Published online Apr 15, 2019. doi: 10.4251/wjgo.v11.i4.270
Peer-review started: January 4, 2019
First decision: January 21, 2019
Revised: January 29, 2019
Accepted: March 27, 2019
Article in press: March 28, 2019
Published online: April 15, 2019
Processing time: 103 Days and 8.4 Hours
Gastrointestinal (GI) cancers prevail and account for an extremely high number of cancer deaths worldwide. The traditional treatment strategies, including surgery, chemotherapy, radiotherapy, and targeted therapy, have a limited therapeutic effect for advanced GI cancers. Recently, immunotherapy has shown promise in treating various refractory malignancies, including the GI cancers with mismatch repair deficiency (dMMR) or microsatellite instability (MSI). Thus, immunotherapy could be a promising treatment approach for GI cancers. Unfortunately, only a small proportion of GI cancer patients currently respond to immunotherapy. Therefore, it is important to discover predictive biomarkers for stratifying GI cancer patients response to immunotherapy. Certain genomic features, such as dMMR/MSI, tumor mutation burden (TMB), and tumor aneuploidy have been associated with tumor immunity and im-munotherapy response and may serve as predictive biomarkers for cancer immunotherapy. In this review, we examined the correlations between tumor immunity and three genomic features: dMMR/MSI, TMB, and tumor aneuploidy. We also explored their correlations using The Cancer Genome Atlas data and confirmed that the dMMR/MSI status, high TMB, and low tumor aneuploidy are associated with elevated tumor immunity in GI cancers. To improve the immunotherapeutic potential in GI cancers, more genetic or genomic features associated with tumor immune response need to be identified. Furthermore, it is worth exploring the combination of different immunotherapeutic methods and the combination of immunotherapy with other therapeutic approaches for cancer therapy.
Core tip: The traditional treatment strategies have a limited effect on advanced gastrointestinal (GI) cancers. Immunotherapy has shown improved effectiveness in treating diverse malignancies, including the GI cancers with mismatch repair deficiency (dMMR) or microsatellite instability (MSI). However, only a small subset of GI cancers can benefit from immunotherapy. Hence, it is crucial to identify predictive biomarkers for GI cancer patients responsive to immunotherapy. We reviewed the associations between three genomic features (dMMR/MSI, tumor mutation burden, and aneuploidy) and tumor immunity. These genomic features have significant correlations with antitumor immune response and are useful biomarkers for immunotherapy of GI cancers.
- Citation: He Y, Liu ZX, Jiang ZH, Wang XS. Identification of genomic features associated with immunotherapy response in gastrointestinal cancers. World J Gastrointest Oncol 2019; 11(4): 270-280
- URL: https://www.wjgnet.com/1948-5204/full/v11/i4/270.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v11.i4.270
Gastrointestinal (GI) cancers, including malignancies of the esophagus, stomach, biliary system, pancreas, small intestine, large intestine, rectum, and anus are the most prevalent malignant carcinomas globally and account for a large number of cancer deaths[1]. Traditional treatment strategies for GI cancers include surgery, chemotherapy, radiotherapy, and targeted therapy[2]. However, for the refractory or metastatic GI malignancies these traditional treatment strategies often have a limited therapeutic effect[3]. Recently, immunotherapy has demonstrated rapid success in treating various refractory malignancies, such as melanoma[4], non-small cell lung cancer (NSCLC)[5], head and neck cancer[6], renal cell carcinoma[7], leukemia[8], and lymphoma[9]. In particular, the immune checkpoint blockade (ICB) that targets molecules involved in mediating antitumor immunosuppression has been used clinically for treating diverse cancers[10]. Several immune checkpoint inhibitors have been approved by the Food and Drug Administration in clinically treating cancer, including ipilimumab (anti-CTLA4), nivolumab and pembrolizumab (anti-PD1), and atezolizumab and avelumab (anti-PD-L1). However, there is currently no immunotherapeutic drug specifically used for treating a GI cancer, although pembrolizumab is being used for treating DNA mismatch repair-deficient GI cancers.
Abundant evidence shows that the response to ICB is associated with certain genomic features[4,11,12]. These include DNA mismatch repair deficiency[13], tumor mutation burden (TMB)[14] or neoantigen load[15], and tumor aneuploidy[16]. Several studies have revealed that the colorectal cancers (CRCs) with mismatch repair deficiency (dMMR) were more sensitive to anti-PD-1 therapy than those with mismatch repair proficiency (pMMR)[17,18]. Similar results have been demonstrated in other cancer types including ovarian[19], endometrial[20], and gastric cancers[21-23]. In addition, previous studies found that higher TMB was associated with a more favorable response to ICB in diverse cancer types, indicating the potential role of TMB in predicting ICB efficacy[14]. A recent study revealed an inverse correlation between tumor aneuploidy and immunotherapy response[16]. These prior studies suggest that it is significant to identify the genomic features associated with immunotherapy response for treating diverse refractory malignancies, including a large number of GI cancers.
In this review, we examined the associations between three genomic features (dMMR or microsatellite instability (MSI) status, TMB, and tumor aneuploidy) and tumor immunity in GI cancers.
DNA mismatch repair is a highly conserved system for repairing the errors of deletion, insertion, and mismatch occurring in DNA replication and recombination[24]. It plays a key role in maintaining genomic stability[25]. dMMR is associated with genome-wide instability and can lead to tumorigenesis and cancer development[26]. dMMR may cause a high increase in the frequency of insertion and deletion mutations in simple repeat (microsatellite) sequences, a phenomenon known as MSI[27,28]. Plentiful evidence shows that MSI can trigger hyperimmunity in a tumor that may promote response to ICB therapy. First, tumors with MSI are hypermutated and thus generate many neoantigens to incite the tumor immune response[13]. In fact, MSI is associated with the increased infiltration of tumor-infiltrating lymphocytes (TILs) in the tumor[29,30]. It has been shown that MSI CRCs exhibited high infiltration of activated CD8+ cytotoxic T lymphocytes and activated Th1 cells[31]. Second, MSI tumors often exhibit the elevated expression of immune checkpoint molecules such as PD-L1 that may increase the sensitivity to immunotherapy[32].
Several GI cancers harbor a comparatively high proportion of MSI-high (MSI-H) tumors, including gastric cancer (GC) (22%), hepatocellular carcinoma (16%), CRC (13%), and esophageal adenocarcinoma (ESCA) (7%)[33]. A number of studies have revealed that MSI-H GI tumors are more responsive to ICB[13,17,21]. In the clinical trial study (KEYNOTE-012)[21], two out of four MSI-H GC patients responded to pembrolizumab. In a phase 2 clinical study of CRC treatment with pembrolizumab[13], the objective response rate (ORR) in dMMR CRCs was 40% vs 0% in pMMR CRCs[13]. Pancreatic cancer generally has a poor response to immunotherapy[34]. However, a study showed that six pancreatic cancer patients with dMMR or MSI exhibited an objective response rate of 83% to pembrolizumab[17]. The high TMB, neoantigen load, and TIL infiltration in MSI-H GI cancers could explain why this subtype has a favorable response to immunotherapy. Furthermore, because the expression of PD-L1 in this subtype is common[35], and anti-PD-1/PD-L1 therapy may achieve a higher response rate in PD-L1-positive tumors than in PD-L1-negative tumors[36], MSI-H GI cancer patients are likely to respond to immunotherapy. Therefore, dMMR/MSI is an important predictive biomarker in GI cancer immunotherapy.
