Published online Aug 15, 2021. doi: 10.4251/wjgo.v13.i8.970
Peer-review started: June 10, 2021
First decision: June 15, 2021
Revised: June 18, 2021
Accepted: July 5, 2021
Article in press: July 5, 2021
Published online: August 15, 2021
Processing time: 65 Days and 2.3 Hours
Early detection, early diagnosis, and early treatment are currently accepted methods that can effectively improve the efficacy of colorectal cancer (CRC) treatment. Exosomes were demonstrated to be potential tumor molecular mar
To evaluate the diagnostic value of CRC by detecting four exosomal microRNAs (miRNAs) (miR-15b, miR-16, miR-21, and miR-31) that were demonstrated to have potential diagnostic value in serum.
Relative expression levels of miR-15b, miR-16, miR-21, and miR-31 in 123 CRC, 117 colorectal adenoma, and 150 healthy controls were detected, and single and panel models were evaluated. The 2-ΔΔCt method was used to calculate the re
Compared to the healthy control group, the best indicator of the four miRNAs was miR-15b, and the sensitivity and specificity were 81.33% and 91.80%, re
We built and validated a diagnostic model containing miR-15b, miR-21, and miR-31 expression levels to discriminate the healthy control group and CRC group, and its sensitivity and specificity were 95.06% and 94.44%, respectively. The miR-15b, miR-16, and miR-21 panel was used to discriminate the colorectal adenoma group and CRC group with a sensitivity and specificity of 85.19% and 82.09%, respectively.
Core Tip: In this study, we aimed to evaluate the diagnostic value of colorectal cancer by detecting the exosome four microRNAs (miR-15b, miR-16, miR-21, miR-31). The diagnostic model may serve as a novel diagnostic model for colorectal cancer.
- Citation: Han L, Shi WJ, Xie YB, Zhang ZG. Diagnostic value of four serum exosome microRNAs panel for the detection of colorectal cancer. World J Gastrointest Oncol 2021; 13(8): 970-979
- URL: https://www.wjgnet.com/1948-5204/full/v13/i8/970.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v13.i8.970
Colorectal cancer (CRC) is a common malignant tumor and the third most common malignant tumor in China. The 5-year survival rate of patients with early-stage CRC after effective treatment exceeds 90%. However, 25% of patients have local or distant metastasis at the time of initial diagnosis, and the 5-year survival rate is only 12%[1]. Early detection, early diagnosis, and early treatment are currently accepted methods that can effectively improve the efficacy of CRC treatment. At present, commonly used clinical laboratory diagnostic tests mainly include fecal occult blood detection and blood marker detection, but their sensitivity and specificity are still insufficient. Imaging examination can effectively assess the extent of the lesion and stage the tumor, but it is of limited value for the diagnosis of early lesions. Endoscopy combined with tissue biopsy is currently the gold standard for the early diagnosis of CRC, but it has the disadvantages of cumbersome operation, poor patient compliance, and invasiveness.
At present, the available diagnostic methods for early CRC are insufficient[2]. A minimally invasive or noninvasive, sensitive, and accurate early diagnostic method is urgently needed in clinical practice. Liquid biopsy technology is highly valued due to its noninvasive, comprehensive, real-time, and repeated monitoring features[3]. Liquid biopsy technology is mainly aimed at body fluid components such as peripheral blood, saliva, and urine to provide tumor heterogeneity and genetic information[4,5], and it can provide strategies for the early diagnosis of CRC and optimal individualized treatment.
With the advent of the era of precision medicine, liquid biopsy has become an important branch of the field of oncology and testing. Circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), and exosomes are currently the main targets of liquid biopsy. CTCs refer to cells shed from the primary tumor or metastasis that enter the peripheral blood and are highly heterogeneous. The detection of CTCs mainly depends on capturing trace CTCs in the peripheral blood and then monitoring the change trend of the type and number of CTCs. CTCs play an important role in the metastasis and recurrence of metastatic solid tumors, the evaluation of therapeutic effects, and prognosis evaluation[6,7].
ctDNA refers to double-stranded DNA fragments released into the blood circulation system after shedding or apoptosis of tumor cells, and their length is approximately 150-250 bp. Studies have shown that the tumor-related information carried by ctDNA has the same biological characteristics as the tumor tissue and can provide relevant information about the genetic changes in the tumor (including mutations, deletions, insertions, gene copy number variation, microsatellite changes, methylation, etc.) It can provide important clues for the diagnosis and treatment of tumors. The results of one study found that the later the stage of CRC patients, the more significant the increase in serum ctDNA content. However, due to its removal by macrophages, the content of free DNA in body fluids is extremely low, and ctDNA only accounts for a very small part of free DNA, which requires detection equipment with extremely high sensitivity[8,9].
