Published online Aug 15, 2022. doi: 10.4251/wjgo.v14.i8.1574
Peer-review started: May 26, 2022
First decision: June 21, 2022
Revised: July 2, 2022
Accepted: July 22, 2022
Article in press: July 22, 2022
Published online: August 15, 2022
Processing time: 76 Days and 3.7 Hours
Studies have validated the potential of methylated cell-free DNA as a biomarker in various tumors, and methylated DNA in plasma may be a potential biomarker for cancer.
To evaluate the diagnostic value of RASSF1A methylation in plasma for colorectal cancer (CRC) and hepatocellular carcinoma (HCC).
A total of 92 CRC patients, 67 colorectal polyp (CRP) patients, 63 HCC patients, and 66 liver cirrhosis (LC) patients were enrolled. The plasma DNA was subjected to DNA extraction, double-strand DNA concentration determination, bisulfite conversion, purification, single-strand DNA concentration determination, and digital polymerase chain reaction (PCR) detection. The methylation rate was calculated. The diagnostic value was evaluated by the area under the curve (AUC).
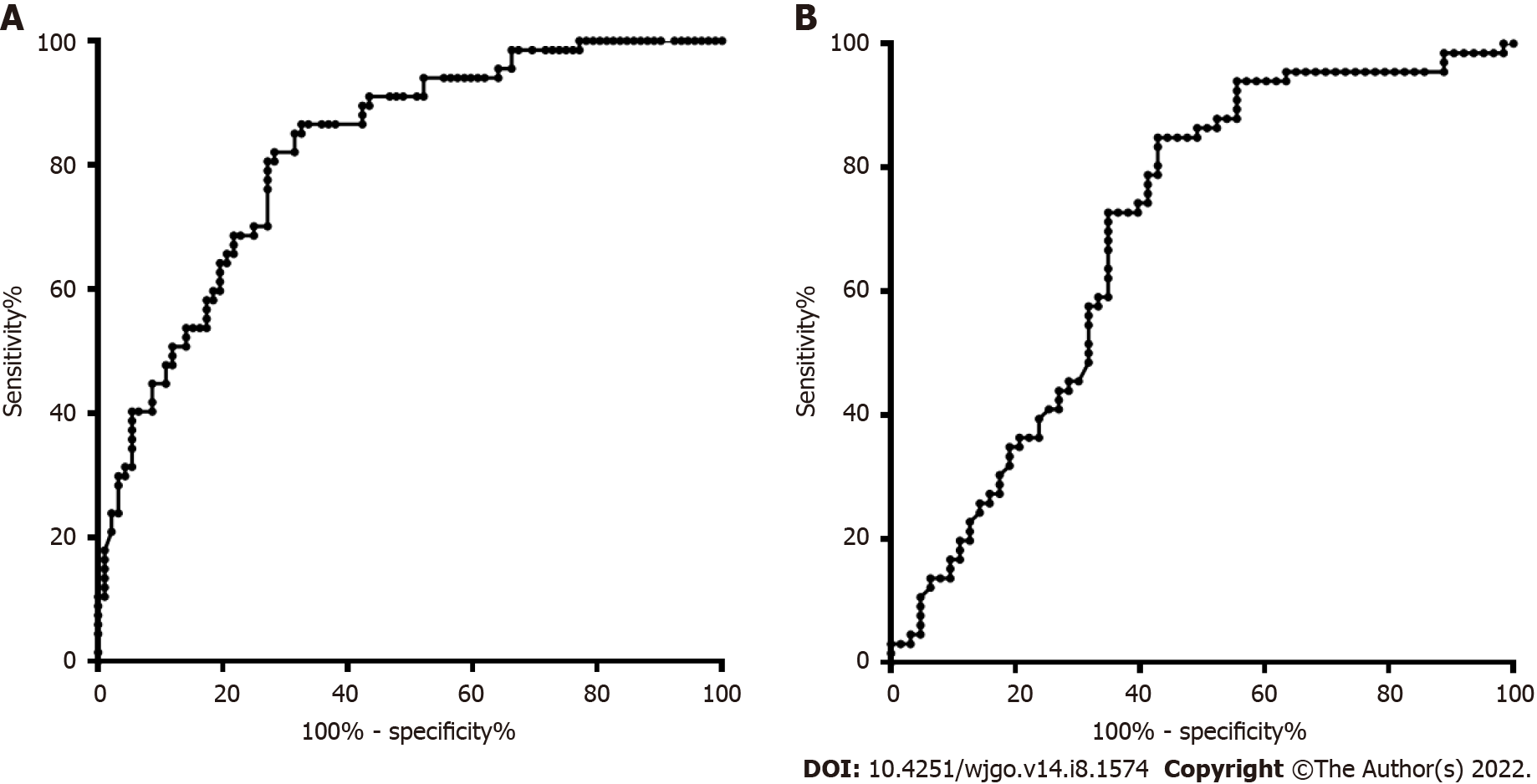
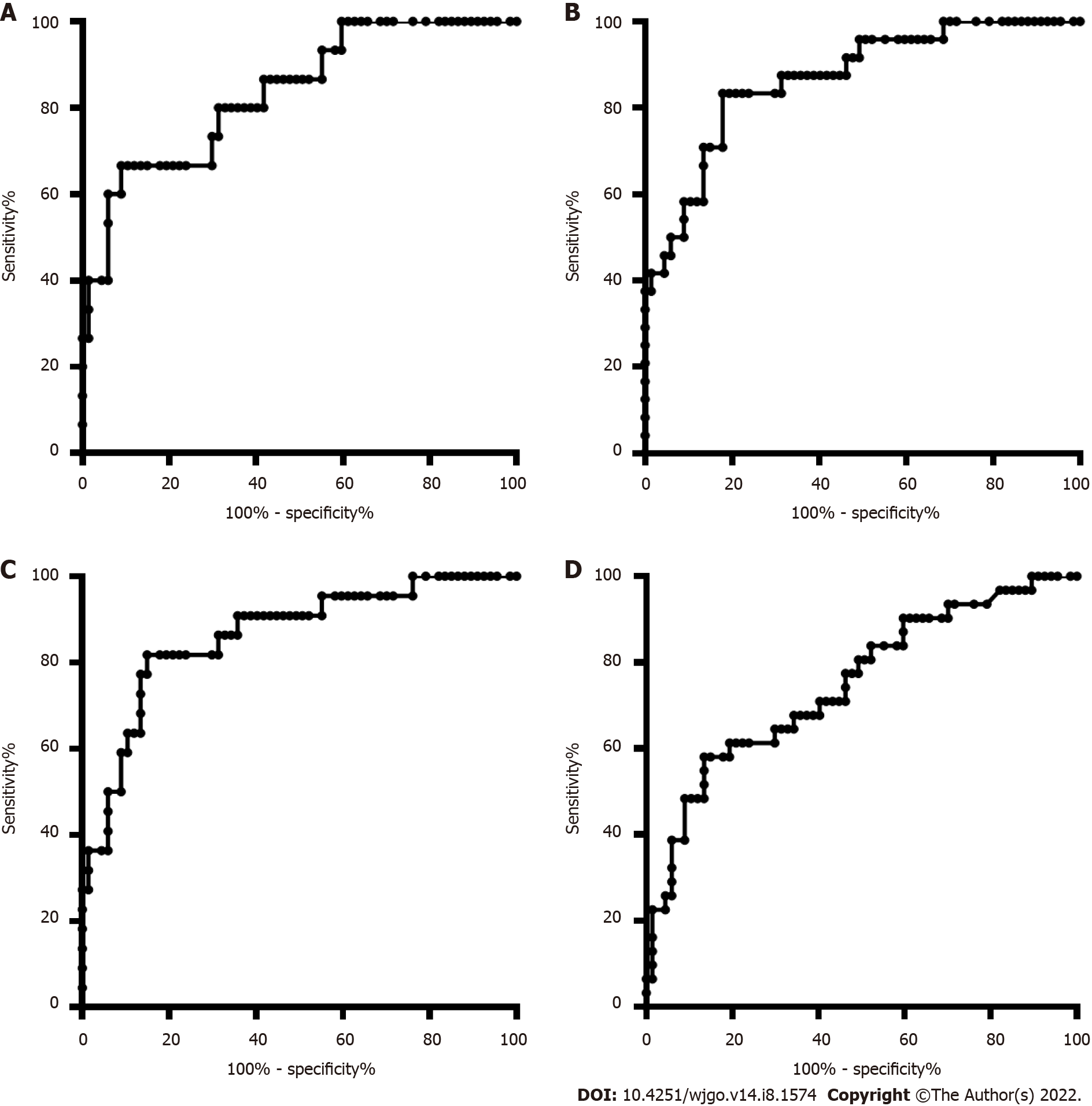
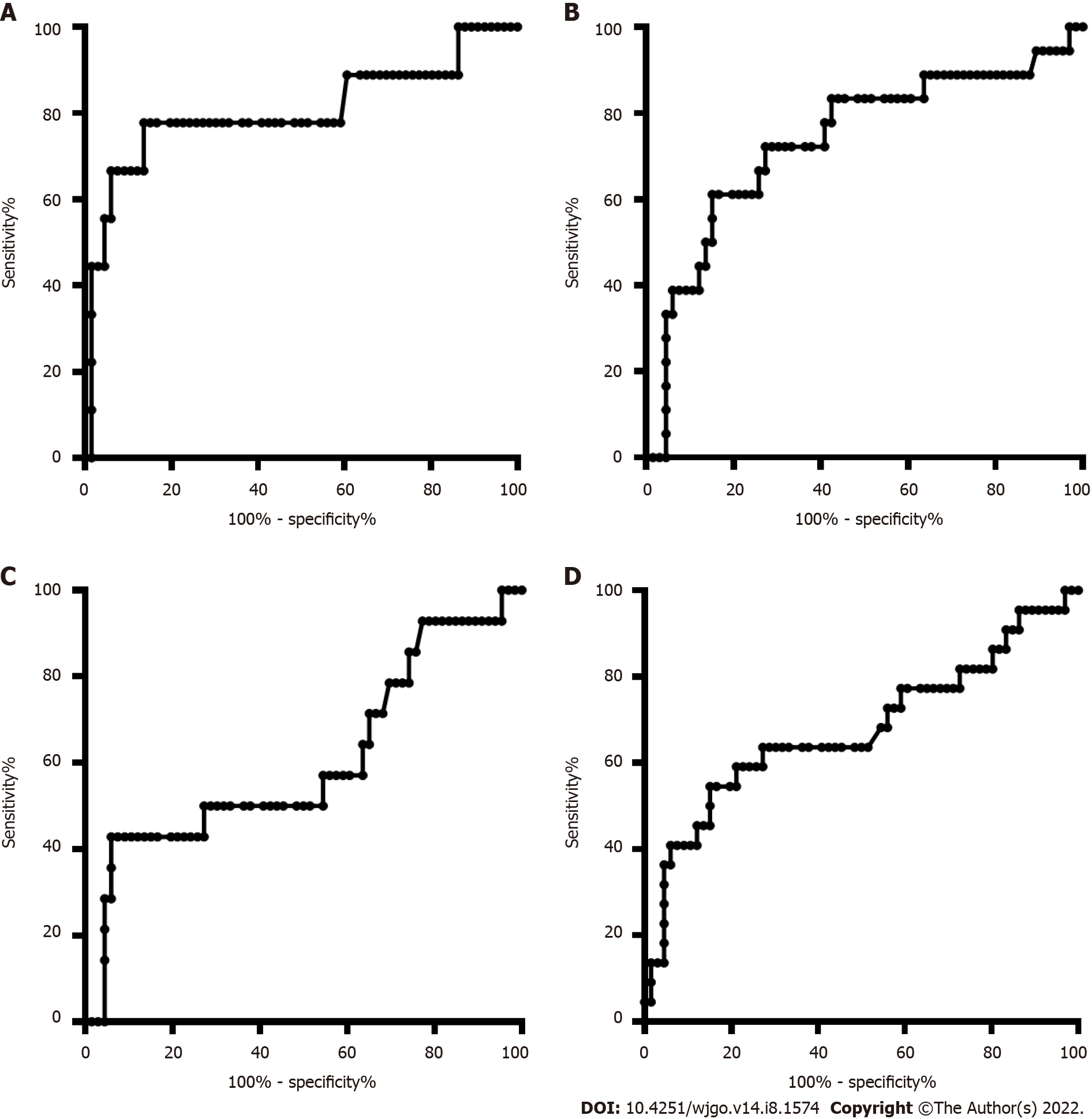
The age and sex in the CRC and CRP groups and the HCC and LC groups were also matched. The DNA methylation rate of RASSF1A in plasma in the CRC group was 2.87 ± 1.80, and that in the CRP group was 1.50 ± 0.64. DNA methylation of RASSF1A in plasma showed a significant difference between the CRC and CRP groups. The AUC of RASSF1A methylation for discriminating the CRC and CRP groups was 0.82 (0.76-0.88). The AUCs of T1, T2, T3 and T4 CRC and CRP were 0.83 (0.72-0.95), 0.87 (0.78-0.95), 0.86 (0.77-0.95), and 0.75 (0.64-0.85), respectively. The DNA methylation rate of RASSF1A in plasma in the HCC group was 4.45 ± 2.93, and that in the LC group was 2.46 ± 2.07. DNA methylation of RASSF1A in plasma for the HCC and LC groups showed a significant difference. The AUC of RASSF1A methylation for discriminating the HCC and LC groups was 0.70 (0.60-0.79). The AUCs of T1, T2, T3 and T4 HCC and LC were 0.80 (0.61, 1.00), 0.74 (0.59-0.88), 0.60 (0.42-0.79), and 0.68 (0.53-0.82), respectively.
