©The Author(s) 2022.
World J Gastrointest Oncol. Sep 15, 2022; 14(9): 1856-1873
Published online Sep 15, 2022. doi: 10.4251/wjgo.v14.i9.1856
Published online Sep 15, 2022. doi: 10.4251/wjgo.v14.i9.1856
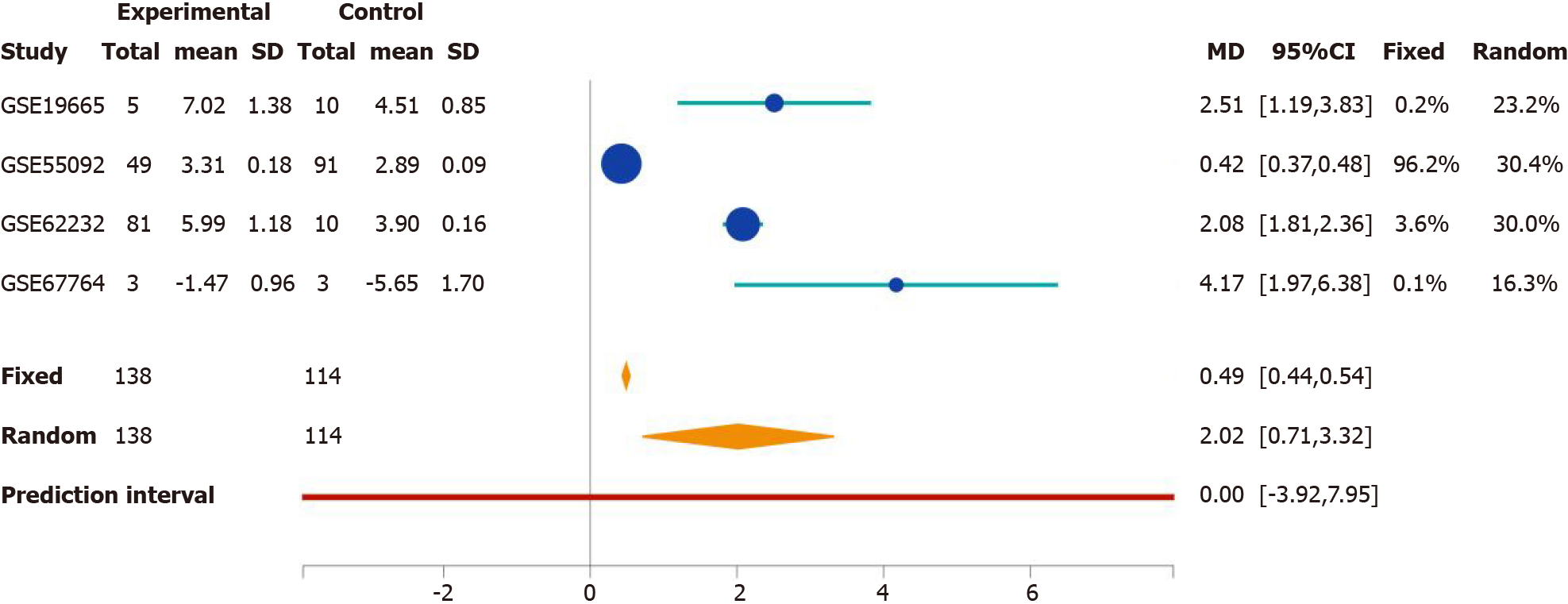
Figure 1 Screenshot from http://stargeo.org/analysis/669/ detailing the expression patterns of PDZ-binding kinase across different studies as shown. Fixed and random treatment effects are illustrated. MD: Mean difference; CI: Confidence interval. Citation: Figures produced from IPA are available under an open-access CC-BY 4.0 license for purposes of publication. The authors have obtained the permission for figure using from the QIAGEN Digital Insights (Supplementary material).
