©The Author(s) 2025.
World J Gastrointest Oncol. Jul 15, 2025; 17(7): 108455
Published online Jul 15, 2025. doi: 10.4251/wjgo.v17.i7.108455
Published online Jul 15, 2025. doi: 10.4251/wjgo.v17.i7.108455
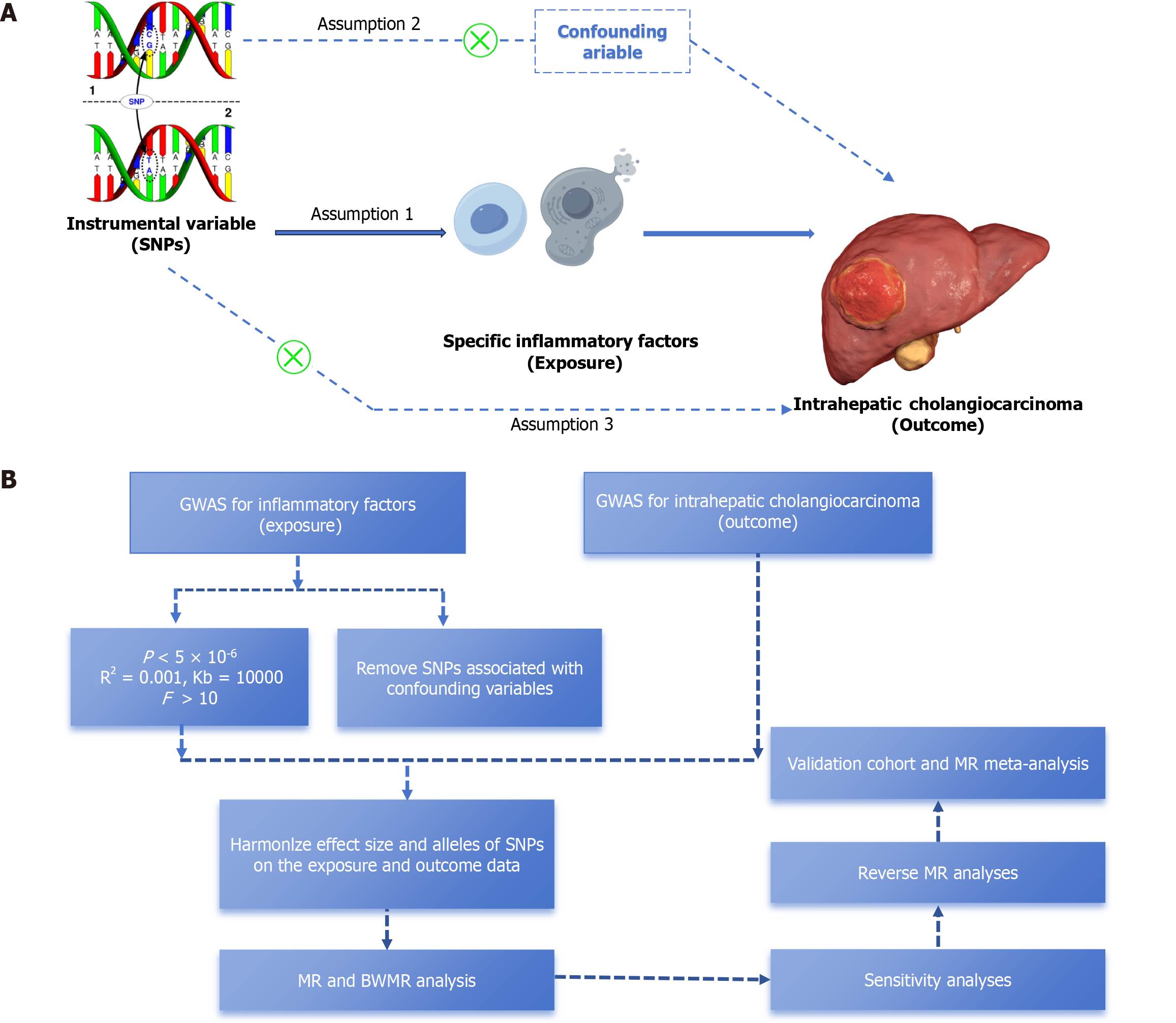
Figure 1 An overview of the current research process and the foundational assumptions of Mendelian randomization analyses.
A: Diagram of the three core assumptions in Mendelian randomization; B: Overall flowchart of the study process. SNPs: Single nucleotide polymorphisms; MR: Mendelian randomization; BWMR: Bayesian weighted Mendelian randomization.