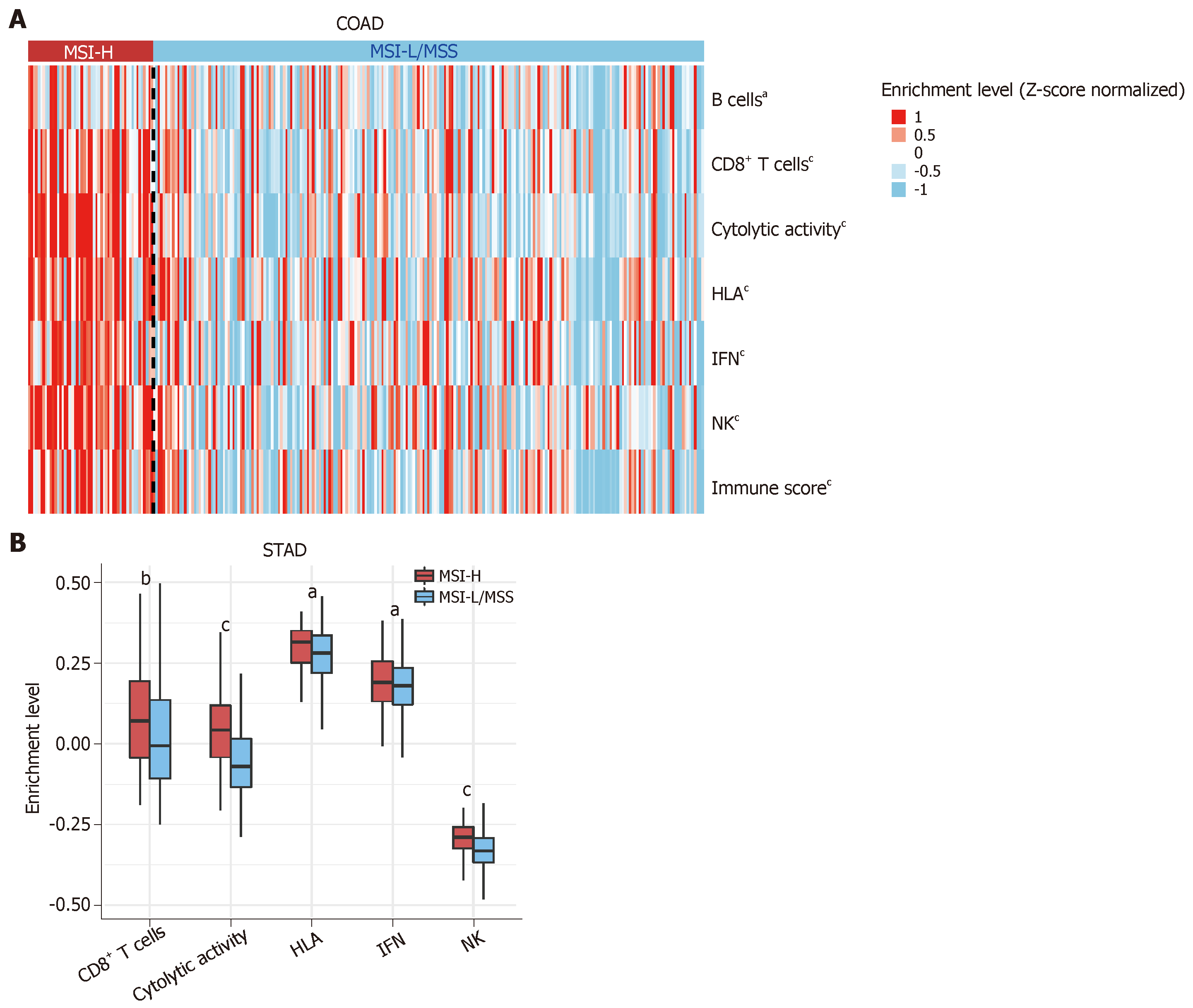
To further investigate the relationship between MSI status and tumor immunity in GI cancers, we downloaded RNA-Seq gene expression (Level 3) and clinical data from The Cancer Genome Atlas (TCGA) project (https://portal.gdc.cancer.gov/). We first quantified the enrichment levels of six immune signatures (B cells, CD8+ T cells, cytolytic activity, human leukocyte antigen (HLA), interferon response, and natural killer (NK) cells) (Table 1)[37] in each GI sample using the single-sample gene-set enrichment analysis score[38]. We compared the enrichment levels between MSI-H GI cancers and MSI low (MSI-L) or microsatellite stability (MSS) GI cancers. We observed a significant upregulation of the six immune signatures in the MSI-H colon adenocarcinoma (COAD) vs the MSI-L/MSS COAD (Mann-Whitney U test, P < 0.05) (Figure 1A). Similar results were observed for stomach adenocarcinoma (STAD) (Figure 1B). Next, we used the ESTIMATE algorithm[39] to evaluate the immune score for each GI sample, which represents the degree of immune cell infiltration in the tumor. We found that the MSI-H COAD had significantly higher immune scores than the MSI-L/MSS COAD (Mann-Whitney U test, P < 0.05) (Figure 1A). Collectively, these results confirmed that MSI-H GI tumors tend to have stronger tumor immunity compared to MSI-L/MSS GI tumors, suggesting that the MSI-H GI cancer subtype could be more responsive to immunotherapy.