RASSF1A methylation in plasma detected by digital PCR may be a potential biomarker for CRC and HCC.
Core Tip: Accurate quantitative polymerase chain reaction (qPCR) quantification relies on a standard curve and good amplification efficiency and is sensitive to factors affecting amplification efficiency. Digital PCR technology is an emerging PCR technology. In this study, we evaluated the diagnostic value of RASSF1A methylation in plasma by digital polymerase chain reaction.
- Citation: Li J, Li H, Run ZC, Wang ZL, Jiang T, An Y, Li Z. RASSF1A methylation as a biomarker for detection of colorectal cancer and hepatocellular carcinoma. World J Gastrointest Oncol 2022; 14(8): 1574-1584
- URL: https://www.wjgnet.com/1948-5204/full/v14/i8/1574.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i8.1574
As a very important epigenetic modification, DNA methylation is closely related to the occurrence and development of tumors[1]. Most of the DNA methylation studies of cancer currently focus on tumor tissue; however, due to its invasive characteristics, it is difficult to use for cancer screening and early diagnosis. Noninvasive biological samples (such as blood) have the advantages of being minimally invasive or noninvasive, having a simple operation and being suitable for multiple collections. Blood DNA mainly includes plasma or serum DNA and blood cell DNA. It is generally believed that DNA in plasma or serum mainly comes from tumor cell necrosis or apoptosis[2-5]. The concentration of free DNA in normal human plasma is ng/mL, and the concentration of DNA in the plasma of benign and malignant lesions can be increased by 5-15 times. Cell-free DNA (cfDNA) in plasma can provide a new method for tumor diagnosis and prognosis[6].
