Published online Aug 15, 2022. doi: 10.4251/wjgo.v14.i8.1528
Peer-review started: March 30, 2022
First decision: June 2, 2022
Revised: June 14, 2022
Accepted: July 19, 2022
Article in press: July 19, 2022
Published online: August 15, 2022
Processing time: 133 Days and 2.9 Hours
Approximately half of all new cases of gastric cancer (GC) and related deaths occur in China. More than 80% of patients with GC are diagnosed at an advanced stage, which results in poor prognosis. Although HER2-directed therapy and immune checkpoint inhibitors have been somewhat successful, new drugs are still needed for the treatment of GC. Notably, several gene fusion-targeted drugs have been approved by the United States Food and Drug Administration for solid tumors, including GC, such as larotrectinib for NTRK fusion-positive cancers and zenocutuzumab for NRG1 fusion-positive cancers. However, gene fusions involving targetable genes have not been well characterized in Chinese patients with GC.
To identify the profile of fusions involving targetable genes in Chinese patients with GC using clinical specimens and determine the distribution of patients with gene fusion variants among the molecular subtypes of GC.
We retrospectively analyzed gene fusion events in tumor tissue samples from 954 Chinese patients with GC. Clinicopathological characteristics were obtained from their medical records. Genetic alterations, such as single nucleotide variants, indels, amplifications, and gene fusions, were identified using a targeted sequencing panel containing 825 genes. Fusions were validated by fluorescence in situ hybridization (FISH) using break-apart probes. The microsatellite instability (MSI) status was evaluated using MSIsensor from the targeted sequencing panel data. Tumor mutational burden (TMB) was calculated using the total number of nonsynonymous mutations divided by the total genomic targeted region. Chi-square analysis was used to determine the enrichment of gene fusions associated with the molecular subtypes of GC.
We found that 1.68% (16/954) of patients harbored 20 fusion events involving targetable genes. RARA fusions (n = 5) were the most common, followed by FGFR2, BRAF, MET, FGFR3, RET, ALK, EGFR, NTRK2, and NRG1 fusions. Two of the RARA fusions, EML4-ALK (E6:E20) and EGFR-SEPTIN14 (E7:E10), have been identified in other tumors but not in GC. Surprisingly, 18 gene fusion events were previously not reported in any cancer types. Twelve of the eighteen novel gene fusions included complete exons encoding functional domains of targetable genes, such as the tyrosine kinase domain of receptor tyrosine kinases and the DNA- and ligand-binding domains of RARA. Consistent with the results of detection using the targeted sequencing fusion panel, the results of FISH (fluorescence in situ hybridization) confirmed the rearrangement of FGFR2 and BRAF in tumors from patients 04 and 09, respectively. Genetic analysis indicated that the fusion genes were significantly enriched in patients with ERBB2 amplification (P = 0.02); however, there were no significant differences between fusion-positive and fusion-negative patients in age, sex, MSI status, and TMB.
We characterized the landscape of fusions involving targetable genes in a Chinese GC cohort and found that 1.68% of patients with GC harbor potential targetable gene fusions, which were enriched in patients with ERBB2 amplification. Gene fusion detection may provide a potential treatment strategy for patients with GC with disease progression following standard therapy.
Core Tip: The proportion of patients with gene fusions in Chinese patients with gastric cancer (GC) has not yet been characterized. In our analysis, we found that 1.68% of such patients harbor fusions involving targetable genes. Moreover, these fusion genes were enriched in patients with ERBB2 amplification. Our study indicates that gene fusion detection may provide a novel approach for GC therapy.
- Citation: Liu ZH, Zhu BW, Shi M, Qu YR, He XJ, Yuan HL, Ma J, Li W, Zhao DD, Liu ZC, Wang BM, Wang CY, Tao HQ, Ma TH. Profiling of gene fusion involving targetable genes in Chinese gastric cancer. World J Gastrointest Oncol 2022; 14(8): 1528-1539
- URL: https://www.wjgnet.com/1948-5204/full/v14/i8/1528.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i8.1528
Gastric cancer (GC) is the fifth most frequent cancer and the third leading cause of cancer deaths worldwide, with more than one million new cases and approximately 769000 deaths in 2020[1]. The overall survival rate of patients with early stage disease is around 90% after surgical resection[2]; however, more than 80% of patients with GC are diagnosed at an advanced stage in China, which limits the effectiveness of the treatment[3]. Although chemotherapy has improved the survival of advanced-stage patients with GC, the objective response rate remains less than 40%, and the median overall survival is less than 12 mo[4]. Nevertheless, new targeted therapies are capable of improving the objective response rate and overall survival of patients with GC expressing certain targets[5].
Approximately 13%-22% of GCs exhibit HER2 overexpression or amplification[6-8]. The College of American Pathologists, the American Society for Clinical Pathology, and the American Society of Clinical Oncology recommend that all patients with advanced gastric adenocarcinoma should be tested for HER2 overexpression[9]. Trastuzumab was approved by the United States Food and Drug Administration (FDA) in 2010 as first-line treatment in combination with chemotherapy for patients with HER2-positive GC. Microsatellite instability-high (MSI-H) tumors are considered a molecular subtype of gastric adenocarcinoma by The Cancer Genome Atlas (TCGA)[10]. The incidence of MSI-H GC is 10%-20%[11]. The NCCN guidelines recommend MSI testing as a standard test for all patients with GC. Regarding targeted therapy, the FDA has approved pembrolizumab (PD1 monoclonal antibody) for the treatment of all unresectable or metastatic solid tumors with MSI-H/dMMR (deficient DNA mismatch repair), including GC. Although drug treatments have shown success to some extent, the development of more targeted drugs is required.
