Published online Feb 18, 2016. doi: 10.4254/wjh.v8.i5.282
Peer-review started: October 1, 2015
First decision: November 4, 2015
Revised: December 3, 2015
Accepted: January 16, 2016
Article in press: January 19, 2016
Published online: February 18, 2016
Processing time: 139 Days and 16.5 Hours
AIM: To address the effect of heat-shock protein 90 (HSP90) inhibitors on the release of the hepatitis C virus (HCV), a cell culture-derived HCV (JFH1/HCVcc) from Huh-7 cells was examined.
METHODS: We quantified both the intracellular and extracellular (culture medium) levels of the components (RNA and core) of JFH-1/HCVcc. The intracellular HCV RNA and core levels were determined after the JFH1/HCVcc-infected Huh-7 cells were treated with radicicol for 36 h. The extracellular HCV RNA and core protein levels were determined from the medium of the last 24 h of radicicol treatment. To determine the possible role of the HSP90 inhibitor in HCV release, we examined the effect of a combined application of low doses of the HSP90 inhibitor radicicol and the RNA replication inhibitors cyclosporin A (CsA) or interferon. Finally, we statistically examined the combined effect of radicicol and CsA using the combination index (CI) and graphical representation proposed by Chou and Talalay.
RESULTS: We found that the HSP90 inhibitors had greater inhibitory effects on the HCV RNA and core protein levels measured in the medium than inside the cells. This inhibitory effect was observed in the presence of a low level of a known RNA replication inhibitor (CsA or interferon-α). Treating the cells with a combination of radicicol and cyclosporin A for 24 h resulted in significant synergy (CI < 1) that affected the release of both the viral RNA and the core protein.
CONCLUSION: In addition to having an inhibitory effect on RNA replication, HSP90 inhibitors may interfere with an HCV replication step that occurs after the synthesis of viral RNA, such as assembly and release.
Core tip: Hepatitis C virus (HCV) is a major causative agent of hepatocellular carcinoma. Several non-structural proteins of HCV physically and functionally interact with heat-shock protein 90 (HSP90). Although HSP90 inhibitors, which inhibit the chaperone function of HSP90, have been shown to inhibit HCV replication by several groups, a recent report using a reporter system for HCV RNA replication (replicon) suggests that the effect is nonspecific. Thus, the inhibitory mechanism of HSP90 inhibitors remains controversial. Here, we address the effect of HSP90 inhibitors on the release of JFH1/cell culture-derived HCV from Huh-7 cells, and suggested that, HSP90 inhibitors may also interfere with an HCV replication step that occurs after the synthesis of viral RNA, such as assembly and release.
- Citation: Kubota N, Nomoto M, Hwang GW, Watanabe T, Kohara M, Wakita T, Naganuma A, Kuge S. Hepatitis C virus inhibitor synergism suggests multistep interactions between heat-shock protein 90 and hepatitis C virus replication. World J Hepatol 2016; 8(5): 282-290
- URL: https://www.wjgnet.com/1948-5182/full/v8/i5/282.htm
- DOI: https://dx.doi.org/10.4254/wjh.v8.i5.282
Chronic infection with hepatitis C virus (HCV) frequently causes liver cirrhosis and hepatocellular carcinoma[1]. Approximately 170 million individuals have been infected by HCV[2] and are at risk for developing liver disease[3]. HCV has a positive-sense single-stranded RNA genome that encodes a 3000 amino acid polyprotein and also contains an internal entry site for translation and a non-coding region for genome replication at the 5’- and 3’- flanking region. Translation of the polyprotein is followed by cleavage by the host and viral proteases, which yields three structural proteins (core, E1 and E2) and seven nonstructural (NS) proteins (p7, NS2, NS3, NS4A, NS4B, NS5A and NS5B)[4]. Establishment of the cell culture-derived HCV (HCVcc) system (i.e., human hepatoma Huh-7 cells propagating the highly infectious clone of HCV genotype 2a, JFH-1[5]) provides a native infection-cycle system, which is suitable for determination of the precise function of HCV proteins and thus the mechanism for replication and secretion of viral particles. Accumulating evidence has indicated that NS3, NS4B and NS5A are required for not only genome RNA replication but also virus assembly[6], whereas P7 and NS2 are dispensable for RNA replication but are required for virus production[7].
