Published online Oct 28, 2016. doi: 10.4254/wjh.v8.i30.1287
Peer-review started: April 25, 2016
First decision: June 6, 2016
Revised: July 30, 2016
Accepted: August 27, 2016
Article in press: August 29, 2016
Published online: October 28, 2016
Processing time: 184 Days and 9.8 Hours
To investigate how Tregs are regulated in chronic hepatitis C virus (HCV) patients via assessment of Tregs markers (granzyme 2, CD69 and FoxP3), Teffs markers [TNFRSF4 (OX40), INFG] and CD4, CD25 genes.
A prospective study was conducted on 120 subjects divided into 4 groups: Group I (n = 30) treatment naïve chronic HCV patients; Group II (n = 30) chronic HCV treated with Peg/Riba; Group III (n = 30) chronic HCV associated with non-organ specific autoantibody and Group IV (n = 30) healthy persons as a control group. Tregs and Teffs markers were assessed in peripheral blood mononuclear cells by quantitative real time reverse transcriptase-polymerase chain reaction.
Chronic HCV patients exhibited significant higher levels of both Teffs and Tregs in comparison to healthy control group. Tregs markers were significantly decreased in Peg/Riba treated HCV patients in comparison to treatment naïve HCV group. In HCV patients with antinuclear antibody (ANA) +ve, Tregs markers were significantly decreased in comparison to all other studied groups. Teffs markers were significantly elevated in all HCV groups in comparison to control and in HCV group with ANA +ve in comparison to treatment naïve HCV group.
Elevated Tregs cells in chronic HCV patients dampen both CD4+ and CD8+ autologous T cell immune response. Interferon-α and ribavirin therapy suppress proliferation of Tregs. More significant suppression of Tregs was observed in HCV patients with autoantibodies favoring pathological autoimmune response.
Core tip: A prospective study conducted on 120 subjects divided into: Treatment naïve hepatitis C virus (HCV) patients, HCV patients treated with old standard of care, HCV associated with antinuclear antibody (ANA) and healthy control group. Teffs/Tregs imbalance was evaluated. Results showed that HCV patients exhibited significant higher levels of both Teffs and Tregs markers. Interferon-α and ribavirin therapy suppresses proliferation of Tregs. More significant suppression of Tregs was observed in HCV patients with autoantibodies favoring pathological autoimmune response. Teffs markers were significantly elevated in HCV treated group and in HCV group with ANA +ve in comparison to treatment naïve HCV group.
- Citation: Fouad H, El Raziky M, Hassan EM, Aziz GMA, Darweesh SK, Sayed AR. Regulatory and activated effector T cells in chronic hepatitis C virus: Relation to autoimmunity. World J Hepatol 2016; 8(30): 1287-1294
- URL: https://www.wjgnet.com/1948-5182/full/v8/i30/1287.htm
- DOI: https://dx.doi.org/10.4254/wjh.v8.i30.1287
Over 200 million people worldwide are suffering from chronic hepatitis C virus (HCV) infection and liver cirrhosis will be developed in about a quarter of these patients[1]. The prevalence rate of HCV genotype 4 in high risk populations in Egypt ranges from 73% to 90% and it was also found to be highly prevalent in sub-Saharan Africa and in the Middle East[2,3].
T lymphocytes play a major role in cell mediated immunity[4,5]. The several subsets of T cells have distinct functions and the majority is part of the adaptive immune system. Other subtypes can effectively present antigens to other T cells and are considered to be part of the innate immune system[6].
HCV is accompanied with different autoimmune manifestations[7], and could be a stimulator for the autoimmune reactions causing production of autoantibodies[8]. More recently, Acay et al[9] stated that the auto-antibodies in chronic HCV infection are highly incident. The authors stated that high percentages of patients with chronic hepatitis C had anti-mitochondrial antibodies, anti-smooth muscle antibodies, antinuclear antibody (ANA), thyroid antibody and anti-liver kidney microsomal antibodies.
