Published online Jun 15, 2019. doi: 10.4251/wjgo.v11.i6.459
Peer-review started: February 12, 2019
First decision: March 15, 2019
Revised: April 6, 2019
Accepted: April 19, 2019
Article in press: April 20, 2019
Published online: June 15, 2019
Processing time: 124 Days and 3.3 Hours
Circular RNA (circRNA) is a newly discovered non-coding RNA with special structure, which is widely expressed in eukaryotic organisms and mainly located in the cytoplasm. circRNAs participate in gene regulation by working as miRNA sponges that block the inhibitory effect of miRNA on its target genes. In addition, circRNAs can bind to RNA binding proteins to regulate gene expression. Based on characteristics of stability, expression specificity and participation in gene regulation, circRNAs are expected to be biomarkers for early diagnosis of cancer or potential targets for cancer therapy. With the help of bioinformatics analysis, circRNA microarray analysis and high-throughput sequencing technology, more circRNAs were discovered to participate in the progression of gastric cancer (GC) over the past three years. This article gives an overview of these recent research focusing on the roles of circRNAs in GC and highlights the advances.
Core tip: Gastric cancer (GC) is a common, worldwide malignant tumor with a poor prognosis. An increasing number of circRNAs was discovered to participate in the progression of GC. Therefore, exploring the function of circRNAs will help to achieve a better understanding of the pathogenesis of GC and identify new diagnostic biomarkers and therapeutic targets. This article gives an overview of the recent research focusing on the roles of circRNAs in GC and highlights the advances that were made over the past three years.
- Citation: Wang KW, Dong M. Role of circular RNAs in gastric cancer: Recent advances and prospects. World J Gastrointest Oncol 2019; 11(6): 459-469
- URL: https://www.wjgnet.com/1948-5204/full/v11/i6/459.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v11.i6.459
Gastric cancer (GC) is one of the most common human cancers. The number of new cases in 2018 was 1033701, which accounted for 5.7% of all new cancers, and the number of deaths from GC was 782685, which accounted for 8.2% of all cancer deaths; only behind lung cancer[1]. Although diagnostic and therapeutic techniques have been developing rapidly, the prognosis of GC remains poor[2]. The poor prognosis is partly due to an incomplete understanding of the molecular mechanisms of GC occurrence and development. Thus, it is critical to identify some new biomarkers and therapeutic targets to improve the diagnosis and treatment of GC.
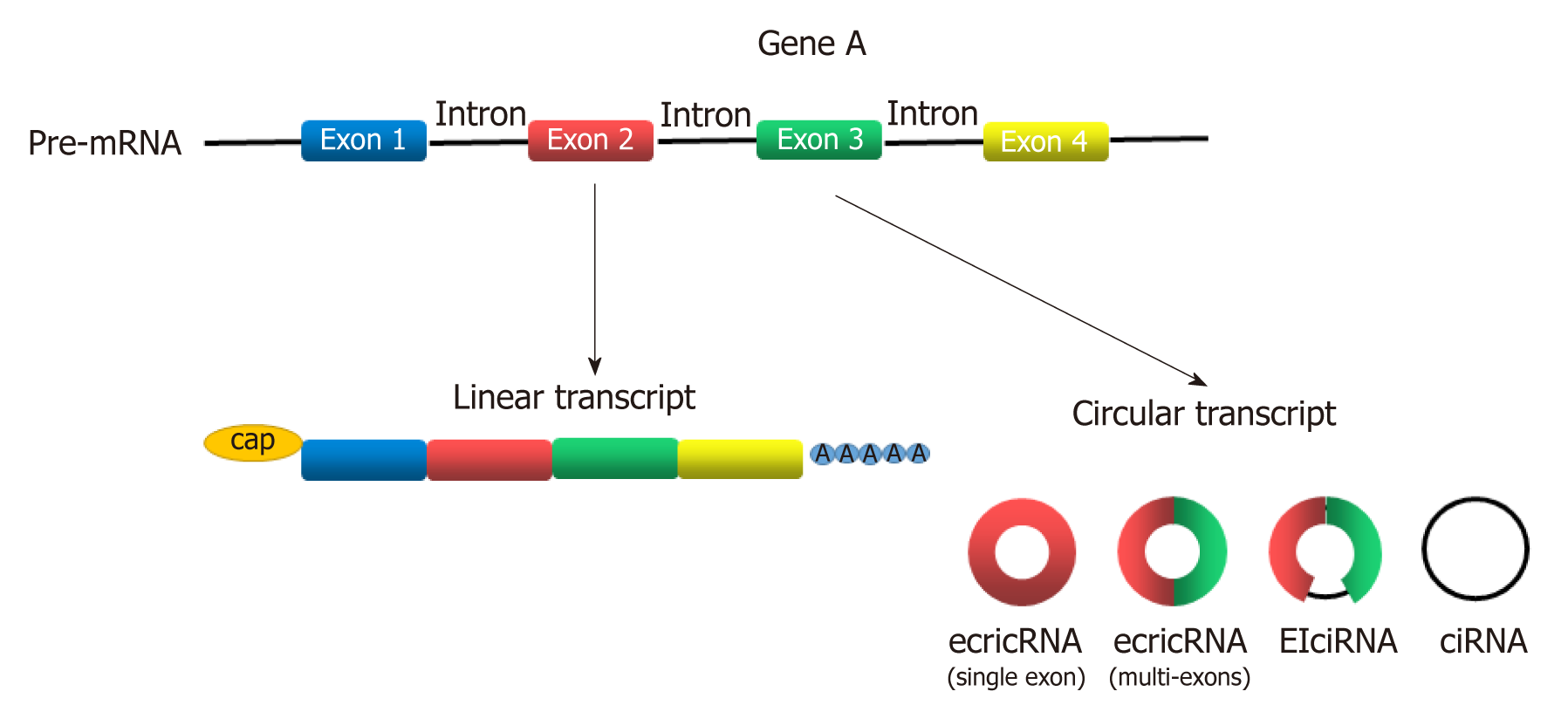
Circular RNAs (circRNAs) are newly discovered endogenous non-coding RNAs (ncRNAs) that form covalently closed continuous loops with neither 5’ to 3’ polarity nor a polyadenylated tail[3]. They are generated from backsplicing of exons, introns, or both, and called exonic circRNAs, intronic RNAs and exon-intron circRNAs according to their components (Figure 1). Although circRNAs have been investigated for almost 40 years[4], significant attention has not been received until recent years[5]. circRNAs were found to participate in gene regulation by working as miRNA sponges that block the inhibitory effect of miRNA on its target genes[6], splicing of target genes[7] or interacting with RNA-binding proteins (RBPs)[8]. In addition, some circRNAs can even encode peptides[9,10]. circRNAs have been widely investigated in recent years because they play many important roles in proliferation, apoptosis and metastasis of cancer cells[11-15]. With the help of bioinformatics analysis, circRNA microarray analysis and high-throughput sequencing technology, more circRNAs was discovered to participate in the progression of GC[16-21]. Gu et al[20] conducted a circRNA microarray analysis to explore the difference in circRNAs expression between tumor and adjacent nontumorous tissues from six patients with GC. They showed that 440 circRNAs were expressed differently in tumor samples, including 176 upregulated and 264 downregulated circRNAs[20]. Exploration of GC-related circRNAs may provide a new insight into the diagnosis and treatment of GC. In addition, circRNAs are hardly degraded by RNA exonuclease or ribonuclease R[22], making them more stable in tissue or plasma. This feature makes circRNAs a potential use for biomarkers, prognosis predictors, and even therapeutic targets of GC.