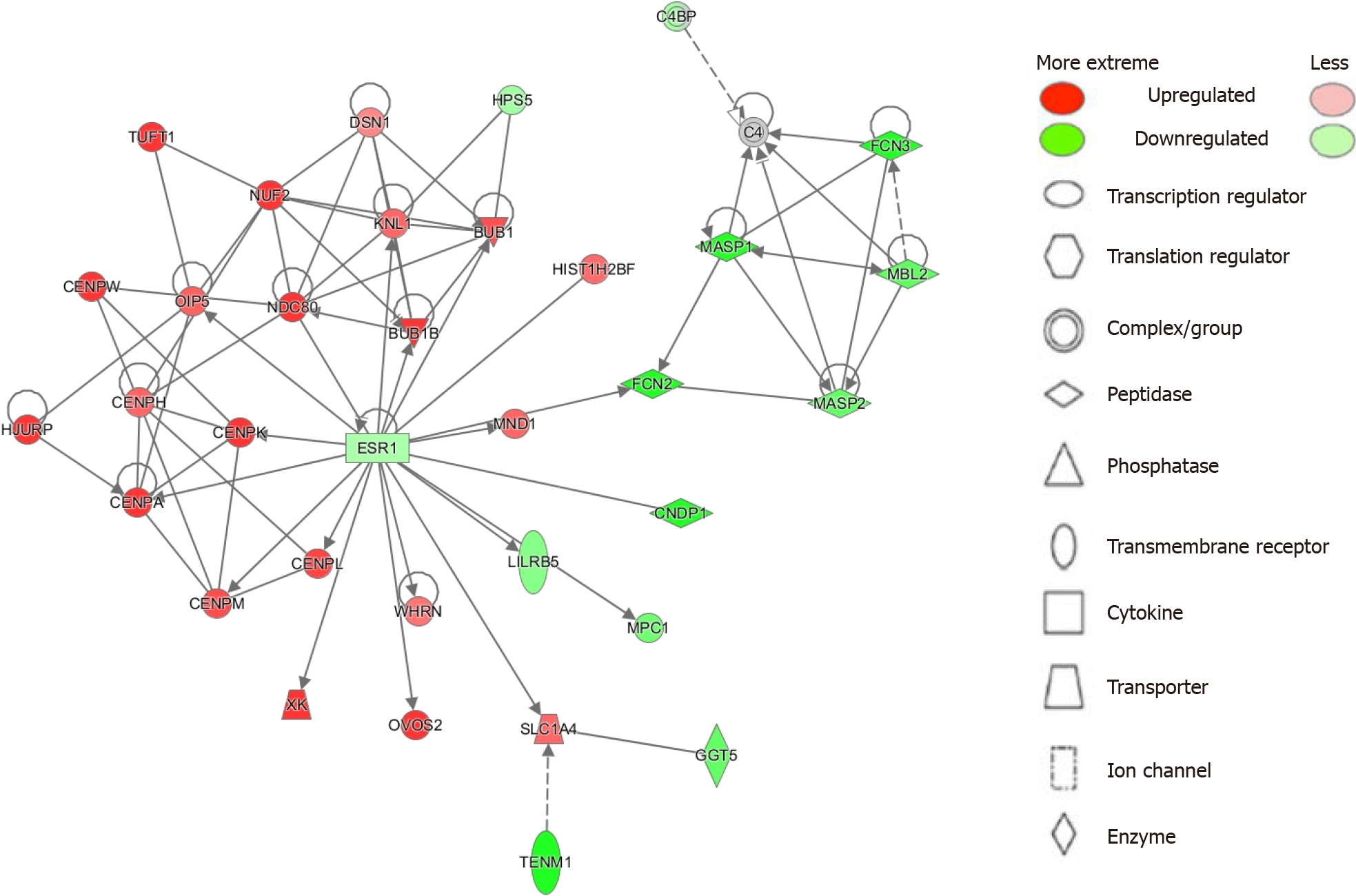
Figure 2 Top network (cell cycle; cellular assembly and organization; DNA replication, recombination and repair) identified by ingenuity pathway analysis Network analysis.