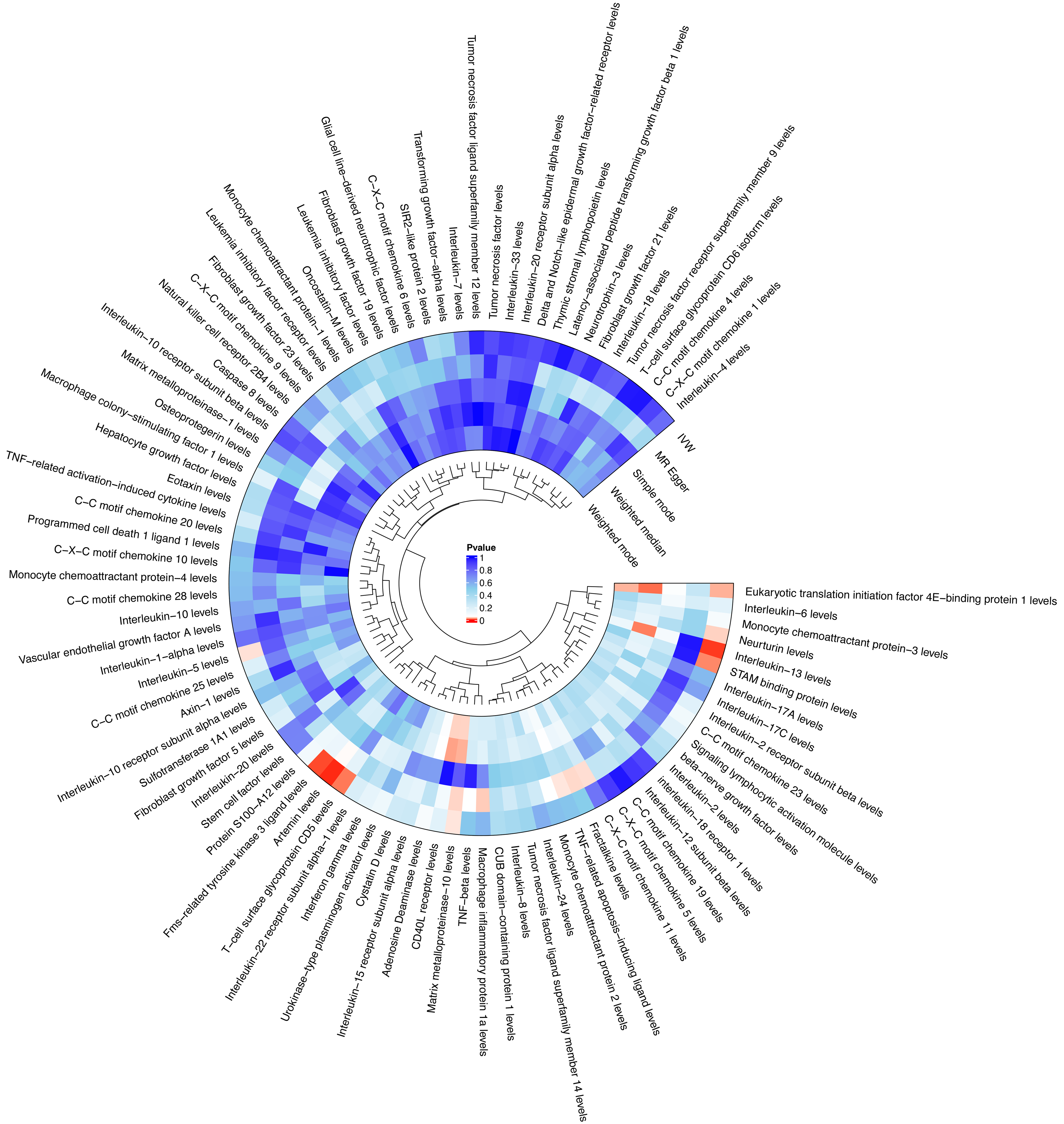
Figure 2
A circular heatmap illustrating the causal impact of inflammatory factors on intrahepatic cholangiocarcinoma risk, analyzed using five statistical methods: Inverse variance weighted, weighted median, Mendelian randomization-Egger, Simple mode, and weighted mode.
