©The Author(s) 2025.
World J Gastrointest Oncol. Nov 15, 2025; 17(11): 111670
Published online Nov 15, 2025. doi: 10.4251/wjgo.v17.i11.111670
Published online Nov 15, 2025. doi: 10.4251/wjgo.v17.i11.111670
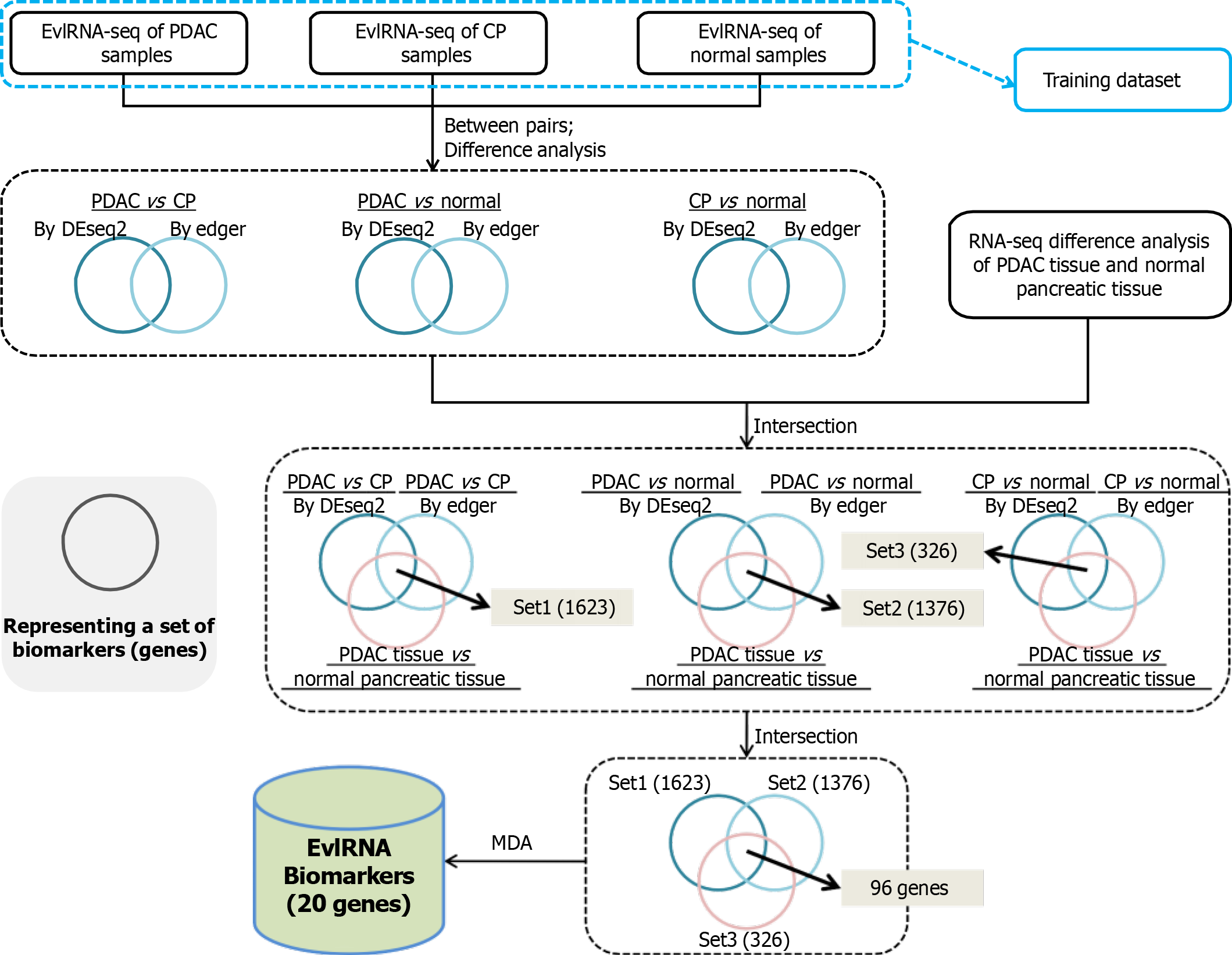
Figure 1 Feature selection method flowchart.
EvlRNA: Extracellular vesicle long RNA; PDAC: Pancreatic ductal adenocarcinoma; CP: Chronic pancreatitis.
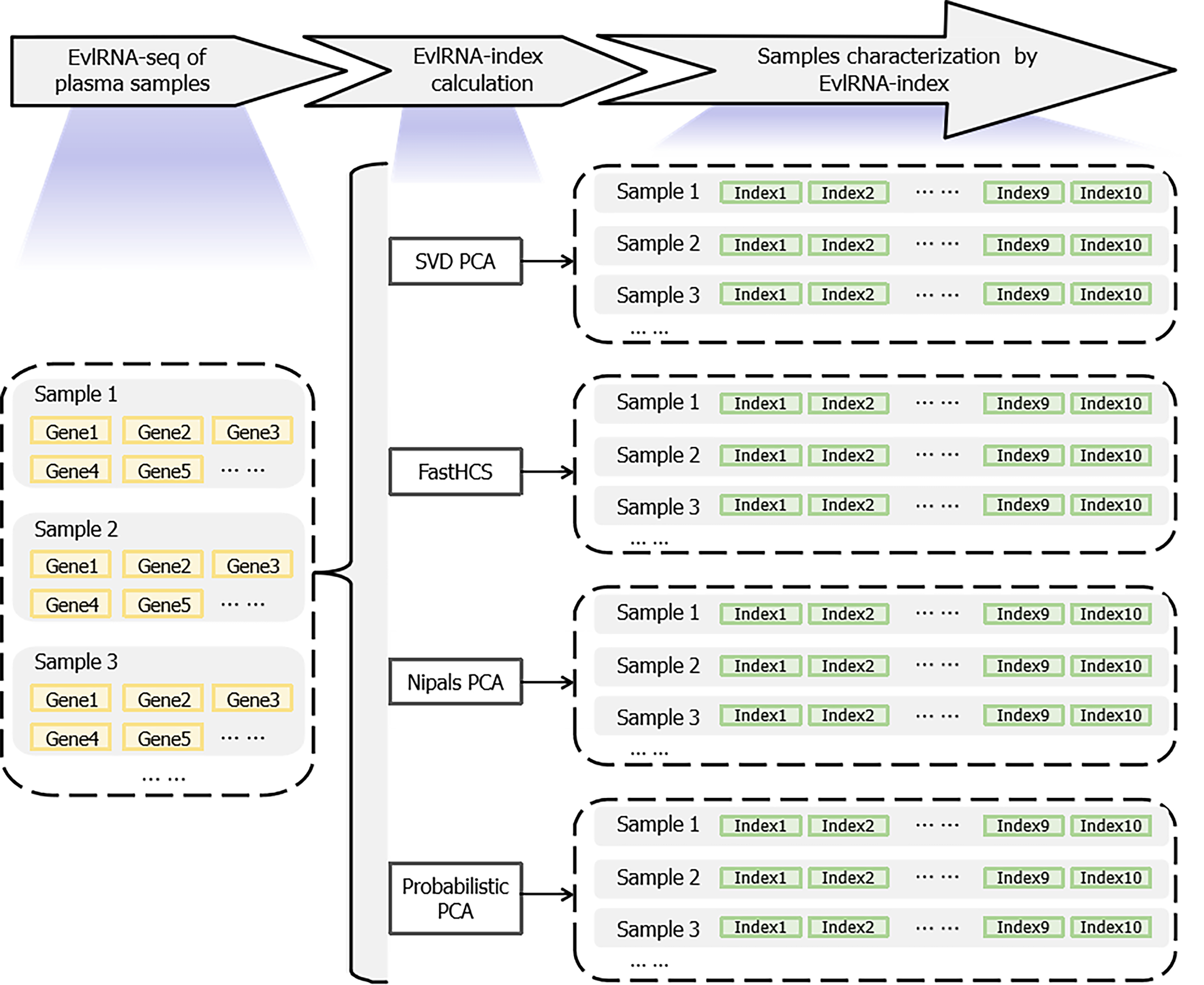
Figure 2 Extracellular vesicle long RNA-index calculation of plasma extracellular vesicle long RNA samples.