Legend illustrates class of the gene. Red indicates upregulation and green downregulation, with shade depicting magnitude of change. Solid and dashed lines depict direct and indirect, respectively, relationship between genes. Figure was generated using ingenuity pathway analysis. TENM1: Teneurin transmembrane protein 1; GGT5: Gamma-glutamyltransferase5; SLC1A4: Solute carrier family 1 member 4; OVOS2: Alpha-2-macroglobulin like 1 pseudogene; XK: X-linked Kx blood group; WHRN: Whirlin; MPC1: Mitochondrial pyruvate carrier 1; LILRB5: Leukocyte immunoglobulin like receptor B5; CNDP1: Carnosine dipeptidase 1; CENPM: Centromere protein M; CENPL: Centromere protein L; ESR1: Estrogen receptor 1; C4BP: C4b-binding protein; TUFT1: Tuftelin 1; DSN1: MIND kinetochore complex component; KNL1: Kinetochore scaffold 1; BUB1: Mitotic checkpoint serine/threonine-protein kinase BUB1; HPS5: Hermansky-Pudlak syndrome 5 protien; FCN3: Ficolin 3; MASP1: MBL associated serine protease 1; MBL2: Mannose binding lectin 2; HIST1H2BF: Histone H2B type 1; OIP5: Opa interacting protein 5; NDC80: Kinetochore protein NDC80; BUB1B: BUB1 mitotic checkpoint serine/threonine kinase B; MND1: Meiotic nuclear divisions 1; CENPW: Centromere protein W; CENPH: Centromere protein H; CENPK: Centromere protein K; CENPA: Centromere protein A. Citation: Figures produced from IPA are available under an open-access CC-BY 4.0 license for purposes of publication. The authors have obtained the permission for figure using from the QIAGEN Digital Insights (Supplementary material).
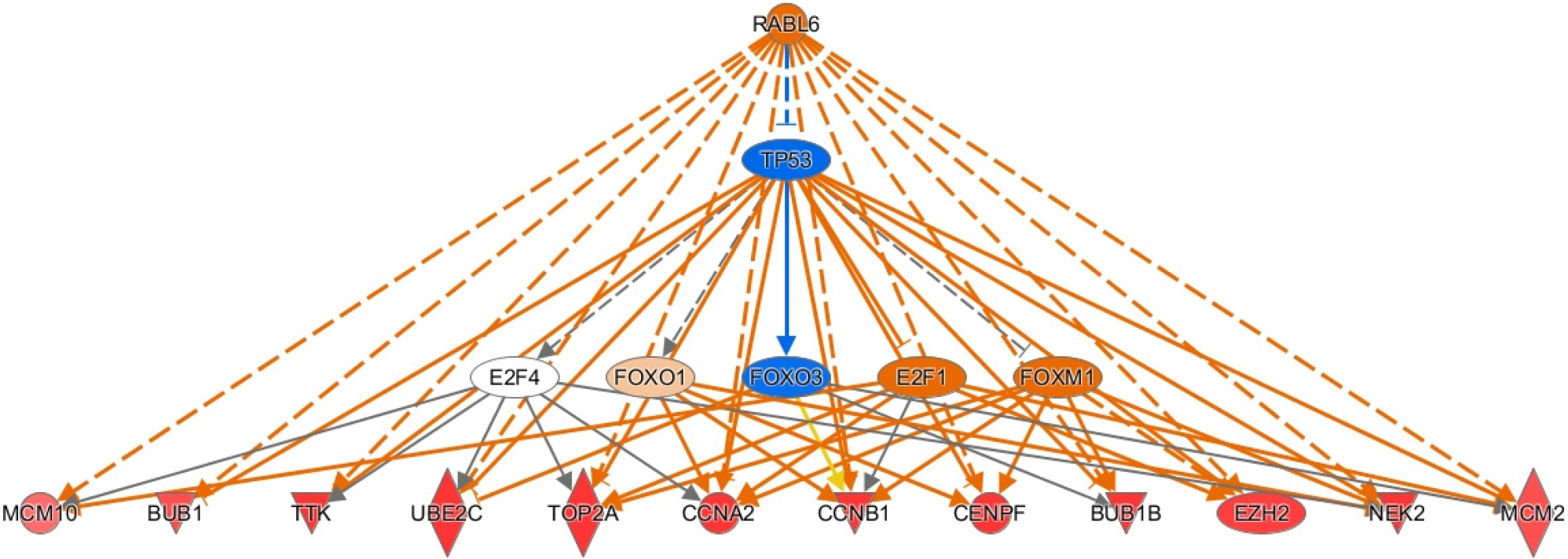
Figure 3 Ingenuity pathway analysis of rab-like protein 6 signaling in hepatitis B-related hepatocellular carcinoma tumors.