Heat-shock protein 90 (HSP90) functions as a molecular chaperone for various client proteins and interacts with a cohort of co-chaperones that modulate the HSP90 ATPase cycle. The ATPase activity of HSP90 is inhibited by HSP90 inhibitors, which compete with ATP for binding and thereby eliminate HSP90 chaperone activity[8]. HSP90 clients include not only host proteins but also some virus proteins. Thus, some of the replication steps are required for HSP90 activity[9]. In fact, activities of multiple HCV proteins are affected by inhibition of HSP90 by specific inhibitors of HSP90, such as radicicol and geldanamycin and its derivatives. HSP90 inhibitors restrict the activity of NS2/3 protease[10], the stability of NS3[11], and the RNA replication of HCV in cells harboring the HCV replicon by inhibiting a complex consisting of HSP90, NS5A and a human FK506-binding protein (FKBP8)[12]. The host factors that are required for HCV replication are also affected by inhibition of HSP90[13,14]. Apart from these positive results, Beran et al[15] have shown that an HSP90 inhibitor elicits effects similar to those of known cytostatic compounds, abrogating propagation of the mini-genome (subgenomic) replicon of HCV indirectly, through slowing cell growth. HCV subgenomic replicons are generally composed of a selection marker gene, such as G418 resistance, a reporter gene such as luciferase, and a whole HCV genome, except for the regions encoding the structural proteins and NS2 protease, which are deleted[16]. The replicons propagating in Huh-7 cells are widely used for studying RNA replication as well as for screening for anti-HCV drugs. However, it is possible that the replicon system does not fully represent the native function of HCV proteins, because mutations that accumulate in NS3 and NS5A in the replicon during enhancement of the RNA replication interfere with virus assembly when the mutation is introduced in the HCVcc construct[17]. Hence, the HCVcc system may be more suitable than the subgenomic replicon system to evaluate the anti-viral effect of HSP90 inhibitors. In addition, HSP90 is required for NS3 stability[11] and for formation of the complex composing NS4A[12]. Because NS3 and NS4A can act on both RNA replication and HCV assembly, it is possible that HSP90 activity may contribute to post-RNA replication steps, such as virus assembly and release, which is represented by HCVcc but not the replicon system.
In this study, we used JFH1/HCVcc to demonstrate the effect of HSP90 inhibition on HCVcc release from infected Huh-7 cells. Our results showed that the HSP90 inhibitor radicicol preferentially reduced the levels of the core and the HCV RNA released from cells in the medium compared with those in the cells. The HSP90 inhibitor had a more potent effect on viral release in the medium than that of the inhibitor for RNA replication.
Radicicol (Sigma-Aldrich, St. Louis, MO, United States) was dissolved in methanol (1 mg/mL). Cyclosporin A [cyclosporin A (CsA); Wako Pure Chemical, Osaka, Japan], 17-AAG [17-(allylamino)-17-demethoxygeldanamycin, Sigma-Aldrich] and 17-DMAG [17-(dimethylaminoethylamino)-17-demethoxygeldanamycin; BIOMOL, Plymouth Meeting, PA, United States] were dissolved in ethanol (1 mg/mL). Interferon-α (IFN-α; PeproTec EC, London, United Kingdom) was dissolved in water (0.1 mg/mL).
Human hepatoma Huh-7 cells (purchased from Human Science Resources Bank, Osaka, Japan) were cultured in DMEM (Nissui Pharmaceuticals, Tokyo, Japan) supplemented with 10% fetal calf serum, 0.06% glutamine, 0.35% glucose and 1% penicillin-streptomycin (Life Technologies, Carlsbad, CA, United States). The cells were maintained at 37 °C in an atmosphere of 5% CO2. We synthesized the HCV genomic RNA of genotype 2a (JFH1) in vitro using a MEGAscript™ T7 kit (Ambion, Austin, TX, United States) and introduced the RNA into Huh-7 cells by electroporating the cells with the GenePulser II electroporation system (Bio-Rad, Hercules, CA, United States) as previously described[5]. The cytotoxic effects of the reagents were examined with Alamar Blue cell viability reagent (Serotec, Raleigh, NC, United States), which allows an estimation of the oxidation levels in the cellular electron-transport pathways with a fluorescent indicator. Alamar Blue was used as described by the manufacturer.