Exosomes are small vesicles secreted and released by cells under normal or pa
Analysis of the protein profile expression of serum exosomes revealed that 22 patients with CRC had a downregulation of protein-peptide profiles. Bioinformatics analysis found that the downregulated protein-peptide profiles mainly participated in the occurrence and development of CRC and had potential clinical application value. Exosomes, as tumor molecular markers, have the advantages of protecting nucleic acids and having good specificity, but their extraction methods and identification methods have low efficiency[11].
In our study, we aimed to evaluate the diagnostic value of CRC by detecting four exosomal miRNAs (miR-15b, miR-16, miR-21, miR-31) that were demonstrated to have potential diagnostic value in serum.
The expression levels of serum exosome miR-15b, miR-16, miR-21, and miR-31 in CRC patients, colorectal adenoma patients, and healthy controls were detected. With patient consent, the samples were collected in Beijing Daxing District People’s Hos
The CRC inclusion criteria included: (1) Diagnosed by surgery and pathology; and (2) No radiotherapy, chemotherapy, or other tumor-related treatments were accepted. The exclusion criteria included: Patients with liver, kidney, and other organ dysfunc
Colorectal adenoma patient inclusion criteria included: Diagnosed by enteroscopy pathology. The exclusion criteria included: No history of tumors, no other intestinal diseases, and no T3, liver, kidney, or other organ dysfunction.
The healthy control group inclusion criteria included follow-up. After enteroscopy, there could be no abnormality found in the intestine. The exclusion criteria included: History of a tumor and dysfunction of the heart, liver, kidney or other organs.
The blood of all subjects was collected on an empty stomach in the morning. We collected 5 mL of peripheral venous blood in a test tube without anticoagulant, centrifuged it at 3500 g for 7 min at room temperature to remove the cells or cell debris. The supernatant was subjected to 4 °C, 17000 g centrifugation for 10 min to produce the serum, which was transferred to a new centrifuge tube and then stored at -80 °C until analysis.
The serum was centrifuged at 3000 g for 15 min at room temperature. According to the ExoQuick kit instruction manual, the vortexed serum and exosome reagent mixture was precipitated at 4 °C for 30 min. After centrifuging at 1500 g for 30 min, we discarded the supernatant and centrifuged it again at 1500 g for 5 min to obtain the precipitate, resuspended it in 200 μL phosphate buffered saline, and filtered it through a 0.22 μm sterile filter to prepare the desired size and concentration of the exosomes. After extracting the serum exosomes, nanoparticle tracking analysis technology and western blotting, which were used to detect the expression of exosome marker pro
Total RNA from the serum exosomes was extracted by the TRIzol method. Briefly, TRIzol reagent was added to the exosome suspension for 5 min to allow the exosomes to be fully lysed and release their internal nucleic acid substances, and then chloroform reagent was added at room temperature for 15 min. After centrifugation, the upper transparent water phase was moved to a new centrifuge tube, and then isopropanol was added. After mixing and centrifugation, ethanol was added to the centrifuge tube, and then diethyl pyrocarbonate water was added to the centrifuge tube to obtain total RNA. A NanoDrop was applied to detect the concentration and purity of the total RNA from the serum exosomes.
The RNA reverse transcription reaction system included 12 μL of total RNA, 2.5 U/μL PolyA polymerase 1 μL, Mix 1 μL, 5× Reaction Buffer 5 μL, and ddH2O (RNase/ DNase free) 6 μL. U6 small nuclear RNA was used as an internal control, and the relative expression of miR-15b, miR-16, miR-21, and miR-31 was detected by an ABI 7500 instrument. The three-step real-time polymerase chain reaction amplification reaction conditions were 95 °C for 10 min; followed by 95 °C for 10 s, 60 °C for 20 s, and 72 °C for 10 s for 40 cycles. The 2-ΔΔCt method was used to calculate the relative expression of miRNA compared to the internal control (U6).