The abnormal methylation changes of different cancers are specific, and the cfDNA of different stages of cancer is also different[7]. By detecting the level and methylation of cfDNA, tumor diagnosis and staging can be achieved. Studies have validated the potential of methylated cfDNA as biomarkers in various tumors, and genes such as DCLK1 were found in the plasma of lung cancer patients[8]. Abnormal hypermethylation occurred in cfDNA; SOX17 promoter hypermethylation was found in cfDNA of patients with early breast cancer, primary breast cancer, and metastatic breast cancer[9]; SEPT9 gene promoter methylation in plasma cfDNA was found to have good sensitivity and specificity in the diagnosis of early colorectal cancer (CRC)[10-12]; quantitative methylation detection of the NEUROG1 gene in serum has also been proven to be an early screening method for CRC[13]; and there are also studies that simultaneously detect multiple genetic loci of cfDNA to establish a combined methylation diagnostic model. At present, liquid biopsy technology to detect cfDNA methylation is gradually becoming a new type of cancer screening and diagnosis.
Currently, real-time quantitative PCR (qPCR) is the main quantitative detection technology for the detection of methylation in nucleic acid samples. However, it is a relatively quantitative technique[14]. Accurate qPCR quantification relies on a standard curve and good amplification efficiency and is sensitive to factors affecting amplification efficiency (such as method design and PCR inhibitors). Digital PCR (dPCR) technology is an emerging PCR technology. Compared with qPCR, dPCR does not require a standard curve, can achieve absolute quantification, has higher sensitivity and specificity and is resistant to background sequences and reaction inhibitors. dPCR has obvious advantages in the detection of rare mutations and rare methylated alleles[15], its lower limit of detection and improved detection accuracy, and the absolute quantification of the nucleic acid to be detected[16].
In this study, using a dPCR detection method, we aimed to evaluate the diagnostic value of RASSF1A methylation in plasma for CRC and hepatocellular carcinoma (HCC).