We washed the JFH1/HCVcc cells with PBS and lysed them in lysis buffer (20 mmol/L Tris-Cl, pH 7.5, 0.1% SDS, 1% Triton X-100, 1% deoxycholate, 0.1 mmol/L EDTA, 0.1 mmol/L phenylmethanesulfonyl fluoride, 50 μmol/L N-p-tosyl-L-phenylalanine chloromethyl ketone, 5 μmol/L Nα-tosyl-L-lysine chloromethyl ketone hydrochloride, 5 μg/mL aprotinin and 5 μg/mL leupeptin). We quantified the level of core protein present in the lysates and spent culture medium using the Ortho HCV antigen ELISA test (Ortho Clinical Diagnostics, Rochester, NY, United States). We used a QIAamp™ Viral RNA Mini kit (Qiagen, Hilden, Germany) to isolate the HCV genomic RNA from the medium. We used an RNeasy™ mini kit (Qiagen) to isolate total RNA from the HCV-infected cells. We quantified the HCV genomic RNA using TaqMan™ EZ RT-PCR Core Reagents (Applied Biosystems, Foster City, CA, United States) and the iCycler™ iQ real-time detection system (Bio-Rad) as previously described[18].
The JFH1/HCVcc cells (5 × 104 cells in one well of a 24-well culture dish) were treated with radicicol, CsA and/or IFN-α for 12 h, at which point the medium was replaced by fresh medium that contained the same level(s) of drug(s). After culturing for another 24 h, the core and the HCV RNA in each cell lysate and culture medium were quantified as described above. The synergism between CsA and radicicol was evaluated by the combination index (CI) equation and the Chou and Talalay method[19,20] using CalcuSyn software (BIOSOFT, Cambridge, United Kingdom). The CI equation is based on the multiple drug-effect equation of Chou et al[20], which is derived from enzyme-kinetic models. Assuming that CsA and radicicol were mutually non-exclusive drugs that have totally independent modes of action, we used the following equation:
Equation 1 dictates that the combination of drug 1 (D)1 and drug 2 (D)2 inhibits a reaction (or phenomenon) by x% in an actual experiment. (Dx)1 and (Dx)2 are the doses of drug 1 and drug 2 alone that inhibit the same reaction by x%.
CI = [(D)1/(Dx)1] + [(D)2/(Dx)2] + [(D)1(D)2/(Dx)2(Dx)2] (1)
We used Student’s t-test to examine statistical significance (P < 0.05). All of the experiments were performed with multiple independent replicates, and all of the data are presented as the mean results of three independent experiments with the standard error of the mean. The statistical methods of this study were reviewed by professor Kotaro Tanahashi from Mathematics, Tohoku Pharmaceutical University.
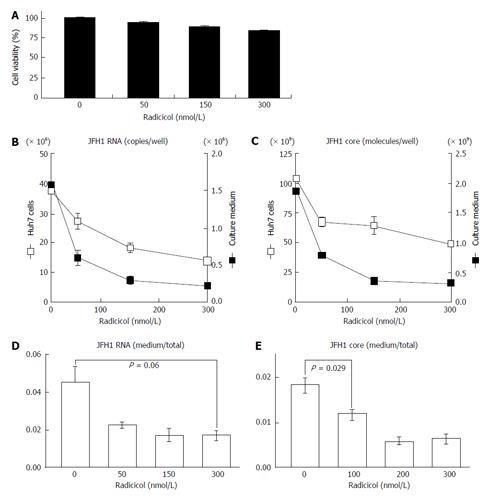
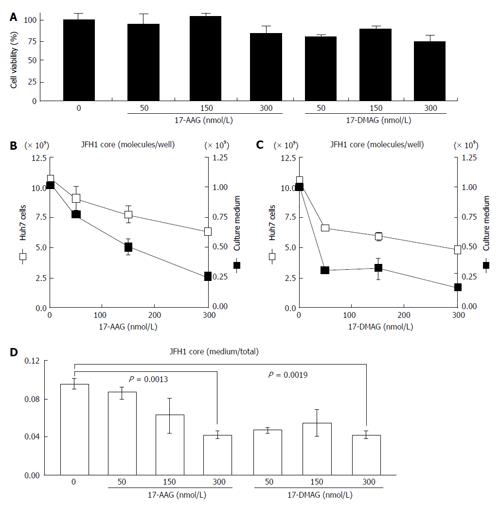
To examine the effects of HSP90 inhibitor on the release of HCV, we quantified both the intracellular and extracellular (culture medium) levels of the components (RNA and core) of JFH-1/HCVcc. The intracellular HCV RNA and core levels were determined after the cells were treated with radicicol for 36 h. The extracellular HCV RNA and the core were determined from the medium of the last 24 h of radicicol treatment. The radicicol treatment (50-300 nmol/L) exhibited no apparent cytotoxic effect (Figure 1A), reduced both the intracellular and extracellular (medium) levels of the HCV RNA (Figure 1B) and the core (Figure 1C) in a dose-dependent manner. Interestingly, the RNA level in the culture medium relative to the total RNA level was apparently reduced by radicicol even at a low concentration (50 nmol/L) (Figure 1D). Similarly, the core level in the medium relative to the total core level was also significantly decreased (P = 0.029) in the presence of 50 nmol/L radicicol (Figure 1E). Furthermore, two derivatives of the geldanamycin HSP90 inhibitor, 17-AAG and 17-DMAG, also inhibited the release of the HCV RNA and core more effectively than they decreased the intracellular HCV RNA and core levels (Figure 2).
