Published online Apr 15, 2022. doi: 10.4251/wjgo.v14.i4.935
Peer-review started: December 19, 2021
First decision: February 21, 2022
Revised: February 21, 2022
Accepted: April 3, 2022
Article in press: April 3, 2022
Published online: April 15, 2022
Processing time: 116 Days and 20.4 Hours
DNA methylation is a part of epigenetic modification, that is closely related to the growth and development of colorectal cancer (CRC). Specific methylated genes and methylated diagnostic models of tumors have become current research focuses. The methylation status of circulating DNA in plasma might serve as a potential biomarker for CRC.
To investigate genome-wide methylation pattern in early CRC using the Illumina Infinium Human Methylation 850K BeadChip.
The 850K Methylation BeadChip was used to analyze the genome-wide methylation status of early CRC patients (n = 5) and colorectal adenoma patients (n = 5). Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment analyses were performed on the selected differentially methylated sites to further discover candidate methylation biomarkers in plasma.
A total of 1865 methylated CpG sites with significant differences were detected, including 676 hypermethylated sites and 1189 hypomethylated sites. The distribution of these sites covered from the 1st to 22nd chromosomes and are mainly distributed on the gene body and gene promoter region. GO and KEGG enrichment analysis showed that the functions of these genes were related to biological regulation, molecular binding, transcription factor activity and signal transduction pathway.
The study demonstrated that the Illumina Infinium Human Methylation 850K BeadChip can be used to investigate genome-wide methylation status of plasma DNA in early CRC and colorectal adenoma patients.
Core Tip: DNA methylation is associated with the growth and development of colorectal cancer (CRC). CRC usually develops from its precancerous lesion, colorectal adenoma. The study demonstrated that the Illumina Infinium Human Methylation 850K BeadChip can be used to investigate genome-wide methylation status of plasma DNA in early CRC and colorectal adenoma patients.
- Citation: Wu YL, Jiang T, Huang W, Wu XY, Zhang PJ, Tian YP. Genome-wide methylation profiling of early colorectal cancer using an Illumina Infinium Methylation EPIC BeadChip. World J Gastrointest Oncol 2022; 14(4): 935-946
- URL: https://www.wjgnet.com/1948-5204/full/v14/i4/935.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i4.935
Colorectal cancer (CRC) is a common malignancy with high morbidity and mortality. Tumors are highly associated with aberrant DNA methylation alterations, which are involved in the regulation of genomic function[1]. DNA methylation in promoter regions leads to gene silencing, which plays an important role in the development and progression of CRC. The incidence of CRC is insidious, and its early manifestation is not obvious, thus, early effective screening is of great significance to reduce morbidity and mortality[2]. CRC usually develops from its precancerous lesion, colorectal adenoma. The evolution from colorectal adenoma to CRC is associated with hypermethylation of tumor-related genes and genome-wide hypomethylation of CpG sites[3,4]. Various histological subtypes of colorectal adenoma also have different methylation patterns[5]. Genome-wide methylation profiling of early CRC and colorectal adenoma will contribute to the discovery of tumor biomarkers, thus improving the early screening rate of CRC.
Circulating tumor DNA (ctDNA), a free DNA released by tumor cells, was found in plasma. The genetic alterations between ctDNA and tumor cell DNA were consistent. A variety of tumor-specific alterations could be detected through ctDNA, including abnormal methylation[6]. Therefore, the detection of ctDNA methylation in plasma can facilitate the cancer diagnosis. Some research had studied genomic methylation of CRC in plasma or serum[7-10], but few focused on the ctDNA methylation differences between early CRC and colorectal adenoma. The Illumina Infinium Human Methylation 850K BeadChip has technical superiority in high-throughput detection for genomic methylation status. The 850K chip contains 91% of the 450K chip and 413745 additional sites (a total of 866895 CpG sites), covering gene promoter regions, gene coding regions, CpG islands and enhancers in the ENCODE and FANTOM5 programs[11-13]. The research efficiency has been greatly improved.
In the present study, an Illumina Infinium Human Methylation 850K BeadChip was used to detect methylation differences of plasma DNA between CRC patients and colorectal adenoma patients, gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses was also performed to investigate the biological functions of differentially methylated sites (DMSs). This study aimed to investigate the different methylation pattern in plasma DNA between early CRC and colorectal adenoma using the Human Methylation 850K BeadChip.
In this study, the Human Methylation 850K BeadChip assay included early CRC patients (n = 5) and colorectal adenoma patients (n = 5) hospitalized in the Chinese PLA General Hospital from January 2018 to January 2020 (Table 1). Their ages ranged from 52 to 79 years old, with an average age of 63 years old. Inclusion criteria: None of the patients had received any treatment when their peripheral blood samples were taken, including surgical resection, radiotherapy, chemotherapy, and targeted therapy; all patients who underwent colonoscopy and endoscopic biopsy, which were confirmed as colorectal adenoma (without cancerization) by pathology, were included in the adenoma group, which were confirmed as CRC by pathology and at stage I according to the American Joint Committee on Cancer tumor-node-metastasis staging system for CRC, were included in the tumor group; all patients had no other malignant tumors or serious diseases. Later clinical follow-up was required for all patients. If the postoperative pathological results of the tumor tissue were inconsistent with those of endoscopic biopsy, postoperative histopathology prevailed, and cases that did not meet the inclusion criteria were excluded. All participants signed written informed consent for the collection of samples and subsequent analyses before inclusion.