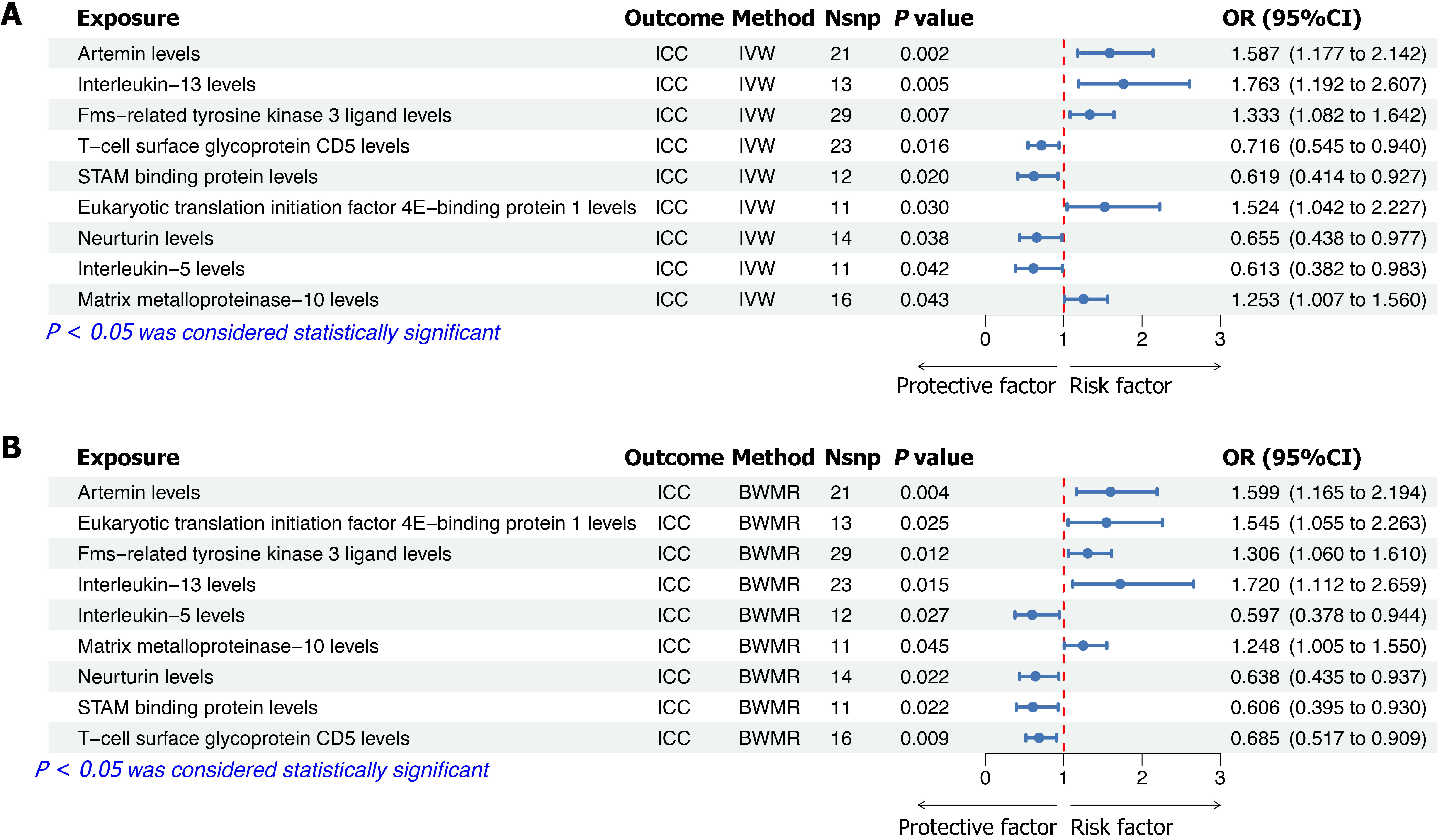
Figure 3 Forest plots illustrating the causal effects of specific inflammatory factors on intrahepatic cholangiocarcinoma risk.
A: Mendelian randomization (MR) analysis results using the Inverse-Variance weighted method, showing odds ratios with corresponding 95%CI and P values for each inflammatory factor; B: Bayesian weighted MR analysis results, validating the causal associations identified by MR. BWMR: Bayesian weighted Mendelian randomization; IVW: Inverse-Variance weighted method; OR: Odds ratio; Nsnp: Number of SNPs.