We searched MEDLINE and PubMed before January 2019 using the following keywords: circular RNA, circRNA and gastric cancer. The inclusion criteria were as follows: (1) Studies associating circRNAs with GC samples or cancer cells, and discussing their potential use as biomarkers for diagnosis of the disease; (2) Studies associating circRNAs with biological functions or potential pathways in GC; and (3) Studies associating circRNAs with clinical significance of GC. This review highlights recent advances in circRNA in GC, especially focusing on their deregulation, biological function and clinical significance.
As mentioned before, circRNAs primarily act as miRNA sponges to regulate gene expression. Most circRNAs contain a miRNA response element, which can bind with miRNAs and regulate their expression. At present, there are 13 circRNAs that act as miRNA regulators in GC. Some of these are downregulated and serve as tumor suppressors, while others are upregulated during carcinogenesis and serve as oncogenes. All these circRNAs are summarized in Table 1.
| circRNA | Deregulation | Function/clinical association | Gene/pathway affected | Ref. |
| hsa_circ_0000993 | Downregulation | Inhibits proliferation, migration and invasion | miR-214-5p | [23] |
| has_circ_0001461 | Downregulation | Inhibits proliferation, migration and invasion; correlates with the clinical stage | miR-548g, RUNX1 in the cytoplasm; YBX1 in the nucleus | [24] |
| has_circ_0002320 | Downregulation | Inhibits proliferation and invasion; correlates with TMN stage and survival time | miR-367-5p, p27 | [25] |
| hsa_circ_0027599 | Downregulation | Inhibits proliferation and migration; correlates with TNM stage | miR-101, PHLDA1 | [26] |
| circRNA_100269 | Downregulation | Inhibits proliferation; correlates with histological subtype,node invasive number and overall survival time | miR-630 | [30] |
| circRNA_101057 | Downregulation | Inhibits proliferation and invasion; correlates with tumor size and lymphatic metastasis and overall survival time | miR-424, LATS1 | [31] |
| circZFR | Downregulation | Inhibits proliferation and promotes apoptosis | miR-130a/miR-107, PTEN | [35] |
| hsa_circ_0017639 | Upregulation | Promotes proliferation; correlates with TNM stage | miR-182-5p, CREB1 | [38] |
| circRNA_0000284 | Upregulation | Promotes proliferation; correlates with T stage | miR-124 and miR-29b, COL1A1, COL4A1 and CDK6 | [42] |
| circRNA_001569 | Upregulation | Promotes proliferation and inhibits apoptosis; correlates with tumor size, depth of invasion and clinical stage | miR-145, NR4A2 | [44] |
| circPDSS1 | Upregulation | Promotes proliferation and inhibits apoptosis;correlates with worse overall survival time | miR-186-5p, NEK2 | [45] |
| circNF1 | Upregulation | Promotes proliferation | miR-16, MAP7 and AKT3 | [49] |
| ciRS-7 | Upregulation | Promotes proliferation and inhibits apoptosis; correlates with TNM stage and poor overall survival time | miR-7, PTEN/PI3K/AKT pathway | [53] |
hsa_circ_0000993 is downregulated in GC. It can act as a miRNA sponge for miR-214-5p and inhibit the proliferation, invasion and migration of GC cells[23]. Has_circ_000146, a sponge for miR-548g, is significantly downregulated in GC cell lines and tissues, and negatively correlated with survival time of GC patients. Overexpression of has_circ_0001461 can inhibit proliferation, migration and invasion of GC cells. These effects can be reversed by overexpression of miR-548g, which can downregulate expression of runt-related transcription factor 1 (RUNX1). These results suggest that has_circ_0001461 acts as a tumor suppressor in GC cells by regulating the miR-548g/RUNX1 pathway[24]. Liu et al[25] reported that has_circ_0002320 level was significantly lower in GC tissues than in paired adjacent nontumorous tissues, and the survival time was shorter in GC patients with lower has_circ_0002320 level. By using fluorescence in-situ hybridization (FISH) in GC tissues, they found that has_circ_0002320 and miR-367-5p were colocalized in the cytoplasm. Overexpression of has_circ_0002320 upregulated expression of p27 Kip1 in GC cells and inhibited their growth and invasion, and these effects could be reversed by miR-367-5p mimics. These results demonstrate that has_circ_0002320 is a tumor suppressor in GC cells by targeting the miR-367-5p/p27 Kip1 pathway and provides a prediction of survival time in GC patients[25]. Hsa_circ_0027599 was significantly downregulated in GC patients and cells, and its overexpression inhibited proliferation and metastasis of GC cells. Moreover, hsa_circ_0027599 was verified to be a sponge of miR-101-3p.1 (miR-101) by bioinformatic technology and luciferase reporter assays. miR-101 can inhibit the expression of its target gene PHLDA1 and promote proliferation of cancer cells. Conversely, overexpression of PHLDA1 decreases the growth and migration of MKN-28 and HGC-27 GC cells. These results suggest that PHLDA1 is regulated by circ_0027599/miR-101, which inhibits the growth and metastasis of GC cells[26]. Another study, which had different conclusions from the above, has shown that miR-101-3p is a tumor suppressor and overexpression of miR-101-3p inhibits proliferation and invasion of AGS GC cells[27]. Therefore, the functions of miR-101 needs more investigation. miR-630 is one of the newly discovered miRNAs, and its role in cancer has attracted increased attention. miR-630 is dysregulated in many tumors[28,29]. Direct interaction of miR-630 and circRNA_100269 was confirmed by dual-luciferase reporter assays. The level of miR-630 decreased significantly by circRNA_100269 