| Gender | Age | Group | TNM stage | |
| C1 | Male | 52 | CRC (I) | I |
| C2 | Male | 62 | CRC (I) | I |
| C3 | Female | 62 | CRC (I) | I |
| C4 | Male | 64 | CRC (I) | I |
| C5 | Female | 79 | CRC (I) | I |
| A1 | Female | 69 | Adenoma | |
| A2 | Male | 69 | Adenoma | |
| A3 | Male | 69 | Adenoma | |
| A4 | Male | 49 | Adenoma | |
| A5 | Male | 74 | Adenoma |
Peripheral blood was collected from the veins of patients and centrifuged within 30 min (4℃, 3000 rpm, 10 min). The plasma was collected and stored at 4℃ for DNA extraction within 3 days, according to the instructions of the QIAamp MinElute ccfDNA Mini Kit (QIAGEN, Germany, 55204). The DNA was treated with bisulfite to convert cytosine to uracil according to the instructions of the EZ DNA Methylation-Gold Kit (Zymo Research, USA, D5001/D5003). Subsequently Qubit ssDNA HS Assay Kit (Invitrogen, USA, Q32854) as well as Qubit 3.0 fluorescence quantizer (Thermo Fisher, USA) were used for DNA quantitative analysis after bisulfite conversion. The bisulfite-converted DNA was stored at -20℃ until use.
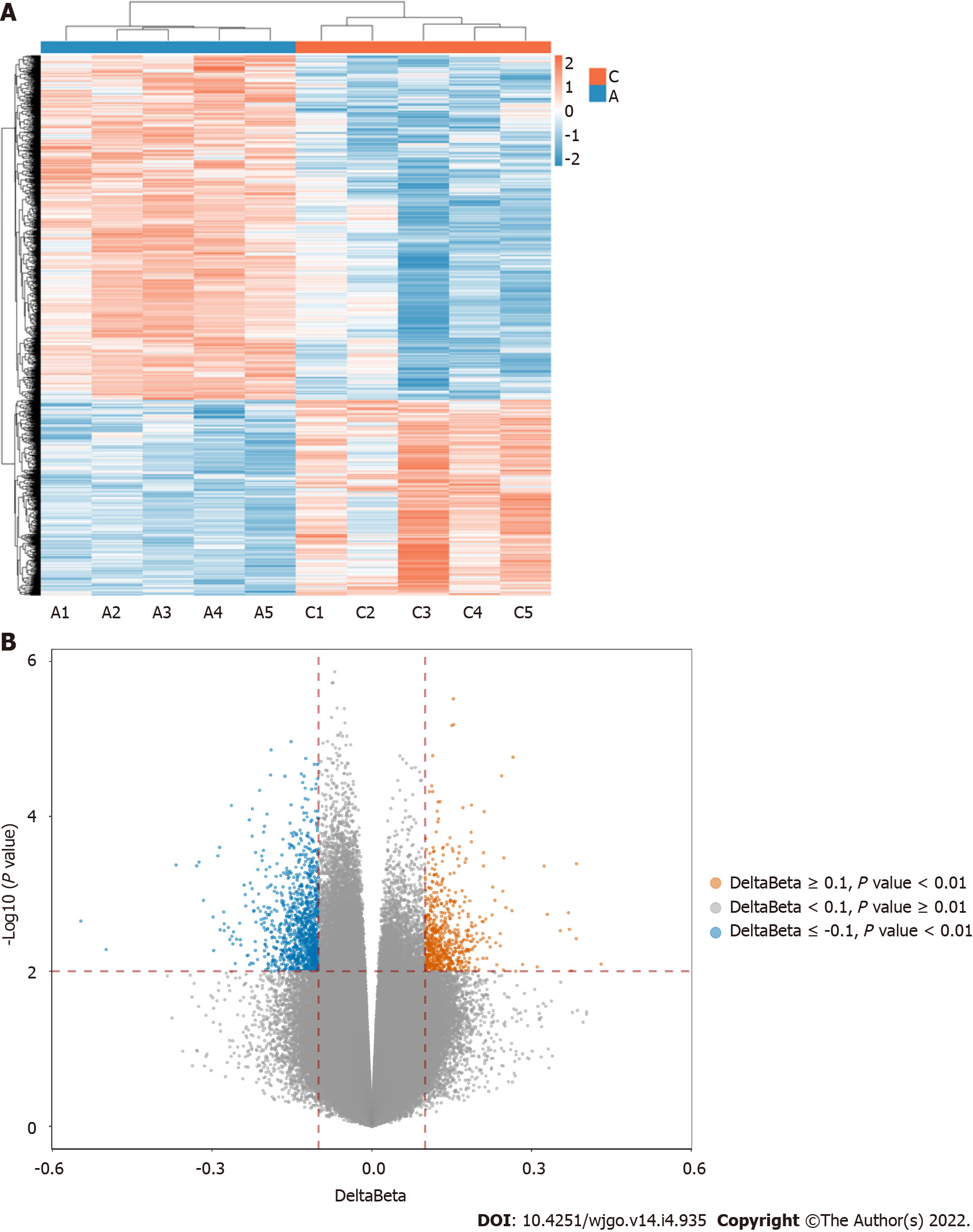
Genome-wide methylation profiling was perfrormed by Illumina Infinium Human Methylation 850K BeadChip (Sinotech Genomics, China), in accordance with Illumina's standard protocol. Bisulfite-converted DNA was used for alkali denaturation and genomic amplification. Then, the amplified DNA was used for hybridization, washing, fluorescent staining, single-base extension and scanning on an 850K BeadChip. An IScan software analysis system (Illumina, USA) was used for signal acquisition and analysis. The data (IDAT files) were mainly analyzed using the ChAMP package in R. We used β values (range, 0-1) to represent the DNA methylation level, wAhich were calculated from the intensity ratio of the methylated signals to the total (methylated and unmethylated) signals for each site. Then we compared the β values of each probe site in the samples between the CRC group and adenoma group, calculating the average β value of each site and the difference in β values between groups (Δβ). For plasma samples, a CpG site with |Δβ| > 0.1 and a P value < 0.01 was considered as a DMS (Δβ > 0.1 was considered a hypermethylated site, and Δβ < -0.1 was considered a hypomethylated site). The Benjamini and Hochberg method was applied to reduce the false-positive rate[14].