The data were analyzed by SPSS 22.0 statistical software (Armonk, NY, United States). The relative expression of miR-15b, miR-16, miR-21, and miR-31 are expressed as the median (25%, 75%). The significance of the four serum exosome miRNAs was compared by analysis of variance. The diagnostic value of miR-15b, miR-16, miR-21, and miR-31 was analyzed by receiver operating characteristic curve analysis. The area under the curve (AUC) was used to evaluate the diagnostic value of single and panel detection. After calculating the cutoff value, the sensitivity and specificity were determined. Binary logistic regression analysis was used to evaluate the serum exo
There were 123 newly diagnosed patients with CRC, including 59 men and 64 women, with an average age of 51.60 ± 11.41 years. A total of 117 patients who were diagnosed with colorectal adenoma by video colonoscopy during the same period were enrolled, including 59 men and 58 women, with an average age of 51.55 ± 10.17 years. A total of 150 healthy controls in our hospital were enrolled as the healthy control group, including 76 men and 74 women, with an average age of 52.30 ± 11.25 years. The age and gender were group matched and showed no significant difference. In addition, 81 newly diagnosed patients with CRC, 67 patients who were diagnosed with colorectal adenoma, and 90 healthy controls were enrolled for the validation of the model.
The relative expression levels of miR-15b, miR-16, miR-21, and miR-31 in 123 newly diagnosed patients with CRC, 117 patients who were diagnosed with colorectal ade
| RE | HC, n = 123 | CA, n = 117 | CC, n = 150 |
| miR-15b | 1.21 (0.96, 1.49) | 2.45 (1.84, 3.03) | 3.94 (3.47, 4.19) |
| miR-16 | 0.79 (0.66, 0.94) | 1.17 (1.03, 1.35) | 0.83 (0.72, 1.01) |
| miR-21 | 1.27 (1.12, 1.38) | 2.14 (1.40, 2.24) | 3.59 (1. 25, 4.02) |
| miR-31 | 1.22 (1.08, 1.48) | 1.38 (0.91, 2.26) | 2.16 (0.98, 2.23) |
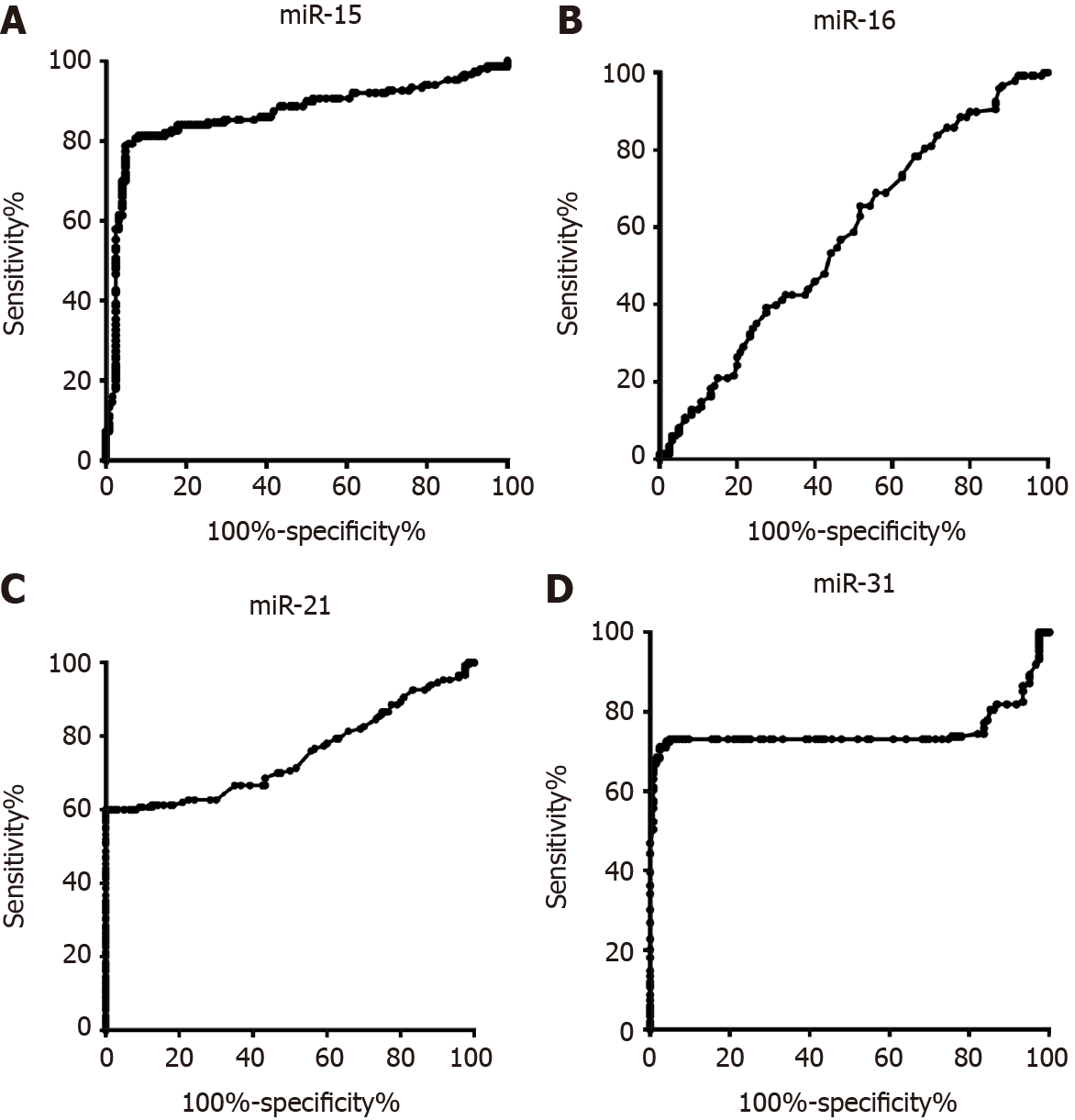
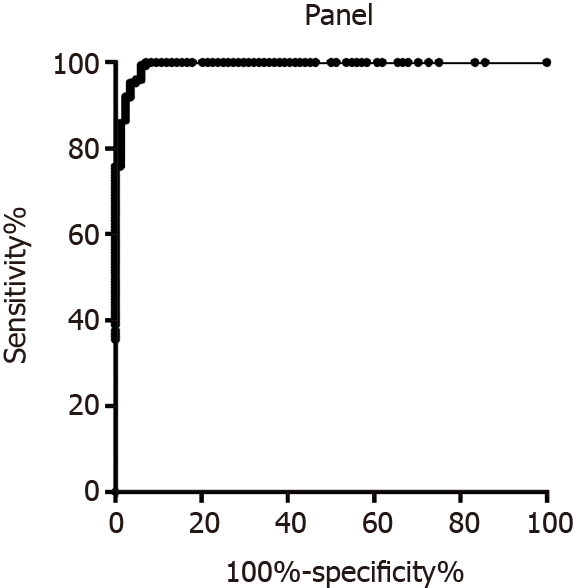
To discriminate the healthy control group and CRC group, the diagnostic value of miR-15b, miR-16, miR-21, and miR-31 was evaluated. As shown in Figure 1A, the AUC of miR-15b was 0.86 (0.82, 0.91). The AUCs of miR-16, miR-21, and miR-31 were 0.58 (0.51, 0.65), 0.75 (0.69, 0.81), and 0.75 (0.68, 0.82), respectively, as shown in Figure 1B, 1C, and 1D. When each miRNA was used alone for evaluation, the best indicator among the four miRNAs was miR-15b, and the sensitivity and specificity were 81.33% and 91.80%, respectively. After evaluating the diagnostic value of a single indicator, binary logistic regression analysis was used to evaluate the combined miR-15b, miR-21, and miR-31 panel, and the sensitivity and specificity were 91.95% and 97.62%, respectively, as shown in Figure 2.