With rapid advancements in the field of oncogenomics, gene fusions in cancer have received increasing attention. The FDA has approved larotrectinib (Vitrakvi) and entrectinib (Rozlytrek) for the first- or subsequent-line treatment of solid tumors with NTRK fusions, including GC[12,13]. In 2021, the FDA accelerated the approval of the NRG1 inhibitor, zenocutuzumab (MCLA-128), in patients with pan-cancer harboring an NRG1 fusion. Apart from these fusion genes with approved drugs in pan-cancer, ALK fusions, such as EML4-ALK, TFG-ALK, and STRN-ALK, have been identified in the majority of tumors, including lung adenocarcinoma and colorectal cancer[14-16]. For lung cancer and mesenchymal tumors, patients harboring an ALK fusion are highly responsive to crizotinib and ceritinib[17,18]. Recently, a RAB10-ALK fusion was identified in a patient with GC[19], which indicates the possibility of future applications of ALK-TKIs (tyrosine kinase inhibitors) in these patients. Recent advances in next-generation sequencing (NGS) have contributed to a surge in the discovery of fusion genes, including BRAF; EGFR; FGFR1, 2, and 3; RET; and ROS1[20]. Gene fusion detection can guide the development of targeted therapeutic strategies for patients with GC with disease progression after standard therapy. Notably, there is a lack of comprehensive data characterizing gene fusions involving targetable genes in GC, particularly in the Chinese population.
This multicenter retrospective study included 1341 patients with GC admitted to Fujian Provincial Hospital (Fuzhou, China) and Zhejiang Provincial People’s Hospital (Hangzhou, China) between October 2015 and December 2021. The clinicopathological characteristics of the patients were retrieved from their medical records. Additionally, MSI status and tumor mutational burden (TMB) scores were extracted for statistical analysis. This study was approved by the Ethics Committee of the Fujian Provincial Hospital.
Mutational profiling of the Onco PanScan panel was performed by Genetron Health (Beijing) Co., Ltd. The coding regions of 825 cancer-related genes were analyzed. Genomic DNA was isolated from formalin-fixed paraffin-embedded (FFPE) tissue specimens with a minimum of 20% viable tumor nuclei. For sequencing, paired tumor and white blood cell DNA libraries were prepared using KAPA HyperPrep Kits (Roche, Germany). Libraries were quantified using Qubit fluorometer (Thermo Fisher Scientific, Waltham, MA, United States), and their quality was evaluated using Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, United States). High-throughput sequencing was performed on Novaseq6000 platform (Illumina, United States). Paired-end reads from Illumina sequencing were processed using script bcl2fastq (v. 2.17.1.14) and aligned against the human genome reference build, GRCh37, using Burrows-Wheeler Aligner (BWA, version 0.7.13). Duplicate removal, local realignment, and base quality recalibration were performed using PICARD (http://broadinstitute.github.io/picard/) and the Genome Analysis Toolkit. Variant calling was performed using an in-house developed pipeline. Variants identified as germline variants were excluded, while single nucleotide variants (SNVs) and indels with allelic fractions of more than 5% and supported by more than 4 unique reads, amplification with a fold-change greater than 2.5 in more than 25% of regions covered, and gene fusions supported by more than 3 unique reads were included.
TMB was calculated using the total number of nonsynonymous mutations divided by the total genomic target region (2.13 Mb). MSI status was determined using MSIsensor from paired tumor-normal targeted sequence data, and 309 MSI sites were included in the panel of 825 cancer-related genes. An MSIsensor score below 10 defines microsatellite stability (MSS) status, while that above 50 defines MSI status. The prevalence of gene fusions involving a targetable gene and driver mutations was compared with the OrigiMed2020 and TCGA cohorts[21]. Clinicopathological and genomic data were retrieved from the cBioPortal (https://www.cbioportal.org).