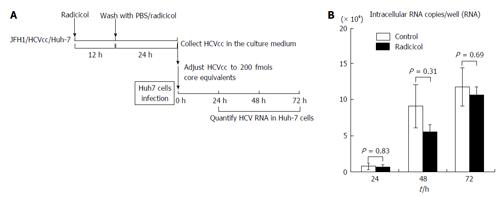
We next examined whether the integrity of HCV was affected by the radicicol treatment during production of HCV from JFH1/HCVcc. The infectivity of the HCV that had been released into the medium in the presence of radicicol was compared to the infectivity of HCV released in the absence of radicicol. As shown in Figure 3, there was no significant difference (P > 0.3) in the infectivity between the HCV produced in the presence and that produced in the absence of radicicol. These results suggested that even though radicicol preferentially reduced HCV release, radicicol did not affect its infectivity.
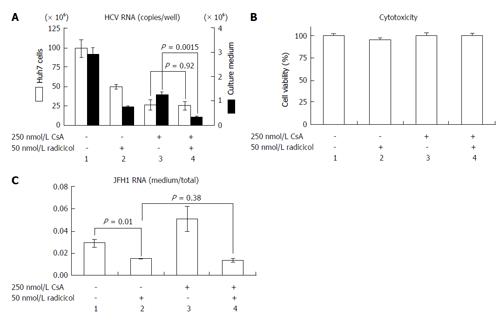
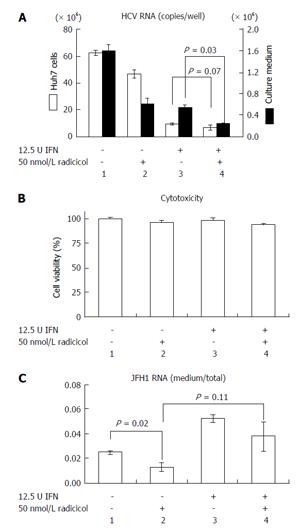
NS3 and NS5A, which are known targets of HSP90[11,12], are required for both RNA replication and virus assembly[6,21]. Thus, it is difficult to distinguish whether the inhibitory step of the HSP90 inhibitor affects the RNA replication or the assembly. If the HSP90 inhibitor were to preferentially inhibit the post-RNA replication steps, the HSP90 inhibitor might enhance the RNA-replication inhibitor-dependent inhibitory effect on HCV release. CsA an immunosuppressant, inhibits interaction between a CsA’s target cyclophilin A and NS5A and the interaction of the cyclophilin A with the NS5B polymerase/RNA complex, thereby inhibiting RNA replication[22]. This inhibitory effect is distinct from CsA’s immunosuppressive activity[23]. To determine the possible role of the HSP90 inhibitor on HCV release, we examined the effect of a combined application of low doses of radicicol and CsA. By studying the dose-dependent inhibition of HCV RNA production in Huh-7 cells (data not shown), we determined the doses showing the partial effect of CsA (250 nmol/L) and radicicol (50 nmol/L) on the viral RNA production in cells after 36 h of treatment (Figure 4A, column 1, 2 and 3, open bars). CsA alone, radicicol alone and simultaneous treatment with both drugs did not affect the cytotoxicity in JFH1-infected Huh-7 cells (Figure 4B). When the HCV-infected cells were treated with the low concentration of both drugs simultaneously, the level of HCV RNA released in the medium after 24 h fell significantly (Figure 4A, comparison of the closed bars in columns 3 and 4, P = 0.0015), whereas the level of HCV RNA in the cells was the same as that in the cells treated with CsA alone (Figure 4A, comparison of the open bars in columns 3 and 4). Intriguingly, the RNA level in the culture medium relative to the RNA levels in the infected cells fell in the presence of radicicol (Figure 4C, comparison of columns 1 and 2). However, this medium-to-total ratio was not affected by simultaneous treatment of CsA with radicicol (Figure 4C, column 2 and 4). Thus, radicicol could have more of an inhibitory effect on viral release from infected cells than CsA.