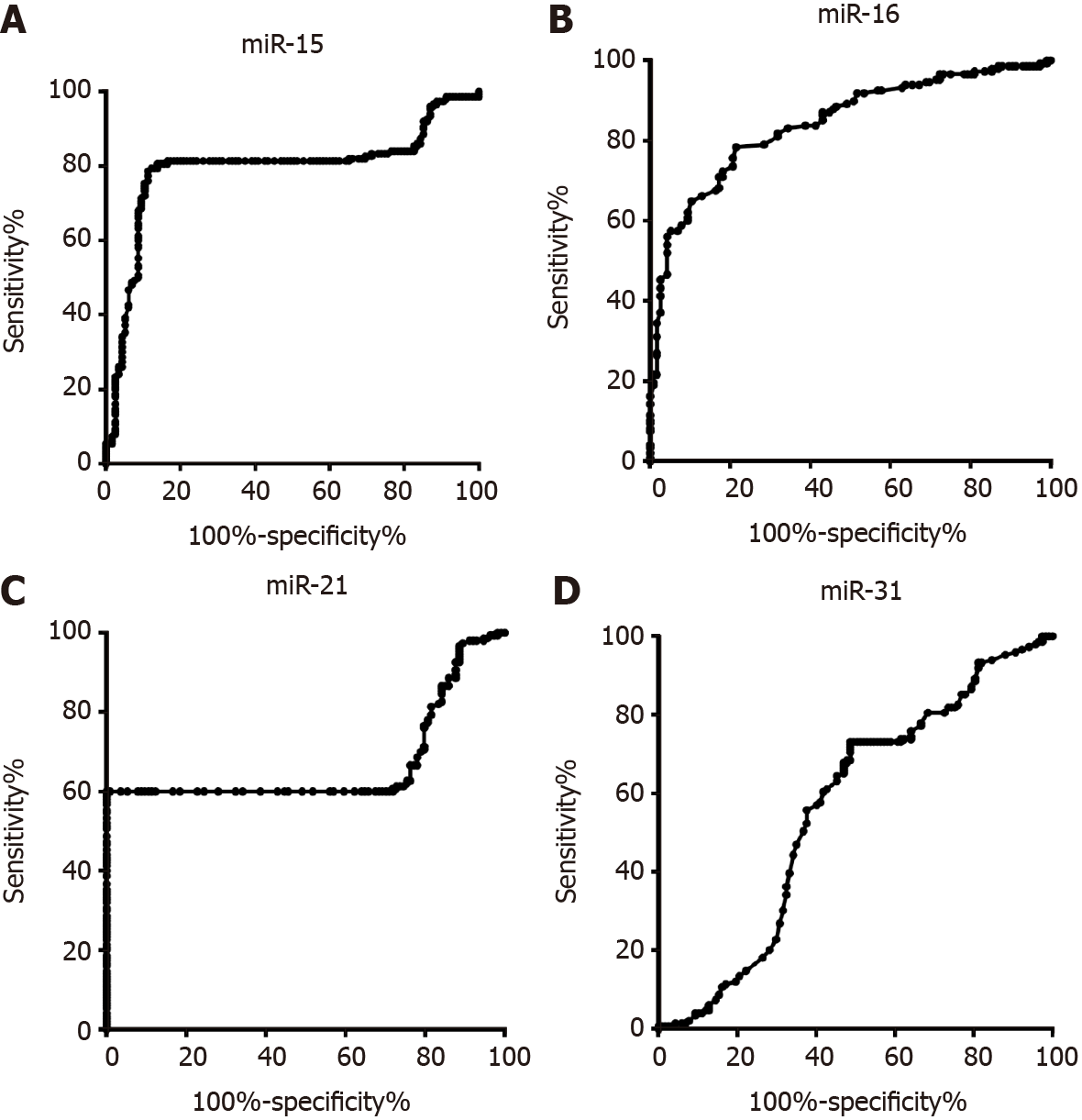
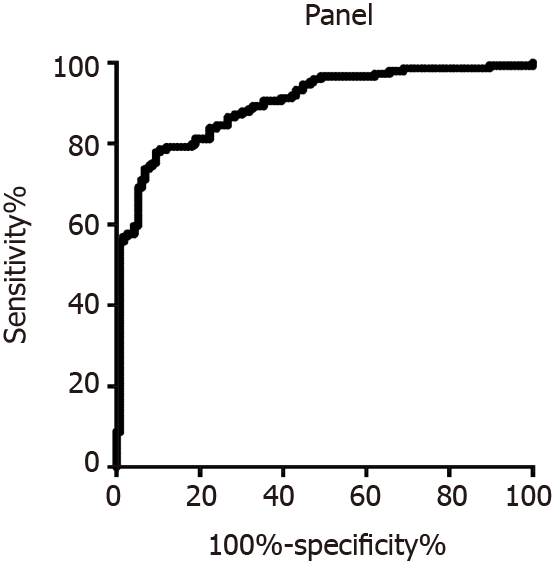
To discriminate the colorectal adenoma group and CRC group, the diagnostic value of miR-15b, miR-16, miR-21, and miR-31 was evaluated. As shown in Figure 3A, the AUC of miR-15b was 0.79 (0.73, 0.85). The AUCs of miR-16, miR-21, and miR-31 were 0.84 (0.80, 0.88), 0.66 (0.59, 0.73), and 0.55 (0.48, 0.63), respectively, as shown in Figure 3B, 3C, and 3D. When the miRNA was used alone for evaluation, the best indicator among the four miRNAs was miR-16, and the sensitivity and specificity were 79.05% and 71.55%, respectively. For the panel that included miR-15b, miR-16, and miR-21 together, the sensitivity and specificity were 81.21% and 81.03%, respectively, as shown in Figure 4.
After building the diagnostic panel model for discriminating the healthy control group and CRC group and the colorectal adenoma group and CRC group, 81 newly diag
Circulating miRNAs are an important part of “liquid biopsy”, including free and vesicle miRNAs. Studies have confirmed that free miRNAs account for only a small proportion of miRNAs and are unstable, and the miRNAs wrapped in vesicles (es
Many studies have shown miRNAs are involved in tumor cell proliferation, apop
Detecting the type and content of miRNAs in tumor-derived exosomes is of great significance for clinical diagnosis and treatment. There are many ways to obtain exosomes. They can not only be extracted from the supernatant of cultured cells but can also be easily obtained from body fluids such as blood, urine, pleural and ascites, cerebrospinal fluid, saliva, and milk[14]. Compared with traditional tumor markers, exosome-derived miRNAs have high sensitivity and specificity[12]. The formation process of exosomes and the characteristics of their shedding in body fluids are being studied for the development of clinical disease monitoring methods[11].