overexpression, which inhibited proliferation of GC cells. These results suggest that the circRNA_100269/miR-630 axis plays an important role in the growth of GC cells[30]. A novel circRNA circ_101057, also termed as circLARP4, was shown downregulated in GC tissues by FISH analysis, and lower circLARP4 expression was associated with poor prognosis. Furthermore, circLARP4 inhibited biological behavior of GC cells[31]. These effects have also been seen in ovarian cancer[32]. circLARP4 was found to sponge miR-424-5p by bioinformatics analysis. miR-424-5p promotes proliferation and invasion of GC cells by targeting LATS1 gene, and positively correlates with higher clinical stage and worse prognosis of GC patients[31]. However, the function of miR-424-5p is the opposite in breast cancer and esophageal squamous cell carcinoma. Wang et al[33] have reported that miR-424-5p acts as a tumor suppressor to regulate proliferation, invasion and migration of breast cancer cells by binding to the functional target Doublecortin Like Kinase 1[33]. Upregulation of miR-424-5p may prevent tumor invasion or metastasis[34]. circ-ZFR is a new circRNA that is markedly downregulated in tumor tissues compared with pair-matched adjacent nontumorous tissues. Moreover, expression of circ-ZFR is significantly lower in GC cell lines HGC-27, AZ521, and AGS than in gastric epithelial cell line GES1. circ-ZFR promotes cell cycle arrest and apoptosis in GC cells by sponging miR-107/miR-130a, and miR-107/miR-130a could bind to the 3’ untranslated region (UTR) of phosphatase and tensin homolog (PTEN)[35]. Many studies have demonstrated that PTEN could be targeted and regulated by miR-107 and miR-130a to influence activities of cancer cells[36,37]. All these results suggest that the circ-ZFR-miR-107/miR-130a-PTEN pathway plays an important role in the progression of GC.
One circRNA hsa_circ_0017639 that is derived from gene SFMBT2, also named circ-SFMBT2, shows higher expression level in GC tissues compared with adjacent nontumorous tissues, and is linked to higher tumor stages. The proliferation of GC cells is significantly suppressed when circ-SFMBT2 is knocked down. Luciferase reporter assay revealed that miR-182-5p mimics induced a lower luciferase level in circ-SFMBT2 WT group than in the normal control group. Furthermore, it has been demonstrated that circ-SFMBT2 acts as a sponge of miR-182-5p to regulate expression of cAMP response element binding protein (CREB)1 and promotes proliferation of GC cells[38]. circHIPK3 (circRNA_0000284) that is derived from the homeodomain-interacting protein kinase-3 (HIPK3) gene sponges multiple miRNAs and serves as an oncogene in multiple cancers[39-41]. In GC tissues, circHIPK3 level is significantly higher than it in paired adjacent nontumorous tissues. Moreover, it negatively regulates expression of miR-29b/miR-124 and is associated with T stage of GC. Three candidate genes (CDK6, COL1A1 andCOL4A1 ) can be regulated by miR-29b and miR-124, suggesting that these genes may play important roles in GC though circHIPK3-miR-29b/miR-124 axes[42]. circRNA_001569 was firstly discovered to act as a positive regulator in cell proliferation and invasion of colorectal cancer[43]. Recently, it was found upregulated in tissues and cells of GC. circRNA_001569 overexpression significantly decreases expression of miR-145, while circRNA_001569 knockdown has the opposite effect. Moreover, circRNA_001569 knockdown decreases cell viability dramatically and promotes apoptosis, but these effects of circRNA_001569 knockdown are reversed when cells are cotransfected with miR-145 inhibitor. The online microRNA.org predicted that miR-145 could bind with NR4A2 3’ UTR. miR-145 overexpression significantly decreased NR4A2 expression and cell viability, and promoted apoptosis. However, cotransfection with NR4A2 abolished the above effects[44]. All these results indicate that circRNA_001569 serves as an oncogene by regulating expression of the miR-145/NR4A2 axis. circPDSS1 was recently discovered to be highly expressed in GC tissue and cell lines. Patients with higher circPDSS1 expression have worse overall survival. CircPDSS1 knockdown significantly inhibits cell proliferation[45]. The expression of miR-186-5p, a tumor suppressor gene[46], is decreased by circPDSS1 overexpression. In luciferase reporter assays, luciferase activity was decreased by cotransfection of miR-186-5p mimics and wt-NEK2. This suggests that NEK2, an oncogene[47,48], is a target of miR-186-5p. Moreover, miR-186-5p inhibits NEK2 expression, while miR-186-5p inhibitor reverses this effect[45]. In summary, circPDSS1/miR-186-5p/NEK2 pathway may play an important role in GC cancer progression, and may be a target for gene therapy. circNF1 is upregulated in GC tissues and cell lines. Functional studies have demonstrated that circNF1 serves as an oncogene and significantly promotes cell proliferation. Furthermore, luciferase reporter assays have shown that circNF1 acts as a sponge to miR-16, thereby affecting its downstream target mRNAs, AKT3 and MAP7[49]. ciRS-7 is a well-known circRNA due to its promotion of carcinogenesis in a variety of cancers[50-52]. In GC, the ciRS-7 level is significantly higher than in nontumorous tissues, and higher ciRS-7 is associated with worse survival. miR-7 overexpression increases expression of PTEN, decreases PI3K and Akt phosphorylation, and inhibits tumor growth, while ciR-7 attenuates these effects[53]. These results indicate that ciRS-7 might be a promising therapeutic target through modulation of mir-7/PTEN/PI3K/AKT pathway in GC.