Genes are implicated in several disease potential disease processes including inflammation, cell division, Akt signaling, and more. Legend illustrates relationship between genes. See Figure 2 legend for identification of shapes. RABL6: RAB, member RAS oncogene family like 6; TP53: Tumor protein p53; E2F4: E2F transcription factor 4; FOXO1: Forkhead box O1; FOXM1: Forkhead box M1; MCM10: Mini-chromosome maintenance 10; BUB1: Mitotic checkpoint serine/threonine-protein kinase BUB1; TTK: TTK protein kinase; UBE2C: Ubiquitin-conjugating enzyme E2 C; TOP2A: DNA topoisomerase IIα; CCNA2: Cyclin A2; CENPF: Centromere protein F; EZH2: Enhancer of zeste homolog 2; NEK2: Serine/threonine-protein kinase Nek2. Citation: Figures produced from IPA are available under an open-access CC-BY 4.0 license for purposes of publication. The authors have obtained the permission for figure using from the QIAGEN Digital Insights (Supplementary material).
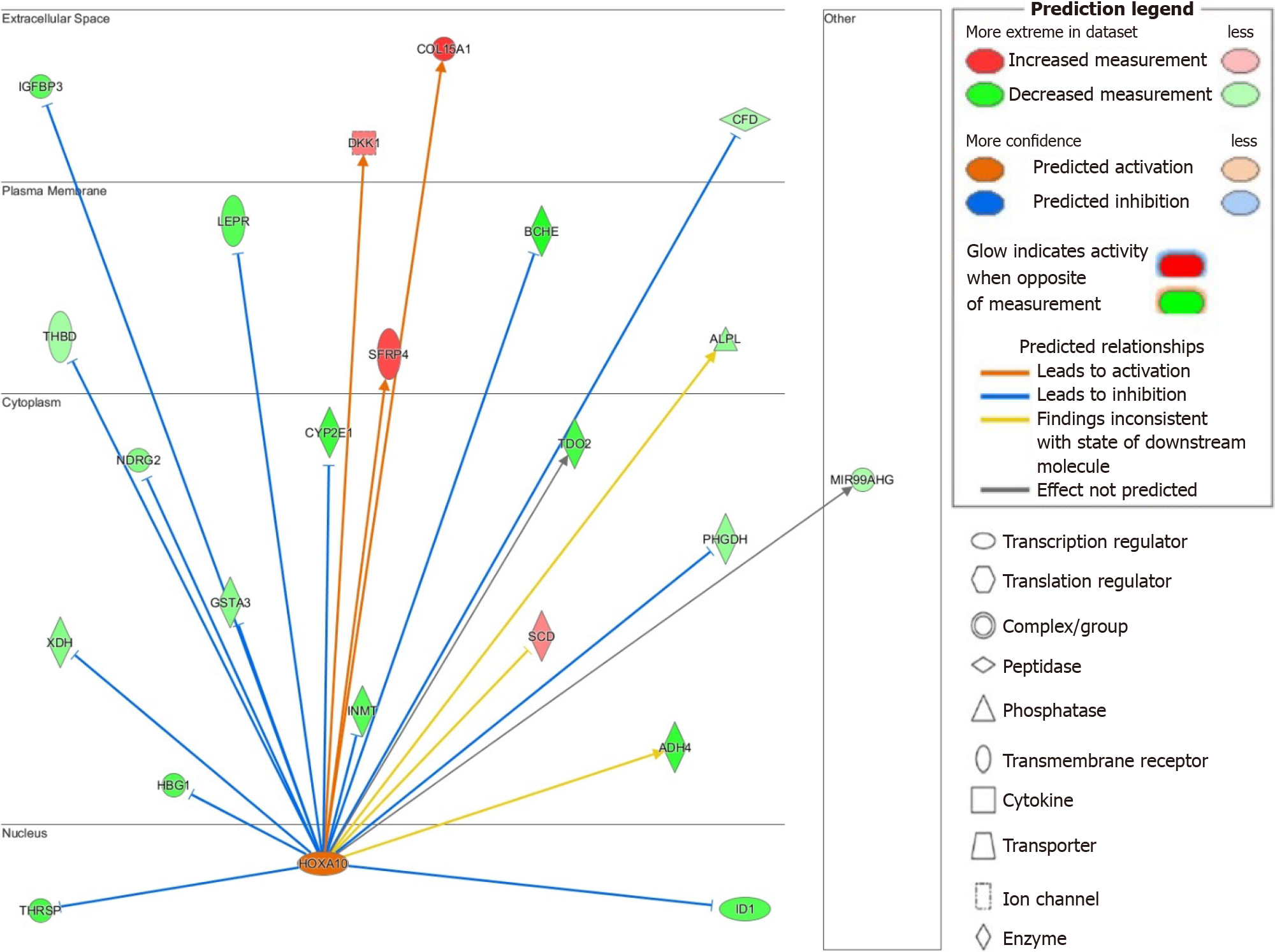
Figure 4 Ingenuity pathway analysis of homeobox A10 activity in hepatitis B-related hepatocellular carcinoma.
