32% of 162 patients developed mutations in RAS. Mutations were found to primarily emerge in KRAS codons 2, 3, and 4 and less frequently in exon 2 of NRAS[48]. In contrast to previous studies, no significant differences were detected in patients with emerging RAS mutations in terms of PFS, overall survival (OS), or objective response rate. Subsequently, in the same cohort of patients, the authors found that the allelic frequency of resistance mutations in EGFR pathway genes, including KRAS, may be more closely associated with worse prognosis in panitumumab-treated patients[49]. These results are consistent with those of another study examining patients with wt KRAS CRC undergoing treatment with cetuximab or panitumumab; the emergence of mutations in KRAS, NRAS, or BRAF resulted in worse OS when compared with patients without mutations in these genes, as determined by analyzing CTC [hazard ratio (HR): 0.60, 95% confidence interval (CI): 0.40–0.91, P = 0.0028], but not when ctDNA liquid biopsy was used to analyze the same cohort (HR: 0.80, 95%CI: 0.59–1.33, P = 0.088)[50]. In summary, growing evidence indicates that the detection of mutations, as well as allelic frequency, can be linked to the prognosis of mCRC.
It has been suggested that once disease progression is detected during anti-EGFR treatment, liquid biopsy can be used to evaluate the timing of reintroducing therapy[51]. This concept is known as rechallenge, whereby a period without treatment (such as anti-EGFR therapy) is followed by re-initiation of prior therapy, despite knowledge regarding the potential emergence of resistance mutations[8]. In a meta-analysis of patients who exhibited prior evidence of anti-EGFR benefits and rechallenge with anti-EGFR treatment (with a strategy of assessing RAS status by ctDNA liquid biopsy), up to 46% of patients converted from wt to mutant RAS following exposure to anti-EGFR treatment. Patients who maintained wt RAS before rechallenge had a better prognosis than those with a de novo RAS mutation[52]. Therefore, based on evidence suggesting a potential benefit in patients who maintain wt RAS prior to rechallenge, strategies have been proposed for patients who exhibit acquired resistance mutations in RAS following anti-EGFR treatment. Growing evidence indicates that resistance mutations decay over time after withdrawing anti-EGFR treatment; thus, withdrawing drug therapy eliminates the selective pressure on clones harboring resistance mutations[44]. An exploratory study of patients with wt KRAS/BRAF who acquired RAS or EGFR mutations during the course of anti-EGFR treatment showed that the frequency of mutant alleles decayed exponentially after discontinuing anti-EGFR treatment, with a mean of 4.4 mo[53]. In a retrospective cohort of 80 patients rechallenged after a longer interval, the authors reported a superior prognosis in terms of overall response[53]. Thus, considering the dynamics of the decay of clones with resistance mutations after treatment suspension, clinical studies have been proposed to corroborate the clinical utility of rechallenge therapies. For instance, it has been speculated that patients who previously progressed to chemotherapy and anti-EGFR antibodies could undergo second-line chemotherapy without anti-EGFR; if they progress, anti-EGFR rechallenge could then be performed based on KRAS allele frequency measurement[54]. This has been proposed in the REMARRY and PURSUIT phase II clinical trials; these studies suggested the reintroduction of FOLFIRI and panitumumab (which have an allelic frequency < 0.1% for mutated KRAS), allowing at least 4 mo without anti-EGFR administration[55]. Therefore, biopsies are not only useful for detecting resistance mutations, but could help determine the timing of treatment reintroduction once resistance-inducing mutations have declined.