CRC is one of the most common malignant tumors. The incidence and mortality due to CRC in China rank third and fifth among cancers, respectively, and the overall trend is increasing because of a lack of sensitivity and specificity of the methods used for its early diagnosis.
A study analyzed 746 CRC patients and 476 healthy people, and the sensitivity and specificity of miR-21 for diagnosing CRC were 72% and 85%, respectively. It has good potential as a diagnostic marker in CRC. A miRNA panel, as a combination of different miRNAs, can improve its detection efficiency. A meta-analysis included 24 studies, including 1558 CRC patients and 1085 healthy individuals, and the diagnostic sen
Serum exosomes have been used not only for cancer detection but also for detection of recurrences and drug therapy effect evaluation. MiR-29c is expected to be a risk monitoring indicator for early recurrence, and miR-203 and miR-19a are also closely related to the poor prognosis of metastatic CRC. Studies have found that circulating miR-106a, miR-484, miR-130, miR-27b, miR-148a, miR-326, and other molecules may be used as predictors of response to oxaliplatin chemotherapy regimens.
miR-15b belongs to the miR-15 family and is located in the 3q25.33 chromosomal region. Studies have shown that miR-15b is abnormally expressed in many tumors. MiR-15b is upregulated in tongue cancer, cervical cancer, and pancreatic cancer cells and acts as an oncogene. The abnormal expression of miR-15b could be used in the diagnosis of various tumors and can also be used as an important biomarker for recurrence detection[15-17]. A study demonstrated that miR-15b expression in liver cancer cells was significantly increased.