FFPE tissue sections (5 μm) were prepared on positively charged slides. After deparaffinizing and rehydrating, the slides were incubated with prewarmed 8% sodium thiocyanate in dH2O at 80 °C and incubated for 30 min. FGFR2 (10q26) or BRAF (7q34) break-apart probes were placed on the slide, covered with a glass coverslip, and sealed with rubber cement. Hybridization was performed overnight at 37 °C. The slides were washed twice in 50% formamide at 47 °C for 2 min and then twice in 2X standard saline citrate at room temperature for 2 min. Nuclei were stained with DAPI as a counterstain. The slides were scanned using a 90i Nikon fluorescent microscope. For each probe, 200 nuclei were evaluated. The 5′ (red) and 3′ (green) signals separated by ≥ 2 signal diameters were considered split as positive.
All statistical analyses were performed using SPSS 24.0 software (IBM, Chicago, IL, United States). χ2 or Fisher’s exact test was used to analyze the association between fusion alterations and driver mutations. A P value of < 0.05 was considered statistically significant.
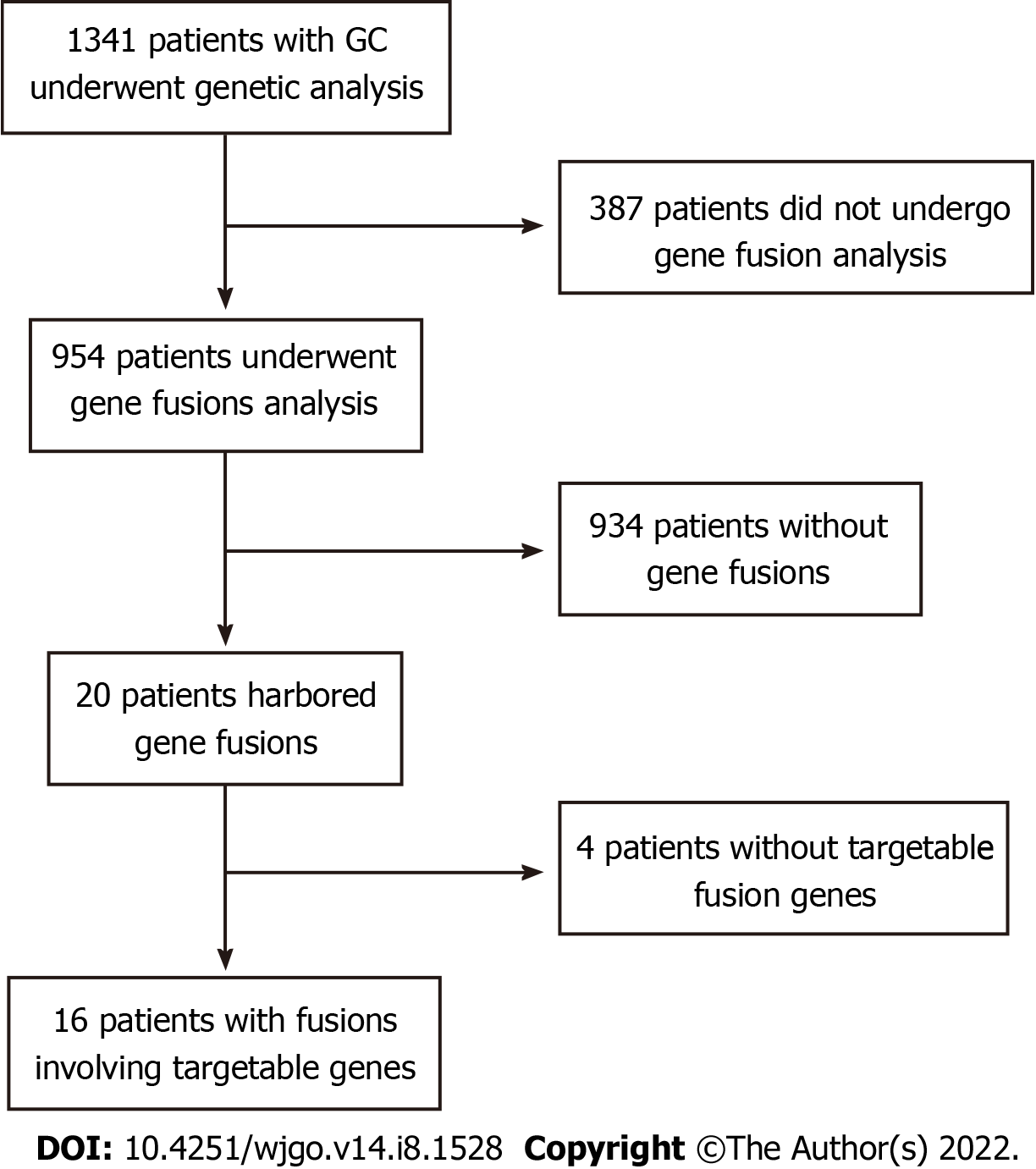
We retrospectively analyzed 1341 Chinese patients with GC who underwent genetic analysis from multiple centers in China. Of these, 387 patients were excluded because gene fusion detection was not performed with the Onco PanScan panel using tumor tissue samples (Figure 1). Gene fusion events were detected in 20 patients; however, 4 patients without any gene fusions involving targetable genes were excluded. Finally, 16 patients with 20 fusion events involving targetable genes were included for further analysis. The clinical characteristics of 954 patients with GC are shown in Table 1. Of these patients, 310 (32.56%) were women and 644 (67.44%) were men, with a median age of 57 and 62, respectively, at diagnosis. There was no significant difference between targetable gene fusion-positive and -negative patients in age (P = 0.293), sex (P = 0.463), MSI status (P = 0.551), or TMB (P = 0.217) (Table 1).
| Variables | Total, n | Fusion involving targetable genes | P value | |
| Positive, n (%) | Negative, n (%) | |||
| Sex | 0.293 | |||
| Female | 310 | 3 (0.97) | 307 (99.03) | |
| Male | 644 | 13 (2.02) | 631 (97.98) | |
| Age, yr | 0.463 | |||
| ≤ 60 | 451 | 6 (1.56) | 445 (98.44) | |
| > 60 | 503 | 10 (1.98) | 493 (98.01) | |
| MSI status | 0.551 | |||
| MSI-H | 46 | 1 (2.17) | 45 (97.93) | |
| MSS | 908 | 15 (1.65) | 893 (98.35) | |
| TMB | 0.217 | |||
| Median TMB score | 2.92 | 5.63 | 2.83 | |
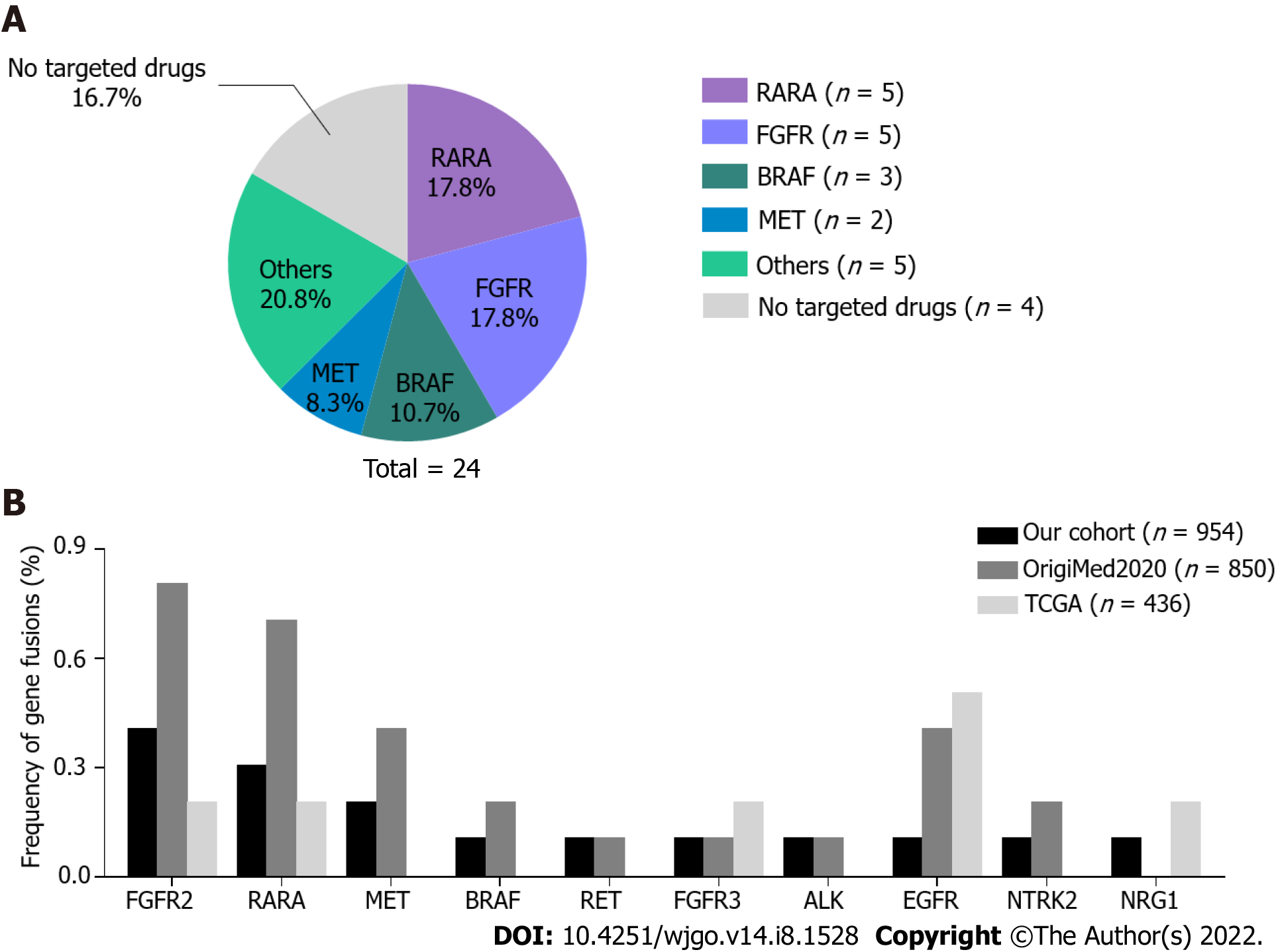
To gain insight into fusion events in GC, we evaluated 954 patients with GC undergoing gene fusion analysis. In total, 20 patients harbored 24 gene fusions, 2 patients had double fusions (patient 01 and 02), and 1 patient (09) harbored triple fusions. RARA fusions (5/24, 17.8%) and FGFR family gene fusions (5/24, 17.8%) occurred most frequently in the cohort, followed by BRAF (3/24, 10.7%) and MET (2/24, 8.3%) (Figure 2A). ALK, RET, NTRK2, NRG1, and EGFR fusions were identified in one patient each. Remarkably, 20 of 24 (83.3%) fusions involved targetable genes (Table 2). RARA has been frequently reported as a 3′ fusion partner in acute promyelocytic leukemia[22]. RARA was identified as a 5′ fusion partner in 4 patients and as a 3′ fusion partner in 1 patient; however, only the KRPAT9-RARA fusion was detected in patient 01 as the 3′ fusion partner including exons 3-9, which encodes a DNA-binding and a ligand-binding domain required for RARA transcription factor activity[22]. Three BRAF fusions were identified in patient 09 as the 3′ fusion partner containing the complete tyrosine kinase domain, which was coded by exons 11-18. All FGFR2 and FGFR3 fusions were detected as 5′ fusion partners. Four FGFR2 fusions were consistent with other known activating FGFR2 fusions[23], which frequently occur with a breakpoint after exon 17 at the 3′ end of FGFR2 with a 3′ fusion partner. The kinase domain was retained in these fusion genes. In patient 10, the MET fusion involved the 5′ end of MET exon 7, thus retaining an intact MET kinase domain.