Resistance mutations to anti-EGFR treatment are frequent, particularly in KRAS, estimated to range between 30 and 89% (Table 1) in patients with mCRC. Although resistance mutations in KRAS are most frequent, mutations or amplification of other genes in the EGFR pathway, such as ERBB2, MEK, BRAF, and MAP2K, could also cause or contribute to anti-EGFR treatment resistance (Figure 1). Basic studies using patient-derived xenograft models, where the acquisition of natural resistance by chronic cetuximab exposure is reproduced, have reported the emergence of driver mutations in EGFR, KRAS, MEK1, and MEK2[56]. These results have been documented in real-world clinical settings, where patients were prospectively followed up by liquid biopsy. For instance, acquisition of MET ampli
HER2 is a tyrosine kinase receptor and member of the HER/ERBB receptor family that includes EGFR (HER1), HER3, and HER4[61]. HER2/ERBB2 activation induces cellular proliferation and activation of the RAS/RAF/ERK and PI3KCA/PTEN/AKT pathways[62]. Mutations or amplification of HER2/ERBB2 has been detected in various tumors. Although most HER2-based studies have primarily focused on breast cancer, the role of this receptor in mCRC has recently been described[63,64]. Previous in vitro and prospective patient studies have suggested that both the presence of mutations related to the active site of the receptor and HER2 amplification are associated with a poor response to anti-EGFR therapy[62,63,65]. In addition, the acquisition of mutations in HER2 may be an underlying mechanism of secondary resistance that can be detected early using liquid biopsy. In a liquid biopsy study, 1 of 11 patients who progressed on anti-EGFR treatment showed HER2 amplification and simultaneous mutation of KRAS[41]. In a study evaluating ctDNA by NGS, one case of HER2 amplification was identified in a series of 15 patients treated with cetuximab[66]. Nonetheless, a case-control study revealed that the presence of HER2 amplification in patients with wt KRAS CRC (prospectively measured by ddPCR of ctDNA) was not associated with a worse prognosis when compared with those without HER2 mutations. However, the number of cases of amplified HER2 was markedly low (five cases) to establish meaningful conclusions[67]. A phase IB clinical study has proposed the use of neratinib (pan-ERBB kinase inhibitor) and cetuximab in patients who have progressed to anti-EGFR therapy[68]. This trial was based on the hypothesis that HER2-negative tumors acquire HER2 ampli
Liquid biopsies for monitoring anti-EGFR resistance mutations have not been performed in routine medical practice. Real-world studies on liquid biopsy programs indicate that the application of these techniques can effectively alter the management of patients with colon cancer[43]. However, im
Therefore, it is necessary to establish protocols for the frequency of taking liquid biopsies, as well as their implications for clinical patient management. Clinical studies are currently being conducted to standardize the frequency of sampling and interpretation of results. Two prospective studies have attempted to establish the prognostic value of liquid biopsy protocols; both studies including periodic three-monthly ctDNA analyses and clinical follow-up in CRC wt KRAS patients exposed to 5-fluorouracil regimens plus anti-EGFR antibodies[72,73]. Finally, current international guidelines, such as ESMO, have concluded that although there is insufficient evidence to recommend follow-up with liquid biopsy, such analysis could be useful for detecting secondary resistance to anti-EFGR[4]. In contrast, the Japanese Society of Medical Oncology clinical guidelines recommend the use of liquid biopsy because of its usefulness in monitoring anti-EGFR therapy[74].
Based on current evidence, liquid biopsy could be developed as an innovative tool for managing patients with mCRC who receive anti-EGFR therapy. De novo KRAS mutations are one of the most commonly described mechanisms of acquired resistance and are associated with poor outcomes. However, establishing panels beyond KRAS, including genes related to the EGFR pathway, is crucial, given that such genes also potentially contribute to anti-EGFR resistance. Adequate strategies are needed to integrate liquid biopsy for the early detection of clinical progression of mCRC in patients undergoing anti-EGFR therapy. Future clinical studies will advance the routine use of liquid biopsy as a tool for reaching clinical decisions that benefit patients.
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 68689] [Article Influence: 13737.8] [Reference Citation Analysis (201)] |