The old HCV therapeutic protocol recommended by National Institutes of Health[10] was pegylated interferon (PEG-IFN) and ribavirin. Either endogenous or exogenous IFN-α leads to down regulation of CD4+ FoxP3hiIFN-γneg activated T regulatory cells (aTregs) while at the same time induces induction of of CD4+ FoxP3low/negIFN-γpos T-activated cells (aTeffs). IFN-α play an essential role in suppression of Tregs via inhibition of interleukin-2 secretion[11].
Together, these observations support the fact that in early antiviral response there is a production of IFN-α which enhances CD4 effector functions by inhibiting Tregs activation, whereas sustained elevation of IFN-α reverses Tregs/Teffs balance towards Teffs activation, generation of auto antibody and development of autoimmunity.
The objective of the present study is to evaluate the extent of Teffs/Tregs imbalance in chronic HCV and its association with old standard of care as well as the presence of ANA.
Our research hypothesis was that HCV with or without IFN-α and ribavirin is usually associated with Tregs/Teffs imbalance with subsequent generation of autoantibodies.
The primary outcome for this study was to evaluate Teffs/Tregs balance and regulation in chronic HCV through assessment of Tregs markers (granzyme 2, CD69 and FoxP3), Teffs markers (TNFRSF4, INFγ) and CD4, CD25 genes. Assessment of the effect of IFN-α and ribavirin on Teffs/Tregs balance as well as the association of Teffs/Tregs balance with the presence of antinuclear antibody were also conducted.
This was a prospective study conducted in Biochemistry and Molecular Biology Unit, Cairo University, Faculty of Medicine. The study included one hundred and twenty subjects categorized into 4 groups: Group I (30 patients) treatment naïve chronic HCV patients; Group II (30 patients) chronic HCV patients treated with the old standard of care therapy; Peg-IFN-α and ribavirin (Peg/Riba), group III (30 patients) chronic HCV patients associated with non-organ specific autoantibody and group IV, 30 healthy persons served as a control group. The patients attended the Internal Medicine Department at Beni-Sueif General Hospital. Healthy controls matched the age and sex of other patients. Cairo University Institutional review board in Faculty of Medicine approved the study. Informed written consent was signed by all subjects of the study.
The eligibility of selected patients included: (1) age between 18 and 65 years old; (2) anti-HCV positive serum; (3) positive HCV RNA detected by reverse-transcription/polymerase chain reaction (RT/PCR); (4) non-organ specific autoantibody by positive ANA test (titer > 1/32) in group III only and < 1/16 in all other groups; and (5) white blood cell > 3.500/mm3.
A signed informed consent was got in accordance with Declaration of Helsinki ethics guidelines.
Exclusion criteria include patients with: Hepatocellular carcinoma, HBV co-infection, severe psychiatric disease, HIV-positive patients, co-morbid serious conditions, schistosomiasis mansoni, past history of alcohol abuse or long use of hepatotoxic drugs.
All HCV-infected patients in the treated group had a 48 wk course of old standard of care (Peg/Riba therapy) and achieved sustained virologic response. The T cells markers were analyzed after more than 6 mo of the end of the Peg/Riba course.
Whole blood was obtained from all subjects of the study. The mononuclear cell layer was isolated using Ficoll (Sigma, St. Louis, MO, United States) and centrifugation was conducted for 30 min at 400 g in cooling centrifuge.
RNA extraction: Total RNA was isolated from mononuclear cell layer using Qiagen purification reagent (Qiagen, CA, United States). The extracted RNA was quantified and checked for purity using a spectrophotometer (260/280 w.l.).
