©The Author(s) 2025.
World J Gastrointest Oncol. Feb 15, 2025; 17(2): 97125
Published online Feb 15, 2025. doi: 10.4251/wjgo.v17.i2.97125
Published online Feb 15, 2025. doi: 10.4251/wjgo.v17.i2.97125
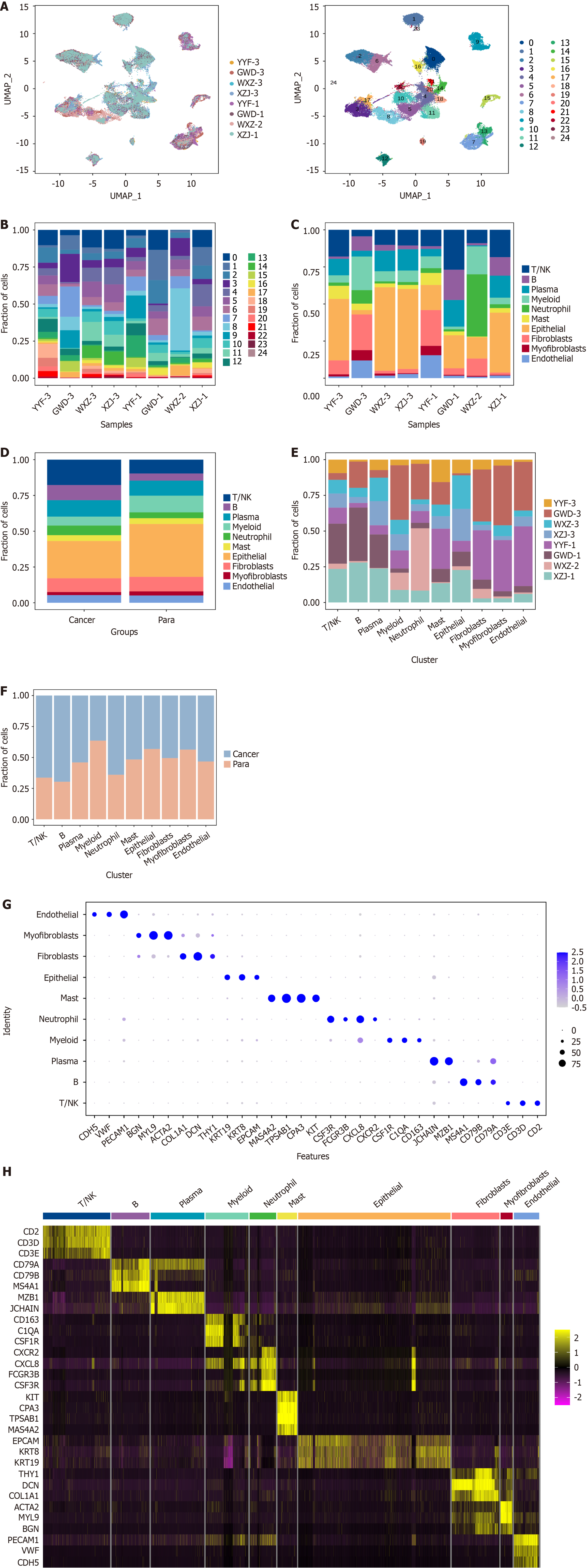
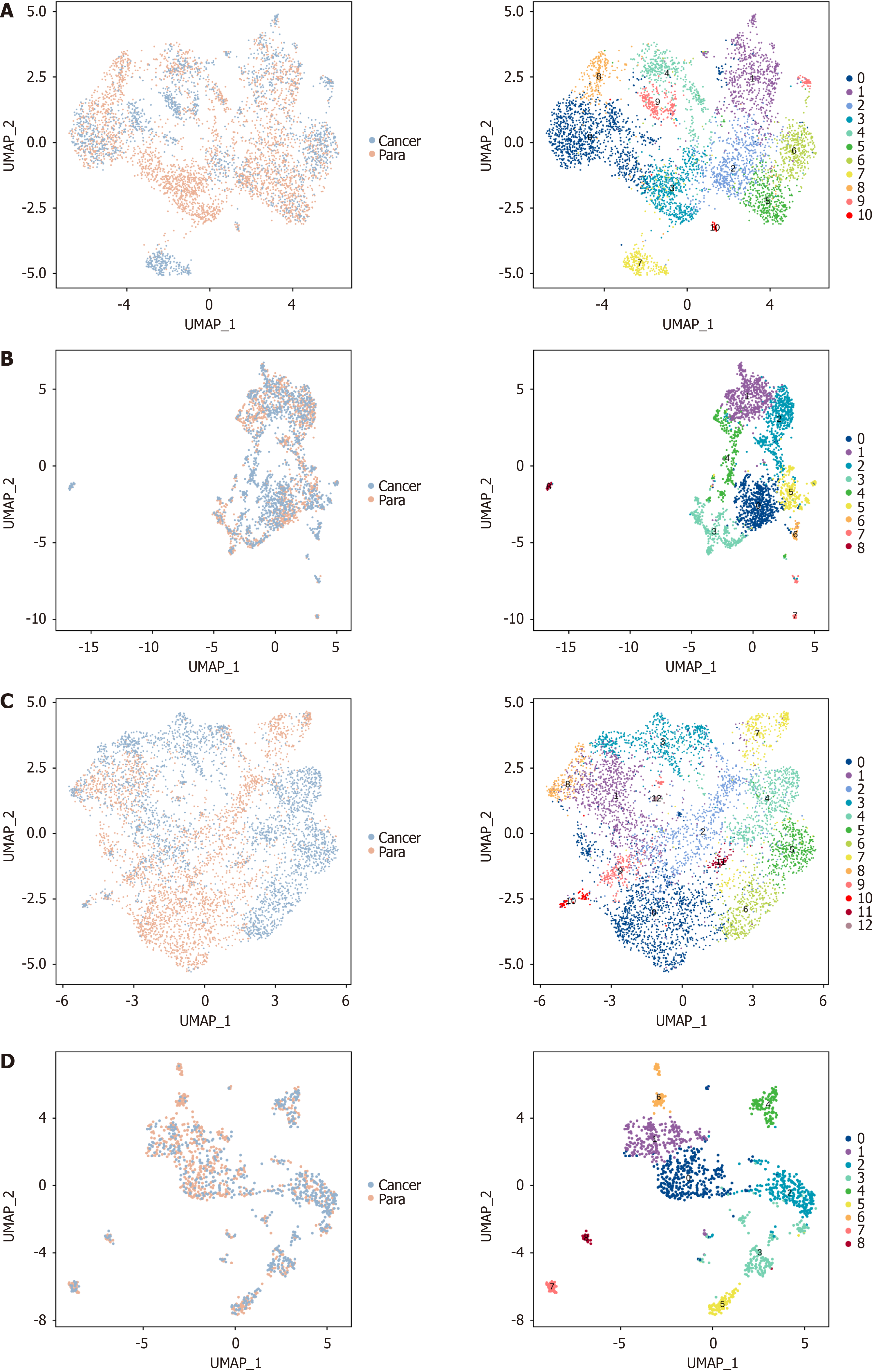
Figure 1 Global single-cell atlas of four different stages of gastric cancer.
A: Single-cell atlas of 73645 cells captured from 25 cell clusters. Tissue samples are shown on the left, and cell clusters on the right, with colors indicating cell types; B and C: Composition of cell clusters and cell types within various tissue samples (n = 8); D: Proportion of different cell types in cancer (n = 4) and para cancerous tissues (n = 4); E: Tissue sample distribution in different cell types; F: Distribution of cancer and para cancerous tissues across varying cell types; G and H: Dot Plot and Heatmap of the corresponding marker genes identifying the cell types.