| Immune signature | Gene set |
| B cells | BACH2, BANK1, BLK, BTLA, CD79A, CD79B, FCRL1, FCRL3, HVCN1, RALGPS2 |
| CD8+ T cells | CD8A |
| Cytolytic activity | PRF1, GZMA |
| HLA | HLA-A, HLA-B, HLA-C, HLA-DMA, HLA-DMB, HLA-DOA, HLA-DOB, HLA-DPA1, HLA-DPB1, HLA-DPB2, HLA-DQA1, HLA-DQA2, HLA-DQB1, HLA-DQB2, HLA-DRA, HLA-DRB1, HLA-DRB5, HLA-DRB6, HLA-E, HLA-F, HLA-G, HLA-J |
| IFN | DDX4, IFIT1, IFIT2, IFIT3, IRF7, ISG20, MX1, MX2, RSAD2, TNFSF10, GPR146, SELP, AHR |
| NK | KLRC1, KLRF1 |
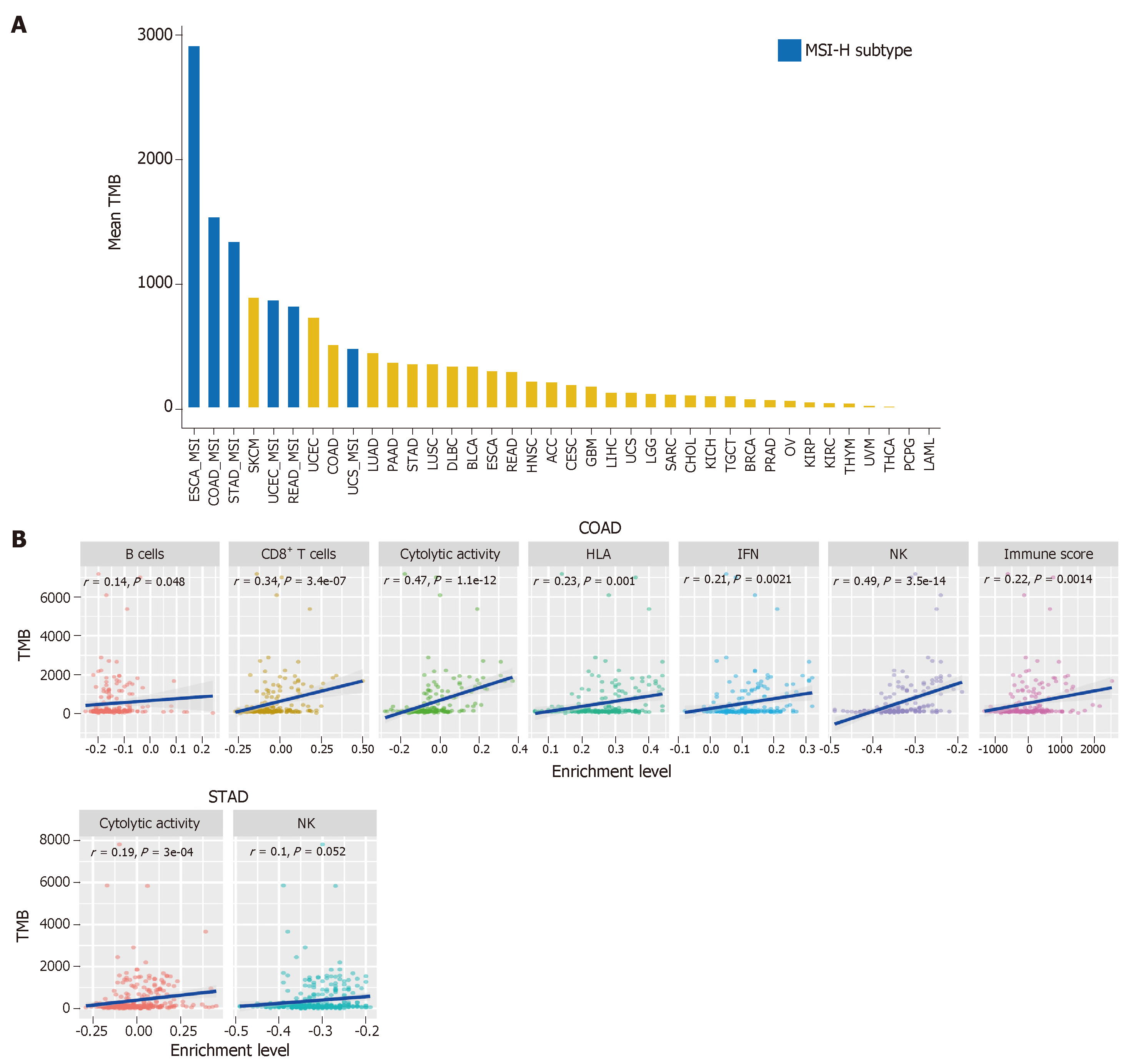
TMB represents the overall load of somatic mutations in the tumor. Tumor somatic mutations may produce neoantigens to drive antitumor immune responses[40]. Therefore, the increase in TMB would result in elevated tumor immunogenicity as well as antitumor immunity[41,42]. Previous studies have shown that TMB varies with cancer type[43,44]. Some cancer types generally have high TMB, such as skin cutaneous melanoma, lung cancer, esophageal carcinoma, and bladder urothelial carcinoma, and some have low TMB, such as leukemia. Most GI cancers have a medium level of TMB (defined as total somatic mutation counts) (Figure 2A). However, some GI cancer subtypes, such as the MSI-H subtype, often have high TMB (Figure 2A).
The high TMB cancer types, such as melanoma, NSCLC, and MSI-H cancers, are more sensitive to the ICB therapy because they can produce more neoantigens[17,45-47]. As a result, high TMB is associated with long-term clinical benefit to anti-CTLA-4[46,48] and anti-PD-1/PD-L1[49-51] therapy. In GI cancers, the high TMB in ESCA was associated with clinical benefit of the ICB therapy[52], the EBV+ (Epstein–Barr virus+) and MSI+ molecular subsets of GC with high TMB showed increased immune cell infiltration and PD-1/PD-L1 pathway activation[53], and the high TMB CRC patients were commonly responsive to PD-1/PD-L1 blockade[14]. Interestingly, a high TMB in pancreatic cancer was negatively associated with T cell activity and a worse overall survival[54].
We evaluated the correlation between TMB and tumor immunity in GI cancers based on the TCGA data. We found that the six immune signatures showed significant positive correlations with TMB in COAD (Spearman's correlation test, P < 0.05) (Figure 2B). In STAD, the cytolytic activity and NK cell signatures were significantly positively associated with TMB (Figure 2B). These data confirmed that TMB is likely to be positively associated with tumor immunity in GC cancers.
Tumor cells often display a high degree of chromosomal instability (CIN)[55]. CIN refers to distortions in the number of chromosome (aneuploidy) or the chromosomal structure (translocation, inversion, and duplication)[56]. Aneuploidy, also known as somatic copy number alterations (SCNAs), is a common characteristic present in 88% of solid tumors[57] and plays a key role in tumor development[58-61]. Numerous studies have revealed a significant correlation between tumor aneuploidy and tumor immunity[16,57]. Davoli et al[16] found that tumors with a high level of aneuploidy inversely correlated with cytotoxic immune infiltration and that the tumor patients with high aneuploidy had a poor survival prognosis. Taylor et al[57] demonstrated that aneuploidy was negatively associated with tumor immune activity. Moreover, tumor aneuploidy is likely to increase intratumor heterogeneity[62,63], which may inhibit tumor immunity[64,65].
In GI cancers, the genomic feature of ESCA resembles the CIN subtype of GC[66]. The CIN subtype, characterized with a high degree of SCNAs[53,67,68], accounts for nearly half of all GC cases[53]. Our previous study showed that immune signatures were significantly downregulated in the CIN subtype versus the genomically stable subtype in GC and COAD[69].