We also conducted GO and KEGG enrichment analyses for DMSs using the R package. In the functional enrichment analysis, the P values represented the likelihood that the gene associated with the GO term would be enriched in the input gene list, and a P value less than 0.05 was considered significant.
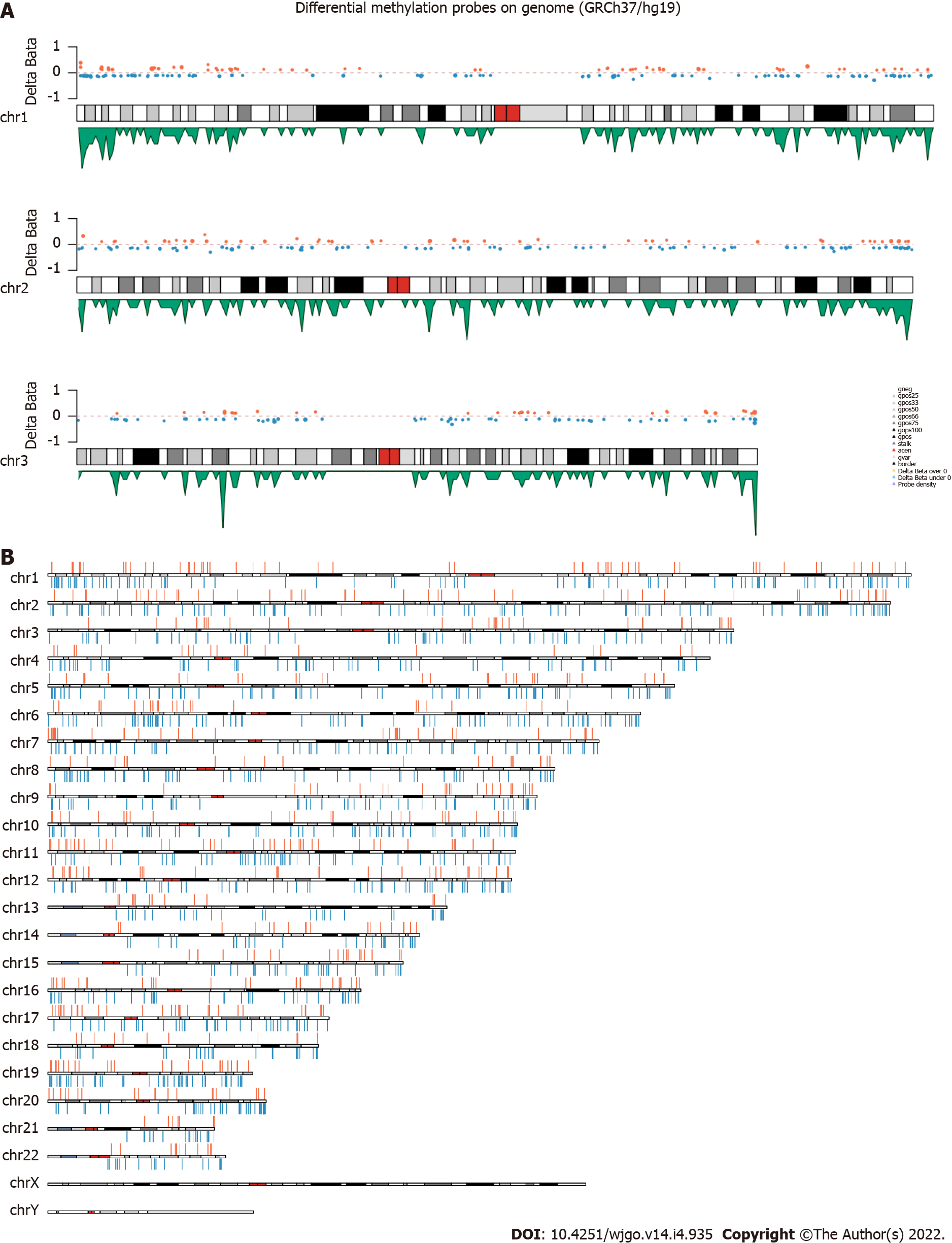
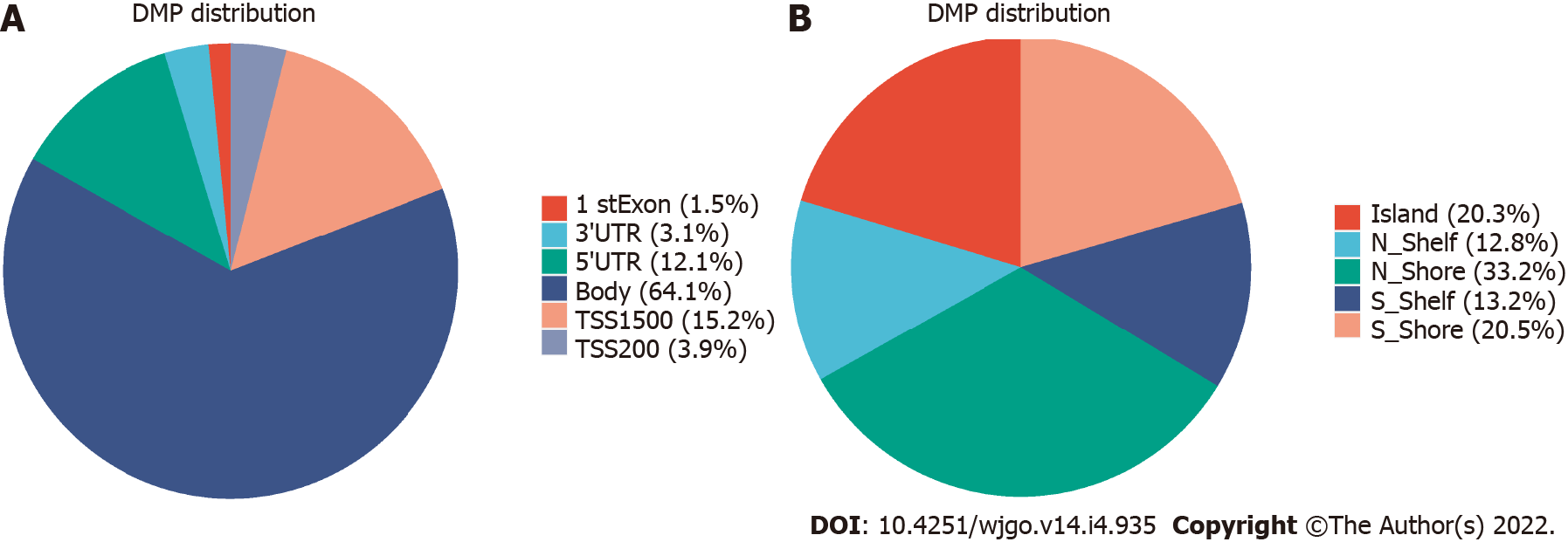
The 850K methylation BeadChip included a total of 866895 CpG sites, screening out 1865 differentially methylated CpG sites between CRC and adenoma plasma samples (|Δβ| > 0.1, P < 0.01), including 676 hypermethylation sites and 1189 hypomethylation sites (Figure 1). These DMSs were widely distributed on all chromosomes, among which, chromosomes 1 and 2 had the most sites and chromosomes 21 and 22 had the least sites (Table 2). Figure 2A and B shows the distribution of DMSs on the chromosomes of the genome. There were also differences in the distribution of the gene structure elements, mainly in the gene body (64.1%) and gene promoter region (32.7%, including TSS1500, TSS200, 5'UTR, and 1stExon). The distribution of DMSs in and around CpG islands was also different, mainly in the CpG Shore (53.7%) and CpG Island regions (20.3%) (Figure 3A and B).