Previous results have indicated that the combined effect of IFN and CsA on RNA replication is mostly additive[23] and have suggested that both CsA and IFN target a similar point in HCV replication. However, Robida et al[24] have indicated that CsA resistant mutants maintain their sensitivity to IFN-α. Thus, although both IFN-α and CsA inhibit RNA replication, the inhibitory effects of IFN-α and CsA seems to be different. As shown in Figure 5A, we observed that radicicol efficiently reduced the level of HCV in the medium even in the presence of IFN-α. Although the combined treatment of radicicol and IFN-α and the treatment of IFN-α alone exhibited a similar level of HCV RNA in cells (columns 3 and 4, open bars), HCV RNA in the medium was significantly reduced (columns 3 and 4, closed bars). Again, HCV RNA in the medium was significantly suppressed in the presence of radicicol (Figure 5C). All of these results suggested that HSP90 might be responsible for a post-replication step such as viral release. It should be noted that a similar synergism has been previously reported: A combined administration of an HSP90 inhibitor and polyethylene glycol-conjugated interferon (PEG-IFN) in HCV-infected chimeric mice with humanized livers was more effective at reducing the HCV genomic RNA levels in mouse serum than a single PEG-IFN treatment[25].
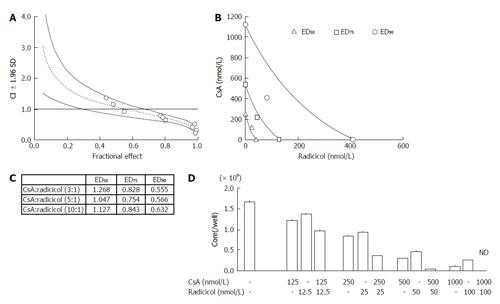
Our results suggested that radicicol has a greater inhibitory effect on the release of HCVcc into the medium and that the point of inhibition of HCV production by the HSP90 inhibitor may be different from that of CsA. Thus, we statistically examined the combined effect of radicicol and CsA using the CI and the graphical representation proposed by Chou et al[19,20]. We examined the effects of various CsA (125, 250, 750 and 1500 nmol/L) and radicicol (25, 50, 150 and 300 nmol/L) concentrations on the release of the core into the medium after 24 h (data not shown). Next, we examined the effects of a fixed molar ratio of these drugs (5:1 CsA to radicicol). The results (Figure 6A) indicated that CsA and radicicol had a synergistic effect (CI < 1) above a fractional effect of 0.5 and close to a strong synergism (CI ≤ 0.3) at a fractional effect of 1. The combined effect was additive near a fractional effect of 0.5 and antagonistic at a fractional effect less than 0.4. Figure 6B shows the conservation isobologram (CI = 1) for the different effective doses of the combination treatment that yielded 50% (ED50), 75% (ED75) and 90% (ED90) inhibition of the core release (constructed using actual experimental data). The combined effect at ED50 (CI = 1.05) was additive, whereas the combined effects at ED75 (CI = 0.75) and ED90 (CI = 0.57) were synergistic (Figure 6). The combined use of 408 nmol/L CsA and 82 nmol/L radicicol yielded ED90 (Figure 6B; if a CI = 1 was expected, an estimated 605 nmol/L CsA and 121 nmol/L radicicol would be required to yield ED90). We obtained similar results for the 3:1 and 10:1 molar ratios of CsA to radicicol (Figure 6C). One example, shown in Figure 6D, indicated that treating the cells with both 1000 nmol/L CsA and 100 nmol/L radicicol for 36 h caused the HCV core to be undetectable in the medium for the last 24 h of the combined treatment.