| Patients ID | Fusion gene | 5' partner gene | 3' partner gene | Variant frequency, % | Functional domain is included or not | Targeted drugs | ||||||
| Gene name | Chromosome | Last observed exon | Breakpoint | Gene name | Chromosome | First observed exon | Breakpoint | |||||
| Patient 01 | RARA-PGAP3 | RARA | 17 | 3 | 38504951 | PGAP3 | 17 | 8 | 37828020 | 24.1 | Partially include | Tamibarotene targeting RARA fusion2 |
| Patient 01 | KRTAP9-7-RARA | KRTAP9-7 | 17 | downstream | 39437039 | RARA | 17 | 3 | 38499547 | 56.9 | Completely include | Tamibarotene targeting RARA fusion2 |
| Patient 02 | RARA-KRT13 | RARA | 17 | 2 | 38491648 | KRT13 | 17 | 8 | 39657269 | 29.2 | Partially include | Tamibarotene targeting RARA fusion2 |
| Patient 02 | RARA-ETV4 | RARA | 17 | 2 | 38499726 | ETV4 | 17 | 5 | 41621243 | 76 | Partially include | Tamibarotene targeting RARA fusion2 |
| Patient 03 | RARA-IKZF3 | RARA | 17 | 2 | 38504120 | IKZF3 | 17 | 2 | 38009555 | 18.4 | Partially include | Tamibarotene targeting RARA fusion2 |
| Patient 04 | FGFR2-PDE2A | FGFR2 | 10 | 17 | 123241248 | PDE2A | 11 | 7 | 72307251 | 1.4 | Completely include | Pemigatinib; Erdafitinib targeting FGFR fusion1 |
| Patient 05 | FGFR2-intergenic | FGFR2 | 10 | 17 | 123242196 | intergenic | 10 | - | 123394107 | 16.6 | Completely include | Pemigatinib; Erdafitinib targeting FGFR fusion1 |
| Patient 06 | FGFR2-intergenic | FGFR2 | 10 | 17 | 123240841 | intergenic | 10 | - | 122793842 | 4.2 | Completely include | Pemigatinib; Erdafitinib targeting FGFR fusion1 |
| Patient 07 | FGFR2-SHTN1 | FGFR2 | 10 | 17 | 123242528 | SHTN1 | 10 | 6 | 118709305 | 5.1 | Completely include | Pemigatinib; Erdafitinib targeting FGFR fusion1 |
| Patient 08 | FGFR3-PHTF2 | FGFR3 | 4 | 18 | 1808927 | PHTF2 | 7 | 11 | 77567982 | 3.3 | Completely include | Pemigatinib; Erdafitinib targeting FGFR fusion1 |
| Patient 09 | STIM2-BRAF | STIM2 | 4 | 11 | 27012641 | BRAF | 7 | 9 | 140487929 | 12.7 | Completely include | Selumetinib targeting BRAF fusion1 |
| Patient 09 | STIM2-BRAF | STIM2 | 4 | 11 | 27013243 | BRAF | 7 | 10 | 140486103 | 1.1 | Completely include | Selumetinib targeting BRAF fusion1 |
| Patient 09 | TBC1D19-BRAF | TBC1D19 | 4 | 4 | 26629603 | BRAF | 7 | 10 | 140486782 | 6.5 | Completely include | Selumetinib targeting BRAF fusion1 |
| Patient 10 | TES-MET | TES | 7 | 1 | 115867013 | MET | 7 | 2 | 116332227 | 0.7 | Completely include | Crizotinib targeting MET fusion1 |
| Patient 11 | MET-TES | MET | 7 | 21 | 116436166 | TES | 7 | 4 | 115889445 | 24.6 | Not include | Crizotinib targeting MET fusion1 |
| Patient 12 | EML4-ALK | EML4 | 2 | 6 | 29447382 | ALK | 2 | 20 | 42498662 | 3.5 | Completely include | Crizotinib; ceritinib targeting ALK fusion1 |
| Patient 13 | OPALIN-RET | OPALIN | 10 | 6 | 98104545 | RET | 10 | 11 | 43610099 | 5.46 | Completely include | Pralsetinib targeting RET fusion1 |
| Patient 14 | ARHGAP10-NTRK2 | ARHGAP10 | 4 | 1 | 148716754 | NTRK2 | 9 | 16 | 87476645 | 15.3 | Completely include | Larotrectinib targeting NTRK2 fusion1 |
| Patient 15 | NRG1-FDFT1 | NRG1 | 8 | 12 | 32617907 | FDFT1 | 8 | 8 | 11685375 | 8.9 | Partially include | MCLA-128 targeting NRG1 fusion2 |
| Patient 16 | EGFR-SEPTIN14 | EGFR | 7 | 25 | 55269173 | SEPTIN14 | 7 | 10 | 55871179 | 9.6 | Completely include | Afatinib targeting EGFR fusion2 |
The frequency of fusion events involving the abovementioned 10 targetable genes in the TCGA GC cohort and another Chinese GC cohort (OrigiMed2020 cohort) were analyzed and compared with our patient data (Figure 2B). Neither our cohort nor the OrigiMed2020 cohort showed significant differences in the incidence of these gene fusions in Chinese patients. In two Chinese cohorts, EGFR fusions occurred less frequently. Fusions in MET, BRAF, RET, ALK, and NTRK2 were only identified in two Chinese cohorts; however, the differences in the incidence of these genes were not statistically significant.