The 5-year survival rate of early GC can exceed 92%[54,55]. However, if GC develops to a late stage, the survival rate is significantly decreased[56]. Therefore, stable and effective diagnostic markers for the early diagnosis of GC need to be identified. Over the past three years, many circRNAs have been found to have specific differences between GC and normal gastric tissue, and these differences have helped circRNAs to become potential markers of early diagnosis or predictors of prognosis[38,57-72]. All these circRNA are summarized in Table 2. We will cover in detail those circRNA with an area under the curve (AUC) > 0.75.
| circRNA | Deregulation | Cut-off value (ΔCt) | AUC | Sensitivity | Specificity | Clinical association | Ref. |
| hsa_circ_00000 96 | Downregulation | 12.9 | 0.82 | - | - | Gender, invasion and TNM stage | [57] |
| hsa_circ_00001 81 | Downregulation | 9.4 | 0.756 | 85.2% | 53.9% | Tumor diameter, lymphatic metastasis, distal metastasis, and CA19-9 (tissue) | [58] |
| 7.27 | 0.582 | 20.6% | 99% | CEA and differentiation (plasma) | |||
| hsa_circ_00001 90 | Downregulation | 6.83 | 0.75 | 72.1% | 68.3% | Tumor diameter, TNM stage and CA19-9 (tissue) | [59] |
| 3.07 | 0.6 | 41.4% | 87.5% | CEA (plasma) | |||
| hsa_circ_00005 20 | Downregulation | - | 0.6129 | 53.57% | 85.71% | TNM stage (tissue) | [60] |
| - | 0.8967 | 82.35% | 84.44% | CEA (plasma) | |||
| hsa_circ_00007 45 | Downregulation | - | 0.683 | 85.5% | 45% | Tumor differentiation (tissue) and TNM stage (plasma) | [61] |
| hsa_circ_00018 95 | Downregulation | 9.53 | 0.792 | 67.8% | 85.7% | Tumor differentiation, Borrmann type, and tissue CEA | [62] |
| hsa_circ_00001649 | Downregulation | 0.227 | 0.834 | 71.1% | 81.6% | Tumor differentiation | [63] |
| hsa_circ_002059 | Downregulation | 12.9 | 0.73 | 81% | 62% | TMN stage, distal metastasis, gender and age | [64] |
| hsa_circ_00031 59 | Downregulation | 12.31 | 0.75 | 85.2% | 56.5% | Gender, distal metastasis, and TMN stage | [65] |
| hsa_circ_00066 33 | Downregulation | 8.17 | 0.741 | 60% | 81% | Distal metastasis and CEA | [66] |
| hsa_circ_00147 17 | Downregulation | 12.14 | 0.696 | 59.38% | 81.25% | Tumor stage; distal metastasis; CEA; CA199 | [67] |
| has_circ_00667 79 | Downregulation | - | 0.6726 | 90.3% | 56.4% | TNM stage overall survival time | [68] |
| hsa_circ_00743 62 | Downregulation | 12.17 | 0.63 | 84.3% | 36.2% | CA19–9 and lymphatic metastasis | [69] |
| hsa_circ_01308 10 | Downregulation | 1.443 | 0.7481 | 77.42% | 68% | TNM stage and overall survival time | [70] |
| hsa_circ_00004 67 | Upregulation | - | 0.79 | 70.5% | 64.8% | TNM stage | [71] |
| hsa_circ_00176 39 | Upregulation | 11.46 | 0.7585 | 80.56% | 63.89% | TNM stage | [38] |
| hsa_circ_00664 44 | Upregulation | - | 0.7328 | 70.75% | 68.87% | Lymphatic metastasis | [72] |
Hsa_circ_0000096 level was found to be lower in GC tissues and cell lines than paired adjacent nontumorous tissues and normal gastric epithelial cells. Furthermore, the cutoff value (ΔCt value) of hsa_circ_0000096 was 12.9 with an AUC of 0.82. Hsa_circ_0000096 was also linked to several clinicopathological features such as invasion and TNM stage[57]. Hsa_circ_0000181 levels in plasma from GC patients and tissues were significantly decreased compared with those from healthy individuals and paired adjacent nontumorous tissues. In addition, its level in plasma of GC patients was associated with differentiation and carcinoembryonic antigen (CEA) level. The AUC of hsa_circ_0000181 in plasma was 0.582 with a specificity of 20.6% and sensitivity of 99.0%. Moreover, hsa_circ_0000181 levels in GC tissues were associated with tumor diameter, lymphatic metastasis, distant metastasis, and carbohydrate antigen (CA)19- 9 level. The AUC of hsa_circ_0000181 in tissues was 0.756 with a specificity of 85.2% and sensitivity of 53.9%[58]. Hsa_circ_0000190 was firstly discovered to be downregulated in plasma and tissues samples from GC patients. Its levels in tissue were significantly associated with TNM stage and CA19-9 level. The AUC of hsa_circ_0000190 in tissue was 0.75. The sensitivity and specificity were 72.1% and 68.3%, respectively[59]. hsa_circ_0000520 expression was significantly downregulated in GC tissue, plasma and cell lines (BGC-823, MKN-45, AGS and MGC-803). In plasma, the AUC was 0.8967, and the sensitivity and specificity were 82.35% and 84.44%, respectively[60]. The limitation of this study was the small number of samples. There were only 56 paired GC tissues, 45 preoperative GC plasma and 17 healthy plasma samples used for analysis, thus indicating the need and necessity to expand the sample size to verify the efficacy of hsa_circ_0000520 as a biomarker for GC. Hsa_circ_0001895 levels were lower in 69.8% of GC tissues than in paired adjacent nontumorous tissues and were also downregulated in five GC cell lines (HGC-27, BGC-823, AGS, SGC7901 and MGC-803). In addition, its level was linked to tissue CEA expression, Borrmann type and cell differentiation. The AUC of hsa_ circ_0001895 in tissue was 0.792. When the optimal cutoff value of hsa_circ_0001895 was set at 9.53, the sensi-tivity and specificity were 67.8% and 85.7%, respectively[62]. Hsa_circ_0001649 is a well-known prognostic biomarker or tumor suppressor in multiple cancers[73-77]. Hsa_circ_0001649 levels in GC tissues were significantly lower than those in paired nontumorous tissues. The AUC was 0.834 with a sensitivity of 71.1% and specificity of 81.6%. Compared with plasma collected preoperatively, has_circ_0001649 level was significantly upregulated in plasma samples collected postoperatively[63]. This suggests that has_circ_0001649 could be used as an index of postoperative follow-up. Whether the nonelevation or redecline of has_circ_0001649 level is related to poor prognosis or recurrence of GC needs further exploration. Compared to paired adjacent nontumorous tissues, hsa_circ_0003159 expression was recently found to be significantly downregulated in GC tissues. Moreover, its levels were negatively related to gender, distant metastasis, and TNM stage. The cutoff value was 12.31 with an AUC of 0.75. The sensitivity and specificity were 85.2% and 56.5%, respectively[65].