Collectively, our results suggest that HSP90 inhibitors might affect both the RNA replication and the post-RNA replication stage of viral propagation. Previous reports have indicated that HSP90 is required for NS3 stability[11] and for the formation of a complex consisting of NS5A and FKBP8[12]. Thus, it is possible that HSP90 may affect the post RNA-replication step, such as assembly, through affecting the activity of NS3 and NS5A. Elucidating the precise mechanism for the HSP90 on HCV assembly may provide an alternative drug target for HCV clearance.
Although heat-shock protein 90 (HSP90) inhibitors, which inhibit the chaperone function of HSP90, have been shown to inhibit hepatitis C virus (HCV) replication by several groups, a recent report using a reporter system for HCV RNA replication (replicon) suggests that the effect is nonspecific. Thus, the inhibitory mechanism of HSP90 inhibitors remains controversial.
The authors found that the HSP90 inhibitors had greater inhibitory effects on the HCV RNA and core protein levels measured in the medium than inside the cells. This inhibitory effect was observed in the presence of a low level of a known RNA replication inhibitor [cyclosporin A (CsA) or interferon-α].
The authors’ results suggested that, HSP90 inhibitors may also interfere with an HCV replication step that occurs after the synthesis of viral RNA, such as assembly and release.
This study would benefit the effort to explore new targets for the treatment of HCV infection.
HSP90 inhibitors inhibit the chaperone function of HSP90. HSP90 clients include not only host proteins but also multiple HCV proteins including NS2/3 protease, NS3 and the RNA replication complex consisting of NS5A.
The authors suggested that HSP90 inhibitors may interfere with an HCV replication step that occurs after the synthesis of viral RNA such as assembly and release. The results of this study were quite interesting. The data were appropriately presented and interpreted. This manuscript also was well prepared.
| 1. | Saito I, Miyamura T, Ohbayashi A, Harada H, Katayama T, Kikuchi S, Watanabe Y, Koi S, Onji M, Ohta Y. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci USA. 1990;87:6547-6549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 826] [Cited by in RCA: 840] [Article Influence: 23.3] [Reference Citation Analysis (1)] |
| 2. | Alter MJ. Epidemiology of hepatitis C virus infection. World J Gastroenterol. 2007;13:2436-2441. [PubMed] [DOI] [Full Text] |
| 3. | Bosch FX, Ribes J, Borràs J. Epidemiology of primary liver cancer. Semin Liver Dis. 1999;19:271-285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 671] [Cited by in RCA: 672] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 4. | Bartenschlager R, Frese M, Pietschmann T. Novel insights into hepatitis C virus replication and persistence. Adv Virus Res. 2004;63:71-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 213] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 5. | Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Kräusslich HG, Mizokami M. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791-796. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2323] [Cited by in RCA: 2279] [Article Influence: 108.5] [Reference Citation Analysis (0)] |
| 6. | Jones DM, McLauchlan J. Hepatitis C virus: assembly and release of virus particles. J Biol Chem. 2010;285:22733-22739. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 86] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 7. | Jones CT, Murray CL, Eastman DK, Tassello J, Rice CM. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J Virol. 2007;81:8374-8383. [PubMed] |
| 8. | Prodromou C, Pearl LH. Structure and functional relationships of Hsp90. Curr Cancer Drug Targets. 2003;3:301-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 206] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 9. | Geller R, Taguwa S, Frydman J. Broad action of Hsp90 as a host chaperone required for viral replication. Biochim Biophys Acta. 2012;1823:698-706. [PubMed] |