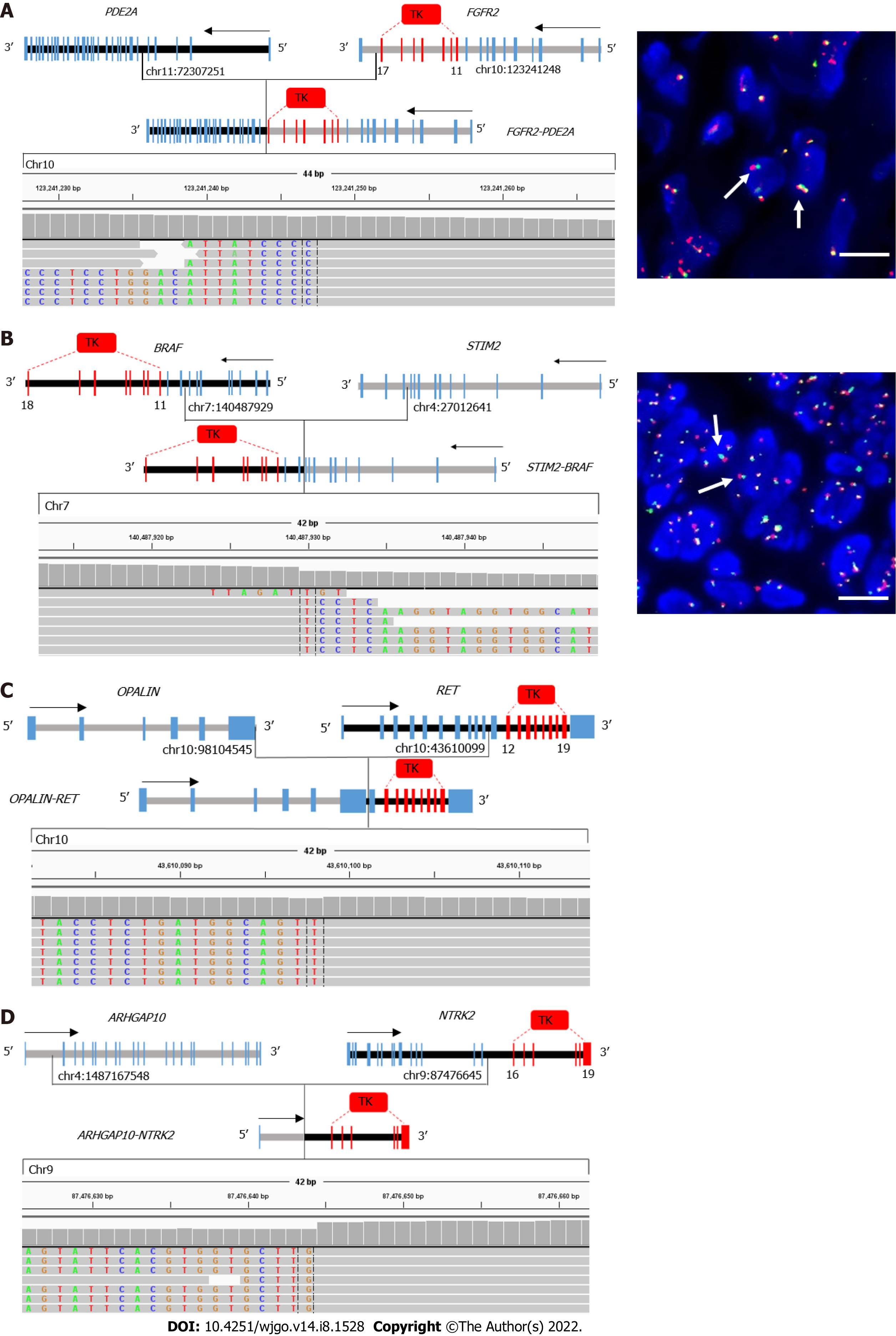
In total, 2 of 20 fusions involving the targetable genes, EML4-ALK and EGFR-SEPTIN14, were reported in other cancers, including non-small-cell lung cancer[24-26]. The remaining 18 gene fusions were not reported in any cancer types. In total, 13 of 18 novel gene fusions contained the exon encoding a tyrosine kinase domain, such as exons 11-17 of FGFR2, exons 11-18 of BRAF, exons 12-19 of MET, and exons 16-21 of NTRK2 (Figure 3). All fusions involving FGFR2, BRAF, RET, and NTRK2 retained the kinase domain (Table 2). Furthermore, reads in Integrative Genomics Viewer plots supported these gene fusions. To verify these novel fusions, fluorescence in situ hybridization (FISH) was performed using break-apart probes. Because only two tumor tissue samples were available, only the FGFR2 and BRAF arrangement in patient 04 and 09, respectively, were confirmed by FISH.