Hsa_circ_0000467 levels were significantly higher in GC tissue compared with adjacent nontumorous tissue. Moreover, its level in tissue was positively related to TNM stage. Similar results of hsa_circ_0000467 expression was obtained in AGS, MGC-803, HGC-27 and NUGC-3 compared with GES-1 cell lines. Furthermore, hsa_circ_0000467 knockdown markedly inhibited the proliferation, invasion and migration, and promoted apoptosis of GC cells in vitro. The AUC of hsa_circ_0000467 in plasma was 0.790. Its levels in plasma of the same patient obviously declined after surgery[71]. However, the small number of samples was a limitation in this study. More samples are needed to increase the accuracy. At present, none of the AUCs that were obtained using a single circRNA as a diagnostic marker for GC was > 0.9. Therefore, some scholars have suggested that the combined application of > 2 circRNAs may help to improve the accuracy of early diagnostic markers for GC. Li et al[78] reported that hsa_circ_0061276 and hsa_circ_0001017 were both downregulated in GC plasma and tissues. Patients with low plasma hsa_circ_0061276 or hsa_circ_0001017 levels had worse overall survival than those with high levels. The AUC of hsa_circ_0061276 and hsa_circ_0001017 in plasma was 85.1% and 84.9%, respectively. When these two plasma biomarkers of GC were used together for analysis, the AUC increased to 0.912, with a sensitivity of 84.7% and specificity of 96.6%[78]. Another similar study also found that the AUC was increased to 0.91 with the combination of hsa_circ_002509 and hsa_circ_0000096[57]. These are the top two highest AUC in all current research.
Studies of circRNAs in GC are just at the beginning compared with coding RNAs, miRNAs and long ncRNAs. Although more functional circRNAs have been discovered and characterized in GC, most of the studies have focused on their relationship with pathological characteristics. For most of these circRNAs, their biogenesis, cellular location, and mechanism of regulation still need to be explored. In recent years, exosomes have been identified to play an important role in the progression of cancer[79]. One recent study showed that ciRS-133 was delivered into preadipocytes by exosomes derived from GC cells, promoting the transformation of preadipocytes into brown-like cells by suppressing miR-133 and activating PRDM16. Additionally, silence of ciRS-133 expression can reduce cachexia in tumor-implanted mice[80]. Therefore, exosome-delivered circRNAs are involved in white adipose tissue browning and play an important role in cancer-related cachexia. In the future, more in-depth studies about the roles of exosome-delivered circRNAs will help to prevent the occurrence of cachexia, improve the prognosis of GC, and prolong the survival time of patients. Some scholars have reported that circRNAs may participate in the process of epithelial–mesenchymal transition (EMT)[81,82], which plays a critical role in cancer metastasis[83]. Further studies on the regulation effect of circRNAs on EMT will be helpful to reveal the mechanism of circRNAs in cancer metastasis. Moreover, how to transfer circRNAs or si-circRNAs efficiently to the accurate lesion site without side effects needs to be resolved urgently for clinical applications. We hope that more basic research about circRNAs will be carried out with the advances in molecular biology and biological informatics technology to reveal the pathological and physiological functions of circRNAs, and to develop circRNA-based therapeutic strategies that can safely and successfully integrate into clinical practice.
Many circRNAs are dysregulated in GC tissues, plasma and cell lines. Moreover, their dysregulation is associated with clinicopathological features and prognosis of GC. By working as miRNA sponges or interacting with RBPs, these circRNAs regulate the expression of miRNAs and target proteins that are associated with cell proliferation, apoptosis, invasion and metastasis. Based on their characteristics of stability, expression specificity and participation in gene regulation, circRNAs are expected to be potential biomarkers for early diagnosis, prognostic predictors, and therapeutic targets of GC.
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 56675] [Article Influence: 7084.4] [Reference Citation Analysis (135)] |
| 2. | Herrero R, Park JY, Forman D. The fight against gastric cancer - the IARC Working Group report. Best Pract Res Clin Gastroenterol. 2014;28:1107-1114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 137] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 3. | Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1045] [Cited by in RCA: 1554] [Article Influence: 155.4] [Reference Citation Analysis (0)] |
| 4. | Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339-340. [PubMed] |
| 5. | Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6253] [Cited by in RCA: 6229] [Article Influence: 479.2] [Reference Citation Analysis (0)] |
| 6. | Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4631] [Cited by in RCA: 6225] [Article Influence: 478.8] [Reference Citation Analysis (0)] |
| 7. | Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1766] [Cited by in RCA: 2435] [Article Influence: 202.9] [Reference Citation Analysis (0)] |
| 8. | Zang J, Lu D, Xu A. The interaction of circRNAs and RNA binding proteins: An important part of circRNA maintenance and function. J Neurosci Res. 2018;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 466] [Article Influence: 58.3] [Reference Citation Analysis (0)] |
| 9. | Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E, Shenzis S, Samson M, Dittmar G, Landthaler M, Chekulaeva M, Rajewsky N, Kadener S. Translation of CircRNAs. Mol Cell. 2017;66:9-21.e7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 988] [Cited by in RCA: 1408] [Article Influence: 156.4] [Reference Citation Analysis (0)] |
| 10. | Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, Laneve P, Rajewsky N, Bozzoni I. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell. 2017;66:22-37.e9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1190] [Cited by in RCA: 1693] [Article Influence: 188.1] [Reference Citation Analysis (1)] |
| 11. | Xu H, Wang C, Song H, Xu Y, Ji G. RNA-Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers. Mol Cancer. 2019;18:8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 151] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 12. | Wang H, Xiao Y, Wu L, Ma D. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-000911/miR-449a pathway in breast carcinogenesis. Int J Oncol. 2018;52:743-754. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 93] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 13. | Tian X, Zhang L, Jiao Y, Chen J, Shan Y, Yang W. CircABCB10 promotes nonsmall cell lung cancer cell proliferation and migration by regulating the miR-1252/FOXR2 axis. J Cell Biochem. 2019;120:3765-3772. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 67] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 14. | Chen Y, Yang F, Fang E, Xiao W, Mei H, Li H, Li D, Song H, Wang J, Hong M, Wang X, Huang K, Zheng L, Tong Q. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2018;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 239] [Article Influence: 29.9] [Reference Citation Analysis (0)] |