miR-15b may bind to MTSS1 to regulate the cell cycle. MTSS1 is a multifunctional protein that plays an important role in tumorigenesis and development: Downregulation of MTSS1 can be used as a marker of stage progression in early adjuvant treatment of lung squamous cell carcinoma. MTTS1 can also be used as a diagnostic marker or therapeutic target for cervical cancer and as a prognostic marker of glioma cancer and pancreatic adenocarcinoma. MiR-15b promotes the proliferation of CRC cells by targeting the downregulation of MTSS1 expression, and inhibiting miR-15b can upregulate the expression of MTSS1, thereby inhibiting the proliferation of cancer cells[16]. In our study, miR-15b was also demonstrated to be a potential biomarker for CRC diagnosis.
miRNAs are located in tumor suppressor genes on the chromosome, and there is a reduction or absence of genes and expression in most patients with chronic lympho
There are still some limitations of our study. First, although we identified serum exosomes, the relatively small sample size limited the study’s application value. Second, the diagnostic value of early and advanced CRC was not evaluated in this study; in future studies, we will evaluate their diagnostic value. Third, although we validated the diagnostic models that were built in our study, this study was performed only in our center, and a multicenter study with a larger sample size should be included in future studies.
In conclusion, we evaluated four serum exosome miRNAs: MiR-15b, miR-16, miR-21, and miR-31. By binary logistic regression analysis, we built and validated a diagnostic model that contained a panel of miR-15b, miR-21 and miR-31 to discriminate the healthy control group and CRC group, and its sensitivity and specificity were 95.06% and 94.44%, respectively. The miR-15b, miR-16, and miR-21 panel could discriminate the colorectal adenoma group and CRC group with a sensitivity and specificity of 85.19% and 82.09%, respectively.
Exosomes, as tumor molecular markers, have the advantages of protecting the nucleic acids in them and having potential diagnostic value for kinds of diseases. Diagnostic indicator panel may improve the diagnostic value when compared to the single indi
The present study evaluated the diagnostic value of colorectal cancer (CRC) by de
This study aimed to evaluate the potential biomarker-exome miRNA in serum for the diagnosis of CRC.
Relative expression of miR-15b, miR-16, miR-21, and miR-31 in CRC, colorectal ade
A diagnostic model including miR-15b, miR-21, and miR-31 panel for discriminating healthy control group and CRC group was built; the sensitivity and specificity were 95.06% and 94.44%. The sensitivity and specificity of miR-15b, miR-16, and miR-21 panel for discriminating colorectal adenoma group and CRC group were 85.19% and 82.09%, respectively.
The diagnostic value of the exosome four miRNAs (miR-15b, miR-16, miR-21, miR-31) were evaluated and were demonstrated to have potential diagnostic value in serum for CRC.
Although we confirmed the diagnostic value of serum exosome miRNAs for CRC, there remains research to be performed, mostly multi-center studies with a larger sample size.
| 1. | Weitz J, Koch M, Debus J, Höhler T, Galle PR, Büchler MW. Colorectal cancer. Lancet. 2005;365:153-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 862] [Cited by in RCA: 956] [Article Influence: 45.5] [Reference Citation Analysis (0)] |
| 2. | Roncucci L, Mariani F. Prevention of colorectal cancer: How many tools do we have in our basket? Eur J Intern Med. 2015;26:752-756. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 81] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 3. | Heinimann K. [Hereditary Colorectal Cancer: Clinics, Diagnostics and Management]. Ther Umsch. 2018;75:601-606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 4. | Chen M, Zhao H. Next-generation sequencing in liquid biopsy: cancer screening and early detection. Hum Genomics. 2019;13:34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 162] [Cited by in RCA: 364] [Article Influence: 52.0] [Reference Citation Analysis (0)] |
| 5. | Poulet G, Massias J, Taly V. Liquid Biopsy: General Concepts. Acta Cytol. 2019;63:449-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 237] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 6. | Alix-Panabières C. EPISPOT assay: detection of viable DTCs/CTCs in solid tumor patients. Recent Results Cancer Res. 2012;195:69-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 122] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 7. | Pantel K, Alix-Panabières C. Liquid biopsy and minimal residual disease - latest advances and implications for cure. Nat Rev Clin Oncol. 2019;16:409-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 440] [Cited by in RCA: 768] [Article Influence: 109.7] [Reference Citation Analysis (0)] |