Primer sequence: PCR primers were got from GenBank RNA sequences cited at the following website: http://www.ncbi.nlm.nih.gov/tools/primer-blast (Table 1).
| TNFRSF4OX 40 | Forward: ′5 GCA ATA GCT CGG ACG CAA TCT 3′ |
| DQ032625.1 | Reverse: ′5 GAG GGT CCC TGT GAG GTT CT 3′ |
| Granzyme 2 | Forward: ′5 TAC CAT TGA GTT GTG CGT GGG 3′ |
| NM_004131.4 | Reverse: ′5 GCC ATT GTT TCG TCC ATA GGA GA 3′ |
| CD69 | Forward: ′5 GGT CAC CCA TGG AAG TGG TC 3′ |
| NM_001781.2 | Reverse: ′5 GAC TTC GGA CCA CAG AGC AG 3′ |
| CD4 NM_001195017.2 | Forward: ′5 CTG CAA GTT CTC ACA CCG TC 3′ |
| Reverse: ′5 CTA GAG TTG CCT GCT CTG CC 3′ | |
| CD25 IL2R NM_000417.2 | Forward: ′5 GCT CTA CAC AGA GGT CCT GC 3′ |
| Reverse: ′5 AGC ACA ACG GAT GTC TCC TG 3′ | |
| FoxP3 | Forward: ′5 CCC ATC CCC AGG AGT CTT G 3′ |
| NG_007392.1 | Reverse: ′5 ACC ATG ACT AGG GGC ACT GTA 3′ |
| Interferon γ | Forward: ′5 ATG GTT GTC CTG CCT GCA AT 3′ |
| NG_015840.1 | Reverse: ′5 CTT GCT TAG GTT GGC TGC CT 3′ |
Real-time quantitative PCR using SYBR Green I: Step One plus real-time PCR system was used in the analysis using software version 3.1 (Applied Biosystems, United States). Optimization of the annealing temperature was conducted for the PCR protocol and for the primer sets.
All cDNA were prepared for all gene markers, glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and for non-template negative control.
Five microliter of total RNA was used to generate cDNA using 20 pmol antisense primer and 0.8 μL AMV reverse transcriptase at 37 °C for 60 min. The relative abundance of mRNA species was evaluated using the SYBR® Green method (Applied Biosystems, CA, United States).
Annealing temperature of 60 °C was optimized for all primer sets. Real time polymerase reaction was performed in 25 μL reaction volume consisting of Mater Mix of SYBR Green, 3 μL of cDNA, 900 nmol/L of every primer. Amplification conditions were conducted according to the manufacturer specifications: 2 min at 50 °C, 10 min at 95 °C, 40 repeated cycles with 15 s denaturation and 10 min of annealing/extension at 60 °C.
The resulting data were expressed in Cycle threshold (Ct). The PCR data results show Ct values of all studied genes (CD69, CD4, granzyme 2, TNFRSF4, FoxP3, CD25 and IFNγ) and the house keeping gene (GAPDH). A negative control sample was no template cDNA was used. Target gene expression was related to GAPDH.
Data were calculated using the Applied Biosystems Step One plus software. Relative gene expressions of all assessed genes were calculated using the comparative Ct method. All values were normalized to GAPDH housekeeping gene and expressed as fold changes relative to the background levels found in the control samples.
Statistical Package of Social Studies (SPSS) version 16.0.1 (SPSS Inc., Chicago, IL, United States) was utilized. Numerical data were presented as mean ± standard deviation. The null hypothesis was calculated for multiple groups by a single-factor ANOVA and for two groups by unpaired t-test. Statistically significant was considered if P value was < 0.05.
This study included 120 subjects divided into four groups. There weren’t any difference between the four groups with statistical significance regarding age, sex distribution, albumin and T.bilirubin values (P-value > 0.05).
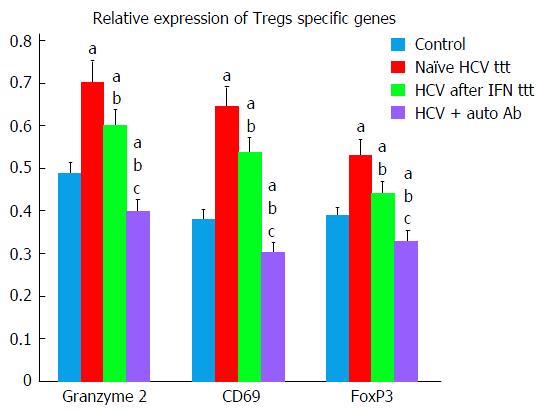
Findings of the present study exhibited that chronic HCV patients exhibited significant higher levels of both Teffs and Tregs markers as compared to healthy control group. Tregs markers (granzyme 2, CD69, FoxP3) were significantly decreased in Peg/Riba treated HCV patients in comparison to treatment naïve HCV group (Tables 2 and 3).