| 15. | Lai Z, Yang Y, Yan Y, Li T, Li Y, Wang Z, Shen Z, Ye Y, Jiang K, Wang S. Analysis of co-expression networks for circular RNAs and mRNAs reveals that circular RNAs hsa_circ_0047905, hsa_circ_0138960 and has-circRNA7690-15 are candidate oncogenes in gastric cancer. Cell Cycle. 2017;16:2301-2311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 54] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 16. | Vidal AF, Ribeiro-Dos-Santos AM, Vinasco-Sandoval T, Magalhães L, Pinto P, Anaissi AKM, Demachki S, de Assumpção PP, Dos Santos SEB, Ribeiro-Dos-Santos Â. The comprehensive expression analysis of circular RNAs in gastric cancer and its association with field cancerization. Sci Rep. 2017;7:14551. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 17. | Sui W, Shi Z, Xue W, Ou M, Zhu Y, Chen J, Lin H, Liu F, Dai Y. Circular RNA and gene expression profiles in gastric cancer based on microarray chip technology. Oncol Rep. 2017;37:1804-1814. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 71] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 18. | Huang YS, Jie N, Zou KJ, Weng Y. Expression profile of circular RNAs in human gastric cancer tissues. Mol Med Rep. 2017;16:2469-2476. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 62] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 19. | Shen Y, Zhang J, Fu Z, Zhang B, Chen M, Ling X, Zou X. Gene microarray analysis of the circular RNAs expression profile in human gastric cancer. Oncol Lett. 2018;15:9965-9972. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 20. | Gu W, Sun Y, Zheng X, Ma J, Hu XY, Gao T, Hu MJ. Identification of Gastric Cancer-Related Circular RNA through Microarray Analysis and Bioinformatics Analysis. Biomed Res Int. 2018;2018:2381680. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 21. | Dang Y, Ouyang X, Zhang F, Wang K, Lin Y, Sun B, Wang Y, Wang L, Huang Q. Circular RNAs expression profiles in human gastric cancer. Sci Rep. 2017;7:9060. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 70] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 22. | Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3481] [Cited by in RCA: 3538] [Article Influence: 272.2] [Reference Citation Analysis (0)] |
| 23. | Zhong S, Wang J, Hou J, Zhang Q, Xu H, Hu J, Zhao J, Feng J. Circular RNA hsa_circ_0000993 inhibits metastasis of gastric cancer cells. Epigenomics. 2018;10:1301-1313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 24. | Fang J, Hong H, Xue X, Zhu X, Jiang L, Qin M, Liang H, Gao L. A novel circular RNA, circFAT1(e2), inhibits gastric cancer progression by targeting miR-548g in the cytoplasm and interacting with YBX1 in the nucleus. Cancer Lett. 2019;442:222-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 136] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 25. | Liu H, Liu Y, Bian Z, Zhang J, Zhang R, Chen X, Huang Y, Wang Y, Zhu J. Circular RNA YAP1 inhibits the proliferation and invasion of gastric cancer cells by regulating the miR-367-5p/p27 <sup>Kip1</sup> axis. Mol Cancer. 2018;17:151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 114] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 26. | Wang L, Shen J, Jiang Y. Circ_0027599/PHDLA1 suppresses gastric cancer progression by sponging miR-101-3p.1. Cell Biosci. 2018;8:58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 27. | Wu X, Zhou J, Wu Z, Chen C, Liu J, Wu G, Zhai J, Liu F, Li G. miR-101-3p Suppresses HOX Transcript Antisense RNA (HOTAIR)-Induced Proliferation and Invasion Through Directly Targeting SRF in Gastric Carcinoma Cells. Oncol Res. 2017;25:1383-1390. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 30] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 28. | Zhang S, Zhang JY, Lu LJ, Wang CH, Wang LH. MiR-630 promotes epithelial ovarian cancer proliferation and invasion via targeting KLF6. Eur Rev Med Pharmacol Sci. 2017;21:4542-4547. [PubMed] |
| 29. | Zhao JJ, Chen PJ, Duan RQ, Li KJ, Wang YZ, Li Y. miR-630 functions as a tumor oncogene in renal cell carcinoma. Arch Med Sci. 2016;12:473-478. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 34] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 30. | Zhang Y, Liu H, Li W, Yu J, Li J, Shen Z, Ye G, Qi X, Li G. CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR-630. Aging (Albany NY). 2017;9:1585-1594. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 217] [Cited by in RCA: 245] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 31. | Zhang J, Liu H, Hou L, Wang G, Zhang R, Huang Y, Chen X, Zhu J. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16:151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 371] [Cited by in RCA: 436] [Article Influence: 48.4] [Reference Citation Analysis (0)] |
| 32. | Zou T, Wang PL, Gao Y, Liang WT. Circular RNA_LARP4 is lower expressed and serves as a potential biomarker of ovarian cancer prognosis. Eur Rev Med Pharmacol Sci. 2018;22:7178-7182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 30] [Reference Citation Analysis (0)] |
| 33. | Wang J, Wang S, Zhou J, Qian Q. miR-424-5p regulates cell proliferation, migration and invasion by targeting doublecortin-like kinase 1 in basal-like breast cancer. Biomed Pharmacother. 2018;102:147-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 73] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 34. | Wang F, Wang J, Yang X, Chen D, Wang L. MiR-424-5p participates in esophageal squamous cell carcinoma invasion and metastasis via SMAD7 pathway mediated EMT. Diagn Pathol. 2016;11:88. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 44] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 35. | Liu T, Liu S, Xu Y, Shu R, Wang F, Chen C, Zeng Y, Luo H. Circular RNA-ZFR Inhibited Cell Proliferation and Promoted Apoptosis in Gastric Cancer by Sponging miR-130a/miR-107 and Modulating PTEN. Cancer Res Treat. 2018;50:1396-1417. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 103] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 36. | Wei H, Cui R, Bahr J, Zanesi N, Luo Z, Meng W, Liang G, Croce CM. miR-130a Deregulates PTEN and Stimulates Tumor Growth. Cancer Res. 2017;77:6168-6178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 53] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 37. | Xiong J, Wang D, Wei A, Lu H, Tan C, Li A, Tang J, Wang Y, He S, Liu X, Hu W. Deregulated expression of miR-107 inhibits metastasis of PDAC through inhibition PI3K/Akt signaling via caveolin-1 and PTEN. Exp Cell Res. 2017;361:316-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 38. | Sun H, Xi P, Sun Z, Wang Q, Zhu B, Zhou J, Jin H, Zheng W, Tang W, Cao H, Cao X. Circ-SFMBT2 promotes the proliferation of gastric cancer cells through sponging miR-182-5p to enhance CREB1 expression. Cancer Manag Res. 2018;10:5725-5734. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 91] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 39. | Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T, Sun H, Pan Y, He B, Wang S. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9:417. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 447] [Cited by in RCA: 492] [Article Influence: 61.5] [Reference Citation Analysis (0)] |