| 2. | Wang J, Li S, Liu Y, Zhang C, Li H, Lai B. Metastatic patterns and survival outcomes in patients with stage IV colon cancer: A population-based analysis. Cancer Med. 2020;9:361-373. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 139] [Cited by in RCA: 223] [Article Influence: 37.2] [Reference Citation Analysis (0)] |
| 3. | Benson AB, Venook AP, Al-Hawary MM, Cederquist L, Chen YJ, Ciombor KK, Cohen S, Cooper HS, Deming D, Engstrom PF, Garrido-Laguna I, Grem JL, Grothey A, Hochster HS, Hoffe S, Hunt S, Kamel A, Kirilcuk N, Krishnamurthi S, Messersmith WA, Meyerhardt J, Miller ED, Mulcahy MF, Murphy JD, Nurkin S, Saltz L, Sharma S, Shibata D, Skibber JM, Sofocleous CT, Stoffel EM, Stotsky-Himelfarb E, Willett CG, Wuthrick E, Gregory KM, Freedman-Cass DA. NCCN Guidelines Insights: Colon Cancer, Version 2.2018. J Natl Compr Canc Netw. 2018;16:359-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 635] [Cited by in RCA: 723] [Article Influence: 90.4] [Reference Citation Analysis (3)] |
| 4. | Van Cutsem E, Cervantes A, Adam R, Sobrero A, Van Krieken JH, Aderka D, Aranda Aguilar E, Bardelli A, Benson A, Bodoky G, Ciardiello F, D'Hoore A, Diaz-Rubio E, Douillard JY, Ducreux M, Falcone A, Grothey A, Gruenberger T, Haustermans K, Heinemann V, Hoff P, Köhne CH, Labianca R, Laurent-Puig P, Ma B, Maughan T, Muro K, Normanno N, Österlund P, Oyen WJ, Papamichael D, Pentheroudakis G, Pfeiffer P, Price TJ, Punt C, Ricke J, Roth A, Salazar R, Scheithauer W, Schmoll HJ, Tabernero J, Taïeb J, Tejpar S, Wasan H, Yoshino T, Zaanan A, Arnold D. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol. 2016;27:1386-1422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2286] [Cited by in RCA: 2541] [Article Influence: 254.1] [Reference Citation Analysis (41)] |
| 5. | Van Cutsem E, Lenz HJ, Köhne CH, Heinemann V, Tejpar S, Melezínek I, Beier F, Stroh C, Rougier P, van Krieken JH, Ciardiello F. Fluorouracil, leucovorin, and irinotecan plus cetuximab treatment and RAS mutations in colorectal cancer. J Clin Oncol. 2015;33:692-700. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 564] [Cited by in RCA: 643] [Article Influence: 58.5] [Reference Citation Analysis (3)] |
| 6. | Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, Rivera F, Kocákova I, Ruff P, Błasińska-Morawiec M, Šmakal M, Canon JL, Rother M, Williams R, Rong A, Wiezorek J, Sidhu R, Patterson SD. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023-1034. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1610] [Cited by in RCA: 1770] [Article Influence: 136.2] [Reference Citation Analysis (0)] |
| 7. | Martins M, Mansinho A, Cruz-Duarte R, Martins SL, Costa L. Anti-EGFR Therapy to Treat Metastatic Colorectal Cancer: Not for All. In: Jordan P, editor. Targeted Therapy of Colorec-tal Cancer Subtypes. Cham: Springer International Publishing, 2018: 113–131. |
| 8. | Rossini D, Germani MM, Pagani F, Pellino A, Dell'Aquila E, Bensi M, Liscia N, Moretto R, Boccaccino A, Prisciandaro M, Manglaviti S, Schirripa M, Vivolo R, Scartozzi M, Santini D, Salvatore L, Pietrantonio F, Loupakis F, Falcone A, Cremolini C. Retreatment With Anti-EGFR Antibodies in Metastatic Colorectal Cancer Patients: A Multi-institutional Analysis. Clin Colorectal Cancer. 2020;19:191-199.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 9. | Sagaert X, Vanstapel A, Verbeek S. Tumor Heterogeneity in Colorectal Cancer: What Do We Know So Far? Pathobiology. 2018;85:72-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 117] [Article Influence: 14.6] [Reference Citation Analysis (1)] |
| 10. | Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda G, Angelino P, Bot BM, Morris JS, Simon IM, Gerster S, Fessler E, De Sousa E Melo F, Missiaglia E, Ramay H, Barras D, Homicsko K, Maru D, Manyam GC, Broom B, Boige V, Perez-Villamil B, Laderas T, Salazar R, Gray JW, Hanahan D, Tabernero J, Bernards R, Friend SH, Laurent-Puig P, Medema JP, Sadanandam A, Wessels L, Delorenzi M, Kopetz S, Vermeulen L, Tejpar S. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350-1356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3408] [Cited by in RCA: 3839] [Article Influence: 349.0] [Reference Citation Analysis (8)] |
| 11. | Valenzuela G, Canepa J, Simonetti C, Solo de Zaldívar L, Marcelain K, González-Montero J. Consensus molecular subtypes of colorectal cancer in clinical practice: A translational approach. World J Clin Oncol. 2021;12:1000-1008. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 12] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (2)] |