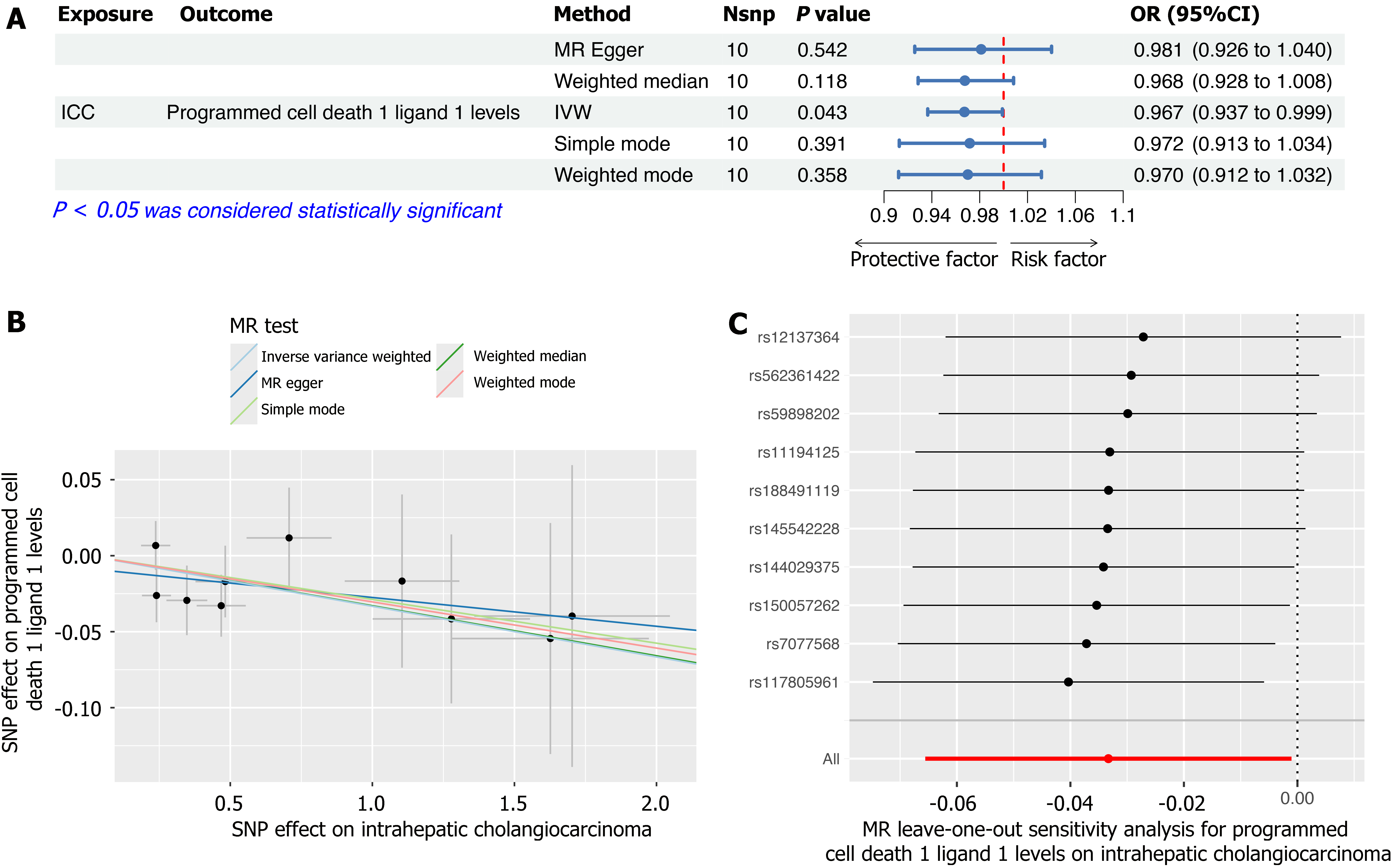
Figure 4 Results of reverse Mendelian randomization analysis examining intrahepatic cholangiocarcinoma as the exposure and specific inflammatory factors as the outcomes.
A: A forest plot displays the significant causal effects of intrahepatic cholangiocarcinoma (ICC) on programmed cell death 1 ligand 1 levels; B: Scatter plots were employed to illustrate the causal relationship between ICC and programmed cell death ligand 1 levels, using five different Mendelian randomization methods; C: Forest plots from "leave-one-out" sensitivity analyses, illustrating the influence of individual single nucleotide polymorphisms on the overall results. ICC: Intrahepatic cholangiocarcinoma; MR: Mendelian randomization; IVW: Inverse-Variance weighted method; OR: Odds ratio; SNPs: Single nucleotide polymorphisms; Nsnp: Number of SNPs.