Homeobox A10 signaling has potential implications on tumor suppression, liver metabolism, and other disease-related activity. Genes and location are shown above. Legend illustrates relationship between genes and gene classification. COL15A1: Collagen alpha-1(XV) chain; IGFBP3: Insulin-like growth factor binding protein 3; DKK1: Dickkopf-related protein 1; CFD: Complement factor D; LEPR: Leptin receptor; BCHE: Butyrylcholinesterase; THBD: Thrombomodulin; SFRP4: Secreted frizzled-related protein 4; ALPL: Alkaline phosphatase; CYP2E1: Cytochrome P450 2E1; TDO2: Tryptophan 2,3-dioxygenase; NDRG2: N-myc downstream-regulated gene family member 2; GSTA3: Glutathione S-transferase A3; PHGDH: Phosphoglycerate dehydrogenase; XDH: Xanthine dehydrogenase; INMT: Indolethylamine N-methyltransferase; SCD: Stearoyl-CoA desaturase; HBG1: Hemoglobin subunit gamma 1; ADH4: Alcohol dehydrogenase 4; HOXA10: Homeobox A10; THRSP: Thyroid hormone-inducible hepatic protein. Citation: Figures produced from IPA are available under an open-access CC-BY 4.0 license for purposes of publication. The authors have obtained the permission for figure using from the QIAGEN Digital Insights (Supplementary material).