| 8. | Pessoa LS, Heringer M, Ferrer VP. ctDNA as a cancer biomarker: A broad overview. Crit Rev Oncol Hematol. 2020;155:103109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 200] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 9. | Gilson P. Enrichment and Analysis of ctDNA. Recent Results Cancer Res. 2020;215:181-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 10. | Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem. 2019;88:487-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 815] [Cited by in RCA: 1990] [Article Influence: 331.7] [Reference Citation Analysis (0)] |
| 11. | Zhang L, Yu D. Exosomes in cancer development, metastasis, and immunity. Biochim Biophys Acta Rev Cancer. 2019;1871:455-468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 737] [Article Influence: 105.3] [Reference Citation Analysis (0)] |
| 12. | Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13:17-24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1443] [Cited by in RCA: 1645] [Article Influence: 149.5] [Reference Citation Analysis (1)] |
| 13. | Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287-314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1357] [Cited by in RCA: 1404] [Article Influence: 108.0] [Reference Citation Analysis (0)] |
| 14. | Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2492] [Cited by in RCA: 3638] [Article Influence: 404.2] [Reference Citation Analysis (0)] |
| 15. | Frampton AE, Krell J, Gall TM, Castellano L, Stebbing J, Jiao LR. miR-15b and miR-17 Are Tumor-derived Plasma MicroRNAs Dysregulated in Colorectal Neoplasia. Ann Surg. 2015;262:e61-e62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 16. | Li J, Chen Y, Guo X, Zhou L, Jia Z, Tang Y, Lin L, Liu W, Ren C. Inhibition of miR-15b decreases cell migration and metastasis in colorectal cancer. Tumour Biol. 2016;37:8765-8773. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 17. | Liu J, Xu H, Wang N, Sun M. miR-15b, a diagnostic biomarker and therapeutic target, inhibits oesophageal cancer progression by regulating the PI3K/AKT signalling pathway. Exp Ther Med. 2020;20:222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 18. | Masuda S, Izpisua Belmonte JC. Re: Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2014;106:djt457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 19. | Okugawa Y, Yao L, Toiyama Y, Yamamoto A, Shigemori T, Yin C, Omura Y, Ide S, Kitajima T, Shimura T, Fujikawa H, Yasuda H, Hiro J, Yoshiyama S, Kobayashi M, Tanaka K, Inoue Y, Araki T, Miki C, Kusunoki M. Prognostic impact of sarcopenia and its correlation with circulating miR-21 in colorectal cancer patients. Oncol Rep. 2018;39:1555-1564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 20. | Sabry D, El-Deek SEM, Maher M, El-Baz MAH, El-Bader HM, Amer E, Hassan EA, Fathy W, El-Deek HEM. Role of miRNA-210, miRNA-21 and miRNA-126 as diagnostic biomarkers in colorectal carcinoma: impact of HIF-1α-VEGF signaling pathway. Mol Cell Biochem. 2019;454:177-189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 109] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 21. | Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013;105:849-859. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 407] [Cited by in RCA: 413] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 22. | Körner C, Keklikoglou I, Bender C, Wörner A, Münstermann E, Wiemann S. MicroRNA-31 sensitizes human breast cells to apoptosis by direct targeting of protein kinase C epsilon (PKCepsilon). J Biol Chem. 2013;288:8750-8761. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 120] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 23. | Mitamura T, Watari H, Wang L, Kanno H, Hassan MK, Miyazaki M, Katoh Y, Kimura T, Tanino M, Nishihara H, Tanaka S, Sakuragi N. Downregulation of miRNA-31 induces taxane resistance in ovarian cancer cells through increase of receptor tyrosine kinase MET. Oncogenesis. 2013;2:e40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 98] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 24. | Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, Nenutil R, Vyzula R. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397-402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 475] [Cited by in RCA: 543] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 25. | Wang CJ, Zhou ZG, Wang L, Yang L, Zhou B, Gu J, Chen HY, Sun XF. Clinicopathological significance of microRNA-31, -143 and -145 expression in colorectal cancer. Dis Markers. 2009;26:27-34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 114] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/Licenses/by-nc/4.0/
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Gerken M, Nguyen Canh B, Shinozaki E S-Editor: Wang JL L-Editor: Filipodia P-Editor: Yuan YY