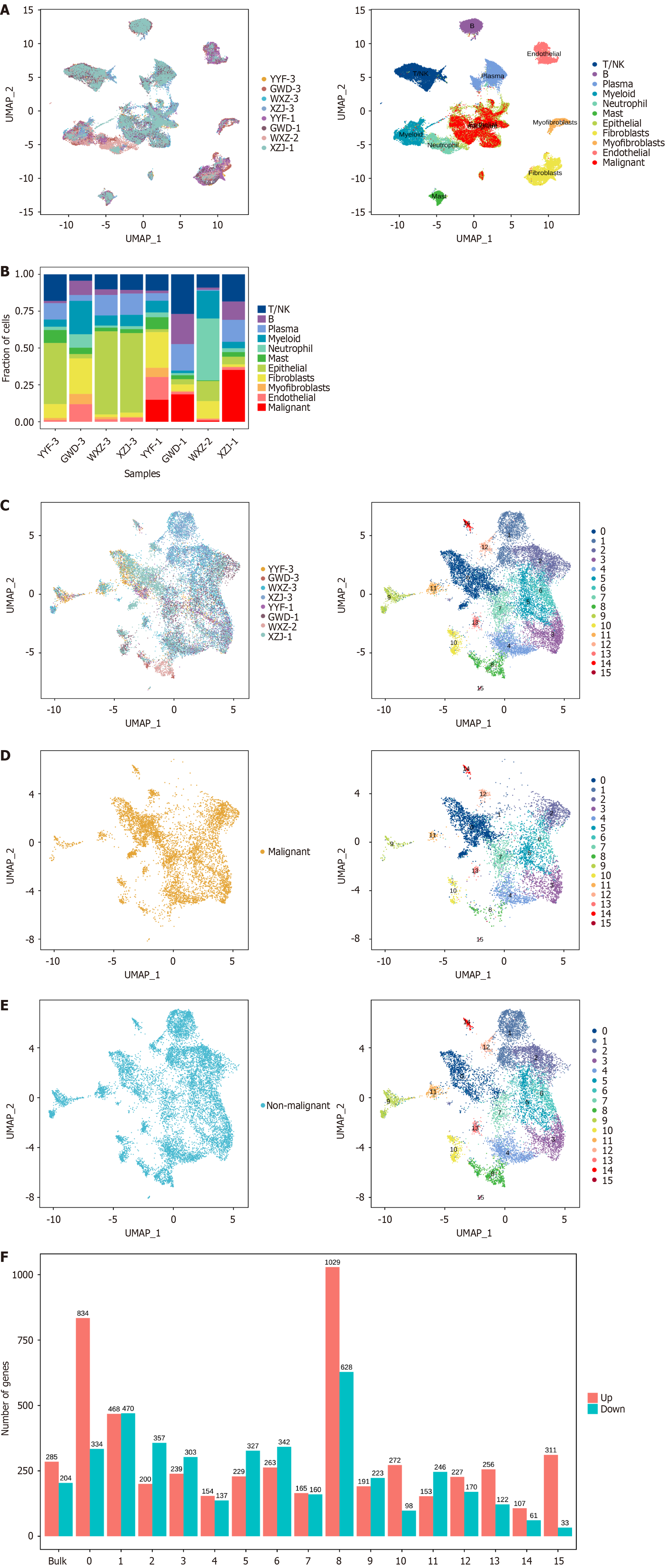
Figure 2 Tumor cell identification and epithelial cell re-clustering.
A: Uniform manifold approximation and projection (UMAP) plot after cancer cell identification, with tissue samples on the left and cell types on the right; B: Proportion of different cell types in tissue samples from patients at various stages after cancer cell identification; C-E: UMAP of epithelial cell re-clustering (C, left: Tissue samples, right: Cell cluster), malignant cell re-clustering (D, left: Malignant cells, right: Cell types), and non-malignant cell re-clustering (E: left: Non-malignant cells, right: Cell types); F: Differential analysis of re-clustering between malignant tumor cells and non-malignant tumor cells.