EvlRNA: Extracellular vesicle long RNA; SVD: Singular value decomposition; PCA: Principal component analysis; FastHCS: High-dimensional congruent subsets; Nipals: Nonlinear iterative partial least squares.
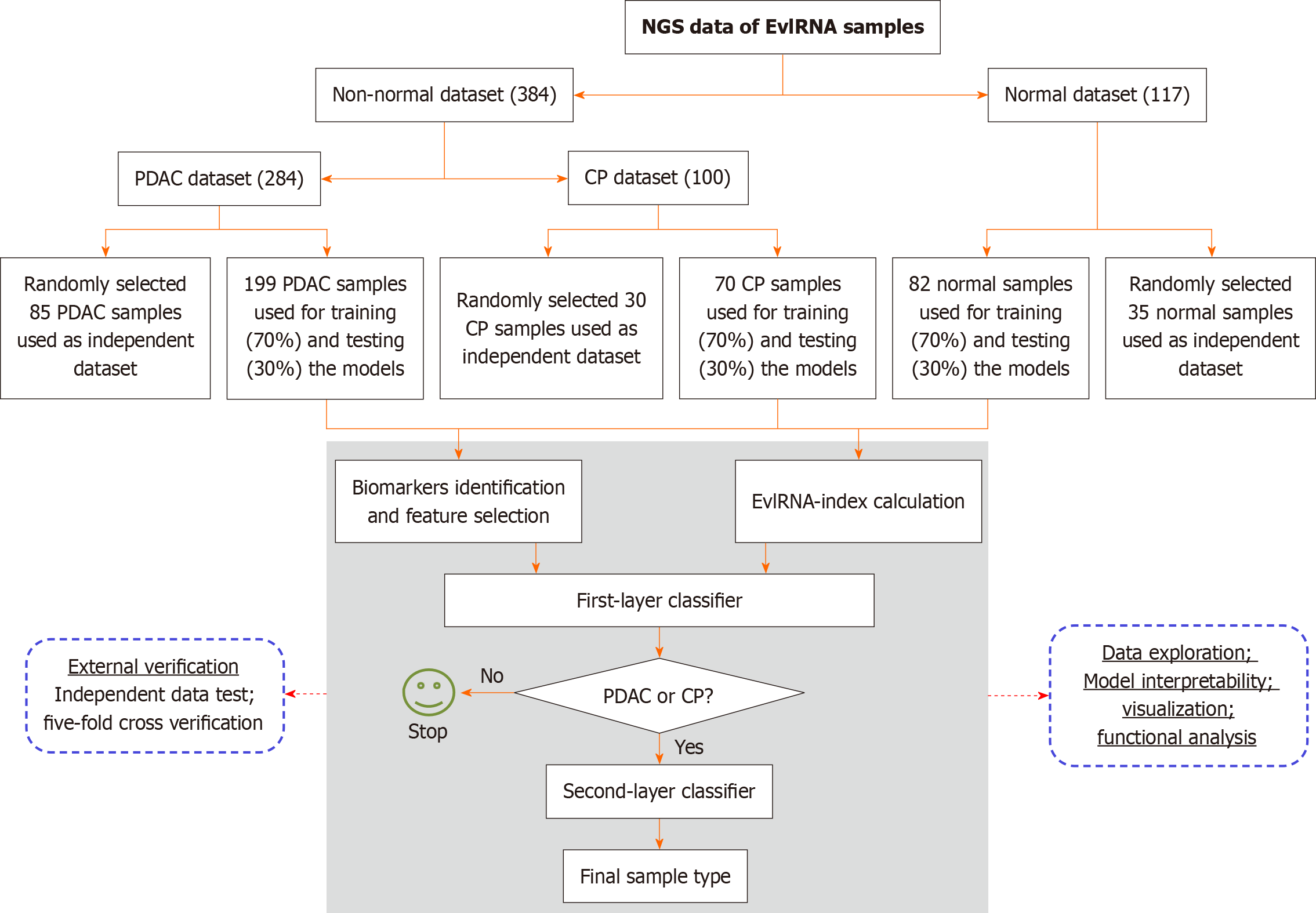
Figure 3 Diagnostic model construction flowchart.
NGS: Next-generation sequencing; EvlRNA: Extracellular vesicle long RNA; PDAC: Pancreatic ductal adenocarcinoma; CP: Chronic pancreatitis.