| Chromosome | DMSs | Hypermethylated | Hypomethylated |
| chr1 | 162 | 62 | 100 |
| chr2 | 158 | 53 | 105 |
| chr3 | 104 | 35 | 69 |
| chr4 | 83 | 22 | 61 |
| chr5 | 112 | 35 | 77 |
| chr6 | 112 | 25 | 87 |
| chr7 | 115 | 55 | 60 |
| chr8 | 89 | 27 | 62 |
| chr9 | 79 | 33 | 46 |
| chr10 | 91 | 29 | 62 |
| chr11 | 104 | 42 | 62 |
| chr12 | 105 | 41 | 64 |
| chr13 | 53 | 18 | 35 |
| chr14 | 50 | 22 | 28 |
| chr15 | 58 | 22 | 36 |
| chr16 | 72 | 34 | 38 |
| chr17 | 76 | 35 | 41 |
| chr18 | 38 | 13 | 25 |
| chr19 | 88 | 33 | 55 |
| chr20 | 60 | 22 | 38 |
| chr21 | 25 | 7 | 18 |
| chr22 | 31 | 11 | 20 |
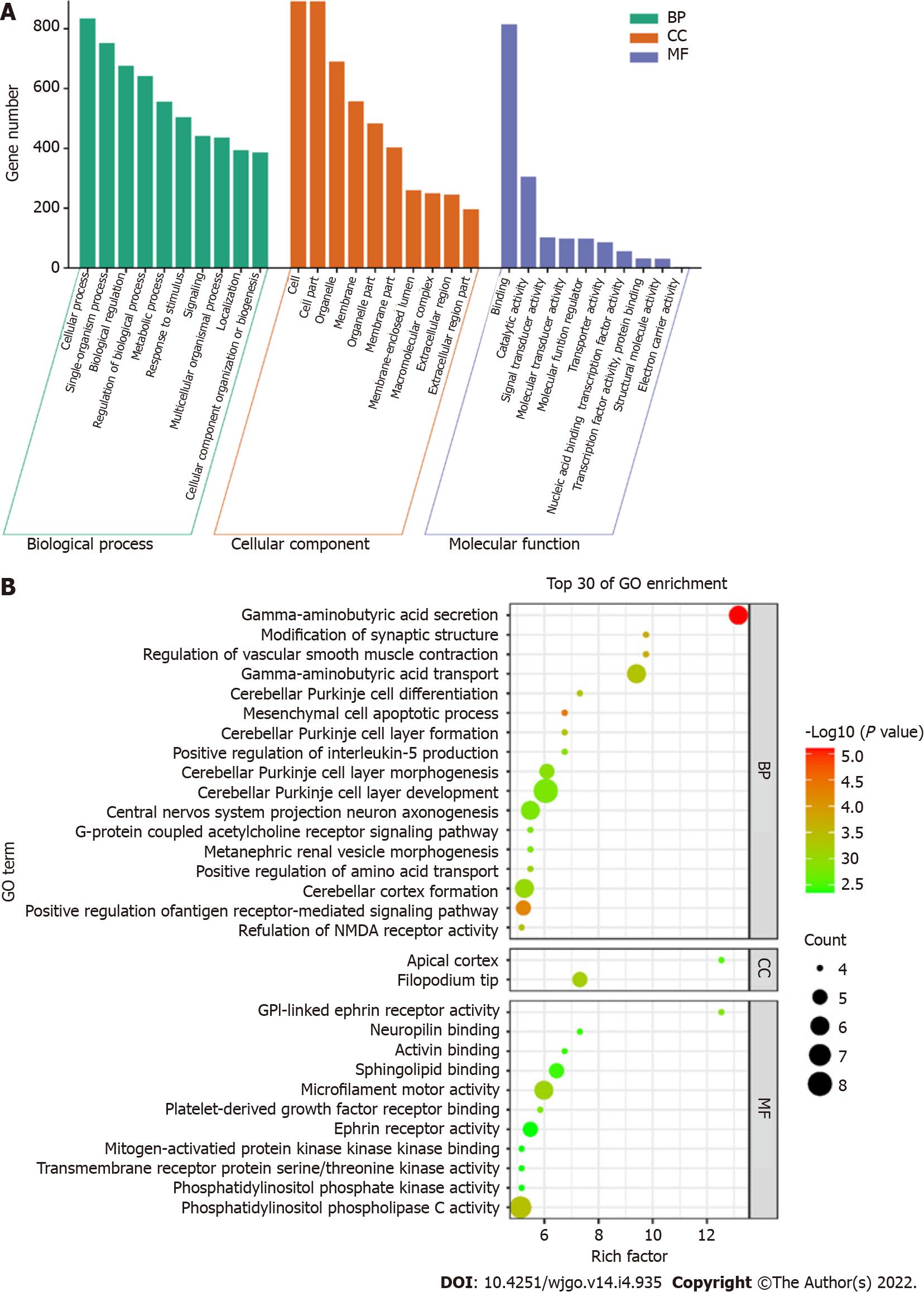
GO enrichment analysis including the biological process, cellular component, and molecular function categories, was performed. A P value of < 0.05 was considered statistically significant. After GO enrichment analysis of the selected DMSs, 783 GO functional annotations were obtained, including 571 terms of biological processes, 107 terms of cell components and 105 terms of molecular functions. These GO functional terms indicated that DMSs in CRC cover a variety of functional communities. In terms of biological process, differentially expressed genes were mainly enriched in biological regulation, cellular process, metabolic process, regulation of biological process, and single organism process. The cell component analysis showed that molecules distributed in the extracellular region, cell membrane, organelle and synapse were significantly enriched. At the molecular level, functional enrichment annotation showed that the highly enriched genes were associated with molecular binding, catalytic activity and signaling pathways (Figure 4A). The bubble diagram in Figure 4B shows the top 30 GO terms with the most significant enrichment effect. Significantly enriched biological processes included gamma−aminobutyric acid depletion and transport and cerebellar Purkinje cell layer development. Significantly enriched cellular components included the filopodium tip and apical cortex. Significantly enriched molecular functions included microfilament motor activity, ephrin receptor activity and phosphatidylinositol phospholipase C and sphingolipid binding (Figure 4B).