After approval by the Ethics Committee of Chinese PLA General Hospital, the research subjects signed informed consent forms. CRC staging was performed according to the American Joint Committee on Cancer tumor node metastasis staging. All patients received no treatment when peripheral blood samples were taken, including surgical resection, radiotherapy, chemotherapy, and targeted therapy; all patients underwent colonoscopy and biopsy and were pathologically diagnosed with colorectal polyps (CRP), CRC, HCC, and liver cirrhosis (LC). All patients needed to undergo follow-up in the later period. If the tumor tissue was obtained by surgery for biopsy and the pathological result was inconsistent with the biopsy under endoscopy, the biopsy result of the tumor tissue would prevail. The healthy control samples were from the physical examination population in the same time period with healthy physical examination results and normal blood biochemical test results.
The samples in this study were peripheral blood collected on an empty stomach in the morning, and EDTA was an anticoagulant. The collected whole blood samples were directly aliquoted into 1.5 mL Eppendorf tubes in 200 μL. The whole blood was centrifuged at 1500 ×g for 10 min to obtain plasma samples. If hemolysis or lipid blood appeared, the samples were discarded. Finally, 1000 μL of plasma was dispensed into Eppendorf tubes for subsequent experiments. Extraction of plasma DNA Extract DNA from 1 mL of plasma samples was performed according to the QIAamp MinElute ccfDNA Mini Kit instructions, and finally 24 μL of ultra-purified water was added to elute the DNA.
The QubitTM double strand DNA (dsDNA) HS assay kit was removed from 4 °C and placed at room temperature for 30 min; 199 μL dsDNA buffer and 1 μL dsDNA reagent were added to specially matched QubitTM assay tubes and mixed by vortexing; 10 μL of the mixture in the two tubes corresponding to the standard were discarded with a pipette, and 1 μL of the mixture in the tube that aspirated the sample was discarded; 10 μL of dsDNA Standard #1 and dsDNA Standard #2 was pipetted into the corresponding tubes, 1 μL was pipetted from each sample to be tested and added to the corresponding tubes, and vortexing was used to mix them well; the Qubit 3.0 instrument was turned on, and the dsDNA high-sensitivity program was selected; the tubes containing standard 1 and standard 2 were inserted into the instrument in turn, the standard curve was drawn, and the samples were placed into the measurement concentration in turn.
Twenty microliters of DNA samples were transformed according to the instructions of the EZ DNA Methylation-Gold Kit, and finally 22 μL of M-Elution Buffer was added to the column matrix to elute the DNA. When the PCR tube was placed in the thermal cycler, the cover temperature of the PCR instrument was set to 105 °C, and the program was changed to (1) 98 °C for 10 min; (2) 64°C with 20 min as a node to set the temperature gradient: 90 min, 110 min, 130 min, and 150 min; and (3) storing at 4 °C.
The sample was then purified again according to the instructions of the Cycle-Pure Kit. After transformation, the DNA was purified again, and 7 μL of elution buffer was added for elution.
The operation steps were the same as those in 2.6.3, except that the dsDNA assay kit reagents were correspondingly replaced with the reagents in the ssDNA assay kit. ssDNA was selected as the assay type to measure.
The dPCR reader QX200 Droplet Reader was turned on and warmed up for 30 min, and the computer and QuantaSoft software were turned on; a 20 μL probe-based quantitative reaction system ddPCR Supermix for Probes, 10 μL, was prepared with methylated upstream primer (10 μM), 1.6 μL; methylation downstream primer (10 μM), 1.6 μL; and methylation probe (10 μM), 0.5 μL. The DNA volume was based on the concentration, and dd water was added according to the reaction system, totaling 20 μL. The RASSF1A methylation primer was synthesized according to previous research[17]. The above reaction system was shaken and mixed and centrifuged briefly to remove air bubbles. The droplet generating card was placed into the metal holder in the direction of the notch. Then, 20 μL of the sample reaction system was added to the 8 wells in the middle row of the droplet generating card, and 70 μL of the droplet generating oil was added to the 8 wells in the bottom row of the droplet generating card. The samples were added slowly to avoid generating air bubbles, as air bubbles in the system would seriously affect the generation of droplets. The disposable rubber pad was hooked to the small holes on both sides of the metal holder, and the droplet-generating card was added. The middle part of the metal holder was held and placed in the droplet generator stably, until droplets started to generate, and whether the droplet generation was completed was judged according to the status of the indicator light. The liquid in the top row of holes of the droplet generation card, generally 40 μL, was aspirated, transferred to the corresponding 96-well plate, and covered with tin foil to prevent the oil from volatilizing. When it was completely transferred, the side marked with the red line was placed on the tin foil film on the 96-well plate. After fixing, the samples were placed in a heat sealer to seal the film. The running program was as follows: 180 °C, 10 s; the sealed film was placed on the C1000 TouchTM Thermal Cycler, and the program was set (95 °C for 10 min, 1 cycle; 94 °C for 30 s and 56 °C for 1 min, 45 cycles; 98 °C for 10 min, 1 cycle; and holding at 12 °C). After amplification, the 96-well plate was placed into the corresponding holder, the button plate was pressed with both hands at the same time, and it was assembled and smoothly placed into the droplet reader. QuantaSoft software was opened, “Flush System” was selected to clean the system, the sample information in the reaction well was set, the program was run after completion, and the data were analyzed after reading.
The number of droplets, copy number, concentration, and copy number ratio of the two channels in each reaction well were obtained in QuantaSoft software for analysis. The area under the curve (AUC) was used to evaluate the diagnostic value, and specificity and sensitivity were listed as the evaluation indicators.