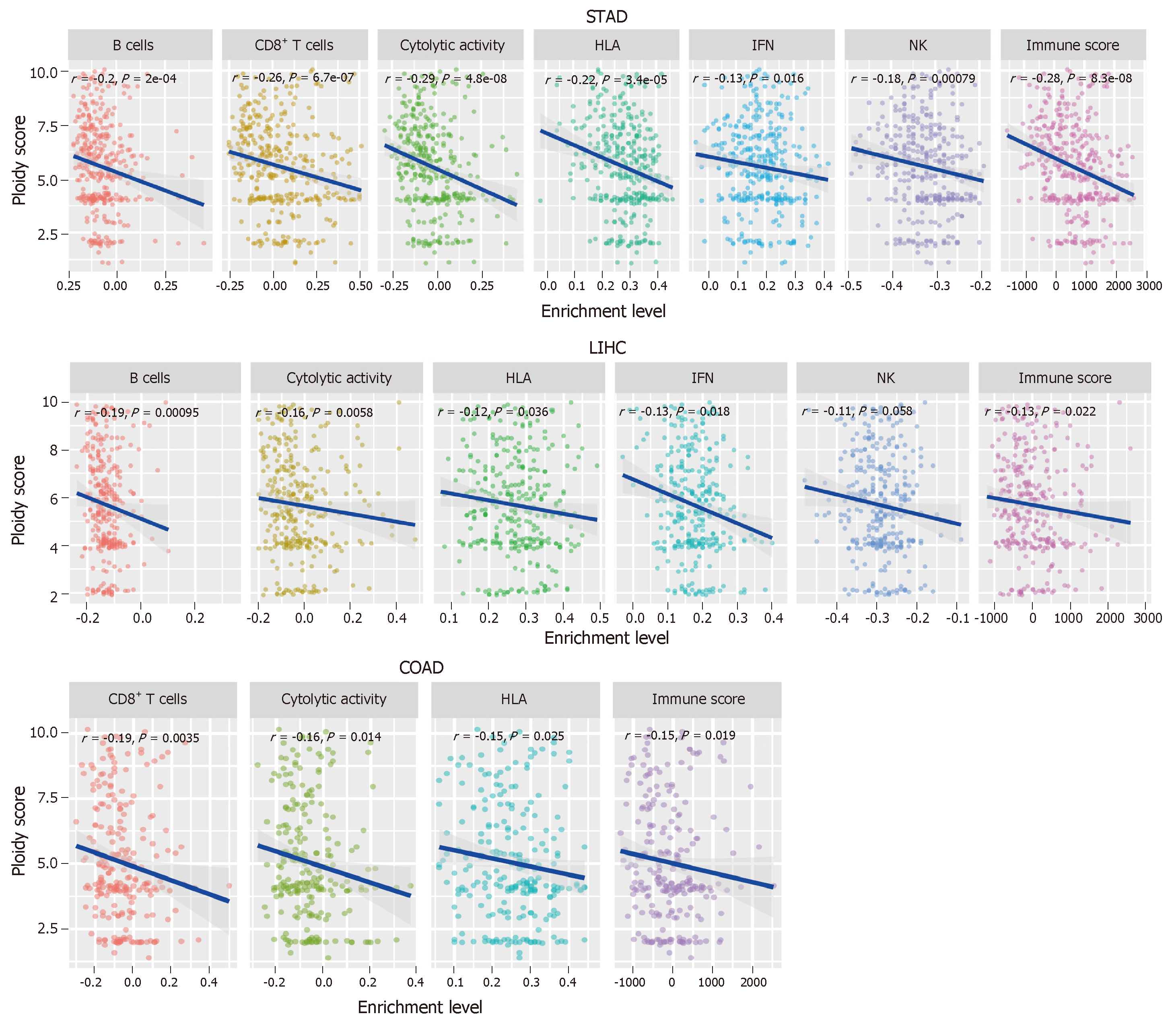
To further explore the relationship between tumor aneuploidy and tumor immunity in GI cancers, we used the absolute algorithm[70] to assess the ploidy score for each GI sample in TCGA, and evaluated the correlations between the six immune signatures and the ploidy scores in GI cancers. We found that diverse immune signatures were significantly inversely correlated with the ploidy scores in GI cancers, including all six immune signatures in STAD, five in liver hepatocellular carcinoma, and four in COAD (Spearman's correlation test, P < 0.05) (Figure 3). We also found that the immune scores were inversely correlated with the aneuploidy in these GI cancer types (Figure 3). Collectively, these results confirmed the negative correlation between tumor aneuploidy and tumor immunity in GI cancers and suggested an important role for aneuploidy in predicting the immunotherapy response in GI cancers.
Due to the limited therapeutic effect of traditional treatment strategies on refractory or metastatic GI cancers, immunotherapy could be an alternative approach for these cancers. Immunotherapy is likely to be effective for the “hot” tumors whose microenvironment has dense T cell infiltration[71]. The tumors with MSI, high TMB, or low aneuploidy are often “hot” tumors that respond to immunotherapy. Nevertheless, immunotherapy often has poor efficiency for the “cold” tumors that lack immune infiltration[71]. To improve the immunotherapy response in “cold” tumors, a combination of different treatment strategies may convert “cold” tumors into “hot” tumors. The combined treatment strategies could be the combination of different immunotherapeutic methods[72-75] or the combination of immunotherapy with other therapeutic approaches[76-79].
Besides the genomic features, mutations in some specific genes may suggest an immunotherapy response. Our previous study showed that TP53 mutations were associated with depressed tumor immunity in STAD and COAD[69], suggesting that the TP53 mutation status may predict the response of STAD and COAD patients to immunotherapy. In addition, KRAS mutations are associated with suppressed immune activity in CRC[80].
Previous studies revealed that ICB was effective in a subset of GI cancer patients, but the response rate in an unselected GI tumor cohort was modest[81]. It suggests that the predictive genetic and genomic features are important for stratifying GI cancer patients responsive to immunotherapy. A large volume of cancer genomics data has been produced through the advancement of next-generation sequencing technology, enabling us to investigate the cancer genomic features associated with tumor immunity and immunotherapy response. The dMMR/MSI status, TMB, and tumor aneuploidy are genomic features associated with tumor immunity and immunotherapy response. Generally speaking, the high TMB tumors are more likely to respond to immunotherapy[14]. However, the association between TMB and immunotherapy response is not absolutely positive. Some responders have a low TMB and some non-responders have a high TMB[12]. One possible explanation is that the intratumor heterogeneity confounds the mutation landscape, affecting tumor immunity[82]. In fact, a previous study has shown that it is clonal neoantigens (generated by mutations identified in distinct regions of a tumor) that associate with immunotherapy response rather than subclonal neoantigens (generated by mutations identified in only a subset of regions of a tumor)[83]. Tumor aneuploidy is another genomic feature that is associated with the antitumor immune response, and could be a stronger predictor of tumor immune infiltration than TMB[16].
In conclusion, dMMR/MSI, TMB, and tumor aneuploidy are the well-recognized genomic features that are associated with the immunotherapy response of GI and other cancers. To improve the potential of immunotherapy for GI cancers, more genomic features need to be identified, and the relevant investigations should remain a high priority.