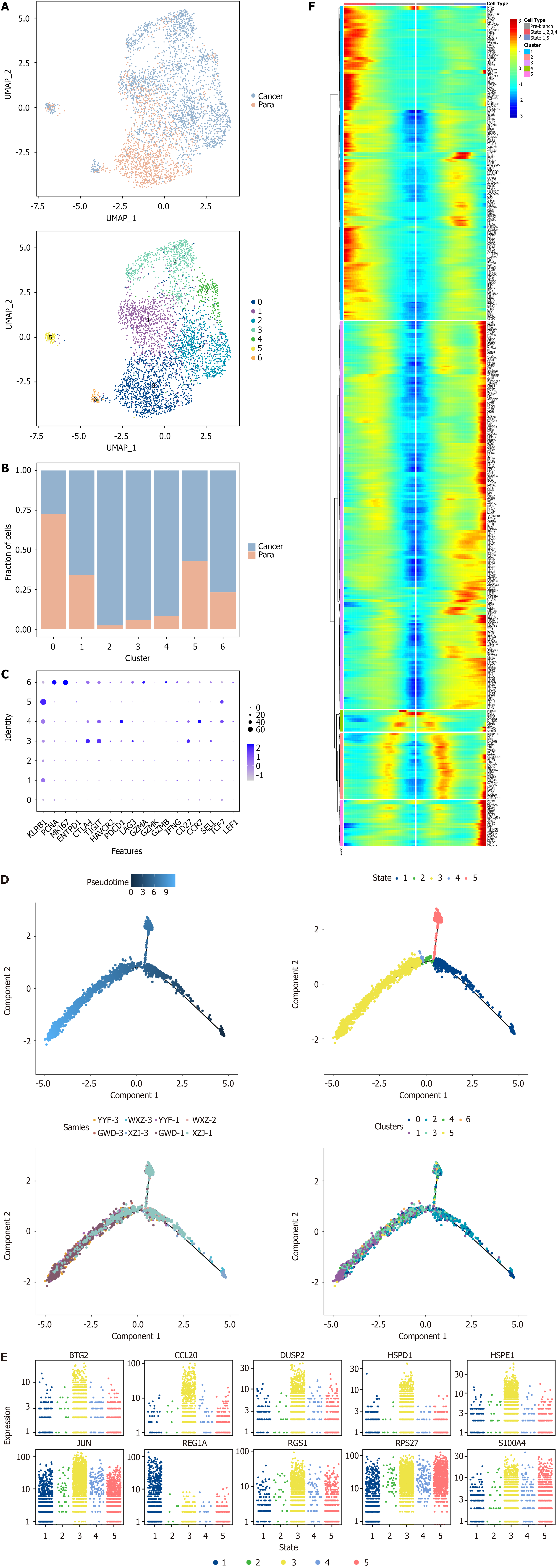
Figure 3 CD4 T cell re-clustering and quasi-temporal analysis.
A: Uniform manifold approximation and projection of CD4 T cell re-clustering, with tissues on the left and cell types on the right; B: Proportion of different CD4 T cell clusters in cancerous tissues and para cancerous tissues; C: Dot Plot illustrating the mean expression levels of marker genes in six main identified cell clusters; D: Quasi-temporal analysis displaying quasi-temporal distribution of cell trajectories (top left), cell trajectories in differentiated states (top right), sample cell trajectories (bottom left), subset cell trajectories (bottom right); E: Scatter plot of differential gene expression in differentiation states; F: Heatmap of hierarchical clustering of branched differential genes.