As shown in Table 1, 92 CRC patients, 67 CRP patients, 63 HCC patients, and 66 LC patients in the training group were enrolled. The age and sex in the CRC and CRP groups of the training and validation groups were matched, and the age and sex in the HCC and LC groups were also matched. The CRC at T1, T2, T3 and T4 were 15, 24, 22, and 31, and the HCC at T1, T2, T3, and T4 were 9, 18, 14, and 22.
| CRC | CRP | HC | LC | |
| n | 92 | 67 | 63 | 66 |
| Age (yr) | ||||
| mean | 57 | 55 | 55 | 53 |
| Range | 42-66 | 41-62 | 35-68 | 34-65 |
| Sex | ||||
| Male | 51 | 38 | 32 | 29 |
| Female | 41 | 29 | 23 | 24 |
| TNM stage | ||||
| T1 | 15 | 9 | ||
| T2 | 24 | 18 | ||
| T3 | 22 | 14 | ||
| T4 | 31 | 22 | ||
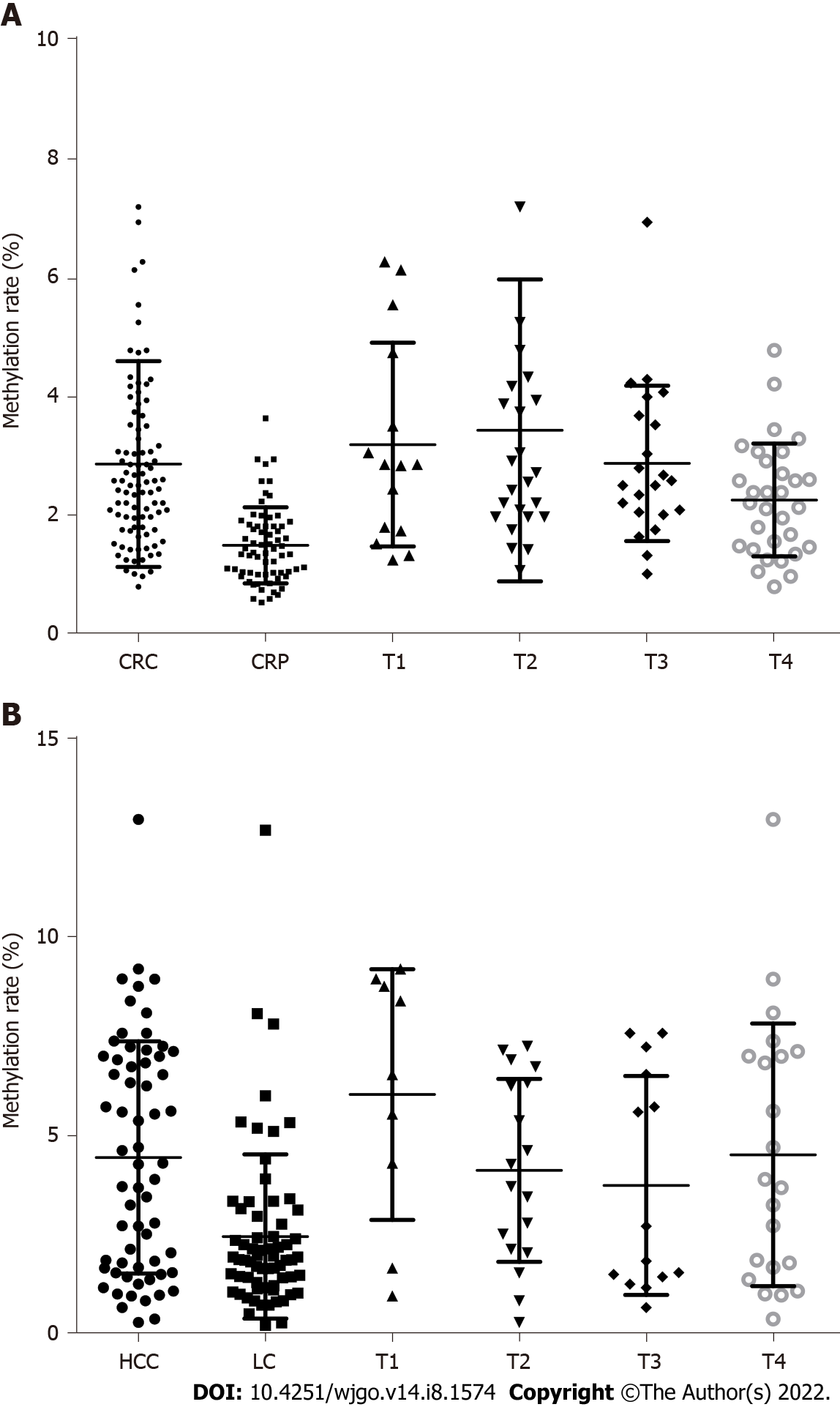
The DNA methylation rate of RASSF1A in plasma in the CRC group was 2.87 ± 1.80, and that in the CRP group was 1.50 ± 0.64 (Figure 1A). DNA methylation of RASSF1A in plasma showed a significant difference between the CRC and CRP groups. The methylation rates of RASSF1A in plasma at T1, T2, T3 and T4 in CRC were 3.20 ± 1.71, 3.45 ± 2.54, 2.88 ± 1.31, and 2.27 ± 0.95, respectively. When CRC at T1, T2, T3 and T4 were compared with the CRP group, all four stages showed significant differences. The AUC of RASSF1A methylation for discriminating the CRC and CRP groups was 0.82 (0.76-0.88) (Figure 2A). The AUCs of T1, T2, T3 and T4 CRC and CRP were 0.83 (0.72-0.95), 0.87 (0.78-0.95), 0.86 (0.77-0.95), and 0.75 (0.64-0.85), respectively (Figure 3A-D).
The DNA methylation rate of RASSF1A in plasma was 4.45 ± 2.93 in the HCC group and 2.46 ± 2.07 in the LC group (Figure 1B). DNA methylation of RASSF1A in plasma for the HCC and LC groups showed a significant difference. The methylation rates of RASSF1A in plasma at T1, T2, T3 and T4 in HCCs were 6.04 ± 3.16, 4.13 ± 2.31, 3.75 ± 2.76, and 4.52 ± 3.31, respectively. When T1, T2, T3 and T4 in the HCC group were compared with those in the LC group, T1, T2 and T4 showed significant differences. T3 showed no significant difference (P = 0.061). The AUC of RASSF1A methylation for discriminating the HCC and LC groups was 0.70 (0.60-0.79) (Figure 2B). The AUCs of T1, T2, T3 and T4 HCC and LC were 0.80 (0.61-1.00), 0.74 (0.59-0.88), 0.60 (0.42-0.79), and 0.68 (0.53-0.82), respectively (Figure 4A-D).