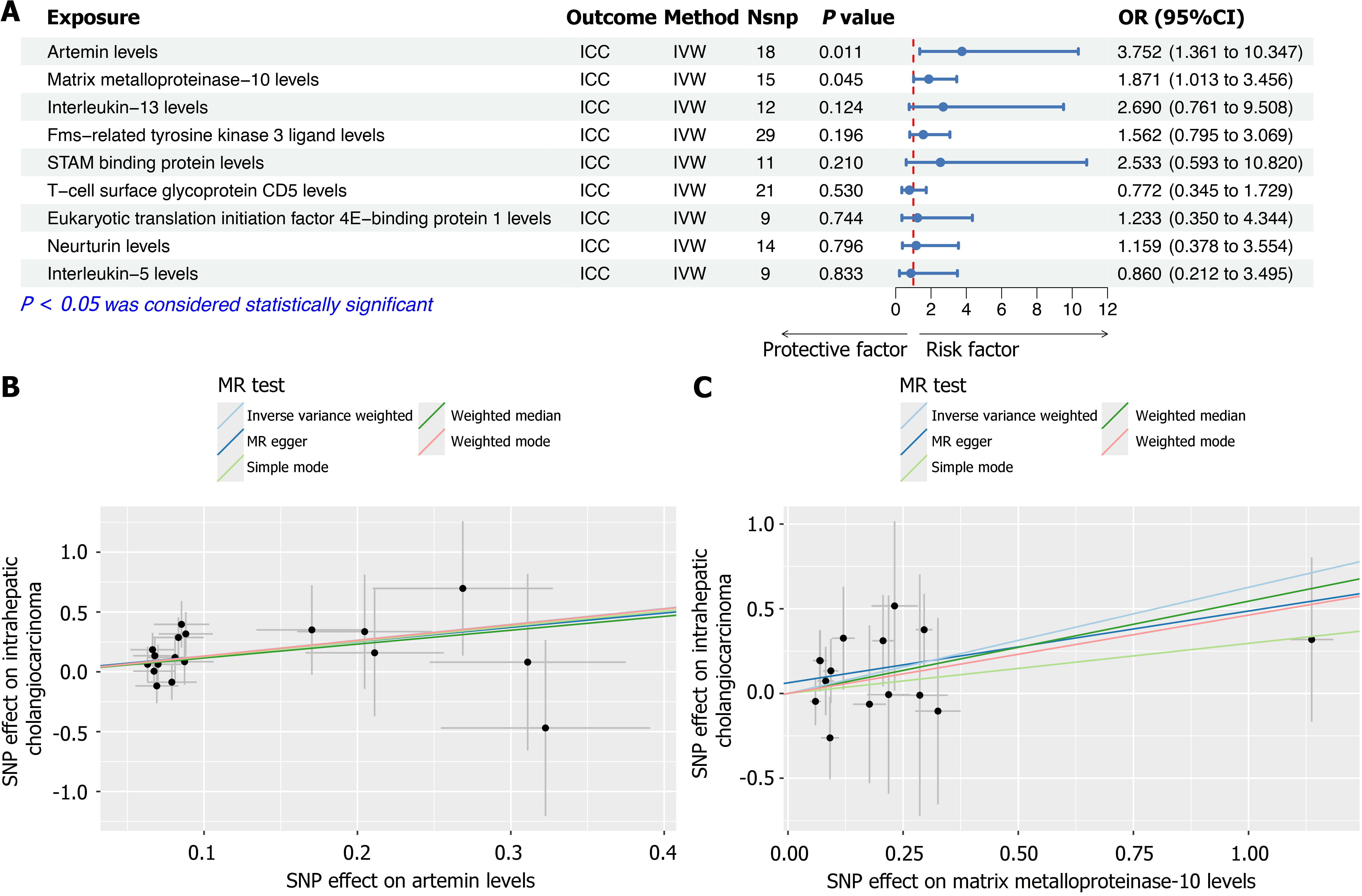
Figure 5 Validation of causal associations between specific inflammatory factors and intrahepatic cholangiocarcinoma risk in an independent cohort.
A: Forest plot showing the odds ratios with 95%CI for inflammatory factors previously identified in the discovery cohort, evaluated in the validation cohort using the Inverse-Variance weighted (IVW) Mendelian randomization (MR) method; B: Scatter plot illustrating the causal association between genetically predicted artemin levels and ICC risk, analyzed using five MR methods (IVW, weighted median, MR-Egger, simple mode, and weighted mode); C: Scatter plot illustrating the causal association between genetically predicted matrix metalloproteinase-10 levels and ICC risk, using the same five MR methods. ICC: Intrahepatic cholangiocarcinoma; MR: Mendelian randomization; IVW: Inverse-Variance weighted method; OR: Odds ratio; SNPs: Single nucleotide polymorphisms; Nsnp: Number of SNPs.