| Group I (naïve) | Group II (Peg/Riba) | Group III (ANA+) | Group IV (control) | |
| Granzyme 2 Tregs | 0.704 ± 0.039 | 0.603 ± 0.046 | 0.400 ± 0.042 | 0.489 ± 0.053 |
| CD69 Tregs | 0.647 ± 0.037 | 0.54 ± 0.049 | 0.306 ± 0.036 | 0.383 ± 0.043 |
| FoxP3 Tregs | 0.531 ± 0.033 | 0.444 ± 0.046 | 0.330 ± 0.039 | 0.39 ± 0.030 |
| TNFRSF4OX 40 | 0.596 ± 0.047 | 0.688 ± 0.057 | 0.707 ± 0.05 | 0.482 ± 0.056 |
| CD4 | 0.584 ± 0.034 | 0.677 ± 0.048 | 0.712 ± 0.042 | 0.528 ± 0.05 |
| CD25 | 0.595 ± 0.039 | 0.684 ± 0.053 | 0.73 ± 0.053 | 0.495 ± 0.041 |
| Interferon γ | 0.466 ± 0.035 | 0.483 ± 0.035 | 0.522 ± 0.044 | 0.357 ± 0.038 |
| Group I naïve | Group II Peg/Riba | Group III ANA+ | Group IV control | |
| FOX3 | ||||
| Group I naïve | aP ≤ 0.05 | dP ≤ 0.001 | bP ≤ 0.01 | |
| Group II Peg/Riba | aP ≤ 0.05 | aP ≤ 0.05 | bP ≤ 0.01 | |
| Group III ANA+ | dP ≤ 0.001 | aP ≤ 0.05 | dP ≤ 0.001 | |
| Group IV control | bP ≤ 0.01 | bP ≤ 0.01 | dP ≤ 0.001 | |
| CD69 | ||||
| Group I naïve | bP ≤ 0.01 | bP ≤ 0.01 | dP ≤ 0.001 | |
| Group II Peg/Riba | bP ≤ 0.01 | bP ≤ 0.01 | dP ≤ 0.001 | |
| Group III ANA+ | bP ≤ 0.01 | bP ≤ 0.01 | dP ≤ 0.001 | |
| Group IV control | dP ≤ 0.001 | dP ≤ 0.001 | dP ≤ 0.001 | |
| Granzyme | ||||
| Group I naïve | dP ≤ 0.001 | dP ≤ 0.001 | dP ≤ 0.001 | |
| Group II Peg/Riba | dP ≤ 0.001 | bP ≤ 0.01 | dP ≤ 0.001 | |
| Group III ANA+ | dP ≤ 0.001 | bP ≤ 0.01 | dP ≤ 0.001 | |
| Group IV control | dP ≤ 0.001 | dP ≤ 0.001 | dP ≤ 0.001 |
In HCV patients with autoantibodies, Tregs markers were significantly decreased in comparison to all the other studied groups (Tables 2 and 3, Figure 1).
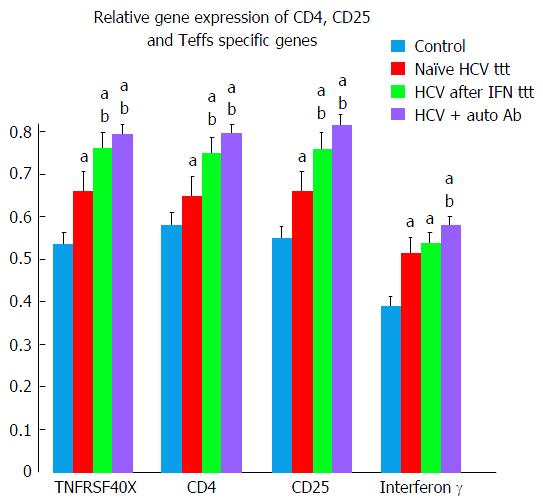
Teffs specific genes (TNFRSF4 and IFN-γ) and CD4, CD25 showed significant elevation in treatment naïve HCV group in comparison to control group (Tables 2 and 4, Figure 2).