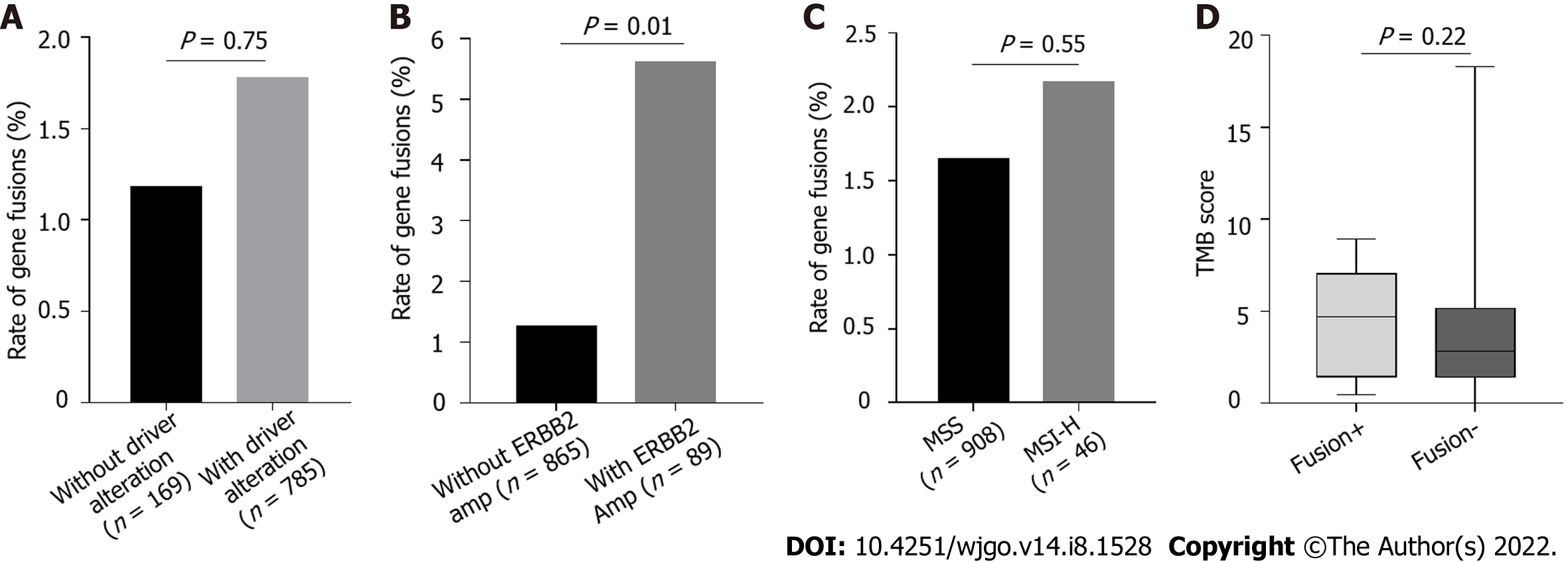
Because of the low frequency of gene fusions in patients with GC, we determined whether gene fusions are enriched in different molecular subtypes of GC, which may indicate the patients that could benefit from gene fusion detection. Fusions are mutually exclusive with other oncogenic mutations and are enriched in patients without driver mutations[27-29]. In our cohort, the frequency of genetic alterations in oncogenic driver genes of GC, such as TP53, ARID1A, CDH1, and PIK3CA mutations and ERBB2 amplification, were comparable with those in the TCGA cohort (Supplementary Figure 1). There was no significant difference in the frequency of fusions involving targetable genes between patients with any alterations in all five driver genes and those without (Figure 4A). Notably, the fusion alteration frequency was significantly higher in patients with ERBB2 amplification than in those without ERBB2 amplification (Figure 4B, P = 0.01). To determine whether fusion alterations were enriched in other driver genes, TP53, ARID1A, CDH1, and PIK3CA were analyzed. There was no enrichment in fusion alterations for these genes (Supplementary Figure 2). Forty-six patients had the MSI-H phenotype. Of these, one patient with fusion genes exhibited MSI-H. There was no obvious difference in the incidence of gene fusions between patients with MSI-H and MSS (Figure 4C). Similarly, TMB scores were evaluated in targetable gene fusion-positive and -negative patients, but the results were not statistically significant (Figure 4D).
Structural gene rearrangements leading to gene fusions are common events that occur in solid tumors. Gene fusions have been considered oncogenic drivers in neoplasia for more than 30 years[30]. Detection and characterization of gene fusions is important for clinical purposes[31]. As the first large-scale study focusing on gene fusion events in Chinese patients with GC, we retrospectively analyzed 954 tumor specimens to identify fusions involving targetable genes and confirmed the occurrence of these fusions in GC.
In this study, 16 of 954 patients harbored 20 fusions involving targetable genes, the majority of which had not been previously reported, including FGFR2-PDE2A, STIM2-BRAF, OPALIN-RET, and ARHGAP10-NTRK2. However, we did not find any significant differences between the Chinese GC cohort (our cohort and OrigiMed2020 cohort) and the TCGA cohort. Fusions in BRAF, RET, ALK, and NTRK2 were detected in two Chinese cohorts but not in the TCGA cohort. This finding may have resulted from the small size of the TCGA cohort, which is prone to bias for gene fusions events because of the low occurrence rate in GC. A comparative study with a larger population is needed to identify differences in fusions involving targetable genes between races.
A major contribution of gene fusions to patients with tumor is the development of drugs that target fusion proteins encoded by these genes. The majority of advances in targeting gene fusions involve kinase domains that constitutively activate downstream signaling pathways[32]. In this study, except RARA and NRG1 fusions, the 14 other fusions involving targetable genes included a receptor tyrosine kinase (RTK) gene, such as FGFR2/3, BRAF, MET, ALK, RET, NTRK2, and EGFR. Furthermore, most of all RTK gene fusions (13/14) completely retained the tyrosine kinase domain, which resulted in functional fusion proteins. We only verified the BRAF rearrangement in patient 09 and the FGFR2 rearrangement in patient 04 using FISH because of insufficient tumor specimens. These fusions were consistent with previously observed fusions[23]; however, only 1 out of 5 RARA fusions contained exons 3-9, which encodes a DNA-binding and ligand-binding domain, which are required for RARA transcription factor activity. These results indicate that most patients with GC with fusions involving targetable genes may benefit from drugs that target fusions. However, patients in this retrospective study had not received targeted drug treatment; thus, we cannot determine whether they would have benefited from fusion-targeted drug therapy.