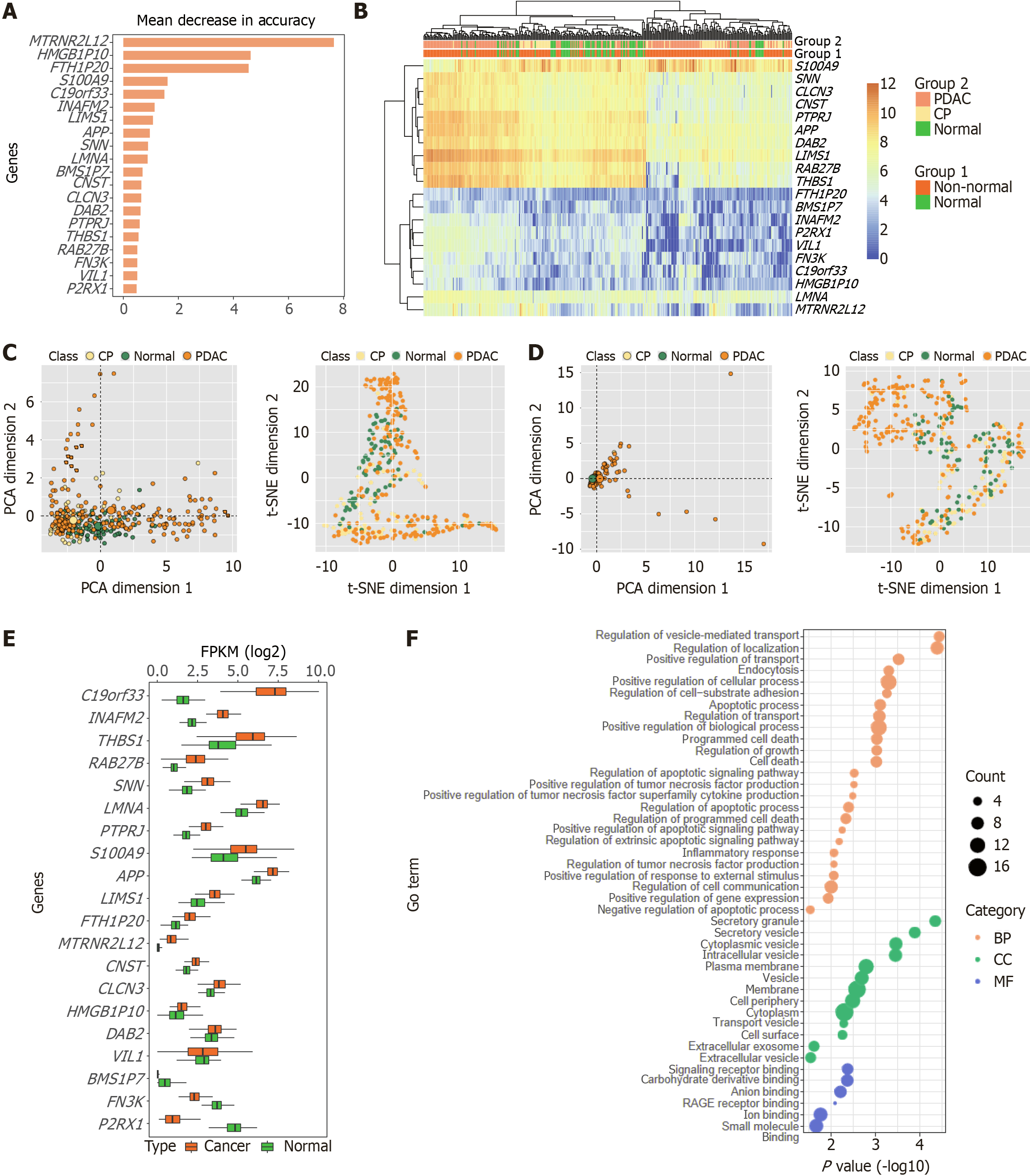
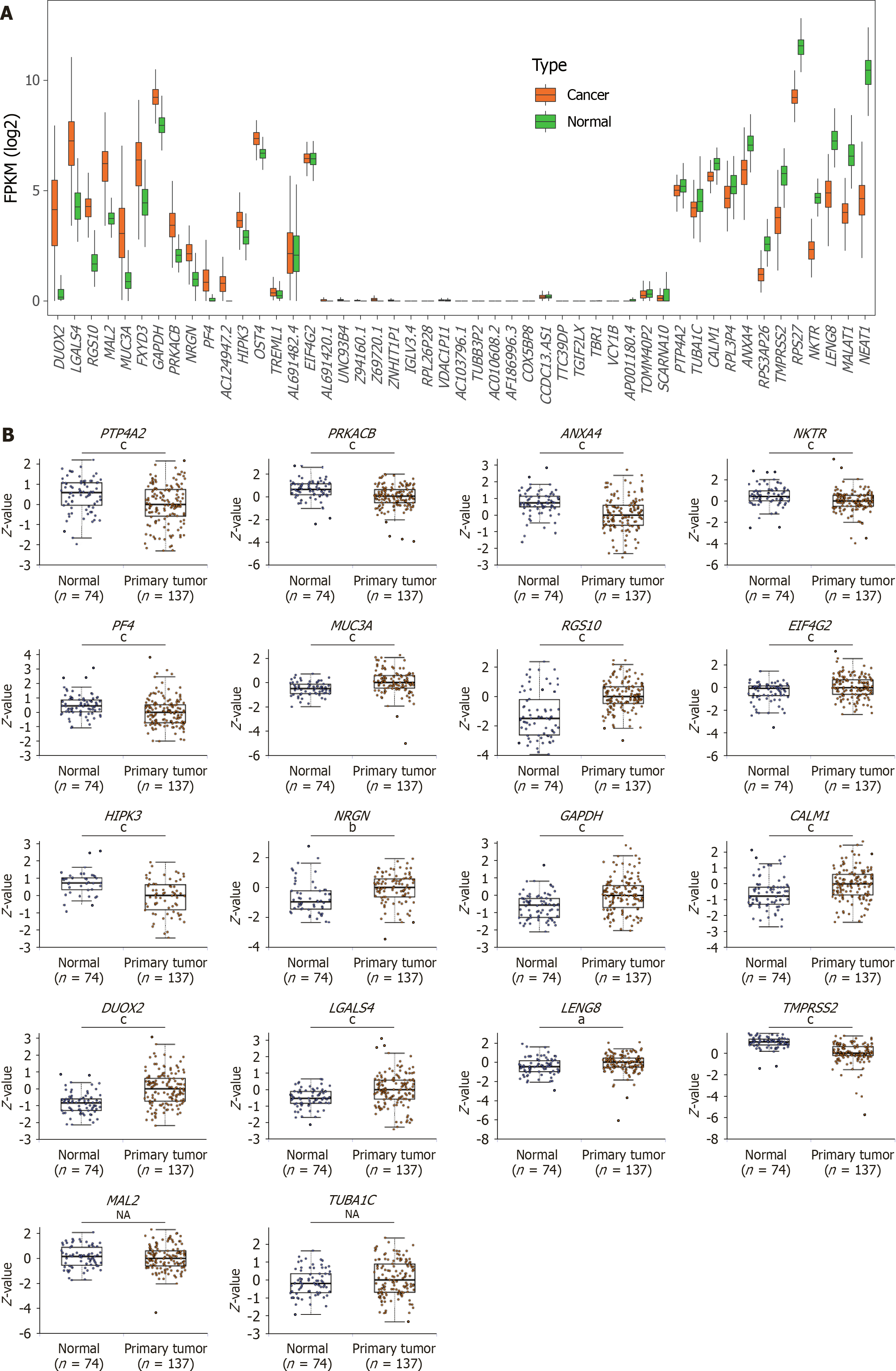
Figure 4 Analysis of potential biomarkers and visualization.