By detecting the methylation level of the PCDH10 promoter region in the tissue and plasma of colorectal cancer patients, the PCDH10 methylation level in the tissue of early colorectal cancer patients was found to be highly correlated with the DNA methylation level in the plasma, suggesting that the PCDH10 promoter in the plasma is highly correlated[18]. The level of regional methylation can be used as a biomarker for the early diagnosis of colorectal cancer. With the advancement of technology, other specific gene methylation levels in plasma, such as RASSF2, sFPR1, SDC2 and other gene promoter methylation, have been confirmed for use as biomarkers for the early diagnosis of colorectal cancer[19-21]. In addition, the determination of Septin 9 methylation in plasma is considered a sensitive and specific biomarker for the early diagnosis of colorectal cancer; however, when it is used for screening in general risk populations of colorectal cancer, it can only be detected. It produces approximately 50% of asymptomatic colorectal cancer patients, with a specificity comparable with the fecal occult blood test. Studies have confirmed that changes in methylation sites in plasma can be used as biomarkers for the early diagnosis of colorectal cancer. However, the methylation sites in plasma for the early diagnosis of colorectal cancer are poorly studied. RASSF1A was the most widely investigated gene in serum or plasma, and it was also demonstrated to be more frequently methylated in cancer patients. Global hypomethylation of RASSF1A was related to increased breast cancer risk[22]. RASSF1A methylation is an attractive biomarker for early cancer detection, which, for most cancers, results in improved clinical outcome. RASSF1A methylation may be used as a diagnostic and prognostic marker in cancer management[23]. RASSF1A hypermethylation is a promising biomarker for the diagnosis of HCC in tissue and blood and is an emerging biomarker for HCC[24-26]. In addition, RASSF1A hypermethylation is an early and potential prognostic biomarker in CRC[21,27,28].
There are many detection methods for DNA methylation sites, mainly including the following methods: (1) Methylation-specific PCR: The basic principle is that after bisulfite treatment, two pairs of primers are designed: one pair amplifies the bisulfite-treated DNA template, and the other pair amplifies the unmethylated fragment. Then, according to whether it can be amplified, it is judged whether methylation has occurred. The disadvantage is that the sequence of the gene to be tested needs to be known in advance, and primers with relatively high specificity are designed[29]; (2) The bisulfite sequencing method: The basic principle is that after bisulfite treatment, PCR amplification is performed, the amplified product is sequenced, and methylation is determined by comparison with the untreated sequence. The disadvantage is that it needs to undergo much cloning, and the process is cumbersome and expensive[30]; (3) Restriction endonuclease analysis method: The basic principle is that after bisulfite treatment and PCR amplification, the amplification product is purified and then digested with restriction enzymes. Then, according to whether it can be cut, it is judged whether methylation has occurred. Its disadvantage is that it can only obtain the methylation status of special enzyme cleavage sites[31]; (4) Methylation-sensitive high-resolution melting curve analysis: The basic principle is that after bisulfite treatment, the difference between methylated sites and unmethylated DNA can be found by melting curve analysis due to the presence of more GCs. The disadvantage is that it is greatly affected by primer design and cannot achieve quantitative detection[32]; (5) Pyrosequencing: The basic principle is that after bisulfite treatment, by accurately quantifying the methylation frequency on a single continuous site, the methylation frequency can be quickly detected, and the methylation sites in the sample can be qualitatively and quantitatively detected. The disadvantage is that there are many steps, so it is difficult to use as a conventional methylation detection method and more often used as a verification method of methylation sites[33]; (6) Sequenom MassArray platform: The basic principle is that after bisulfite treatment, primers are designed for PCR amplification, and the product is subjected to a single-base extension reaction after outpatient substance abuse program treatment. Flight mass spectrometry can detect the molecular weight difference between methylated and unmethylated sites to be detected. The disadvantage is that the experimental operation requirements are high, its detection sensitivity is low, and it is difficult to achieve quantitative detection of methylation sites[34]; and (7) Fluorescence quantitative method: The basic principle is to use TaqMan probes and PCR primers to distinguish methylated and unmethylated DNA after bisulfite treatment. As a highly sensitive relative quantitative detection method for DNA methylation, it is widely used. The disadvantage is that it is difficult to achieve absolute quantitative detection of methylated sites[35]. In addition to the fluorescence quantitative method, the above methylation detection methods have difficulty achieving high-sensitivity detection for trace DNA, the detection result of the fluorescence quantitative method is relatively quantitative, and it is difficult to achieve high-precision and absolute quantitative detection of plasma DNA methylation.
dPCR technology evenly distributes the PCR system into tens of thousands of reaction units. Each reaction unit does not contain or only contains one nucleic acid sequence to be tested. After the number of nucleic acids to be tested conformed to the Poisson distribution, PCR amplification was independently performed in each reaction unit. Finally, the fluorescence signal of each reaction unit was detected, and the copy number of the nucleic acid sequence to be tested was calculated according to the Poisson distribution and the proportion of reaction units with positive fluorescence signals to all reaction units. Compared with other methylation detection methods, ddPCR has the following advantages: (1) High sensitivity: ddPCR turns PCRs into tens of thousands of PCRs that independently detect nucleic acids. Compared with traditional detection methods, the detection sensitivity is greatly improved; (2) High accuracy: ddPCR can accurately detect small changes in the nucleic acid to be detected by calculating the number and proportion of positive reaction units in tens of thousands or even tens of millions of reaction units; (3) High tolerance; and (4) absolute quantification: ddPCR technology can achieve the absolute quantitative detection of the nucleic acid to be detected without relying on the Ct value and the standard curve.