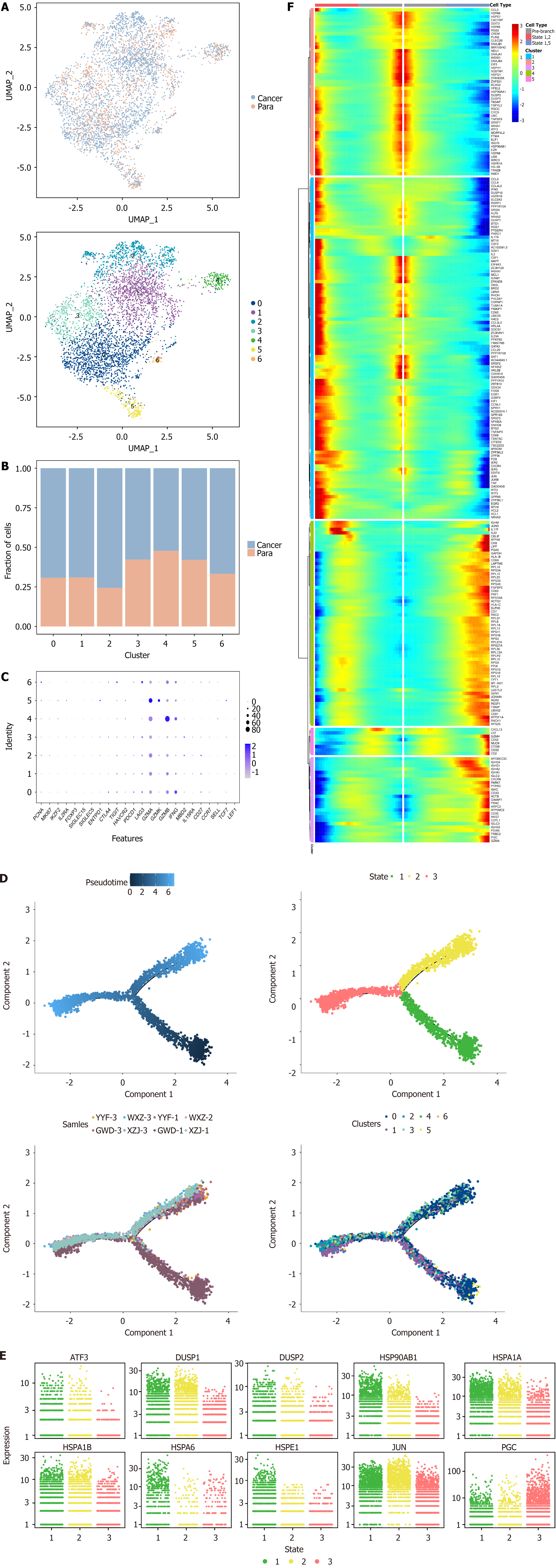
Figure 4 CD8 T cell re-clustering and quasi-timing analysis.
A: Uniform manifold approximation and projection of CD8 T cell re-clustering, with tissues on the left and cell types on the right; B: Proportion of different CD8 T cell clusters in cancerous tissues and para cancerous tissues; C: Dot Plot illustrating the mean expression levels of marker genes across six main identified cell clusters; D: Quasi-temporal analysis depicting quasi-temporal distribution of cell trajectories (top left), cell trajectories in differentiated state (top right), sample cell trajectories (bottom left), and subset cell trajectories (bottom right); E: Scatter plot of differential gene expression in differentiation states; F: Heatmap of hierarchical clustering of branching differential genes.
Figure 5 Reclassification of myeloid cells, endothelial cells, fibroblasts, and myofibroblasts.
Uniform manifold approximation and projection. A: Myeloid cell; B: Endothelial cell; C: Fibroblast; D: Myofibroblast; re-clustering, with tissues on the left and cell types on the right.