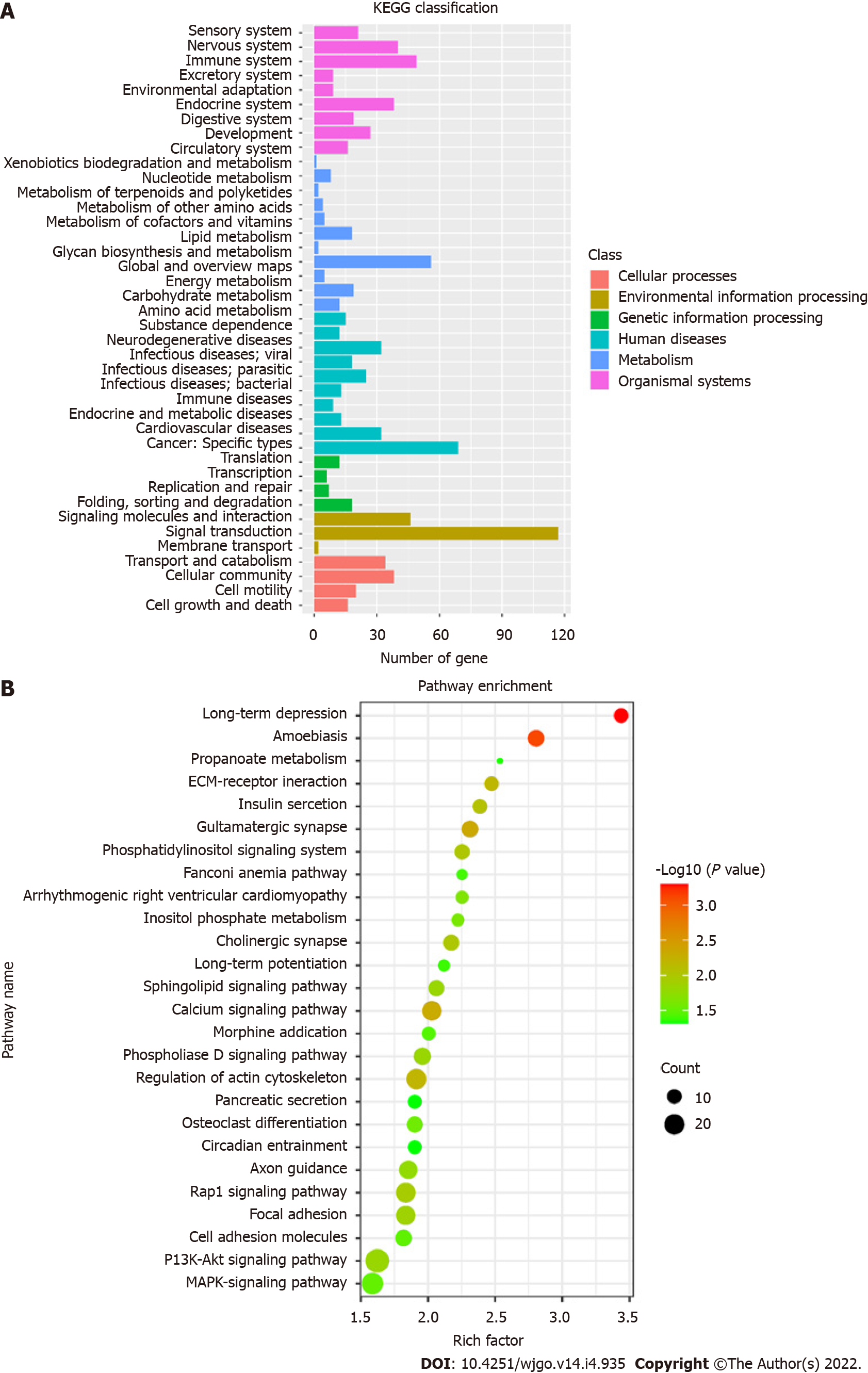
KEGG pathways were divided into cellular processes, environmental information processing, genetic information processing, human disease metabolism, metabolism, and organismal systems. KEGG annotation results of DMSs were classified according to the above six pathways (Figure 5A). A total of 26 KEGG signaling pathways were significantly enriched, which are shown in the bubble diagram (Figure 5B). KEGG analysis showed that DMSs were involved in various cellular pathways in CRC. Significantly enriched pathways included long-term depression, calcium signaling pathway, synapse related pathway, Rap1 signaling pathway, PI3K-Akt signaling pathway and MAPK signaling pathway.
Cancer is associated with a series of abnormal alterations in genetics and epigenetics. Mutations or epigenetic alterations in key pathways involved in cell growth and division may induce the occurrence of tumors[15]. Aberrant methylation modifications in the colon can be observed in early premalignant lesions such as adenoma and normal mucosa of para-carcinoma tissue[16]. And DNA methylation is an important epigenetic alteration that leads to silencing of the expression of tumor-related genes, and many abnormal methylations have been found in the genomic DNA of tumor cells, which are also reflected in plasma ctDNA. Therefore, the detection of ctDNA in plasma is becoming a new method of cancer diagnosis and monitoring[9]. Current ctDNA detection techniques based on gene mutations mainly rely on the number of tumor-derived DNA molecules in the sample, the number of mutated cancer cells, the concentration fraction of ctDNA and the sensitivity of analysis[17]. Using methylation patterns to detect ctDNA in plasma has advantages over mutation-based tests. Epigenetic modifications are more consistent in tumor cells than genetic mutations[18]. Therefore, early CRC can be diagnosed by detecting the methylation pattern of ctDNA.
The detecting DNA for 850K Methylation BeadChip is first treated with bisulfite to convert methylation differences into differences in bases. The Illumina Infinium Human Methylation 850K BeadChip has two detection methods (Infinium I and Infinium II). Infinium I has two specific probes for the bisulfite-converted DNA sequence. The M-type magnetic beads match the methylated sites, and the U-type magnetic beads match the unmethylated sites. Infinium II had only one probe, and only one base is added for single nucleotide extension after pairing. Different bases are added for methylated and unmethylated sequences, producing different fluorescence signals, and the level of methylation is calculated according to the signal ratio[11,19].