| 10. | Waxman L, Whitney M, Pollok BA, Kuo LC, Darke PL. Host cell factor requirement for hepatitis C virus enzyme maturation. Proc Natl Acad Sci USA. 2001;98:13931-13935. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 72] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 11. | Ujino S, Yamaguchi S, Shimotohno K, Takaku H. Heat-shock protein 90 is essential for stabilization of the hepatitis C virus nonstructural protein NS3. J Biol Chem. 2009;284:6841-6846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 65] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 12. | Okamoto T, Nishimura Y, Ichimura T, Suzuki K, Miyamura T, Suzuki T, Moriishi K, Matsuura Y. Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. EMBO J. 2006;25:5015-5025. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 210] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 13. | Ujino S, Nishitsuji H, Sugiyama R, Suzuki H, Hishiki T, Sugiyama K, Shimotohno K, Takaku H. The interaction between human initiation factor eIF3 subunit c and heat-shock protein 90: a necessary factor for translation mediated by the hepatitis C virus internal ribosome entry site. Virus Res. 2012;163:390-395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 14. | Kim MG, Moon JS, Kim EJ, Lee SH, Oh JW. Destabilization of PDK1 by Hsp90 inactivation suppresses hepatitis C virus replication through inhibition of PRK2-mediated viral RNA polymerase phosphorylation. Biochem Biophys Res Commun. 2012;421:112-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 23] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 15. | Beran RK, Sharma R, Corsa AC, Tian Y, Golde J, Lundgaard G, Delaney WE, Zhong W, Greenstein AE. Cellular growth kinetics distinguish a cyclophilin inhibitor from an HSP90 inhibitor as a selective inhibitor of hepatitis C virus. PLoS One. 2012;7:e30286. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 16. | Bartenschlager R, Lohmann V, Penin F. The molecular and structural basis of advanced antiviral therapy for hepatitis C virus infection. Nat Rev Microbiol. 2013;11:482-496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 287] [Cited by in RCA: 296] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 17. | Pietschmann T, Zayas M, Meuleman P, Long G, Appel N, Koutsoudakis G, Kallis S, Leroux-Roels G, Lohmann V, Bartenschlager R. Production of infectious genotype 1b virus particles in cell culture and impairment by replication enhancing mutations. PLoS Pathog. 2009;5:e1000475. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 106] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 18. | Takeuchi T, Katsume A, Tanaka T, Abe A, Inoue K, Tsukiyama-Kohara K, Kawaguchi R, Tanaka S, Kohara M. Real-time detection system for quantification of hepatitis C virus genome. Gastroenterology. 1999;116:636-642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 234] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 19. | Chou TC, Talaly P. A simple generalized equation for the analysis of multiple inhibitions of Michaelis-Menten kinetic systems. J Biol Chem. 1977;252:6438-6442. [PubMed] |
| 20. | Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5258] [Cited by in RCA: 5787] [Article Influence: 137.8] [Reference Citation Analysis (2)] |
| 21. | Paul D, Madan V, Bartenschlager R. Hepatitis C virus RNA replication and assembly: living on the fat of the land. Cell Host Microbe. 2014;16:569-579. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 195] [Cited by in RCA: 206] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 22. | Yang F, Robotham JM, Nelson HB, Irsigler A, Kenworthy R, Tang H. Cyclophilin A is an essential cofactor for hepatitis C virus infection and the principal mediator of cyclosporine resistance in vitro. J Virol. 2008;82:5269-5278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 186] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 23. | Goto K, Watashi K, Murata T, Hishiki T, Hijikata M, Shimotohno K. Evaluation of the anti-hepatitis C virus effects of cyclophilin inhibitors, cyclosporin A, and NIM811. Biochem Biophys Res Commun. 2006;343:879-884. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 114] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 24. | Robida JM, Nelson HB, Liu Z, Tang H. Characterization of hepatitis C virus subgenomic replicon resistance to cyclosporine in vitro. J Virol. 2007;81:5829-5840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 75] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 25. | Nakagawa S, Umehara T, Matsuda C, Kuge S, Sudoh M, Kohara M. Hsp90 inhibitors suppress HCV replication in replicon cells and humanized liver mice. Biochem Biophys Res Commun. 2007;353:882-888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 87] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
P- Reviewer: Chuang WL, Jin B S- Editor: Qiu S L- Editor: A E- Editor: Liu SQ