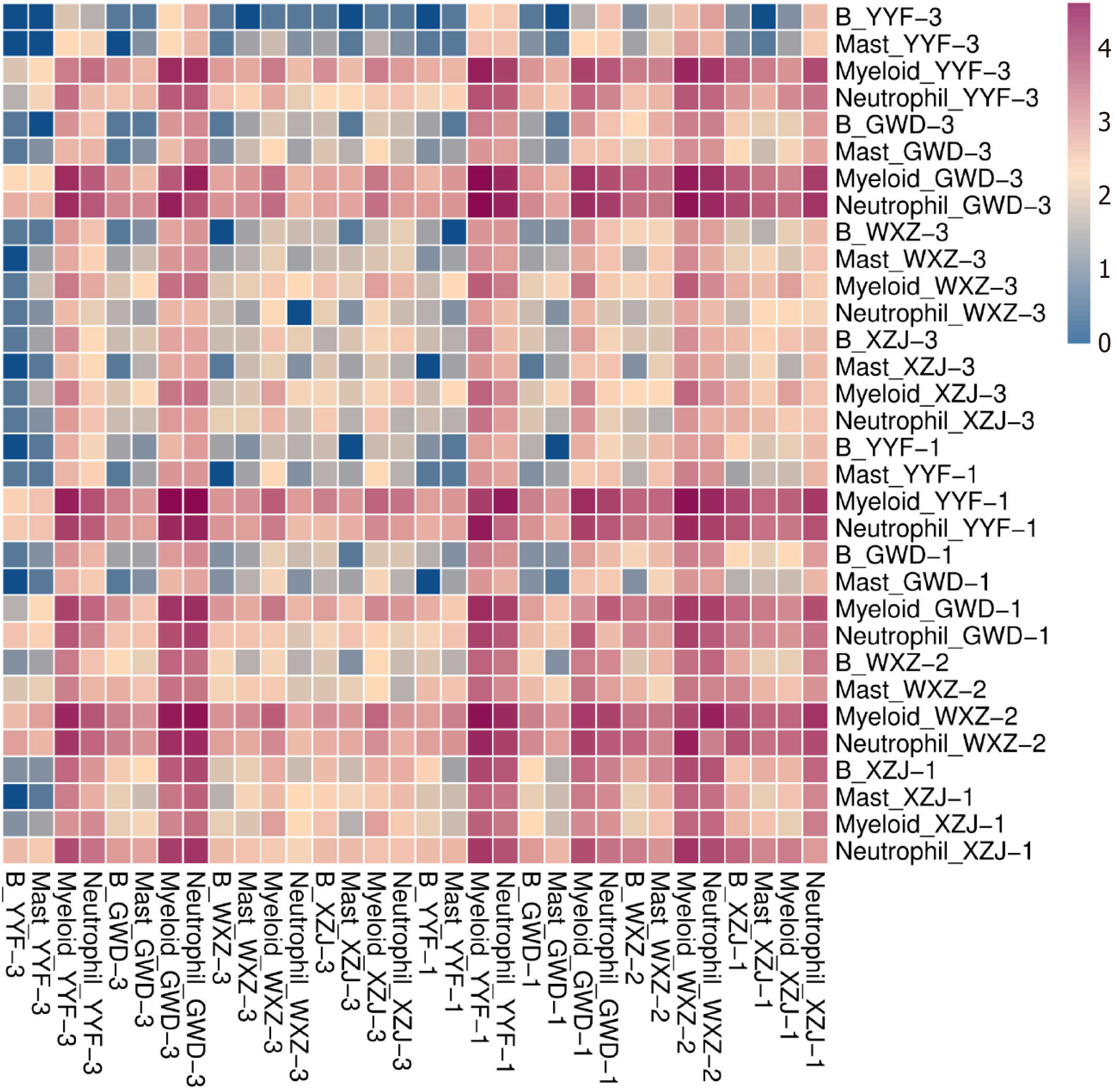
Figure 6 Cell-cell interactions.
The rows and columns in the heatmap representing different cell types across various tissue samples, respectively. The colors indicating the degree of intercellular interaction, ranging from blue (0) to red (4), with higher values denoting stronger intercellular activity.