This study used an Illumina Infinium Human Methylation 850K BeadChip to detect and analyze the genome-wide methylation status of 5 early CRC patients and 5 cases of adenoma. According to the research findings, there were 1865 DMSs, including 676 hypermethylated sites and 1189 hypome
This study showed that the Illumina Infinium Human Methylation 850K BeadChip can be used to investigate genome-wide methylation pattern of plasma DNA in tumors for the discovery of biomarkers. The limitations of this study are the sample size and the deficiency of verification. Further researches are needed on how to reduce the amount of initial DNA for the chip, and how to apply the selected biomarkers to cancer diagnosis.
DNA methylation is closely related to the growth and development of colorectal cancer (CRC). Circulating tumor DNA (ctDNA) in plasma also has tumor-specific methylation patterns, the methylation status of ctDNA might serve as a potential biomarker.
In this study, we investigated the different methylation pattern between early CRC and colorectal adenoma.
This study aimed to analyze the genomic methylation status of CRC and discover potential methylated biomarkers for CRC diagnosis by Illumina Infinium Human Methylation 850K BeadChip.
We used 850K Methylation BeadChip and enrichment analysis to select the differentially methylated sites of early CRC patients (n = 5) and colorectal adenoma patients (n = 5). Then, the gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed.
We found 1865 methylated sites with significant differences (676 hypermethylated sites and 1189 hypomethylated sites). The distribution of these sites covered from the 1st to 22nd chromosomes and are mainly distributed on the gene body and gene promoter region. GO and KEGG enrichment analysis identified that the functions of these genes were related to biological regulation, molecular binding, transcription factor activity and signal transduction pathway.
The Illumina Infinium Human Methylation 850K BeadChip can be used to investigate genome-wide methylation status of plasma DNA to select potential methylated biomarkers for CRC diagnosis.
Genome-wide methylation analysis of early CRC and screening of biomarkers by 850K Methylation BeadChip provided a new possibility for early diagnosis of CRC.
| 1. | Edwards JR, Yarychkivska O, Boulard M, Bestor TH. DNA methylation and DNA methyltransferases. Epigenetics Chromatin. 2017;10:23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 244] [Cited by in RCA: 334] [Article Influence: 37.1] [Reference Citation Analysis (0)] |
| 2. | Brenner H, Stock C, Hoffmeister M. Effect of screening sigmoidoscopy and screening colonoscopy on colorectal cancer incidence and mortality: systematic review and meta-analysis of randomised controlled trials and observational studies. BMJ. 2014;348:g2467. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 663] [Cited by in RCA: 634] [Article Influence: 52.8] [Reference Citation Analysis (2)] |
| 3. | Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683-692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3299] [Cited by in RCA: 3450] [Article Influence: 181.6] [Reference Citation Analysis (0)] |
| 4. | García-Solano J, Turpin MC, Torres-Moreno D, Huertas-López F, Tuomisto A, Mäkinen MJ, Conesa A, Conesa-Zamora P. Two histologically colorectal carcinomas subsets from the serrated pathway show different methylome signatures and diagnostic biomarkers. Clin Epigenetics. 2018;10:141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 5. | Fiedler D, Hirsch D, El Hajj N, Yang HH, Hu Y, Sticht C, Nanda I, Belle S, Rueschoff J, Lee MP, Ried T, Haaf T, Gaiser T. Genome-wide DNA methylation analysis of colorectal adenomas with and without recurrence reveals an association between cytosine-phosphate-guanine methylation and histological subtypes. Genes Chromosomes Cancer. 2019;58:783-797. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 6. | Luo H, Zhao Q, Wei W, Zheng L, Yi S, Li G, Wang W, Sheng H, Pu H, Mo H, Zuo Z, Liu Z, Li C, Xie C, Zeng Z, Li W, Hao X, Liu Y, Cao S, Liu W, Gibson S, Zhang K, Xu G, Xu RH. Circulating tumor DNA methylation profiles enable early diagnosis, prognosis prediction, and screening for colorectal cancer. Sci Transl Med. 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 302] [Article Influence: 50.3] [Reference Citation Analysis (0)] |
| 7. | Rahier JF, Druez A, Faugeras L, Martinet JP, Géhénot M, Josseaux E, Herzog M, Micallef J, George F, Delos M, De Ronde T, Badaoui A, D'Hondt L. Circulating nucleosomes as new blood-based biomarkers for detection of colorectal cancer. Clin Epigenetics. 2017;9:53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 66] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 8. | Barault L, Amatu A, Siravegna G, Ponzetti A, Moran S, Cassingena A, Mussolin B, Falcomatà C, Binder AM, Cristiano C, Oddo D, Guarrera S, Cancelliere C, Bustreo S, Bencardino K, Maden S, Vanzati A, Zavattari P, Matullo G, Truini M, Grady WM, Racca P, Michels KB, Siena S, Esteller M, Bardelli A, Sartore-Bianchi A, Di Nicolantonio F. Discovery of methylated circulating DNA biomarkers for comprehensive non-invasive monitoring of treatment response in metastatic colorectal cancer. Gut. 2018;67:1995-2005. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 181] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 9. | Symonds EL, Pedersen SK, Murray D, Byrne SE, Roy A, Karapetis C, Hollington P, Rabbitt P, Jones FS, LaPointe L, Segelov E, Young GP. Circulating epigenetic biomarkers for detection of recurrent colorectal cancer. Cancer. 2020;126:1460-1469. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 10. | Molnár B, Galamb O, Kalmár A, Barták BK, Nagy ZB, Tóth K, Tulassay Z, Igaz P, Dank M. Circulating cell-free nucleic acids as biomarkers in colorectal cancer screening and diagnosis - an update. Expert Rev Mol Diagn. 2019;19:477-498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 11. | Moran S, Arribas C, Esteller M. Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics. 2016;8:389-399. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 405] [Cited by in RCA: 529] [Article Influence: 48.1] [Reference Citation Analysis (0)] |