| 40. | Chen G, Shi Y, Liu M, Sun J. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. 2018;9:175. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 177] [Cited by in RCA: 247] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 41. | Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, Liang L, Gu J, He X, Huang S. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1230] [Cited by in RCA: 1655] [Article Influence: 165.5] [Reference Citation Analysis (0)] |
| 42. | Cheng J, Zhuo H, Xu M, Wang L, Xu H, Peng J, Hou J, Lin L, Cai J. Regulatory network of circRNA-miRNA-mRNA contributes to the histological classification and disease progression in gastric cancer. J Transl Med. 2018;16:216. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 141] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 43. | Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y, Yang S, Zeng Z, Liao W, Ding YQ, Liang L. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7:26680-26691. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 281] [Cited by in RCA: 373] [Article Influence: 46.6] [Reference Citation Analysis (0)] |
| 44. | Shen F, Liu P, Xu Z, Li N, Yi Z, Tie X, Zhang Y, Gao L. CircRNA_001569 promotes cell proliferation through absorbing miR-145 in gastric cancer. J Biochem. 2019;165:27-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 45. | Ouyang Y, Li Y, Huang Y, Li X, Zhu Y, Long Y, Wang Y, Guo X, Gong K. CircRNA circPDSS1 promotes the gastric cancer progression by sponging miR-186-5p and modulating NEK2. J Cell Physiol. 2019;234:10458-10469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 83] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 46. | Li J, Xia L, Zhou Z, Zuo Z, Xu C, Song H, Cai J. MiR-186-5p upregulation inhibits proliferation, metastasis and epithelial-to-mesenchymal transition of colorectal cancer cell by targeting ZEB1. Arch Biochem Biophys. 2018;640:53-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 86] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 47. | Cappello P, Blaser H, Gorrini C, Lin DC, Elia AJ, Wakeham A, Haider S, Boutros PC, Mason JM, Miller NA, Youngson B, Done SJ, Mak TW. Role of Nek2 on centrosome duplication and aneuploidy in breast cancer cells. Oncogene. 2014;33:2375-2384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 87] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 48. | Chang YY, Yen CJ, Chan SH, Chou YW, Lee YP, Bao CY, Huang CJ, Huang W. NEK2 Promotes Hepatoma Metastasis and Serves as Biomarker for High Recurrence Risk after Hepatic Resection. Ann Hepatol. 2018;17:843-856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 49. | Wang Z, Ma K, Pitts S, Cheng Y, Liu X, Ke X, Kovaka S, Ashktorab H, Smoot DT, Schatz M, Wang Z, Meltzer SJ. Novel circular RNA NF1 acts as a molecular sponge, promoting gastric cancer by absorbing miR-16. Endocr Relat Cancer. 2018;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 50] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 50. | Li RC, Ke S, Meng FK, Lu J, Zou XJ, He ZG, Wang WF, Fang MH. CiRS-7 promotes growth and metastasis of esophageal squamous cell carcinoma via regulation of miR-7/HOXB13. Cell Death Dis. 2018;9:838. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 159] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 51. | Su C, Han Y, Zhang H, Li Y, Yi L, Wang X, Zhou S, Yu D, Song X, Xiao N, Cao X, Liu Z. CiRS-7 targeting miR-7 modulates the progression of non-small cell lung cancer in a manner dependent on NF-κB signalling. J Cell Mol Med. 2018;22:3097-3107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 108] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 52. | Weng W, Wei Q, Toden S, Yoshida K, Nagasaka T, Fujiwara T, Cai S, Qin H, Ma Y, Goel A. Circular RNA ciRS-7-A Promising Prognostic Biomarker and a Potential Therapeutic Target in Colorectal Cancer. Clin Cancer Res. 2017;23:3918-3928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 393] [Article Influence: 43.7] [Reference Citation Analysis (0)] |
| 53. | Pan H, Li T, Jiang Y, Pan C, Ding Y, Huang Z, Yu H, Kong D. Overexpression of Circular RNA ciRS-7 Abrogates the Tumor Suppressive Effect of miR-7 on Gastric Cancer via PTEN/PI3K/AKT Signaling Pathway. J Cell Biochem. 2018;119:440-446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 231] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 54. | Suzuki H, Oda I, Abe S, Sekiguchi M, Mori G, Nonaka S, Yoshinaga S, Saito Y. High rate of 5-year survival among patients with early gastric cancer undergoing curative endoscopic submucosal dissection. Gastric Cancer. 2016;19:198-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 195] [Article Influence: 19.5] [Reference Citation Analysis (1)] |
| 55. | Jeon HK, Kim GH, Lee BE, Park DY, Song GA, Kim DH, Jeon TY. Long-term outcome of endoscopic submucosal dissection is comparable to that of surgery for early gastric cancer: a propensity-matched analysis. Gastric Cancer. 2018;21:133-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 51] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 56. | Lee Y, Min SH, Park KB, Park YS, Kim JW, Ahn SH, Kim JW, Park DJ, Lee KW, Kim HH. Effect of Early Adjuvant Chemotherapy on Survival of Advanced Gastric Cancer Patients: a Propensity Score-matched Analysis. J Gastric Cancer. 2018;18:58-68. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 57. | Li P, Chen H, Chen S, Mo X, Li T, Xiao B, Yu R, Guo J. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br J Cancer. 2017;116:626-633. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 190] [Cited by in RCA: 202] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 58. | Zhao Q, Chen S, Li T, Xiao B, Zhang X. Clinical values of circular RNA 0000181 in the screening of gastric cancer. J Clin Lab Anal. 2018;32:e22333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 88] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 59. | Chen S, Li T, Zhao Q, Xiao B, Guo J. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta. 2017;466:167-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 309] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 60. | Sun H, Tang W, Rong D, Jin H, Fu K, Zhang W, Liu Z, Cao H, Cao X. Hsa_circ_0000520, a potential new circular RNA biomarker, is involved in gastric carcinoma. Cancer Biomark. 