| 12. | Burrell RA, Swanton C. Tumour heterogeneity and the evolution of polyclonal drug resistance. Mol Oncol. 2014;8:1095-1111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 329] [Cited by in RCA: 308] [Article Influence: 25.7] [Reference Citation Analysis (1)] |
| 13. | Parikh AR, Leshchiner I, Elagina L, Goyal L, Levovitz C, Siravegna G, Livitz D, Rhrissorrakrai K, Martin EE, Van Seventer EE, Hanna M, Slowik K, Utro F, Pinto CJ, Wong A, Danysh BP, de la Cruz FF, Fetter IJ, Nadres B, Shahzade HA, Allen JN, Blaszkowsky LS, Clark JW, Giantonio B, Murphy JE, Nipp RD, Roeland E, Ryan DP, Weekes CD, Kwak EL, Faris JE, Wo JY, Aguet F, Dey-Guha I, Hazar-Rethinam M, Dias-Santagata D, Ting DT, Zhu AX, Hong TS, Golub TR, Iafrate AJ, Adalsteinsson VA, Bardelli A, Parida L, Juric D, Getz G, Corcoran RB. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat Med. 2019;25:1415-1421. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 213] [Cited by in RCA: 391] [Article Influence: 55.9] [Reference Citation Analysis (0)] |
| 14. | Stahler A, Heinemann V, Holch JW, von Einem JC, Westphalen CB, Heinrich K, Schlieker L, Jelas I, Alig AHS, Fischer LE, Weiss L, Modest DP, von Weikersthal LF, Decker T, Kiani A, Moehler M, Kaiser F, Kirchner T, Jung A, Stintzing S. Mutational profiles of metastatic colorectal cancer treated with FOLFIRI plus cetuximab or bevacizumab before and after secondary resection (AIO KRK 0306; FIRE-3). Int J Cancer. 2021;149:1935-1943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 15. | Woolston A, Barber LJ, Griffiths B, Pich O, Lopez-Bigas N, Matthews N, Rao S, Watkins D, Chau I, Starling N, Cunningham D, Gerlinger M. Mutational signatures impact the evolution of anti-EGFR antibody resistance in colorectal cancer. Nat Ecol Evol. 2021;5:1024-1032. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 16. | Heitzer E, Haque IS, Roberts CES, Speicher MR. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet. 2019;20:71-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 969] [Cited by in RCA: 1022] [Article Influence: 146.0] [Reference Citation Analysis (0)] |
| 17. | Luo W, Rao M, Qu J, Luo D. Applications of liquid biopsy in lung cancer-diagnosis, prognosis prediction, and disease monitoring. Am J Transl Res. 2018;10:3911-3923. [PubMed] |
| 18. | Vitiello PP, De Falco V, Giunta EF, Ciardiello D, Cardone C, Vitale P, Zanaletti N, Borrelli C, Poliero L, Terminiello M, Arrichiello G, Caputo V, Famiglietti V, Mattera Iacono V, Marrone F, Di Liello A, Martini G, Napolitano S, Caraglia M, Lombardi A, Franco R, De Vita F, Morgillo F, Troiani T, Ciardiello F, Martinelli E. Clinical Practice Use of Liquid Biopsy to Identify RAS/BRAF Mutations in Patients with Metastatic Colorectal Cancer (mCRC): A Single Institution Experience. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 19. | Yi Z, Qu C, Zeng Y, Liu Z. Liquid biopsy: early and accurate diagnosis of brain tumor. J Cancer Res Clin Oncol. 2022;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 20. | Palacín-Aliana I, García-Romero N, Asensi-Puig A, Carrión-Navarro J, González-Rumayor V, Ayuso-Sacido Á. Clinical Utility of Liquid Biopsy-Based Actionable Mutations Detected via ddPCR. Biomedicines. 2021;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (1)] |
| 21. | Vendrell JA, Mau-Them FT, Béganton B, Godreuil S, Coopman P, Solassol J. Circulating Cell Free Tumor DNA Detection as a Routine Tool forLung Cancer Patient Management. Int J Mol Sci. 2017;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 70] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 22. | Moreno-Manuel A, Calabuig-Fariñas S, Obrador-Hevia A, Blasco A, Fernández-Díaz A, Sirera R, Camps C, Jantus-Lewintre E. dPCR application in liquid biopsies: divide and conquer. Expert Rev Mol Diagn. 2021;21:3-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 23. | Perkins G, Lu H, Garlan F, Taly V. Droplet-Based Digital PCR. In: Advances in Clinical Chemistry. Elsevier, 2017: 43–91. |
| 24. | Diehl F, Li M, He Y, Kinzler KW, Vogelstein B, Dressman D. BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions. Nat Methods. 2006;3:551-559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 353] [Cited by in RCA: 385] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 25. | Bai Y, Wang Z, Liu Z, Liang G, Gu W, Ge Q. Technical progress in circulating tumor DNA analysis using next generation sequencing. Mol Cell Probes. 2020;49:101480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 26. | Thress KS, Brant R, Carr TH, Dearden S, Jenkins S, Brown H, Hammett T, Cantarini M, Barrett JC. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer. 2015;90:509-515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 414] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 27. | Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32:579-586. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1413] [Cited by in RCA: 1733] [Article Influence: 144.4] [Reference Citation Analysis (0)] |