A: Feature (gene set) selection with mean decrease in accuracy; B: Heatmap analysis of the selected biomarkers (20 genes); C: Principal component analysis and t-distributed stochastic neighbor embedding analysis based on the extracellular vesicle long RNA expression of the selected biomarkers (20 genes); D: Principal component analysis and t-distributed stochastic neighbor embedding analysis based on the extracellular vesicle long RNA expression of the eight genes screened by Yu et al[25]; E: Gene expression analysis of potential biomarkers was performed using RNA-seq data from the The Cancer Genome Atlas and Genotype-Tissue Expression databases, which included 178 pancreatic ductal adenocarcinoma tissue samples and 171 normal pancreatic tissue samples; F: Gene Ontology analysis of 20 genes (potential biomarkers). PDAC: Pancreatic ductal adenocarcinoma; CP: Chronic pancreatitis; PCA: Principal component analysis; t-SNE: T-distributed stochastic neighbor embedding; FPKM: Fragments per kilobase of exon per million mapped fragments; GO: Gene Ontology; RAGE: Receptor for advanced glycation end products; BP: Biological process; CC: Cellular component; MF: Molecular function.
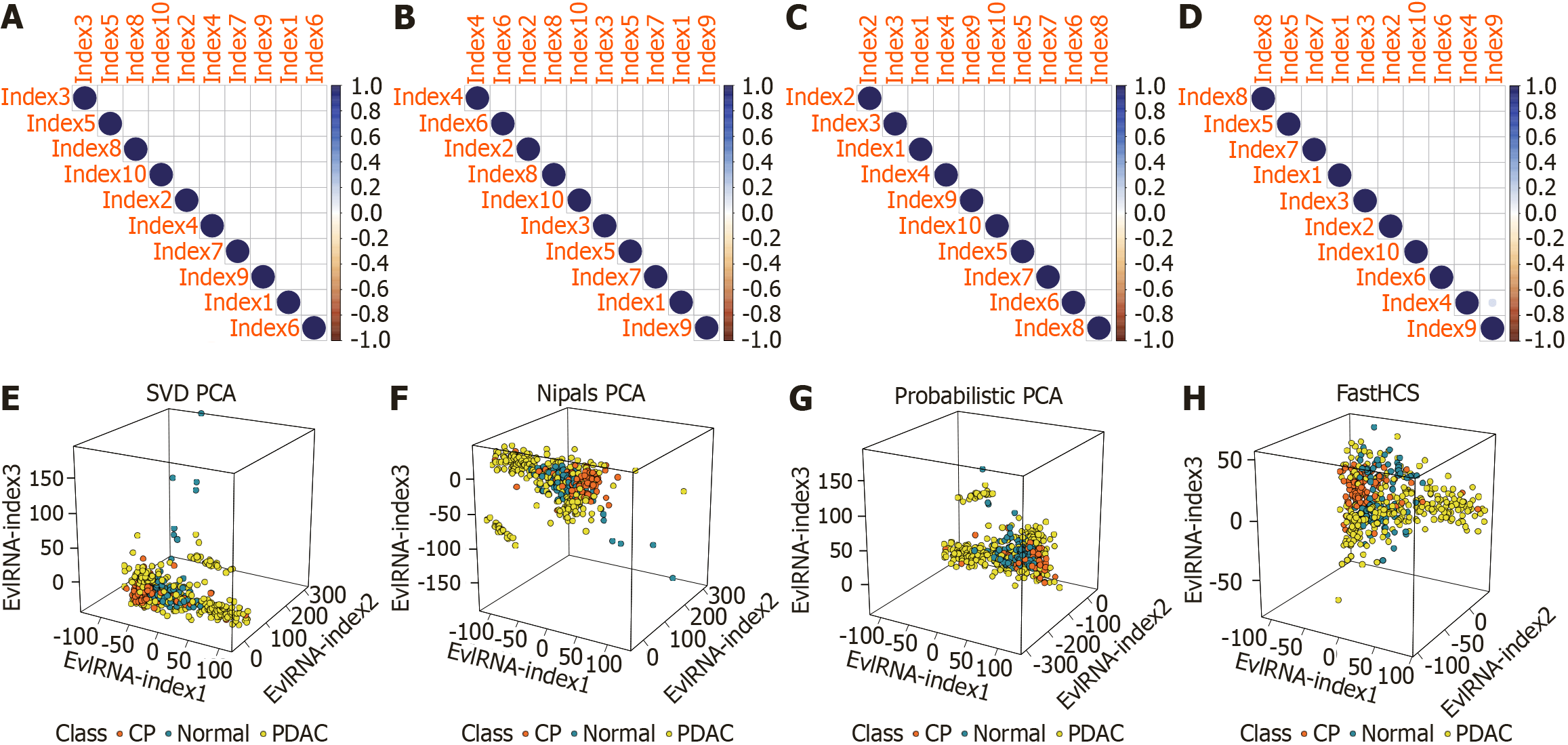
Figure 5 Correlation analysis and visualization of extracellular vesicle long RNA-index calculated by different algorithms.
A: Correlation analysis of extracellular vesicle long RNA (EvlRNA)-index calculated based on singular value decomposition principal component analysis (PCA) algorithm; B: Correlation analysis of EvlRNA-index calculated based on nonlinear iterative partial least squares PCA algorithm; C: Correlation analysis of EvlRNA-index calculated based on Probabilistic PCA algorithm; D: Correlation analysis of EvlRNA-index calculated based on FastHCS algorithm; E: Visualization of EvlRNA-index calculated by singular value decomposition PCA algorithm; F: Visualization of EvlRNA-index calculated by nonlinear iterative partial least squares PCA algorithm; G: Visualization of EvlRNA-index calculated by Probabilistic PCA algorithm; H: Visualization of EvlRNA-index calculated by FastHCS algorithm. SVD: Singular value decomposition; PCA: Principal component analysis; EvlRNA: Extracellular vesicle long RNA; PDAC: Pancreatic ductal adenocarcinoma; CP: Chronic pancreatitis; FastHCS: High-dimensional congruent subsets; Nipals: Nonlinear iterative partial least squares.