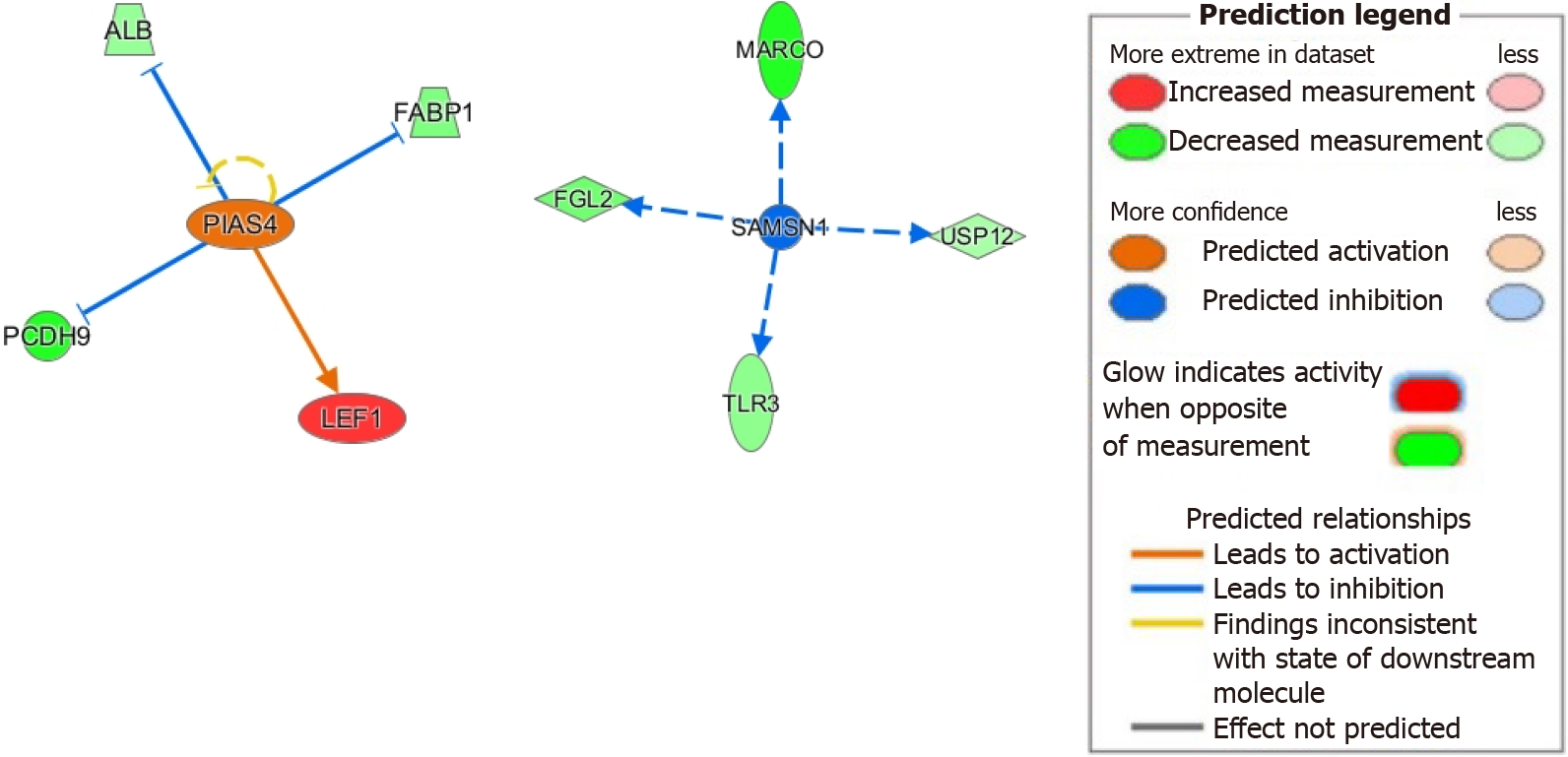
Figure 5 Protein inhibitor of activated STAT 4 and SH3 domain, and nuclear localization signals 1 potential role in homeobox A10 have only been recently described and much remains to be understood.