| 28. | Cabel L, Proudhon C, Mariani P, Tzanis D, Beinse G, Bieche I, Pierga JY, Bidard FC. Circulating tumor cells and circulating tumor DNA: What surgical oncologists need to know? Eur J Surg Oncol. 2017;43:949-962. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 29. | Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14:531-548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1017] [Cited by in RCA: 1460] [Article Influence: 162.2] [Reference Citation Analysis (0)] |
| 30. | De Rubis G, Rajeev Krishnan S, Bebawy M. Liquid Biopsies in Cancer Diagnosis, Monitoring, and Prognosis. Trends Pharmacol Sci. 2019;40:172-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 402] [Article Influence: 57.4] [Reference Citation Analysis (0)] |
| 31. | Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4920] [Cited by in RCA: 5198] [Article Influence: 207.9] [Reference Citation Analysis (1)] |
| 32. | Sforza V, Martinelli E, Ciardiello F, Gambardella V, Napolitano S, Martini G, Della Corte C, Cardone C, Ferrara ML, Reginelli A, Liguori G, Belli G, Troiani T. Mechanisms of resistance to anti-epidermal growth factor receptor inhibitors in metastatic colorectal cancer. World J Gastroenterol. 2016;22:6345-6361. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 86] [Cited by in RCA: 102] [Article Influence: 10.2] [Reference Citation Analysis (2)] |
| 33. | Diaz LA Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, Allen B, Bozic I, Reiter JG, Nowak MA, Kinzler KW, Oliner KS, Vogelstein B. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537-540. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1259] [Cited by in RCA: 1366] [Article Influence: 97.6] [Reference Citation Analysis (0)] |
| 34. | Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761-774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1238] [Cited by in RCA: 1408] [Article Influence: 93.9] [Reference Citation Analysis (0)] |
| 35. | Knickelbein K, Zhang L. Mutant KRAS as a critical determinant of the therapeutic re-sponse of colorectal cancer. Genes Dis. 2015;2:4-12. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 87] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 36. | Lowy DR, Willumsen BM. Function and regulation of ras. Annu Rev Biochem. 1993;62:851-891. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 878] [Cited by in RCA: 861] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 37. | Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, Valtorta E, Schiavo R, Buscarino M, Siravegna G, Bencardino K, Cercek A, Chen CT, Veronese S, Zanon C, Sartore-Bianchi A, Gambacorta M, Gallicchio M, Vakiani E, Boscaro V, Medico E, Weiser M, Siena S, Di Nicolantonio F, Solit D, Bardelli A. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1313] [Cited by in RCA: 1504] [Article Influence: 107.4] [Reference Citation Analysis (0)] |
| 38. | Misale S, Di Nicolantonio F, Sartore-Bianchi A, Siena S, Bardelli A. Resistance to anti-EGFR therapy in colorectal cancer: from heterogeneity to convergent evolution. Cancer Discov. 2014;4:1269-1280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 416] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 39. | Gerlinger M. Targeted drugs ramp up cancer mutability. Science. 2019;366:1452-1453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 40. | Russo M, Crisafulli G, Sogari A, Reilly NM, Arena S, Lamba S, Bartolini A, Amodio V, Magrì A, Novara L, Sarotto I, Nagel ZD, Piett CG, Amatu A, Sartore-Bianchi A, Siena S, Bertotti A, Trusolino L, Corigliano M, Gherardi M, Lagomarsino MC, Di Nicolantonio F, Bardelli A. Adaptive mutability of colorectal cancers in response to targeted therapies. Science. 2019;366:1473-1480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 319] [Article Influence: 45.6] [Reference Citation Analysis (0)] |