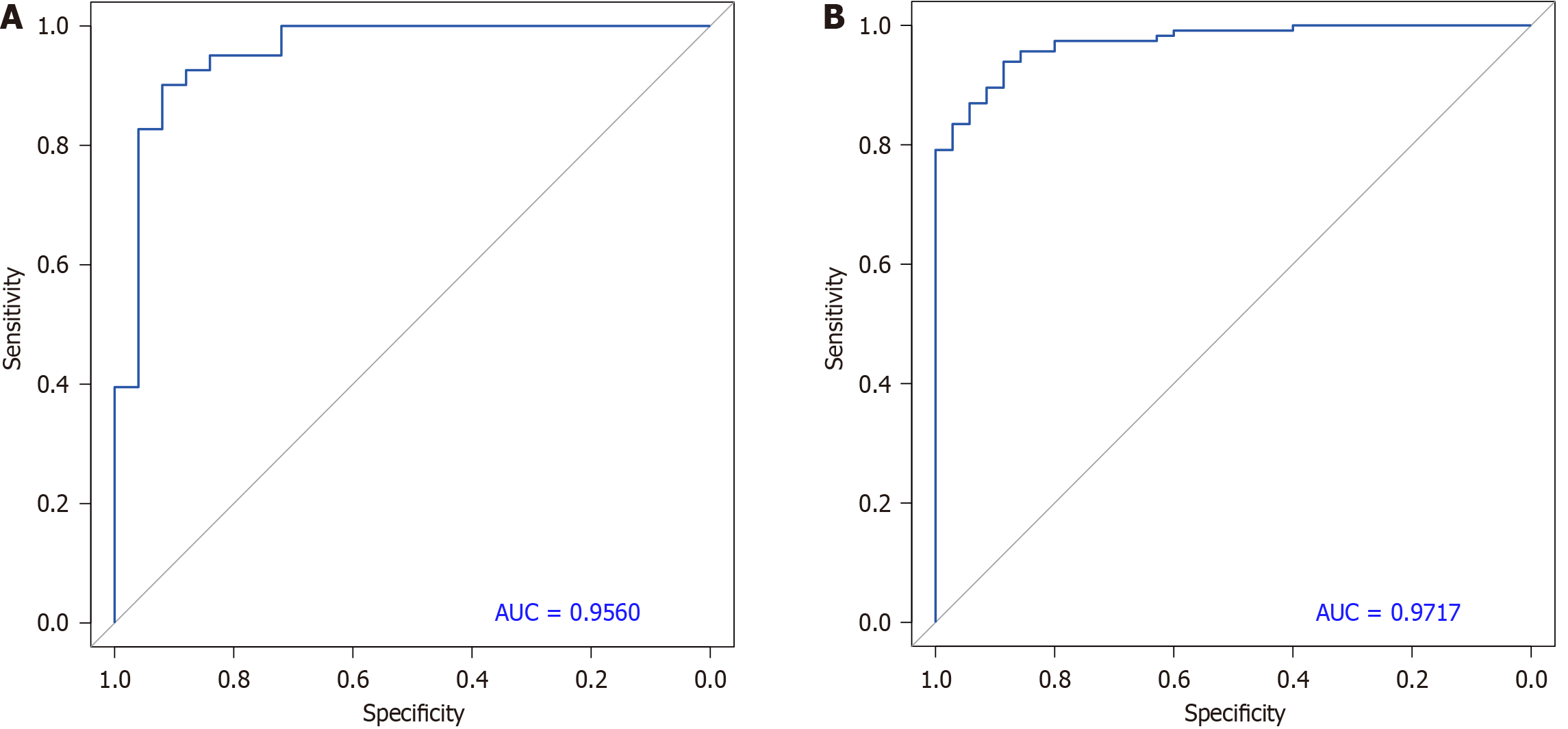
Figure 6 Receiver operating characteristic curve of pancreatic ductal adenocarcinoma and chronic pancreatitis vs health-probabilistic principal component analysis index-support vector machine (1-18).
A: Based on training dataset; B: Based on the independent dataset. AUC: Area under the receiver operating characteristic curve.
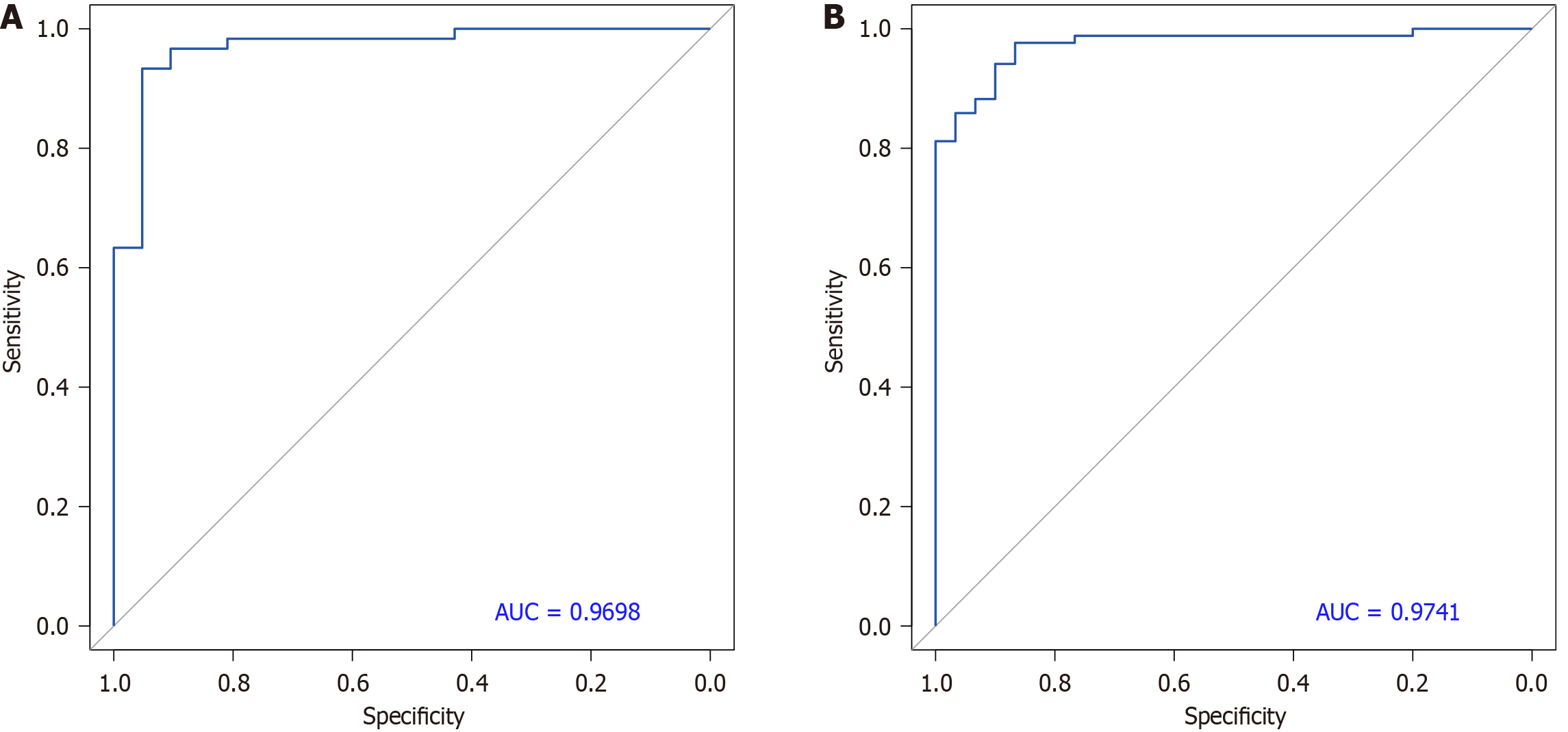
Figure 7 Receiver operating characteristic curve of pancreatic ductal adenocarcinoma vs chronic pancreatitis-probabilistic principal component analysis index-random forest (2-17).