There are some limitations in our study. First, the digital PCR methylation of RASSF1A was not compared with the conventional real-time PCR method. Second, the healthy control group was not detected in our study, and the methylation rate of RASSF1A was not evaluated. Third, the sample size was small, and the results may be affected.
In conclusion, we demonstrate that RASSF1A methylation in plasma detected by digital PCR may be a potential biomarker for CRC and HCC.
DNA methylation in serum or plasma was demonstrated to be a potential biomarker for cancer detection and prognosis.
More sensitive and accurate methods for detecting methylation in plasma are urgently needed in clinical practice.
In this study, we aimed to evaluate RASSF1A methylation in plasma by digital polymerase chain reaction (PCR) for colorectal cancer (CRC) and hepatocellular carcinoma (HCC).
A total of 92 CRC patients, 67 colorectal polyp (CRP) patients, 63 HCC patients, and 66 liver cirrhosis (LC) patients were enrolled. The plasma DNA was detected by digital PCR. The diagnostic value was evaluated by the area under the curve (AUC).
The DNA methylation rate of RASSF1A in plasma in the CRC group was 2.87 ± 1.80, and that in the CRP group was 1.50 ± 0.64. The AUC of RASSF1A methylation for discriminating the CRC and CRP groups was 0.82 (0.76-0.88). The AUCs of T1, T2, T3 and T4 CRC and CRP were 0.83 (0.72-0.95), 0.87 (0.78-0.95), 0.86 (0.77-0.95), and 0.75 (0.64-0.85), respectively. The DNA methylation rate of RASSF1A in plasma in the HCC group was 4.45 ± 2.93, and that in the LC group was 2.46 ± 2.07. The AUC of RASSF1A methylation for discriminating the HCC and LC groups was 0.70 (0.60-0.79). The AUCs of T1, T2, T3 and T4 HCC and LC were 0.80 (0.61-1.00), 0.74 (0.59-0.88), 0.60 (0.42-0.79), and 0.68 (0.53-0.82), respectively.
We demonstrate that RASSF1A methylation in plasma detected by digital PCR may be a potential biomarker for CRC and HCC.
Different methylation detection methods should be compared, and more samples need to be detected.
| 1. | Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 863] [Cited by in RCA: 1167] [Article Influence: 72.9] [Reference Citation Analysis (0)] |
| 2. | Ignatiadis M, Sledge GW, Jeffrey SS. Liquid biopsy enters the clinic - implementation issues and future challenges. Nat Rev Clin Oncol. 2021;18:297-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 242] [Cited by in RCA: 820] [Article Influence: 164.0] [Reference Citation Analysis (0)] |
| 3. | Alix-Panabières C, Pantel K. Liquid Biopsy: From Discovery to Clinical Application. Cancer Discov. 2021;11:858-873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 663] [Article Influence: 132.6] [Reference Citation Analysis (0)] |
| 4. | Mader S, Pantel K. Liquid Biopsy: Current Status and Future Perspectives. Oncol Res Treat. 2017;40:404-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 169] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 5. | Chen D, Xu T, Wang S, Chang H, Yu T, Zhu Y, Chen J. Liquid Biopsy Applications in the Clinic. Mol Diagn Ther. 2020;24:125-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 6. | Li W, Li Q, Kang S, Same M, Zhou Y, Sun C, Liu CC, Matsuoka L, Sher L, Wong WH, Alber F, Zhou XJ. CancerDetector: ultrasensitive and non-invasive cancer detection at the resolution of individual reads using cell-free DNA methylation sequencing data. Nucleic Acids Res. 2018;46:e89. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 136] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 7. | Jung G, Hernández-Illán E, Moreira L, Balaguer F, Goel A. Epigenetics of colorectal cancer: biomarker and therapeutic potential. Nat Rev Gastroenterol Hepatol. 2020;17:111-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 542] [Cited by in RCA: 569] [Article Influence: 94.8] [Reference Citation Analysis (5)] |
| 8. | Duan H, Liu Y, Gao Z, Huang W. Recent advances in drug delivery systems for targeting cancer stem cells. Acta Pharm Sin B. 2021;11:55-70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 148] [Cited by in RCA: 187] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 9. | Fu DY, Wang ZM, Li-Chen, Wang BL, Shen ZZ, Huang W, Shao ZM. Sox17, the canonical Wnt antagonist, is epigenetically inactivated by promoter methylation in human breast cancer. Breast Cancer Res Treat. 2010;119:601-612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 73] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 10. | Wang Y, Chen PM, Liu RB. Advance in plasma SEPT9 gene methylation assay for colorectal cancer early detection. World J Gastrointest Oncol. 2018;10:15-22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 38] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 11. | Tepus M, Yau TO. Non-Invasive Colorectal Cancer Screening: An Overview. Gastrointest Tumors. 2020;7:62-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 72] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 12. | Sun J, Fei F, Zhang M, Li Y, Zhang X, Zhu S, Zhang S. The role of mSEPT9 in screening, diagnosis, and recurrence monitoring of colorectal cancer. BMC Cancer. 2019;19:450. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 103] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 13. | Otero-Estévez O, Gallardo-Gomez M, Cadena MP, Rodríguez-Berrocal FJ, Cubiella J, Ramirez VH, García-Nimo L, Chiara L. Value of Serum NEUROG1 Methylation for the Detection of Advanced Adenomas and Colorectal Cancer. Diagnostics (Basel). 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 14. | Olmedillas-López S, García-Arranz M, García-Olmo D. Current and Emerging Applications of Droplet Digital PCR in Oncology. Mol Diagn Ther. 2017;21:493-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 156] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 15. | Yu M, Heinzerling TJ, Grady WM. DNA Methylation Analysis Using Droplet Digital PCR. Methods Mol Biol. 2018;1768:363-383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 16. | Nell RJ, van Steenderen D, Menger NV, Weitering TJ, Versluis M, van der Velden PA. Quantification of DNA methylation independent of sodium bisulfite conversion using methylation-sensitive restriction enzymes and digital PCR. Hum Mutat. 