| 41. | Pietrantonio F, Vernieri C, Siravegna G, Mennitto A, Berenato R, Perrone F, Gloghini A, Tamborini E, Lonardi S, Morano F, Picciani B, Busico A, Volpi CC, Martinetti A, Battaglin F, Bossi I, Pellegrinelli A, Milione M, Cremolini C, Di Bartolomeo M, Bardelli A, de Braud F. Heterogeneity of Acquired Resistance to Anti-EGFR Monoclonal Antibodies in Patients with Metastatic Colorectal Cancer. Clin Cancer Res. 2017;23:2414-2422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 151] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 42. | Vidal J, Muinelo L, Dalmases A, Jones F, Edelstein D, Iglesias M, Orrillo M, Abalo A, Rodríguez C, Brozos E, Vidal Y, Candamio S, Vázquez F, Ruiz J, Guix M, Visa L, Sikri V, Albanell J, Bellosillo B, López R, Montagut C. Plasma ctDNA RAS mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients. Ann Oncol. 2017;28:1325-1332. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 216] [Cited by in RCA: 279] [Article Influence: 34.9] [Reference Citation Analysis (0)] |
| 43. | Hedtke M, Pessoa Rejas R, Froelich MF, Ast V, Duda A, Mirbach L, Costina V, Martens UM, Hofheinz RD, Neumaier M, Haselmann V. Liquid profiling of circulating tumor DNA in colorectal cancer: steps needed to achieve its full clinical value as standard care. Mol Oncol. 2022;16:2042-2056. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 44. | Morelli MP, Overman MJ, Dasari A, Kazmi SMA, Mazard T, Vilar E, Morris VK, Lee MS, Herron D, Eng C, Morris J, Kee BK, Janku F, Deaton FL, Garrett C, Maru D, Diehl F, Angenendt P, Kopetz S. Characterizing the patterns of clonal selection in circulating tumor DNA from patients with colorectal cancer refractory to anti-EGFR treatment. Ann Oncol. 2015;26:731-736. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 218] [Cited by in RCA: 205] [Article Influence: 18.6] [Reference Citation Analysis (1)] |
| 45. | Strickler JH, Loree JM, Ahronian LG, Parikh AR, Niedzwiecki D, Pereira AAL, McKinney M, Korn WM, Atreya CE, Banks KC, Nagy RJ, Meric-Bernstam F, Lanman RB, Talasaz A, Tsigelny IF, Corcoran RB, Kopetz S. Genomic Landscape of Cell-Free DNA in Patients with Colorectal Cancer. Cancer Discov. 2018;8:164-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 231] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 46. | Yamada T, Matsuda A, Takahashi G, Iwai T, Takeda K, Ueda K, Kuriyama S, Koizumi M, Shinji S, Yokoyama Y, Ohta R, Yoshida H. Emerging RAS, BRAF, and EGFR mutations in cell-free DNA of metastatic colorectal patients are associated with both primary and secondary resistance to first-line anti-EGFR therapy. Int J Clin Oncol. 2020;25:1523-1532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 47. | Price TJ, Peeters M, Kim TW, Li J, Cascinu S, Ruff P, Suresh AS, Thomas A, Tjulandin S, Zhang K, Murugappan S, Sidhu R. Panitumumab versus cetuximab in patients with chemotherapy-refractory wild-type KRAS exon 2 metastatic colorectal cancer (ASPECCT): a randomised, multicentre, open-label, non-inferiority phase 3 study. Lancet Oncol. 2014;15:569-579. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 285] [Cited by in RCA: 345] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 48. | Kim TW, Peeters M, Thomas A, Gibbs P, Hool K, Zhang J, Ang AL, Bach BA, Price T. Impact of Emergent Circulating Tumor DNA RAS Mutation in Panitumumab-Treated Chemoresistant Metastatic Colorectal Cancer. Clin Cancer Res. 2018;24:5602-5609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 49. | Peeters M, Price T, Boedigheimer M, Kim TW, Ruff P, Gibbs P, Thomas A, Demonty G, Hool K, Ang A. Evaluation of Emergent Mutations in Circulating Cell-Free DNA and Clinical Outcomes in Patients with Metastatic Colorectal Cancer Treated with Panitumumab in the ASPECCT Study. Clin Cancer Res. 2019;25:1216-1225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 50. | Sun Q, Liu Y, Liu B. Use of Liquid Biopsy in Monitoring Colorectal Cancer Progression Shows Strong Clinical Correlation. Am J Med Sci. 2018;355:220-227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 51. | Naidoo M, Piercey O, Tie J. Circulating Tumour DNA and Colorectal Cancer: the Next Revolutionary Biomarker? Curr Oncol Rep. 2021;23:140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 52. | Vlachou MS, Mauri D, Zarkavelis G, Ntellas P, Tagkas C, Gkoura S, Pentheroudakis G. Plasma ctDNA RAS status selects patients for anti-EGFR treatment rechallenge in metastatic colorectal cancer: a meta-analysis. Exp Oncol. 2021;43:252-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 53. | Parseghian CM, Loree JM, Morris VK, Liu X, Clifton KK, Napolitano S, Henry JT, Pereira AA, Vilar E, Johnson B, Kee B, Raghav K, Dasari A, Wu J, Garg N, Raymond VM, Banks KC, Talasaz AA, Lanman RB, Strickler JH, Hong DS, Corcoran RB, Overman MJ, Kopetz S. Anti-EGFR-resistant clones decay exponentially after progression: implications for anti-EGFR re-challenge. Ann Oncol. 