A: Based on training dataset; B: Based on the independent dataset. AUC: Area under the receiver operating characteristic curve.
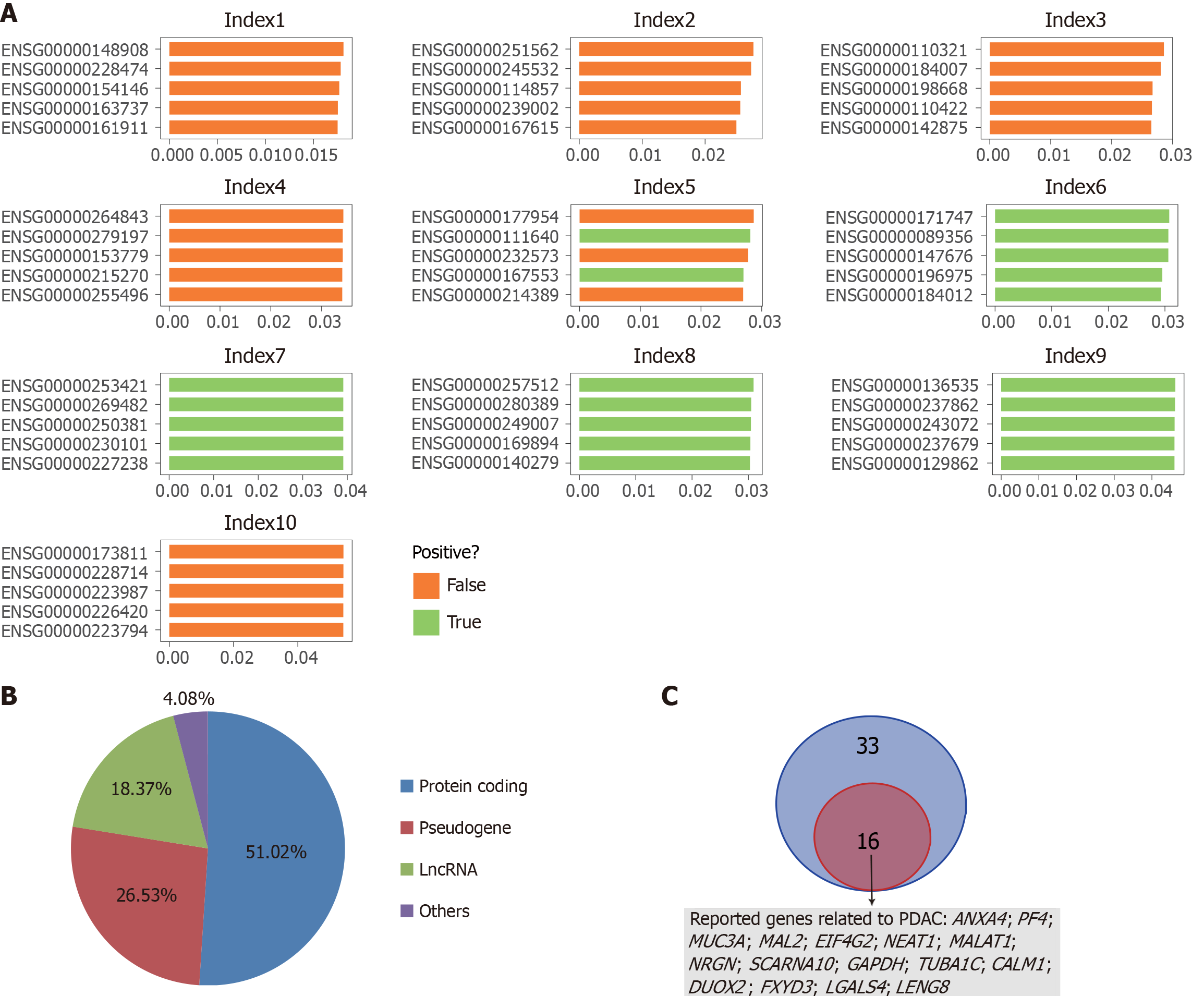
Figure 8 Extraction and classification of important genes.
A: The top five genes in terms of weight in each probabilistic principal component analysis-based extracellular vesicle long RNA index calculation process; B: Classification of gene types; C: The 49 pancreatic ductal adenocarcinoma cancer-related genes identified in this study were compared with those reported in the literature. PDAC: Pancreatic ductal adenocarcinoma; lncRNA: Long non-coding RNA.
Figure 9 Differential expression of selected genes in normal and pancreatic ductal adenocarcinoma tissues.
A: Expression of selected genes was performed using RNA-seq data from The Cancer Genome Atlas and Genotype-Tissue Expression databases; B: Protein expression of encoding protein genes in normal and primary pancreatic ductal adenocarcinoma tissues based on the clinical proteomic tumor analysis consortium dataset. FPKM: Fragments per kilobase of exon per million mapped fragments; NA: Not available. aP < 0.05, bP < 0.01, and cP < 0.001.