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 56682] [Article Influence: 7085.3] [Reference Citation Analysis (135)] |
| 2. | Long J, Lin J, Wang A, Wu L, Zheng Y, Yang X, Wan X, Xu H, Chen S, Zhao H. PD-1/PD-L blockade in gastrointestinal cancers: lessons learned and the road toward precision immunotherapy. J Hematol Oncol. 2017;10:146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 85] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 3. | Kirk R. Gastrointestinal cancer: New drug shows promise in refractory colorectal cancer. Nat Rev Clin Oncol. 2012;9:610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 4. | Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Foppen MHG, Goldinger SM, Utikal J, Hassel JC, Weide B, Kaehler KC, Loquai C, Mohr P, Gutzmer R, Dummer R, Gabriel S, Wu CJ, Schadendorf D, Garraway LA. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2065] [Cited by in RCA: 2253] [Article Influence: 204.8] [Reference Citation Analysis (0)] |
| 5. | Herbst RS, Baas P, Kim DW, Felip E, Pérez-Gracia JL, Han JY, Molina J, Kim JH, Arvis CD, Ahn MJ, Majem M, Fidler MJ, de Castro G, Garrido M, Lubiniecki GM, Shentu Y, Im E, Dolled-Filhart M, Garon EB. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet. 2016;387:1540-1550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4116] [Cited by in RCA: 5181] [Article Influence: 518.1] [Reference Citation Analysis (2)] |
| 6. | Bauml J, Seiwert TY, Pfister DG, Worden F, Liu SV, Gilbert J, Saba NF, Weiss J, Wirth L, Sukari A, Kang H, Gibson MK, Massarelli E, Powell S, Meister A, Shu X, Cheng JD, Haddad R. Pembrolizumab for Platinum- and Cetuximab-Refractory Head and Neck Cancer: Results From a Single-Arm, Phase II Study. J Clin Oncol. 2017;35:1542-1549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 459] [Cited by in RCA: 534] [Article Influence: 59.3] [Reference Citation Analysis (0)] |
| 7. | Tomita Y, Fukasawa S, Shinohara N, Kitamura H, Oya M, Eto M, Tanabe K, Kimura G, Yonese J, Yao M, Motzer RJ, Uemura H, McHenry MB, Berghorn E, Ozono S. Nivolumab versus everolimus in advanced renal cell carcinoma: Japanese subgroup analysis from the CheckMate 025 study. Jpn J Clin Oncol. 2017;47:639-646. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 8. | Maude SL, Laetsch TW, Buechner J, Rives S, Boyer M, Bittencourt H, Bader P, Verneris MR, Stefanski HE, Myers GD, Qayed M, De Moerloose B, Hiramatsu H, Schlis K, Davis KL, Martin PL, Nemecek ER, Yanik GA, Peters C, Baruchel A, Boissel N, Mechinaud F, Balduzzi A, Krueger J, June CH, Levine BL, Wood P, Taran T, Leung M, Mueller KT, Zhang Y, Sen K, Lebwohl D, Pulsipher MA, Grupp SA. Tisagenlecleucel in Children and Young Adults with B-Cell Lymphoblastic Leukemia. N Engl J Med. 2018;378:439-448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2882] [Cited by in RCA: 4317] [Article Influence: 539.6] [Reference Citation Analysis (0)] |
| 9. | Smyth L, Buckstein R, Pennell N, Weerasinghe R, Cheung MC, Imrie K, Spaner D, Piliotis E, Chodirker L, Reis M, Ghorab Z, Zhang L, Boudreau V, Miliken A, Berinstein N. Autologous stem cell transplant and combination immunotherapy of rituximab and interferon-α induces prolonged clinical and molecular remissions in patients with follicular lymphoma. Br J Haematol. 2019;184:469-472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 10. | Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27:450-461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2655] [Cited by in RCA: 3355] [Article Influence: 305.0] [Reference Citation Analysis (0)] |
| 11. | Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, Berent-Maoz B, Pang J, Chmielowski B, Cherry G, Seja E, Lomeli S, Kong X, Kelley MC, Sosman JA, Johnson DB, Ribas A, Lo RS. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell. 2016;165:35-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1695] [Cited by in RCA: 2471] [Article Influence: 247.1] [Reference Citation Analysis (1)] |
| 12. | Braun DA, Burke KP, Van Allen EM. Genomic Approaches to Understanding Response and Resistance to Immunotherapy. Clin Cancer Res. 2016;22:5642-5650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 124] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 13. | Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, Biedrzycki B, Donehower RC, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Duffy SM, Goldberg RM, de la Chapelle A, Koshiji M, Bhaijee F, Huebner T, Hruban RH, Wood LD, Cuka N, Pardoll DM, Papadopoulos N, Kinzler KW, Zhou S, Cornish TC, Taube JM, Anders RA, Eshleman JR, Vogelstein B, Diaz LA. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509-2520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6096] [Cited by in RCA: 7502] [Article Influence: 682.0] [Reference Citation Analysis (2)] |
| 14. | Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, Stephens PJ, Daniels GA, Kurzrock R. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol Cancer Ther. 2017;16:2598-2608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1259] [Cited by in RCA: 1767] [Article Influence: 196.3] [Reference Citation Analysis (0)] |
| 15. | Efremova M, Finotello F, Rieder D, Trajanoski Z. Neoantigens Generated by Individual Mutations and Their Role in Cancer Immunity and Immunotherapy. Front Immunol. 2017;8:1679. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 162] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 16. | Davoli T, Uno H, Wooten EC, Elledge SJ. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science. 2017;355:eaaf8399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 720] [Cited by in RCA: 1013] [Article Influence: 112.6] [Reference Citation Analysis (0)] |
| 17. | Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS, Wong F, Azad NS, Rucki AA, Laheru D, Donehower R, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Greten TF, Duffy AG, Ciombor KK, Eyring AD, Lam BH, Joe A, Kang SP, Holdhoff M, Danilova L, Cope L, Meyer C, Zhou S, Goldberg RM, Armstrong DK, Bever KM, Fader AN, Taube J, Housseau F, Spetzler D, Xiao N, Pardoll DM, Papadopoulos N, Kinzler KW, Eshleman JR, Vogelstein B, Anders RA, Diaz LA. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3799] [Cited by in RCA: 5185] [Article Influence: 576.1] [Reference Citation Analysis (0)] |
| 18. | Lee JJ, Chu E. Recent Advances in the Clinical Development of Immune Checkpoint Blockade Therapy for Mismatch Repair Proficient (pMMR)/non-MSI-H Metastatic Colorectal Cancer. Clin Colorectal Cancer. 2018;17:258-273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 19. | Xiao X, Dong D, He W, Song L, Wang Q, Yue J, Xie L. Mismatch repair deficiency is associated with MSI phenotype, increased tumor-infiltrating lymphocytes and PD-L1 expression in immune cells in ovarian cancer. Gynecol Oncol. 2018;149:146-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 20. | Gargiulo P, Della Pepa C, Berardi S, Califano D, Scala S, Buonaguro L, Ciliberto G, Brauchli P, Pignata S. Tumor genotype and immune microenvironment in POLE-ultramutated and MSI-hypermutated Endometrial Cancers: New candidates for checkpoint blockade immunotherapy? Cancer Treat Rev. 2016;48:61-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 97] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 21. | Muro K, Chung HC, Shankaran V, Geva R, Catenacci D, Gupta S, Eder JP, Golan T, Le DT, Burtness B, McRee AJ, Lin CC, Pathiraja K, Lunceford J, Emancipator K, Juco J, Koshiji M, Bang YJ. Pembrolizumab for patients with PD-L1-positive advanced gastric cancer (KEYNOTE-012): a multicentre, open-label, phase 1b trial. Lancet Oncol. 2016;17:717-726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 878] [Cited by in RCA: 930] [Article Influence: 93.0] [Reference Citation Analysis (1)] |