Interestingly, we also discovered 18 novel fusions with unreported partner genes or with an intergenic space. In other words, screening for known fusions in GC by FISH or polymerase chain reaction will likely miss most of the gene fusions that involve targetable genes. This is not conducive to patients with GC participating in clinical trials of fusion-targeted drugs in pan-cancer. Additionally, we found gene fusions enriched in patients with ERBB2 amplification. We did not confirm all fusions using FISH because of limited tumor tissue samples, nor could we identify gene fusions enriched in distinct molecular subtypes of GC. Moreover, the efficacy of fusion-targeted drugs in GC remains to be further validated in clinical trials. Despite these limitations, for patients who fail standard therapy, NGS-based novel gene fusion detection may provide a new treatment strategy and facilitate participation into clinical trials involving targeted therapy.
As the first large-scale study focusing on gene fusion events in Chinese patients with GC, we determined the frequency (16/954) of targetable gene fusions, and the majority of these fusions, including TES-MET, FGFR2-PDE2A, OPALIN-RET, STIM-BRAF, ARHGAP10-NTRK2, and EGFR-SEPTIN14, had not been previously described. These novel fusions completely retain a kinase domain. Additionally, we found gene fusions that were enriched in patients with ERBB2 amplification. Gene fusion detection may aid in the development of novel treatment strategies for patients with GC.
With rapid advancements in oncogenomics, increasing attention has been focused on gene fusions in cancer. The Food and Drug Administration has approved several fusion-targeted drugs for the treatment of solid tumors, such as larotrectinib for NTRK fusion-positive cancers and Zenocutuzumab for NRG1 fusion-positive cancers. However, targetable gene fusions in Chinese patients with gastric cancer (GC) have not been well characterized.
To investigate the incidence of gene fusions involving targetable genes in Chinese patients with GC and explore a potential treatment strategy for patients with GC.
To explore the types and proportion of targetable gene fusions in Chinese patients with GC and determine the distribution of patients with gene fusions among the molecular subtypes of GC.
This was a multicenter retrospective study that evaluated patients with GC. A total of 954 tumor tissue samples from patients with GC who underwent gene fusion detection were included. Genetic alterations, including SNVs, indels, amplifications, and gene fusions, were analyzed. The enrichment of gene fusions in the molecular subtypes of GC was explored.
Twenty fusions involving targetable genes were detected. Among them, 18 novel gene fusion events were previously not reported in other cancers. Owing to a limited number of tumor tissue samples, only BRAF and FGFR2 fusions were identified by fluorescence in situ hybridization. Additionally, we found that gene fusions were enriched in patients with ERBB2 amplification.
Gene fusions involving targetable genes were characterized in Chinese patients with GC. Testing gene fusions may provide insight for the treatment of GC.
A large study should be performed to further confirm the targetable gene fusions and identify whether gene fusions are enriched in distinct molecular subtypes of GC.
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 68628] [Article Influence: 13725.6] [Reference Citation Analysis (201)] |
| 2. | Sexton RE, Al Hallak MN, Diab M, Azmi AS. Gastric cancer: a comprehensive review of current and future treatment strategies. Cancer Metastasis Rev. 2020;39:1179-1203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 459] [Cited by in RCA: 536] [Article Influence: 89.3] [Reference Citation Analysis (1)] |
| 3. | Jin G, Lv J, Yang M, Wang M, Zhu M, Wang T, Yan C, Yu C, Ding Y, Li G, Ren C, Ni J, Zhang R, Guo Y, Bian Z, Zheng Y, Zhang N, Jiang Y, Chen J, Wang Y, Xu D, Zheng H, Yang L, Chen Y, Walters R, Millwood IY, Dai J, Ma H, Chen K, Chen Z, Hu Z, Wei Q, Shen H, Li L. Genetic risk, incident gastric cancer, and healthy lifestyle: a meta-analysis of genome-wide association studies and prospective cohort study. Lancet Oncol. 2020;21:1378-1386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 185] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 4. | Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23762] [Cited by in RCA: 25605] [Article Influence: 1707.0] [Reference Citation Analysis (11)] |
| 5. | Patel TH, Cecchini M. Targeted Therapies in Advanced Gastric Cancer. Curr Treat Options Oncol. 2020;21:70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 183] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 6. | Grillo F, Fassan M, Sarocchi F, Fiocca R, Mastracci L. HER2 heterogeneity in gastric/gastroesophageal cancers: From benchside to practice. World J Gastroenterol. 2016;22:5879-5887. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 108] [Cited by in RCA: 105] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 7. | Abrahao-Machado LF, Scapulatempo-Neto C. HER2 testing in gastric cancer: An update. World J Gastroenterol. 2016;22:4619-4625. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 155] [Cited by in RCA: 222] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 8. | Rüschoff J, Hanna W, Bilous M, Hofmann M, Osamura RY, Penault-Llorca F, van de Vijver M, Viale G. HER2 testing in gastric cancer: a practical approach. Mod Pathol. 2012;25:637-650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 379] [Cited by in RCA: 450] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 9. | Bartley AN, Washington MK, Colasacco C, Ventura CB, Ismaila N, Benson AB 3rd, Carrato A, Gulley ML, Jain D, Kakar S, Mackay HJ, Streutker C, Tang L, Troxell M, Ajani JA. HER2 Testing and Clinical Decision Making in Gastroesophageal Adenocarcinoma: Guideline From the College of American Pathologists, American Society for Clinical Pathology, and the American Society of Clinical Oncology. J Clin Oncol. 2017;35:446-464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 249] [Cited by in RCA: 336] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 10. | Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202-209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5015] [Cited by in RCA: 5089] [Article Influence: 424.1] [Reference Citation Analysis (4)] |