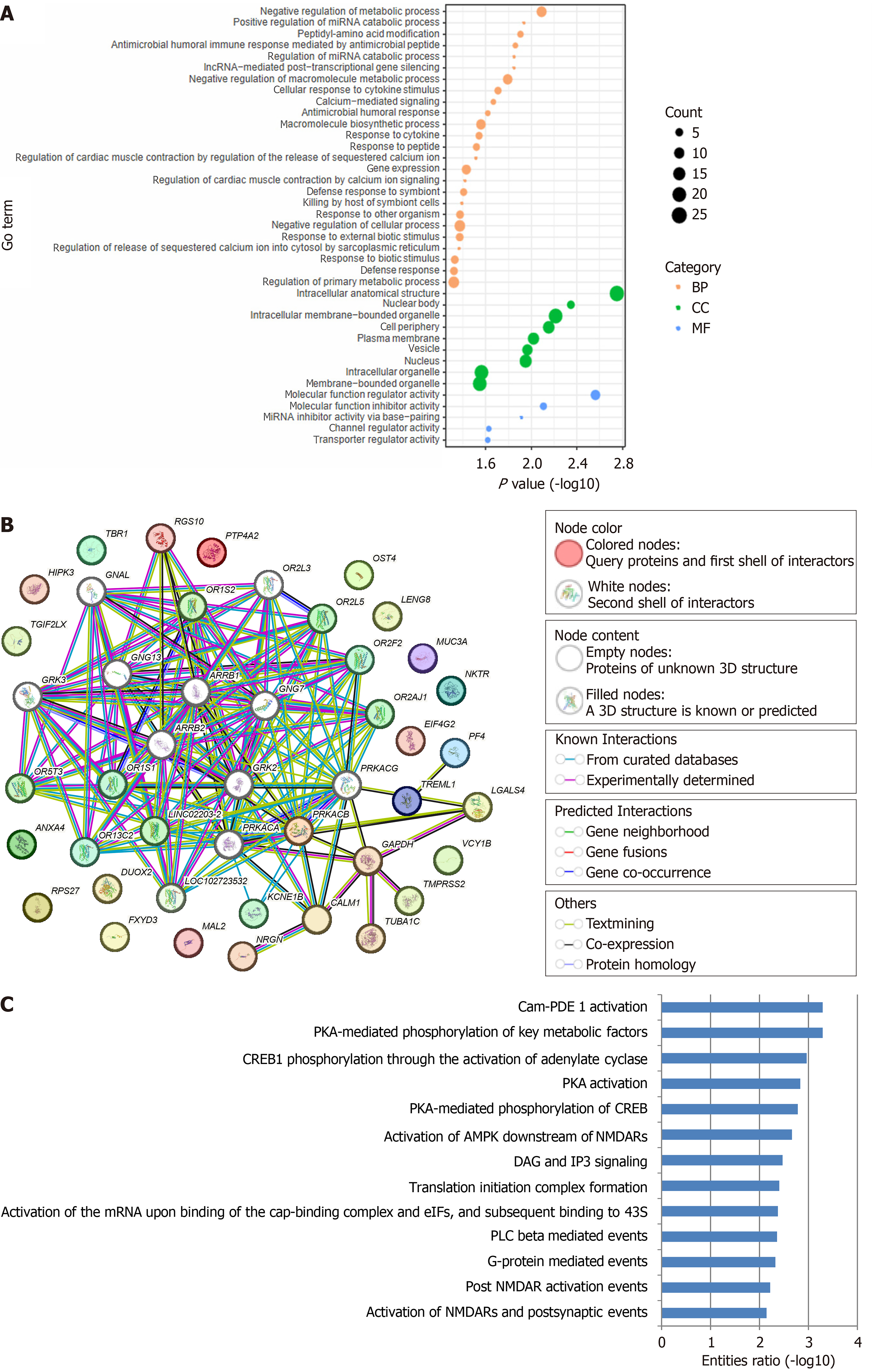
Figure 10 Functional analysis.
A: Gene Ontology analysis; B: Protein-protein interaction analysis (protein-protein interaction enrichment P value: < 1.0e-16), network nodes represent proteins, edges represent protein-protein associations; C: Pathway analysis. GO: Gene Ontology; miRNA: MicroRNA; lncRNA: Long non-coding RNA; Cam-PDE: Calmodulin-dependent phosphodiesterase; PKA: Protein kinase A; CREB1: CAMP response element-binding protein 1; AMPK: AMP-activated protein kinase; NMDARs: N-methyl-D-aspartate receptors; DAG: Diacylglycerol; IP3: Inositol 1,4,5-trisphosphate; eIFs: Eukaryotic initiation factors; PLC: Phospholipase C.
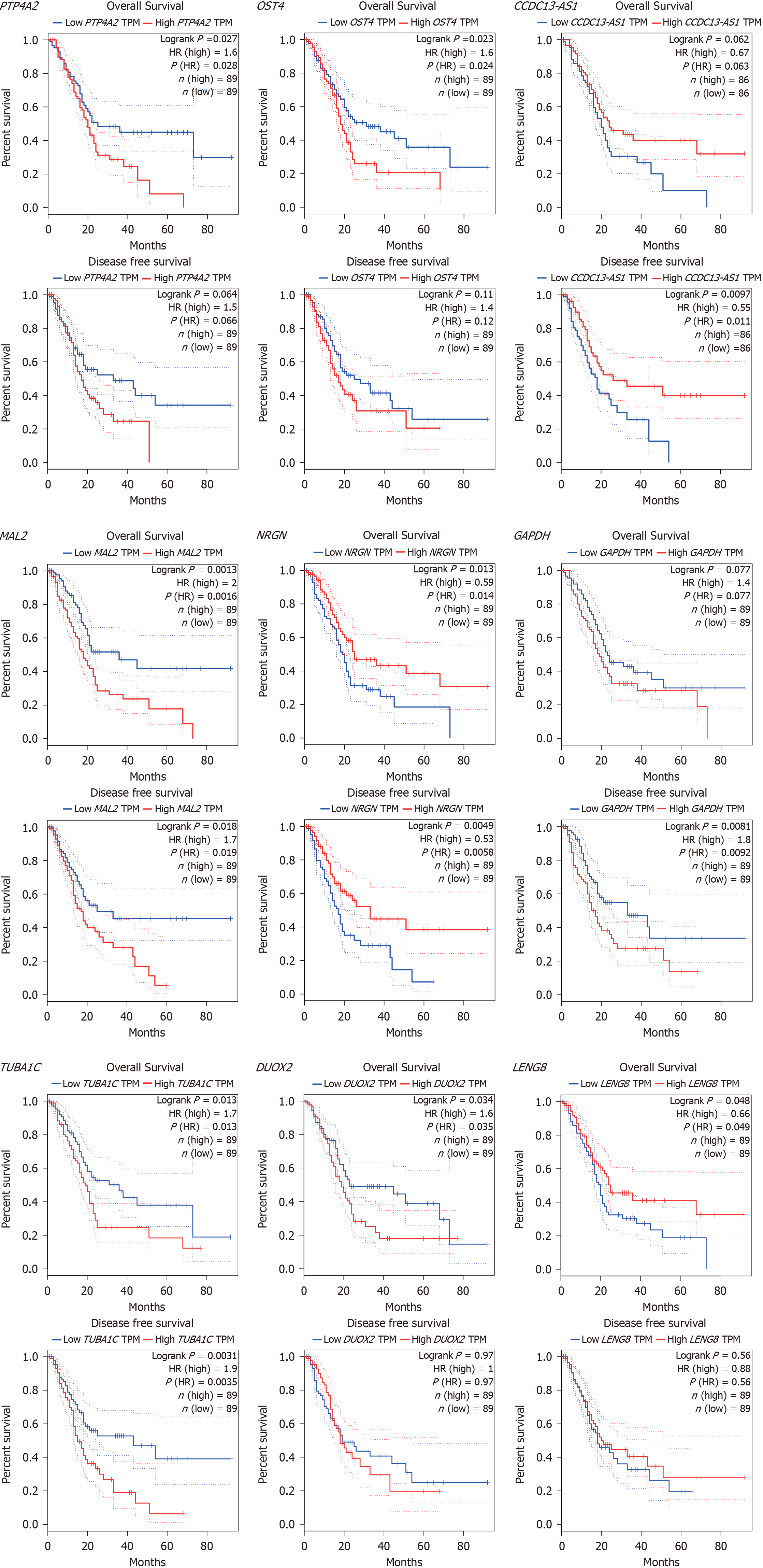
Figure 11 Analysis of overall survival and disease free survival.