| 22. | Kang YK, Boku N, Satoh T, Ryu MH, Chao Y, Kato K, Chung HC, Chen JS, Muro K, Kang WK, Yeh KH, Yoshikawa T, Oh SC, Bai LY, Tamura T, Lee KW, Hamamoto Y, Kim JG, Chin K, Oh DY, Minashi K, Cho JY, Tsuda M, Chen LT. Nivolumab in patients with advanced gastric or gastro-oesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO-4538-12, ATTRACTION-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390:2461-2471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1283] [Cited by in RCA: 1789] [Article Influence: 198.8] [Reference Citation Analysis (0)] |
| 23. | Ott PA, Le DT, Kim JW, Ascierto PA, Sharma P, Bono P, Peltola K, Jager D, Evans TRJ, de Braud F, Chau I, Bendell JC, Tschaika M, Harbison CT, Zhao H, Calvo E, Janjigian Y. Nivolumab (NIVO) in patients (pts) with advanced (adv) chemotherapy-refractory (CT-Rx) esophagogastric (EG) cancer according to microsatellite instability (MSI) status: checkmate 032. Ann Oncol. 2017;28:v209-v268. [RCA] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 24. | Zhang L, Peng Y, Peng G. Mismatch repair-based stratification for immune checkpoint blockade therapy. Am J Cancer Res. 2018;8:1977-1988. [PubMed] |
| 25. | Harfe BD, Jinks-Robertson S. DNA mismatch repair and genetic instability. Annu Rev Genet. 2000;34:359-399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 461] [Cited by in RCA: 459] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 26. | Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 816] [Cited by in RCA: 1028] [Article Influence: 57.1] [Reference Citation Analysis (0)] |
| 27. | Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 873] [Cited by in RCA: 980] [Article Influence: 49.0] [Reference Citation Analysis (0)] |
| 28. | Aaltonen LA, Peltomäki P, Leach FS, Sistonen P, Pylkkänen L, Mecklin JP, Järvinen H, Powell SM, Jen J, Hamilton SR. Clues to the pathogenesis of familial colorectal cancer. Science. 1993;260:812-816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1810] [Cited by in RCA: 1818] [Article Influence: 55.1] [Reference Citation Analysis (1)] |
| 29. | Giampieri R, Maccaroni E, Mandolesi A, Del Prete M, Andrikou K, Faloppi L, Bittoni A, Bianconi M, Scarpelli M, Bracci R, Scartozzi M, Cascinu S. Mismatch repair deficiency may affect clinical outcome through immune response activation in metastatic gastric cancer patients receiving first-line chemotherapy. Gastric Cancer. 2017;20:156-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 66] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 30. | Mlecnik B, Bindea G, Angell HK, Maby P, Angelova M, Tougeron D, Church SE, Lafontaine L, Fischer M, Fredriksen T, Sasso M, Bilocq AM, Kirilovsky A, Obenauf AC, Hamieh M, Berger A, Bruneval P, Tuech JJ, Sabourin JC, Le Pessot F, Mauillon J, Rafii A, Laurent-Puig P, Speicher MR, Trajanoski Z, Michel P, Sesboüe R, Frebourg T, Pagès F, Valge-Archer V, Latouche JB, Galon J. Integrative Analyses of Colorectal Cancer Show Immunoscore Is a Stronger Predictor of Patient Survival Than Microsatellite Instability. Immunity. 2016;44:698-711. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 788] [Cited by in RCA: 791] [Article Influence: 79.1] [Reference Citation Analysis (0)] |
| 31. | Halama N, Michel S, Kloor M, Zoernig I, Benner A, Spille A, Pommerencke T, von Knebel DM, Folprecht G, Luber B, Feyen N, Martens UM, Beckhove P, Gnjatic S, Schirmacher P, Herpel E, Weitz J, Grabe N, Jaeger D. Localization and density of immune cells in the invasive margin of human colorectal cancer liver metastases are prognostic for response to chemotherapy. Cancer Res. 2011;71:5670-5677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 295] [Cited by in RCA: 333] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 32. | Llosa NJ, Cruise M, Tam A, Wicks EC, Hechenbleikner EM, Taube JM, Blosser RL, Fan H, Wang H, Luber BS, Zhang M, Papadopoulos N, Kinzler KW, Vogelstein B, Sears CL, Anders RA, Pardoll DM, Housseau F. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5:43-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 889] [Cited by in RCA: 1206] [Article Influence: 100.5] [Reference Citation Analysis (0)] |
| 33. | Dudley JC, Lin MT, Le DT, Eshleman JR. Microsatellite Instability as a Biomarker for PD-1 Blockade. Clin Cancer Res. 2016;22:813-820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 488] [Cited by in RCA: 695] [Article Influence: 69.5] [Reference Citation Analysis (0)] |
| 34. | Tan E, El-Rayes B. Pancreatic Cancer and Immunotherapy: Resistance Mechanisms and Proposed Solutions. J Gastrointest Cancer. 2019;50:1-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 35. | Ma C, Patel K, Singhi AD, Ren B, Zhu B, Shaikh F, Sun W. Programmed Death-Ligand 1 Expression Is Common in Gastric Cancer Associated With Epstein-Barr Virus or Microsatellite Instability. Am J Surg Pathol. 2016;40:1496-1506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 123] [Article Influence: 13.7] [Reference Citation Analysis (1)] |
| 36. | Patel SP, Kurzrock R. PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol Cancer Ther. 2015;14:847-856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1250] [Cited by in RCA: 1806] [Article Influence: 164.2] [Reference Citation Analysis (1)] |
| 37. | Liu Z, Li M, Jiang Z, Wang X. A Comprehensive Immunologic Portrait of Triple-Negative Breast Cancer. Transl Oncol. 2018;11:311-329. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 206] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 38. | Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7222] [Cited by in RCA: 10496] [Article Influence: 807.4] [Reference Citation Analysis (4)] |
| 39. | Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW, Levine DA, Carter SL, Getz G, Stemke-Hale K, Mills GB, Verhaak RG. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3056] [Cited by in RCA: 6972] [Article Influence: 581.0] [Reference Citation Analysis (2)] |
| 40. | Castle JC, Kreiter S, Diekmann J, Löwer M, van de Roemer N, de Graaf J, Selmi A, Diken M, Boegel S, Paret C, Koslowski M, Kuhn AN, Britten CM, Huber C, Türeci O, Sahin U. Exploiting the mutanome for tumor vaccination. Cancer Res. 2012;72:1081-1091. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 566] [Cited by in RCA: 652] [Article Influence: 46.6] [Reference Citation Analysis (0)] |
| 41. | Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348:69-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2935] [Cited by in RCA: 3695] [Article Influence: 335.9] [Reference Citation Analysis (2)] |
| 42. | Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2009] [Cited by in RCA: 2882] [Article Influence: 262.0] [Reference Citation Analysis (1)] |
| 43. | Sun Q, Li M, Wang X. The Cancer Omics Atlas: an integrative resource for cancer omics annotations. BMC Med Genomics. 2018;11:63. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 44. | Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, Leiserson MDM, Miller CA, Welch JS, Walter MJ, Wendl MC, Ley TJ, Wilson RK, Raphael BJ, Ding L. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333-339. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2931] [Cited by in RCA: 3470] [Article Influence: 266.9] [Reference Citation Analysis (0)] |
| 45. | Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjörd JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Jäger N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, López-Otín C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdés-Mas R, van Buuren MM, van 't Veer L, Vincent-Salomon A, Waddell N, Yates LR; Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain, Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR. Signatures of mutational processes in human cancer. Nature. 2013;500:415-421. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7533] [Cited by in RCA: 7569] [Article Influence: 582.2] [Reference Citation Analysis (3)] |
| 46. | Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, Hollmann TJ, Bruggeman C, Kannan K, Li Y, Elipenahli C, Liu C, Harbison CT, Wang L, Ribas A, Wolchok JD, Chan TA. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189-2199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3101] [Cited by in RCA: 3453] [Article Influence: 287.8] [Reference Citation Analysis (1)] |