| 11. | Cho J, Kang SY, Kim KM. MMR protein immunohistochemistry and microsatellite instability in gastric cancers. Pathology. 2019;51:110-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 12. | Scott LJ. Larotrectinib: First Global Approval. Drugs. 2019;79:201-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 169] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 13. | Frampton JE. Entrectinib: A Review in NTRK+ Solid Tumours and ROS1+ NSCLC. Drugs. 2021;81:697-708. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 14. | Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, Bando M, Ohno S, Ishikawa Y, Aburatani H, Niki T, Sohara Y, Sugiyama Y, Mano H. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561-566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3816] [Cited by in RCA: 4174] [Article Influence: 219.7] [Reference Citation Analysis (0)] |
| 15. | Roskoski R Jr. Anaplastic lymphoma kinase (ALK): structure, oncogenic activation, and pharmacological inhibition. Pharmacol Res. 2013;68:68-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 233] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 16. | Holla VR, Elamin YY, Bailey AM, Johnson AM, Litzenburger BC, Khotskaya YB, Sanchez NS, Zeng J, Shufean MA, Shaw KR, Mendelsohn J, Mills GB, Meric-Bernstam F, Simon GR. ALK: a tyrosine kinase target for cancer therapy. Cold Spring Harb Mol Case Stud. 2017;3:a001115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 138] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 17. | Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou SH, Dezube BJ, Jänne PA, Costa DB, Varella-Garcia M, Kim WH, Lynch TJ, Fidias P, Stubbs H, Engelman JA, Sequist LV, Tan W, Gandhi L, Mino-Kenudson M, Wei GC, Shreeve SM, Ratain MJ, Settleman J, Christensen JG, Haber DA, Wilner K, Salgia R, Shapiro GI, Clark JW, Iafrate AJ. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693-1703. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3613] [Cited by in RCA: 3574] [Article Influence: 223.4] [Reference Citation Analysis (0)] |
| 18. | Camidge DR, Bang YJ, Kwak EL, Iafrate AJ, Varella-Garcia M, Fox SB, Riely GJ, Solomon B, Ou SH, Kim DW, Salgia R, Fidias P, Engelman JA, Gandhi L, Jänne PA, Costa DB, Shapiro GI, Lorusso P, Ruffner K, Stephenson P, Tang Y, Wilner K, Clark JW, Shaw AT. Activity and safety of crizotinib in patients with ALK-positive non-small-cell lung cancer: updated results from a phase 1 study. Lancet Oncol. 2012;13:1011-1019. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 999] [Cited by in RCA: 1017] [Article Influence: 72.6] [Reference Citation Analysis (0)] |
| 19. | Wen Z, Xiong D, Zhang S, Liu J, Li B, Li R, Zhang H. Case Report: RAB10-ALK: A Novel ALK Fusion in a Patient With Gastric Cancer. Front Oncol. 2021;11:645370. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 20. | Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat Commun. 2014;5:4846. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 605] [Cited by in RCA: 773] [Article Influence: 64.4] [Reference Citation Analysis (0)] |
| 21. | Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, Shen R, Taylor AM, Cherniack AD, Thorsson V, Akbani R, Bowlby R, Wong CK, Wiznerowicz M, Sanchez-Vega F, Robertson AG, Schneider BG, Lawrence MS, Noushmehr H, Malta TM; Cancer Genome Atlas Network, Stuart JM, Benz CC, Laird PW. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell. 2018;173:291-304.e6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1839] [Cited by in RCA: 1731] [Article Influence: 216.4] [Reference Citation Analysis (0)] |
| 22. | De Braekeleer E, Douet-Guilbert N, De Braekeleer M. RARA fusion genes in acute promyelocytic leukemia: a review. Expert Rev Hematol. 2014;7:347-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 83] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 23. | Fusco MJ, Saeed-Vafa D, Carballido EM, Boyle TA, Malafa M, Blue KL, Teer JK, Walko CM, McLeod HL, Hicks JK, Extermann M, Fleming JB, Knepper TC, Kim DW. Identification of Targetable Gene Fusions and Structural Rearrangements to Foster Precision Medicine in KRAS Wild-Type Pancreatic Cancer. JCO Precis Oncol. 2021;5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 24. | Koivunen JP, Mermel C, Zejnullahu K, Murphy C, Lifshits E, Holmes AJ, Choi HG, Kim J, Chiang D, Thomas R, Lee J, Richards WG, Sugarbaker DJ, Ducko C, Lindeman N, Marcoux JP, Engelman JA, Gray NS, Lee C, Meyerson M, Jänne PA. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res. 2008;14:4275-4283. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 817] [Cited by in RCA: 828] [Article Influence: 46.0] [Reference Citation Analysis (0)] |
| 25. | Cantile M, Marra L, Franco R, Ascierto P, Liguori G, De Chiara A, Botti G. Molecular detection and targeting of EWSR1 fusion transcripts in soft tissue tumors. Med Oncol. 2013;30:412. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 63] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 26. | Zhu YC, Wang WX, Li XL, Xu CW, Chen G, Zhuang W, Lv T, Song Y. Identification of a Novel Icotinib-Sensitive EGFR-SEPTIN14 Fusion Variant in Lung Adenocarcinoma by Next-Generation Sequencing. J Thorac Oncol. 2019;14:e181-e183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 27. | Chou A, Fraser T, Ahadi M, Fuchs T, Sioson L, Clarkson A, Sheen A, Singh N, Corless CL, Gill AJ. NTRK gene rearrangements are highly enriched in MLH1/PMS2 deficient, BRAF wild-type colorectal carcinomas-a study of 4569 cases. Mod Pathol. 2020;33:924-932. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 28. | Wang Y, Xu Y, Wang X, Sun C, Guo Y, Shao G, Yang Z, Qiu S, Ma K. RET fusion in advanced non-small-cell lung cancer and response to cabozantinib: A case report. Medicine (Baltimore). 2019;98:e14120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 29. | Kobayashi M, Sakakibara T, Inoue A, Fukuhara T, Sasano H, Ichinose M, Nukiwa T. Effective enrichment strategy for EML4-ALK fusion gene screening in patients with non-small cell lung cancer. Respir Investig. 2014;52:49-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 30. | Mertens F, Johansson B, Fioretos T, Mitelman F. The emerging complexity of gene fusions in cancer. Nat Rev Cancer. 2015;15:371-381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 439] [Cited by in RCA: 512] [Article Influence: 46.5] [Reference Citation Analysis (0)] |
| 31. | Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1017] [Cited by in RCA: 1029] [Article Influence: 54.2] [Reference Citation Analysis (0)] |
| 32. | Schram AM, Chang MT, Jonsson P, Drilon A. Fusions in solid tumours: diagnostic strategies, targeted therapy, and acquired resistance. Nat Rev Clin Oncol. 2017;14:735-748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 263] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author's Membership in Professional Societies: American Association for Cancer Research, No. 1070947.
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Goebel WS, United States; Park J, South Korea S-Editor: Gong ZM L-Editor: Filipodia P-Editor: Gong ZM