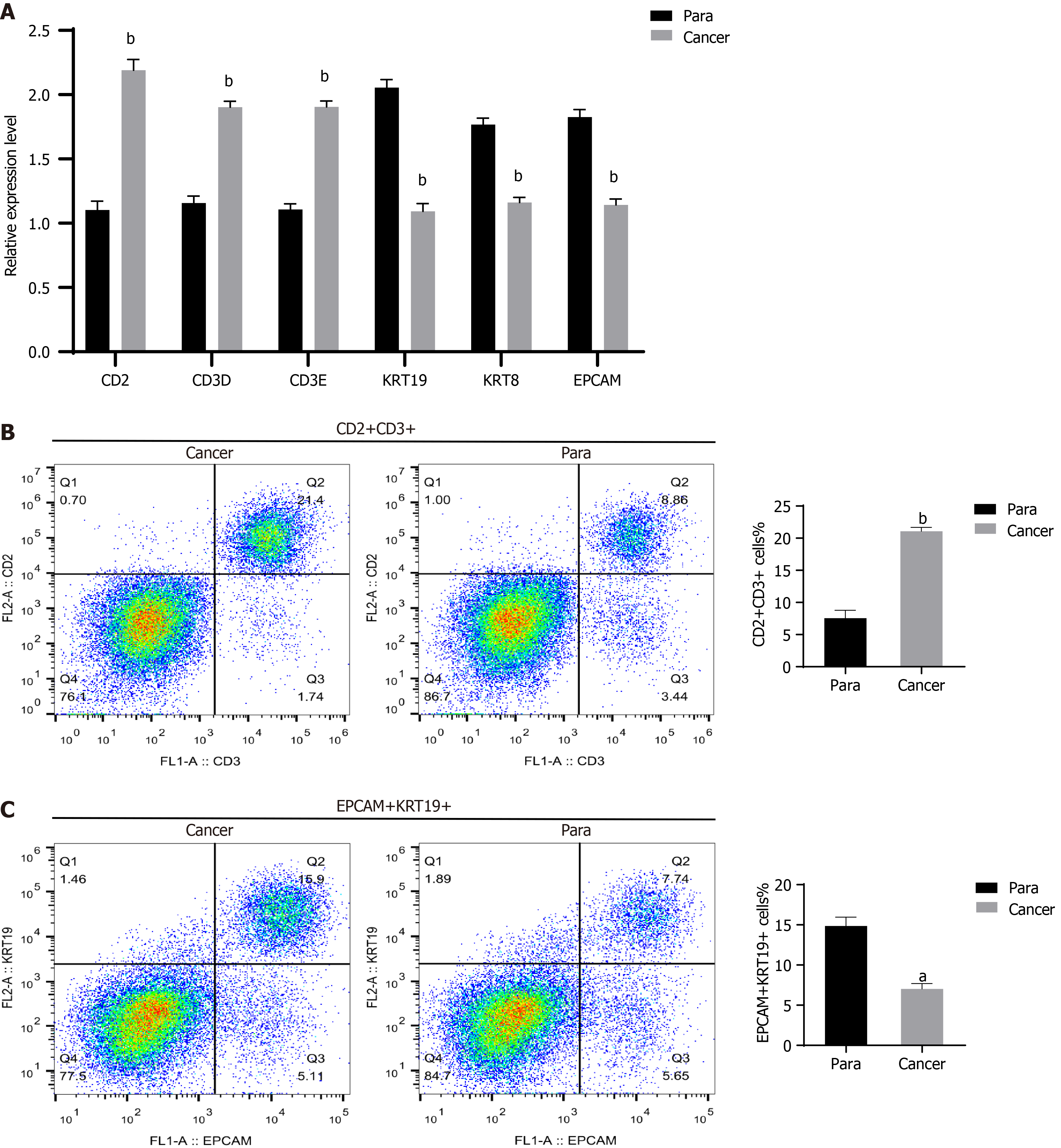
Figure 7 Trends in T cells and epithelial cells in gastric cancer tissues.
A: Quantitative real-time polymerase chain reaction for assessing the marker gene expression levels of T cells and epithelial cells; B and C: Flow cytometry for analyzing the quantities of T cell and epithelial cell marker genes. aP < 0.001, bP < 0.0001 vs para cancerous tissues.
- Citation: Tang XS, Xu CL, Li N, Zhang JQ, Tang Y. Landscape of four different stages of human gastric cancer revealed by single-cell sequencing. World J Gastrointest Oncol 2025; 17(2): 97125
- URL: https://www.wjgnet.com/1948-5204/full/v17/i2/97125.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i2.97125