2018;21:299-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 124] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 61. | Huang M, He YR, Liang LC, Huang Q, Zhu ZQ. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J Gastroenterol. 2017;23:6330-6338. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 133] [Cited by in RCA: 178] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 62. | Shao Y, Chen L, Lu R, Zhang X, Xiao B, Ye G, Guo J. Decreased expression of hsa_circ_0001895 in human gastric cancer and its clinical significances. Tumour Biol. 2017;39:1010428317699125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 74] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 63. | Li WH, Song YC, Zhang H, Zhou ZJ, Xie X, Zeng QN, Guo K, Wang T, Xia P, Chang DM. Decreased Expression of Hsa_circ_00001649 in Gastric Cancer and Its Clinical Significance. Dis Markers. 2017;2017:4587698. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 94] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 64. | Li P, Chen S, Chen H, Mo X, Li T, Shao Y, Xiao B, Guo J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 543] [Cited by in RCA: 640] [Article Influence: 58.2] [Reference Citation Analysis (0)] |
| 65. | Tian M, Chen R, Li T, Xiao B. Reduced expression of circRNA hsa_circ_0003159 in gastric cancer and its clinical significance. J Clin Lab Anal. 2018;32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 123] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 66. | Lu R, Shao Y, Ye G, Xiao B, Guo J. Low expression of hsa_circ_0006633 in human gastric cancer and its clinical significances. Tumour Biol. 2017;39:1010428317704175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 67. | Shao Y, Li J, Lu R, Li T, Yang Y, Xiao B, Guo J. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. 2017;6:1173-1180. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 217] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 68. | Sun HD, Xu ZP, Sun ZQ, Zhu B, Wang Q, Zhou J, Jin H, Zhao A, Tang WW, Cao XF. Down-regulation of circPVRL3 promotes the proliferation and migration of gastric cancer cells. Sci Rep. 2018;8:10111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 71] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 69. | Xie Y, Shao Y, Sun W, Ye G, Zhang X, Xiao B, Guo J. Downregulated expression of hsa_circ_0074362 in gastric cancer and its potential diagnostic values. Biomark Med. 2018;12:11-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 72] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 70. | Tang W, Fu K, Sun H, Rong D, Wang H, Cao H. CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol Cancer. 2018;17:137. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 143] [Cited by in RCA: 206] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 71. | Lu J, Zhang PY, Xie JW, Wang JB, Lin JX, Chen QY, Cao LL, Huang CM, Li P, Zheng CH. Hsa_circ_0000467 promotes cancer progression and serves as a diagnostic and prognostic biomarker for gastric cancer. J Clin Lab Anal. 2019;33:e22726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 61] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 72. | Rong D, Dong C, Fu K, Wang H, Tang W, Cao H. Upregulation of circ_0066444 promotes the proliferation, invasion, and migration of gastric cancer cells. Onco Targets Ther. 2018;11:2753-2761. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 73. | Jiang Y, Wang T, Yan L, Qu L. A novel prognostic biomarker for pancreatic ductal adenocarcinoma: hsa_circ_0001649. Gene. 2018;675:88-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 74. | Wang Y, Sui X, Zhao H, Cong L, Li Y, Xin T, Guo M, Hao W. Decreased circular RNA hsa_circ_0001649 predicts unfavorable prognosis in glioma and exerts oncogenic properties in vitro and in vivo. Gene. 2018;676:117-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 75. | Xing L, Zhang L, Feng Y, Cui Z, Ding L. Downregulation of circular RNA hsa_circ_0001649 indicates poor prognosis for retinoblastoma and regulates cell proliferation and apoptosis via AKT/mTOR signaling pathway. Biomed Pharmacother. 2018;105:326-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 79] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 76. | Zhang X, Qiu S, Luo P, Zhou H, Jing W, Liang C, Tu J. Down-regulation of hsa_circ_0001649 in hepatocellular carcinoma predicts a poor prognosis. Cancer Biomark. 2018;22:135-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 77. | Ji W, Qiu C, Wang M, Mao N, Wu S, Dai Y. Hsa_circ_0001649: A circular RNA and potential novel biomarker for colorectal cancer. Biochem Biophys Res Commun. 2018;497:122-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 102] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 78. | Li T, Shao Y, Fu L, Xie Y, Zhu L, Sun W, Yu R, Xiao B, Guo J. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J Mol Med (Berl). 2018;96:85-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 216] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 79. | Sundararajan V, Sarkar FH, Ramasamy TS. The versatile role of exosomes in cancer progression: diagnostic and therapeutic implications. Cell Oncol (Dordr). 2018;41:223-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 56] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 80. | Zhang H, Zhu L, Bai M, Liu Y, Zhan Y, Deng T, Yang H, Sun W, Wang X, Zhu K, Fan Q, Li J, Ying G, Ba Y. Exosomal circRNA derived from gastric tumor promotes white adipose browning by targeting the miR-133/PRDM16 pathway. Int J Cancer. 2019;144:2501-2515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 188] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 81. | Li J, Zhen L, Zhang Y, Zhao L, Liu H, Cai D, Chen H, Yu J, Qi X, Li G. Circ-104916 is downregulated in gastric cancer and suppresses migration and invasion of gastric cancer cells. Onco Targets Ther. 2017;10:3521-3529. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 66] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 82. | Zhou LH, Yang YC, Zhang RY, Wang P, Pang MH, Liang LQ. CircRNA_0023642 promotes migration and invasion of gastric cancer cells by regulating EMT. Eur Rev Med Pharmacol Sci. 2018;22:2297-2303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 48] [Reference Citation Analysis (0)] |
| 83. | Nieto MA. Epithelial plasticity: a common theme in embryonic and cancer cells. Science. 2013;342:1234850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 683] [Cited by in RCA: 768] [Article Influence: 59.1] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See:
Manuscript source: Invited manuscript
Specialty type: Medicine, research and experimental
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ghorbian S, Sorio C, Ieni A S-Editor: Dou Y L-Editor: A E-Editor: Xing YX