2019;30:243-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 183] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 54. | Ciardiello D, Martini G, Famiglietti V, Napolitano S, De Falco V, Troiani T, Latiano TP, Ros J, Elez Fernandez E, Vitiello PP, Maiello E, Ciardiello F, Martinelli E. Biomarker-Guided Anti-Egfr Rechallenge Therapy in Metastatic Colorectal Cancer. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 55. | Nakajima H, Kotani D, Bando H, Kato T, Oki E, Shinozaki E, Sunakawa Y, Yamazaki K, Yuki S, Nakamura Y, Yamanaka T, Yoshino T, Ohta T, Taniguchi H, Kagawa Y. REMARRY and PURSUIT trials: liquid biopsy-guided rechallenge with anti-epidermal growth factor receptor (EGFR) therapy with panitumumab plus irinotecan for patients with plasma RAS wild-type metastatic colorectal cancer. BMC Cancer. 2021;21:674. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (1)] |
| 56. | Vangala D, Ladigan S, Liffers ST, Noseir S, Maghnouj A, Götze TM, Verdoodt B, Klein-Scory S, Godfrey L, Zowada MK, Huerta M, Edelstein DL, de Villarreal JM, Marqués M, Kumbrink J, Jung A, Schiergens T, Werner J, Heinemann V, Stintzing S, Lindoerfer D, Mansmann U, Pohl M, Teschendorf C, Bernhardt C, Wolters H, Stern J, Usta S, Viebahn R, Admard J, Casadei N, Fröhling S, Ball CR, Siveke JT, Glimm H, Tannapfel A, Schmiegel W, Hahn SA. Secondary resistance to anti-EGFR therapy by transcriptional reprogramming in patient-derived colorectal cancer models. Genome Med. 2021;13:116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 57. | Raghav K, Morris V, Tang C, Morelli P, Amin HM, Chen K, Manyam GC, Broom B, Overman MJ, Shaw K, Meric-Bernstam F, Maru D, Menter D, Ellis LM, Eng C, Hong D, Kopetz S. MET amplification in metastatic colorectal cancer: an acquired response to EGFR inhibition, not a de novo phenomenon. Oncotarget. 2016;7:54627-54631. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 59] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 58. | Rimassa L, Bozzarelli S, Pietrantonio F, Cordio S, Lonardi S, Toppo L, Zaniboni A, Bordonaro R, Di Bartolomeo M, Tomasello G, Dadduzio V, Tronconi MC, Piombo C, Giordano L, Gloghini A, Di Tommaso L, Santoro A. Phase II Study of Tivantinib and Cetuximab in Patients With KRAS Wild-type Metastatic Colorectal Cancer With Acquired Resistance to EGFR Inhibitors and Emergence of MET Overexpression: Lesson Learned for Future Trials With EGFR/MET Dual Inhibition. Clin Colorectal Cancer. 2019;18:125-132.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 59. | Xu JM, Wang Y, Wang YL, Liu T, Ni M, Li MS, Lin L, Ge FJ, Gong C, Gu JY, Jia R, Wang HF, Chen YL, Liu RR, Zhao CH, Tan ZL, Jin Y, Zhu YP, Ogino S, Qian ZR. PIK3CA Mutations Contribute to Acquired Cetuximab Resistance in Patients with Metastatic Colorectal Cancer. Clin Cancer Res. 2017;23:4602-4616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 68] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 60. | Clifton K, Rich TA, Parseghian C, Raymond VM, Dasari A, Pereira AAL, Willis J, Loree JM, Bauer TM, Chae YK, Sherrill G, Fanta P, Grothey A, Hendifar A, Henry D, Mahadevan D, Nezami MA, Tan B, Wainberg ZA, Lanman R, Kopetz S, Morris V. Identification of Actionable Fusions as an Anti-EGFR Resistance Mechanism Using a Circulating Tumor DNA Assay. JCO Precis Oncol. 2019;3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 61. | Hynes NE, MacDonald G. ErbB receptors and signaling pathways in cancer. Curr Opin Cell Biol. 2009;21:177-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 660] [Cited by in RCA: 745] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 62. | Martin V, Landi L, Molinari F, Fountzilas G, Geva R, Riva A, Saletti P, De Dosso S, Spitale A, Tejpar S, Kalogeras KT, Mazzucchelli L, Frattini M, Cappuzzo F. HER2 gene copy number status may influence clinical efficacy to anti-EGFR monoclonal antibodies in metastatic colorectal cancer patients. Br J Cancer. 2013;108:668-675. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 129] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 63. | Takegawa N, Yonesaka K. HER2 as an Emerging Oncotarget for Colorectal Cancer Treatment After Failure of Anti-Epidermal Growth Factor Receptor Therapy. Clin Colorectal Cancer. 2017;16:247-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 40] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 64. | La Salvia A, Lopez-Gomez V, Garcia-Carbonero R. HER2-targeted therapy: an emerging strategy in advanced colorectal cancer. Expert Opin Investig Drugs. 2019;28:29-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 65. | Gharib E, Salmanipour R, Nazemalhosseini Mojarad E, Yaghoob Taleghani M, Sarlak S, Malekzade-Moghani M, Nasrabadi PN, Meiary MA, Asadzadeh Aghdaei H, Zali MR. HER2+ mCRC patients with exon 20 R784G substitution mutation do not respond to the cetuximab therapy. J Cell Physiol. 