| 47. | Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, Miller ML, Rekhtman N, Moreira AL, Ibrahim F, Bruggeman C, Gasmi B, Zappasodi R, Maeda Y, Sander C, Garon EB, Merghoub T, Wolchok JD, Schumacher TN, Chan TA. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6065] [Cited by in RCA: 6499] [Article Influence: 590.8] [Reference Citation Analysis (0)] |
| 48. | Robert C, Thomas L, Bondarenko I, O'Day S, Weber J, Garbe C, Lebbe C, Baurain JF, Testori A, Grob JJ, Davidson N, Richards J, Maio M, Hauschild A, Miller WH, Gascon P, Lotem M, Harmankaya K, Ibrahim R, Francis S, Chen TT, Humphrey R, Hoos A, Wolchok JD. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N Engl J Med. 2011;364:2517-2526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3331] [Cited by in RCA: 3447] [Article Influence: 229.8] [Reference Citation Analysis (11)] |
| 49. | Muller M, Schouten RD, De Gooijer CJ, Baas P. Pembrolizumab for the treatment of non-small cell lung cancer. Expert Rev Anticancer Ther. 2017;17:399-409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 41] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 50. | Robert C, Long GV, Brady B, Dutriaux C, Maio M, Mortier L, Hassel JC, Rutkowski P, McNeil C, Kalinka-Warzocha E, Savage KJ, Hernberg MM, Lebbé C, Charles J, Mihalcioiu C, Chiarion-Sileni V, Mauch C, Cognetti F, Arance A, Schmidt H, Schadendorf D, Gogas H, Lundgren-Eriksson L, Horak C, Sharkey B, Waxman IM, Atkinson V, Ascierto PA. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2015;372:320-330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3964] [Cited by in RCA: 4438] [Article Influence: 403.5] [Reference Citation Analysis (0)] |
| 51. | Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, Barlesi F, Kohlhäufl M, Arrieta O, Burgio MA, Fayette J, Lena H, Poddubskaya E, Gerber DE, Gettinger SN, Rudin CM, Rizvi N, Crinò L, Blumenschein GR, Antonia SJ, Dorange C, Harbison CT, Graf Finckenstein F, Brahmer JR. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med. 2015;373:1627-1639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6945] [Cited by in RCA: 7758] [Article Influence: 705.3] [Reference Citation Analysis (0)] |
| 52. | Hazama S, Tamada K, Yamaguchi Y, Kawakami Y, Nagano H. Current status of immunotherapy against gastrointestinal cancers and its biomarkers: Perspective for precision immunotherapy. Ann Gastroenterol Surg. 2018;2:289-303. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 53. | Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202-209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5015] [Cited by in RCA: 5093] [Article Influence: 424.4] [Reference Citation Analysis (4)] |
| 54. | Bailey P, Chang DK, Forget MA, Lucas FA, Alvarez HA, Haymaker C, Chattopadhyay C, Kim SH, Ekmekcioglu S, Grimm EA, Biankin AV, Hwu P, Maitra A, Roszik J. Exploiting the neoantigen landscape for immunotherapy of pancreatic ductal adenocarcinoma. Sci Rep. 2016;6:35848. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 135] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 55. | Sansregret L, Swanton C. The Role of Aneuploidy in Cancer Evolution. Cold Spring Harb Perspect Med. 2017;7:a028373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 182] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 56. | Bockorny B, Pectasides E. The emerging role of immunotherapy in gastric and esophageal adenocarcinoma. Future Oncol. 2016;12:1833-1846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 57. | Taylor AM, Shih J, Ha G, Gao GF, Zhang X, Berger AC, Schumacher SE, Wang C, Hu H, Liu J, Lazar AJ; Cancer Genome Atlas Research Network, Cherniack AD, Beroukhim R, Meyerson M. Genomic and Functional Approaches to Understanding Cancer Aneuploidy. Cancer Cell. 2018;33:676-689.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 846] [Cited by in RCA: 774] [Article Influence: 96.8] [Reference Citation Analysis (0)] |
| 58. | Watson EV, Elledge SJ. Aneuploidy Police Detect Chromosomal Imbalance Triggering Immune Crackdown! Trends Genet. 2017;33:662-664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 59. | Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623-627. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1367] [Cited by in RCA: 1391] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 60. | Cahill DP, Kinzler KW, Vogelstein B, Lengauer C. Genetic instability and darwinian selection in tumours. Trends Cell Biol. 1999;9:M57-M60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 482] [Cited by in RCA: 430] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 61. | Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 550] [Cited by in RCA: 564] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 62. | Caswell DR, Swanton C. The role of tumour heterogeneity and clonal cooperativity in metastasis, immune evasion and clinical outcome. BMC Med. 2017;15:133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 175] [Article Influence: 19.4] [Reference Citation Analysis (1)] |
| 63. | Oltmann J, Heselmeyer-Haddad K, Hernandez LS, Meyer R, Torres I, Hu Y, Doberstein N, Killian JK, Petersen D, Zhu YJ, Edelman DC, Meltzer PS, Schwartz R, Gertz EM, Schäffer AA, Auer G, Habermann JK, Ried T. Aneuploidy, TP53 mutation, and amplification of MYC correlate with increased intratumor heterogeneity and poor prognosis of breast cancer patients. Genes Chromosomes Cancer. 2018;57:165-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 64. | Safonov A, Jiang T, Bianchini G, Győrffy B, Karn T, Hatzis C, Pusztai L. Immune Gene Expression Is Associated with Genomic Aberrations in Breast Cancer. Cancer Res. 2017;77:3317-3324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 115] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 65. | Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, Ziv E, Culhane AC, Paull EO, Sivakumar IKA, Gentles AJ, Malhotra R, Farshidfar F, Colaprico A, Parker JS, Mose LE, Vo NS, Liu J, Liu Y, Rader J, Dhankani V, Reynolds SM, Bowlby R, Califano A, Cherniack AD, Anastassiou D, Bedognetti D, Rao A, Chen K, Krasnitz A, Hu H, Malta TM, Noushmehr H, Pedamallu CS, Bullman S, Ojesina AI, Lamb A, Zhou W, Shen H, Choueiri TK, Weinstein JN, Guinney J, Saltz J, Holt RA, Rabkin CE; Cancer Genome Atlas Research Network, Lazar AJ, Serody JS, Demicco EG, Disis ML, Vincent BG, Shmulevich L. The Immune Landscape of Cancer. Immunity. 2018;48:812-830.e14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4007] [Cited by in RCA: 4123] [Article Influence: 515.4] [Reference Citation Analysis (5)] |
| 66. | Cancer Genome Atlas Research Network;. Analysis Working Group: Asan University; BC Cancer Agency; Brigham and Women’s Hospital; Broad Institute; Brown University; Case Western Reserve University; Dana-Farber Cancer Institute; Duke University; Greater Poland Cancer Centre; Harvard Medical School; Institute for Systems Biology; KU Leuven; Mayo Clinic; Memorial Sloan Kettering Cancer Center; National Cancer Institute; Nationwide Children’s Hospital; Stanford University; University of Alabama; University of Michigan; University of North Carolina; University of Pittsburgh; University of Rochester; University of Southern California; University of Texas MD Anderson Cancer Center; University of Washington; Van Andel Research Institute; Vanderbilt University; Washington University; Genome Sequencing Center: Broad Institute; Washington University in St. Louis; Genome Characterization Centers: BC Cancer Agency; Broad Institute; Harvard Medical School; Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins University; University of North Carolina; University of Southern California Epigenome Center; University of Texas MD Anderson Cancer Center; Van Andel Research Institute; Genome Data Analysis Centers: Broad Institute; Brown University:; Harvard Medical School; Institute for Systems Biology; Memorial Sloan Kettering Cancer Center; University of California Santa Cruz; University of Texas MD Anderson Cancer Center; Biospecimen Core Resource: International Genomics Consortium; Research Institute at Nationwide Children’s Hospital; Tissue Source Sites: Analytic Biologic Services; Asan Medical Center; Asterand Bioscience; Barretos Cancer Hospital; BioreclamationIVT; Botkin Municipal Clinic; Chonnam National University Medical School; Christiana Care Health System; Cureline; Duke University; Emory University; Erasmus University; Indiana University School of Medicine; Institute of Oncology of Moldova; International Genomics Consortium; Invidumed; Israelitisches Krankenhaus Hamburg; Keimyung University School of Medicine; Memorial Sloan Kettering Cancer Center; National Cancer Center Goyang; Ontario Tumour Bank; Peter MacCallum Cancer Centre; Pusan National University Medical School; Ribeirão Preto Medical School; St. Joseph’s Hospital &Medical Center; St. Petersburg Academic University; Tayside Tissue Bank; University of Dundee; University of Kansas Medical Center; University of Michigan; University of North Carolina at Chapel Hill; University of Pittsburgh School of Medicine; University of Texas MD Anderson Cancer Center; Disease Working Group: Duke University; Memorial Sloan Kettering Cancer Center; National Cancer Institute; University of Texas MD Anderson Cancer Center; Yonsei University College of Medicine; Data Coordination Center: CSRA Inc; Project Team: National Institutes of Health. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541:169-175. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1458] [Cited by in RCA: 1408] [Article Influence: 156.4] [Reference Citation Analysis (0)] |
| 67. | Geigl JB, Obenauf AC, Schwarzbraun T, Speicher MR. Defining 'chromosomal instability'. Trends Genet. 2008;24:64-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 248] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 68. | Tsai CK, Yeh TS, Wu RC, Lai YC, Chiang MH, Lu KY, Hung CY, Ho HY, Cheng ML, Lin G. Metabolomic alterations and chromosomal instability status in gastric cancer. World J Gastroenterol. 2018;24:3760-3769. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 23] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (1)] |
| 69. | Jiang Z, Liu Z, Li M, Chen C, Wang X. Immunogenomics Analysis Reveals that TP53 Mutations Inhibit Tumor Immunity in Gastric Cancer. Transl Oncol. 2018;11:1171-1187. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 93] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 70. | Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, Beroukhim R, Pellman D, Levine DA, Lander ES, Meyerson M, Getz G. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413-421. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1637] [Cited by in RCA: 1592] [Article Influence: 113.7] [Reference Citation Analysis (0)] |
| 71. | Haanen JBAG. Converting Cold into Hot Tumors by Combining Immunotherapies. Cell. 2017;170:1055-1056. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 204] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 72. | Postow MA, Chesney J, Pavlick AC, Robert C, Grossmann K, McDermott D, Linette GP, Meyer N, Giguere JK, Agarwala SS, Shaheen M, Ernstoff MS, Minor D, Salama AK, Taylor M, Ott PA, Rollin LM, Horak C, Gagnier P, Wolchok JD, Hodi FS. Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N Engl J Med. 2015;372:2006-2017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2210] [Cited by in RCA: 2256] [Article Influence: 205.1] [Reference Citation Analysis (0)] |
| 73. | Wang B, Zhang W, Jankovic V, Golubov J, Poon P, Oswald EM, Gurer C, Wei J, Ramos I, Wu Q, Waite J, Ni M, Adler C, Wei Y, Macdonald L, Rowlands T, Brydges S, Siao J, Poueymirou W, MacDonald D, Yancopoulos GD, Sleeman MA, Murphy AJ, Skokos D. Combination cancer immunotherapy targeting PD-1 and GITR can rescue CD8+ T cell dysfunction and maintain memory phenotype. Sci Immunol. 2018;3:eaat7061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 139] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 74. | Ribas A, Dummer R, Puzanov I, VanderWalde A, Andtbacka RHI, Michielin O, Olszanski AJ, Malvehy J, Cebon J, Fernandez E, Kirkwood JM, Gajewski TF, Chen L, Gorski KS, Anderson AA, Diede SJ, Lassman ME, Gansert J, Hodi FS, Long GV. Oncolytic Virotherapy Promotes Intratumoral T Cell Infiltration and Improves Anti-PD-1 Immunotherapy. Cell. 2017;170:1109-1119.e10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 800] [Cited by in RCA: 1145] [Article Influence: 127.2] [Reference Citation Analysis (0)] |
| 75. | Bourgeois-Daigneault MC, Roy DG, Aitken AS, El Sayes N, Martin NT, Varette O, Falls T, St-Germain LE, Pelin A, Lichty BD, Stojdl DF, Ungerechts G, Diallo JS, Bell JC. Neoadjuvant oncolytic virotherapy before surgery sensitizes triple-negative breast cancer to immune checkpoint therapy. Sci Transl Med. 2018;10:eaao1641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 233] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 76. | Song W, Shen L, Wang Y, Liu Q, Goodwin TJ, Li J, Dorosheva O, Liu T, Liu R, Huang L. Synergistic and low adverse effect cancer immunotherapy by immunogenic chemotherapy and locally expressed PD-L1 trap. Nat Commun. 2018;9:2237. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 215] [Cited by in RCA: 321] [Article Influence: 40.1] [Reference Citation Analysis (0)] |
| 77. | Bommareddy PK, Aspromonte S, Zloza A, Rabkin SD, Kaufman HL. MEK inhibition enhances oncolytic virus immunotherapy through increased tumor cell killing and T cell activation. Sci Transl Med. 2018;10:eaau0417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 110] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 78. | Goel S, DeCristo MJ, Watt AC, BrinJones H, Sceneay J, Li BB, Khan N, Ubellacker JM, Xie S, Metzger-Filho O, Hoog J, Ellis MJ, Ma CX, Ramm S, Krop IE, Winer EP, Roberts TM, Kim HJ, McAllister SS, Zhao JJ. CDK4/6 inhibition triggers anti-tumour immunity. Nature. 2017;548:471-475. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 700] [Cited by in RCA: 1147] [Article Influence: 127.4] [Reference Citation Analysis (0)] |
| 79. | Li M, Liu Z, Wang X. Exploration of the Combination of PLK1 Inhibition with Immunotherapy in Cancer Treatment. J Oncol. 2018;2018:3979527. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 80. | Lal N, White BS, Goussous G, Pickles O, Mason MJ, Beggs AD, Taniere P, Willcox BE, Guinney J, Middleton GW. KRAS Mutation and Consensus Molecular Subtypes 2 and 3 Are Independently Associated with Reduced Immune Infiltration and Reactivity in Colorectal Cancer. Clin Cancer Res. 2018;24:224-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 122] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 81. | Lee B, Hutchinson R, Wong HL, Tie J, Putoczki T, Tran B, Gibbs P, Christie M. Emerging biomarkers for immunomodulatory cancer treatment of upper gastrointestinal, pancreatic and hepatic cancers. Semin Cancer Biol. 2018;52:241-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 82. | Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883-892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6102] [Cited by in RCA: 6089] [Article Influence: 434.9] [Reference Citation Analysis (0)] |
| 83. | McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, Jamal-Hanjani M, Wilson GA, Birkbak NJ, Hiley CT, Watkins TB, Shafi S, Murugaesu N, Mitter R, Akarca AU, Linares J, Marafioti T, Henry JY, Van Allen EM, Miao D, Schilling B, Schadendorf D, Garraway LA, Makarov V, Rizvi NA, Snyder A, Hellmann MD, Merghoub T, Wolchok JD, Shukla SA, Wu CJ, Peggs KS, Chan TA, Hadrup SR, Quezada SA, Swanton C. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463-1469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2292] [Cited by in RCA: 2489] [Article Influence: 248.9] [Reference Citation Analysis (14)] |
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See:
Manuscript source: Invited manuscript
Specialty type: Oncology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Mastoraki A, Nakayama Y, Koukourakis GVV S-Editor: Wang JL L-Editor: Filipodia E-Editor: Wu YXJ