2020;41:2205-2216. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 17. | Abe M, Kagara N, Miyake T, Tanei T, Naoi Y, Shimoda M, Shimazu K, Kim SJ, Noguchi S. Highly sensitive detection of sentinel lymph node metastasis of breast cancer by digital PCR for RASSF1A methylation. Oncol Rep. 2019;42:2382-2389. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 18. | Danese E, Minicozzi AM, Benati M, Montagnana M, Paviati E, Salvagno GL, Gusella M, Pasini F, Guidi GC, Lippi G. Epigenetic alteration: new insights moving from tissue to plasma - the example of PCDH10 promoter methylation in colorectal cancer. Br J Cancer. 2013;109:807-813. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 19. | Lyu Z, Chen H, Jiang L, Zheng H, Hu J. [Detection of RASSF2 and sFRP1 promoter region methylation in sporadic colorectal cancer patients]. Zhonghua Wei Chang Wai Ke Za Zhi. 2014;17:41-44. [PubMed] |
| 20. | Barták BK, Kalmár A, Péterfia B, Patai ÁV, Galamb O, Valcz G, Spisák S, Wichmann B, Nagy ZB, Tóth K, Tulassay Z, Igaz P, Molnár B. Colorectal adenoma and cancer detection based on altered methylation pattern of SFRP1, SFRP2, SDC2, and PRIMA1 in plasma samples. Epigenetics. 2017;12:751-763. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 100] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 21. | Zhang L, Dong L, Lu C, Huang W, Yang C, Wang Q, Lei R, Sun R, Wan K, Li T, Sun F, Gan T, Lin J, Yin L. Methylation of SDC2/TFPI2 and Its Diagnostic Value in Colorectal Tumorous Lesions. Front Mol Biosci. 2021;8:706754. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 22. | Tang Q, Cheng J, Cao X, Surowy H, Burwinkel B. Blood-based DNA methylation as biomarker for breast cancer: a systematic review. Clin Epigenetics. 2016;8:115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 126] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 23. | Hesson LB, Cooper WN, Latif F. The role of RASSF1A methylation in cancer. Dis Markers. 2007;23:73-87. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 158] [Cited by in RCA: 156] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 24. | Xu G, Zhou X, Xing J, Xiao Y, Jin B, Sun L, Yang H, Du S, Xu H, Mao Y. Identification of RASSF1A promoter hypermethylation as a biomarker for hepatocellular carcinoma. Cancer Cell Int. 2020;20:547. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 25. | Pasha HF, Mohamed RH, Radwan MI. RASSF1A and SOCS1 genes methylation status as a noninvasive marker for hepatocellular carcinoma. Cancer Biomark. 2019;24:241-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 26. | Zhang C, Li J, Huang T, Duan S, Dai D, Jiang D, Sui X, Li D, Chen Y, Ding F, Huang C, Chen G, Wang K. Meta-analysis of DNA methylation biomarkers in hepatocellular carcinoma. Oncotarget. 2016;7:81255-81267. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 86] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 27. | Hu F, Chen L, Bi MY, Zheng L, He JX, Huang YZ, Zhang Y, Zhang XL, Guo Q, Luo Y, Tang WR, Sheng MM. Potential of RASSF1A promoter methylation as a biomarker for colorectal cancer: Meta-analysis and TCGA analysis. Pathol Res Pract. 2020;216:153009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 28. | Wang HL, Zhang Y, Liu P, Zhou PY. Aberrant promoter methylation of RASSF1A gene may be correlated with colorectal carcinogenesis: a meta-analysis. Mol Biol Rep. 2014;41:3991-3999. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 29. | Yoshioka M, Matsutani T, Hara A, Hirono S, Hiwasa T, Takiguchi M, Iwadate Y. Real-time methylation-specific PCR for the evaluation of methylation status of MGMT gene in glioblastoma. Oncotarget. 2018;9:27728-27735. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 30. | Grunau C, Clark SJ, Rosenthal A. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res. 2001;29:E65-E65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 539] [Cited by in RCA: 561] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 31. | Olszewska MJ, Gernand D, Sakowicz T. Methylation-sensitive restriction endonuclease digestion patterns revealed in Vicia faba L. chromosomes by in situ nick-translation. Folia Histochem Cytobiol. 1999;37:267-274. [PubMed] |
| 32. | White HE, Hall VJ, Cross NC. Methylation-sensitive high-resolution melting-curve analysis of the SNRPN gene as a diagnostic screen for Prader-Willi and Angelman syndromes. Clin Chem. 2007;53:1960-1962. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 62] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 33. | Roesch LF, Fulthorpe RR, Riva A, Casella G, Hadwin AK, Kent AD, Daroub SH, Camargo FA, Farmerie WG, Triplett EW. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007;1:283-290. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1433] [Cited by in RCA: 1005] [Article Influence: 52.9] [Reference Citation Analysis (0)] |
| 34. | Song F, Mahmood S, Ghosh S, Liang P, Smiraglia DJ, Nagase H, Held WA. Tissue specific differentially methylated regions (TDMR): Changes in DNA methylation during development. Genomics. 2009;93:130-139. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 98] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 35. | Rosa F, Osorio JS. Quantitative determination of histone methylation via fluorescence resonance energy transfer (FRET) technology in immortalized bovine mammary alveolar epithelial cells supplemented with methionine. PLoS One. 2020;15:e0244135. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Gupta S, India; Heij LR, Germany; Nardone G, Italy S-Editor: Wang JL L-Editor: A P-Editor: Wang JL