2019;234:13137-13144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 66. | Zhang H, Liu R, Yan C, Liu L, Tong Z, Jiang W, Yao M, Fang W, Chen Z. Advantage of Next-Generation Sequencing in Dynamic Monitoring of Circulating Tumor DNA over Droplet Digital PCR in Cetuximab Treated Colorectal Cancer Patients. Transl Oncol. 2019;12:426-431. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 67. | Liu R, Zhao X, Guo W, Huang M, Qiu L, Zhang W, Zhang Z, Li W, Zhu X, Chen Z. Dynamic monitoring of HER2 amplification in circulating DNA of patients with metastatic colorectal cancer treated with cetuximab. Clin Transl Oncol. 2020;22:928-934. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 68. | Jacobs SA, Lee JJ, George TJ, Wade JL 3rd, Stella PJ, Wang D, Sama AR, Piette F, Pogue-Geile KL, Kim RS, Gavin PG, Lipchik C, Feng H, Wang Y, Finnigan M, Kiesel BF, Beumer JH, Wolmark N, Lucas PC, Allegra CJ, Srinivasan A. Neratinib-Plus-Cetuximab in Quadruple-WT (KRAS, NRAS, BRAF, PIK3CA) Metastatic Colorectal Cancer Resistant to Cetuximab or Panitumumab: NSABP FC-7, A Phase Ib Study. Clin Cancer Res. 2021;27:1612-1622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 69. | Jacobs SA, Lee JJ, George TJ, Yothers G, Kolevska T, Yost KJ, Wade JL, Buchschacher GL, Stella PJ, Shipstone A, Pogue-Geile KL, Srinivasan A, Lucas PC, Allegra CJ. NSABP FC-11: A phase II study of neratinib (N) plus trastuzumab (T) or n plus cetuximab (C) in pa-tients (pts) with ‘quadruple wild-type (WT)’ (KRAS/NRAS/BRAF/PIK3CA WT) metastatic colorectal cancer (mCRC) based on HER2 status—Amplified (amp), non-amplified (non-amp), WT, or mutated (mt). J Clin Oncol. 2019;37:TPS716-TPS716. [RCA] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 70. | IJzerman MJ, de Boer J, Azad A, Degeling K, Geoghegan J, Hewitt C, Hollande F, Lee B, To YH, Tothill RW, Wright G, Tie J, Dawson SJ. Towards Routine Implementation of Liquid Biopsies in Cancer Management: It Is Always Too Early, until Suddenly It Is Too Late. Diagnostics (Basel). 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 71. | Dasari A, Morris VK, Allegra CJ, Atreya C, Benson AB 3rd, Boland P, Chung K, Copur MS, Corcoran RB, Deming DA, Dwyer A, Diehn M, Eng C, George TJ, Gollub MJ, Goodwin RA, Hamilton SR, Hechtman JF, Hochster H, Hong TS, Innocenti F, Iqbal A, Jacobs SA, Kennecke HF, Lee JJ, Lieu CH, Lenz HJ, Lindwasser OW, Montagut C, Odisio B, Ou FS, Porter L, Raghav K, Schrag D, Scott AJ, Shi Q, Strickler JH, Venook A, Yaeger R, Yothers G, You YN, Zell JA, Kopetz S. ctDNA applications and integration in colorectal cancer: an NCI Colon and Rectal-Anal Task Forces whitepaper. Nat Rev Clin Oncol. 2020;17:757-770. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 181] [Cited by in RCA: 277] [Article Influence: 46.2] [Reference Citation Analysis (0)] |
| 72. | Chen SH, Tsai HL, Jiang JK, Sung YC, Huang CW, Yeh YM, Chen LT, Wang JY. Emergence of RAS mutations in patients with metastatic colorectal cancer receiving cetuximab-based treatment: a study protocol. BMC Cancer. 2019;19:640. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 73. | Matsuda A, Yamada T, Takahashi T, Hirata K, Nagasaka T, Ishimaru K, Sakamoto K, Koda K, Ishikawa T, Ishida H, Matsuda K, Kuramochi H, Yoshida Y, Sonoda H, Yoshida H. A Trial Protocol of Precision Medicine for Patients with RAS Wild Metastatic Colorectal Cancer Using Liquid Biopsy (RAS-liquid Study): A Prospective, Multicenter Observational Study. J Anus Rectum Colon. 2022;6:52-57. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 74. | Ebi H, Bando H, Taniguchi H, Sunakawa Y, Okugawa Y, Hatanaka Y, Hosoda W, Kumamoto K, Nakatani K, Yamazaki K. Japanese Society of Medical Oncology Clinical Guidelines: Molecular Testing for Colorectal Cancer Treatment, 4th edition. Cancer Sci. 2020;111:3962-3969. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 75. | Takayama Y, Suzuki K, Muto Y, Ichida K, Fukui T, Kakizawa N, Ishikawa H, Watanabe F, Hasegawa F, Saito M, Tsujinaka S, Futsuhara K, Miyakura Y, Noda H, Konishi F, Rikiyama T. Monitoring circulating tumor DNA revealed dynamic changes in KRAS status in patients with metastatic colorectal cancer. Oncotarget. 2018;9:24398-24413. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: Chile
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B, B, B, B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Herold Z, Hungary; Martin MJ, Argentina; Sun C, United States; Wan XH, China; Ye X, China; Ying HQ, China S-Editor: Fan JR L-Editor: A P-Editor: Fan JR