| 12. | Naumov VA, Generozov EV, Zaharjevskaya NB, Matushkina DS, Larin AK, Chernyshov SV, Alekseev MV, Shelygin YA, Govorun VM. Genome-scale analysis of DNA methylation in colorectal cancer using Infinium HumanMethylation450 BeadChips. Epigenetics. 2013;8:921-934. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 117] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 13. | Sandoval J, Heyn H, Moran S, Serra-Musach J, Pujana MA, Bibikova M, Esteller M. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6:692-702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 763] [Cited by in RCA: 765] [Article Influence: 51.0] [Reference Citation Analysis (0)] |
| 14. | Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811-818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1952] [Cited by in RCA: 2150] [Article Influence: 59.7] [Reference Citation Analysis (0)] |
| 15. | Vrba L, Futscher BW. A suite of DNA methylation markers that can detect most common human cancers. Epigenetics. 2018;13:61-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 51] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 16. | Takane K, Fukuyo M, Matsusaka K, Ota S, Rahmutulla B, Matsushita K, Miyauchi H, Nakatani Y, Matsubara H, Kaneda A. The frequency of promoter DNA hypermethylation is decreased in colorectal neoplasms of familial adenomatous polyposis. Oncotarget. 2018;9:32653-32666. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 17. | Wong FC, Sun K, Jiang P, Cheng YK, Chan KC, Leung TY, Chiu RW, Lo YM. Cell-free DNA in maternal plasma and serum: A comparison of quantity, quality and tissue origin using genomic and epigenomic approaches. Clin Biochem. 2016;49:1379-1386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 62] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 18. | Sun K, Jiang P, Chan KC, Wong J, Cheng YK, Liang RH, Chan WK, Ma ES, Chan SL, Cheng SH, Chan RW, Tong YK, Ng SS, Wong RS, Hui DS, Leung TN, Leung TY, Lai PB, Chiu RW, Lo YM. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci U S A. 2015;112:E5503-E5512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 416] [Cited by in RCA: 566] [Article Influence: 51.5] [Reference Citation Analysis (0)] |
| 19. | Solomon O, MacIsaac J, Quach H, Tindula G, Kobor MS, Huen K, Meaney MJ, Eskenazi B, Barcellos LF, Holland N. Comparison of DNA methylation measured by Illumina 450K and EPIC BeadChips in blood of newborns and 14-year-old children. Epigenetics. 2018;13:655-664. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 80] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 20. | Zhu L, Yan F, Wang Z, Dong H, Bian C, Wang T, Yu E, Li J. Genome-wide DNA methylation profiling of primary colorectal laterally spreading tumors identifies disease-specific epimutations on common pathways. Int J Cancer. 2018;143:2488-2498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 21. | Wong CC, Xu J, Bian X, Wu JL, Kang W, Qian Y, Li W, Chen H, Gou H, Liu D, Yat Luk ST, Zhou Q, Ji F, Chan LS, Shirasawa S, Sung JJ, Yu J. In Colorectal Cancer Cells With Mutant KRAS, SLC25A22-Mediated Glutaminolysis Reduces DNA Demethylation to Increase WNT Signaling, Stemness, and Drug Resistance. Gastroenterology. 2020;159:2163-2180.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 133] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 22. | Kel A, Boyarskikh U, Stegmaier P, Leskov LS, Sokolov AV, Yevshin I, Mandrik N, Stelmashenko D, Koschmann J, Kel-Margoulis O, Krull M, Martínez-Cardús A, Moran S, Esteller M, Kolpakov F, Filipenko M, Wingender E. Walking pathways with positive feedback loops reveal DNA methylation biomarkers of colorectal cancer. BMC Bioinformatics. 2019;20:119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 23. | Nguyen LH, Goel A, Chung DC. Pathways of Colorectal Carcinogenesis. Gastroenterology. 2020;158:291-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 344] [Cited by in RCA: 382] [Article Influence: 63.7] [Reference Citation Analysis (0)] |
| 24. | Warton K, Lin V, Navin T, Armstrong NJ, Kaplan W, Ying K, Gloss B, Mangs H, Nair SS, Hacker NF, Sutherland RL, Clark SJ, Samimi G. Methylation-capture and Next-Generation Sequencing of free circulating DNA from human plasma. BMC Genomics. 2014;15:476. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 62] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 25. | Liu L, Chen Y, Liu T, Yu J, Ma L, Wu H. Genome-wide DNA methylation profiling and gut flora analysis in intestinal polyps patients. Eur J Gastroenterol Hepatol. 2021;33:1071-1081. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kołat D, Poland; Osera S, Japan S-Editor: Wang JL L-Editor: A P-Editor: Wang JL