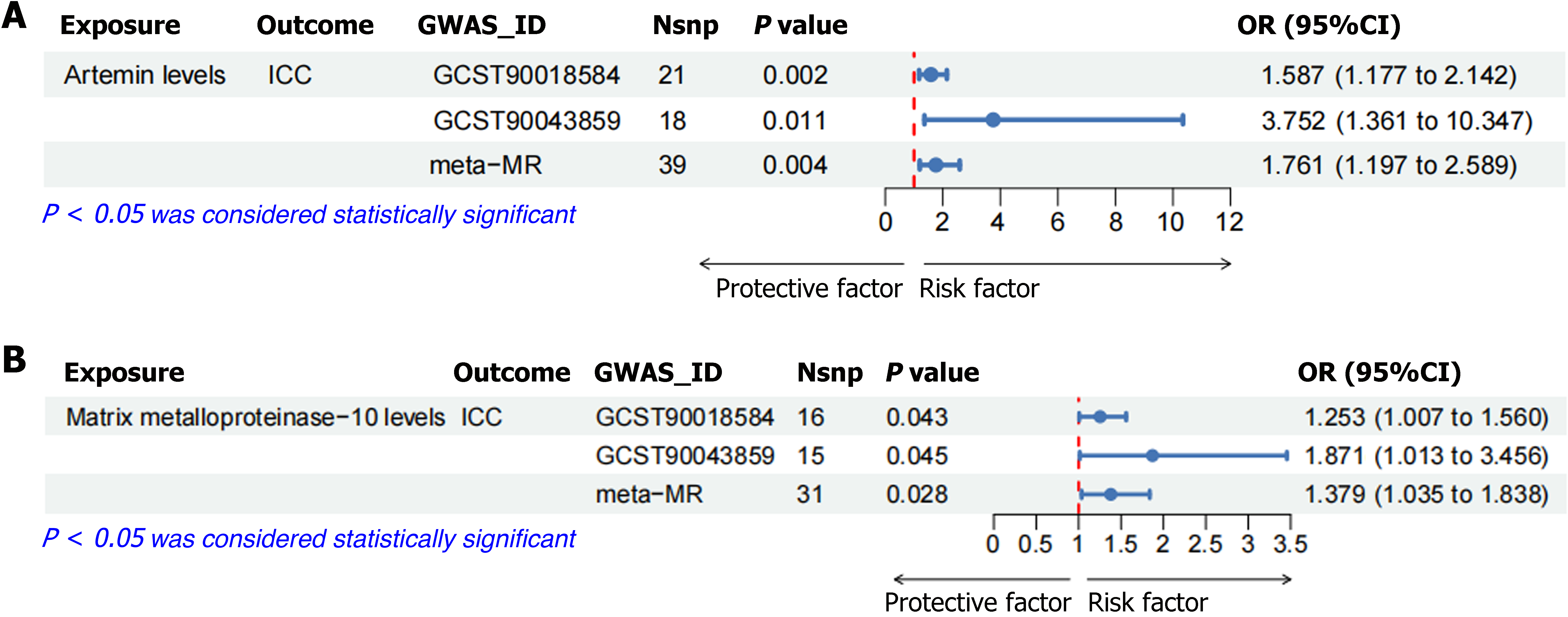
Figure 6 The meta-Mendelian randomization analysis was conducted to examine the association between artemin levels and matrix metalloproteinase-10 levels using data from both the training and validation sets.
A: Meta-Mendelian randomization (MR) analysis of artemin levels was conducted based on intrahepatic cholangiocarcinoma (ICC) data from both the training and validation sets; B: Meta-MR analysis of matrix metalloproteinase-10 levels was conducted based on ICC data from both the training and validation sets. ICC: Intrahepatic cholangiocarcinoma; OR: Odds ratio; GWAS: Genome-wide association studies; Nsnp: Number of SNPs.
- Citation: Chen B, Chen J, Chen ZT, Feng ZP, Lv HB, Jiang GP. Genetic evidence for the causal influence of inflammatory factors on intrahepatic cholangiocarcinoma risk. World J Gastrointest Oncol 2025; 17(7): 108455
- URL: https://www.wjgnet.com/1948-5204/full/v17/i7/108455.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i7.108455