Ingenuity pathway analysis analysis demonstrated activation of protein inhibitor of activated STAT 4, and activation of downstream genes implicated in epithelial-mesenchymal transition through LEF1 and protocadherin 9. Our analysis also demonstrated inhibition of SH3 domain, and nuclear localization signals 1 with downstream effects on viral recognition and regulation of cell survival. Legend illustrates relationship between genes. See legend of Figure 3 for classification of genes. ALB: Albumin; FABP1: Fatty acid binding protein 1; MARCO: Macrophage receptor with collagenous structure; FGL2: Fibrinogen like 2; SAMSN1: SAM domain, SH3 domain and nuclear localization signals 1; USP12: Ubiquitin-specific protease 12; PIAS4: Protein inhibitor of activated STAT protein gamma; PCDH9: Protocadherin 9; LEF1: Lymphoid enhancer-binding factor 1; TLR3: Toll-like receptor 3. Citation: Figures produced from IPA are available under an open-access CC-BY 4.0 license for purposes of publication. The authors have obtained the permission for figure using from the QIAGEN Digital Insights (Supplementary material).
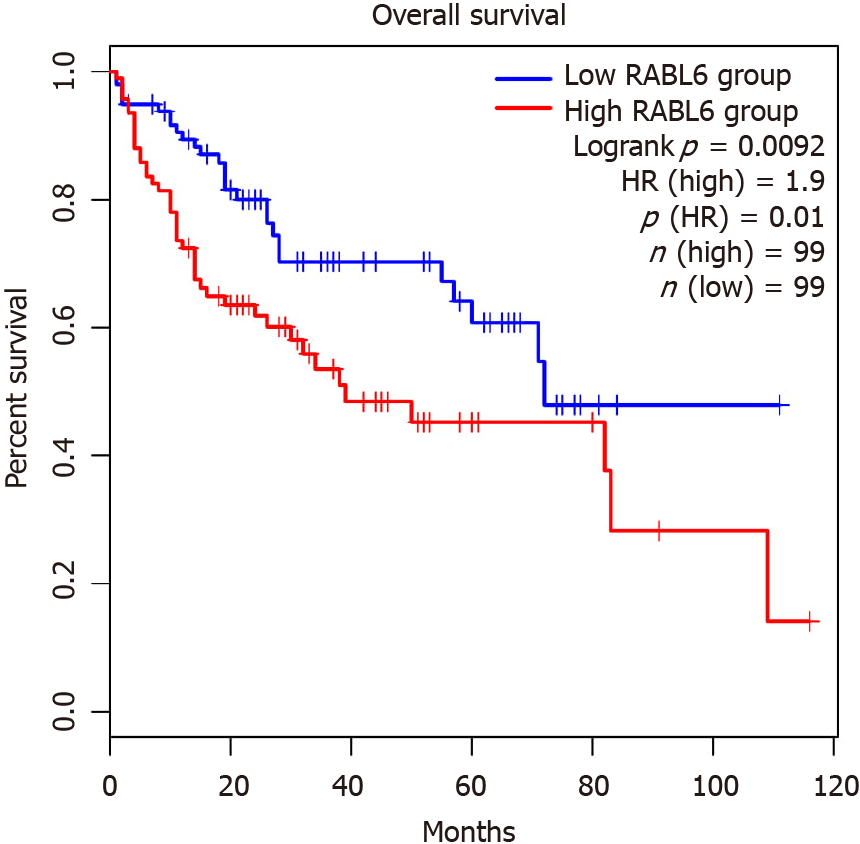
Figure 6 Survival analysis comparing high and low rab-like protein 6 expression in survival of hepatocellular carcinoma patients.
RABL6: Rab-like protein 6; HR: Hazards ratio. Plot generated using GPEIA2. Citation: Figures produced from IPA are available under an open-access CC-BY 4.0 license for purposes of publication. The authors have obtained the permission for figure using from the QIAGEN Digital Insights (Supplementary material).
- Citation: Aljabban J, Rohr M, Syed S, Cohen E, Hashi N, Syed S, Khorfan K, Aljabban H, Borkowski V, Segal M, Mukhtar M, Mohammed M, Boateng E, Nemer M, Panahiazar M, Hadley D, Jalil S, Mumtaz K. Dissecting novel mechanisms of hepatitis B virus related hepatocellular carcinoma using meta-analysis of public data. World J Gastrointest Oncol 2022; 14(9): 1856-1873
- URL: https://www.wjgnet.com/1948-5204/full/v14/i9/1856.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i9.1856