TPM: Transcripts per kilobase of exon model per million mapped reads; HR: Hazard ratio.
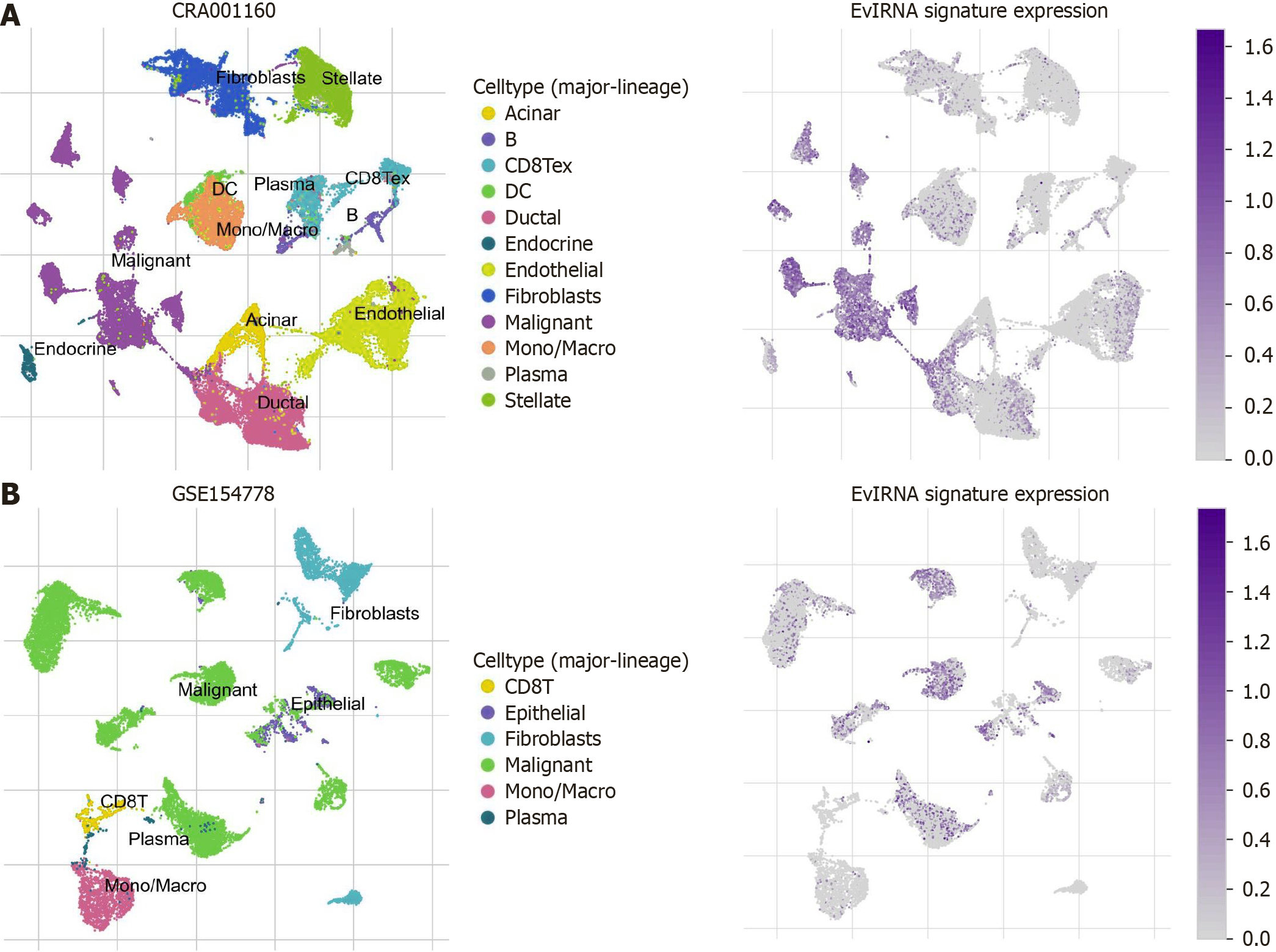
Figure 12 Analysis of single-cell RNA expression in pancreatic ductal adenocarcinoma tumor microenvironment.
A: Based on CRA001160, number of cells: 57443; B: Based on GSE154778, number of cells: 14953. EvlRNA: Extracellular vesicle long RNA; CD8T: CD8+ T cell; DC: Dendritic cell; B: B cell.
- Citation: Liu SC, Zhang H. Early cancer diagnosis via interpretable two-layer machine learning of plasma extracellular vesicle long RNA. World J Gastrointest Oncol 2025; 17(11): 111670
- URL: https://www.wjgnet.com/1948-5204/full/v17/i11/111670.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i11.111670