| Group I naïve | Group II Peg/Riba | Group III ANA+ | Group IV control | |
| TNFRSF4OX 40 | ||||
| Group I naïve | bP ≤ 0.01 | aP ≤ 0.05 | aP ≤ 0.05 | |
| Group II Peg/Riba | bP ≤ 0.01 | NS (P > 0.05) | bP ≤ 0.01 | |
| Group III ANA+ | aP ≤ 0.05 | NS (P > 0.05) | dP ≤ 0.001 | |
| Group IV control | aP ≤ 0.05 | bP ≤ 0.01 | dP ≤ 0.001 | |
| CD4 | ||||
| Group I naïve | bP ≤ 0.01 | aP ≤ 0.05 | aP ≤ 0.05 | |
| Group II Peg/Riba | bP ≤ 0.01 | NS (P > 0.05) | aP ≤ 0.05 | |
| Group III ANA+ | aP ≤ 0.05 | NS (P > 0.05) | bP ≤ 0.01 | |
| Group IV control | aP ≤ 0.05 | aP ≤ 0.05 | bP ≤ 0.01 | |
| CD25 | ||||
| Group I naïve | bP ≤ 0.01 | aP ≤ 0.05 | aP ≤ 0.05 | |
| Group II Peg/Riba | bP ≤ 0.01 | NS (P > 0.05) | bP ≤ 0.01 | |
| Group III ANA+ | aP ≤ 0.05 | NS (P > 0.05) | dP ≤ 0.001 | |
| Group IV control | aP ≤ 0.05 | bP ≤ 0.01 | dP ≤ 0.001 | |
| Interferon-γ | ||||
| Group I naïve | bP ≤ 0.001 | bP ≤ 0.001 | aP ≤ 0.05 | |
| Group II Peg/Riba | bP ≤ 0.001 | NS (P > 0.05) | aP ≤ 0.05 | |
| Group III ANA+ | bP ≤ 0.001 | NS (P > 0.05) | bP ≤ 0.01 | |
| Group IV control | aP ≤ 0.05 | aP ≤ 0.05 | bP ≤ 0.01 |
More significant elevation in Teffs genes was observed in both Peg/Riba treated HCV and HCV with autoantibodies groups as compared to treatment naïve HCV and control groups (Tables 2 and 4, Figure 2).
HCV is reported to suppress immune system to sustain chronic infection. Accumulation of Tregs and activation of inhibitory signaling pathways play essential roles in suppressing antiviral effector T cells (Teffs). The mechanisms by which HCV impairs Teffs include: Induction of Tregs, Th1 deficiency or Th2 dominance, blunted T cell activation, T cell apoptosis and T cell anergy[12].
The objectives of the present study were to assess the extent of upregulation of Tregs in HCV patients whether or not associated with auto-antibodies. In the present study, we have evaluated certain markers of Tregs and Teffs in peripheral blood mononuclear cells.
FOXP3 (forkhead box P3) is a member of forkhead/winged-helix family of transcriptional regulators which are master regulators in the development and function of Tregs[13,14].
Our results exhibited significant upregulation of FOXP3 in all HCV patients groups as compared to healthy controls. Other studies also confirmed accumulation of FOXP3+ Tregs in most chronic viral infections with subsequent suppression of antiviral CD4+ and CD8+ T cell responses[15-18].
Moreover, findings of our study demonstrated more significant decrease in Tregs specific genes (CD69, FoxP3, and granzyme 2) in HCV patients group after Peg/Riba therapy. Similar findings were reported by Langhans et al[19] who stated that ribavirin can inhibit functions of HCV-specific Tregs beside its immuno-stimulatory effects on TH1 cells. Ribavirin can subsequently inhibit Treg-mediated suppression of Teffs in chronic HCV infections pushing the disease towards autoimmune responses.
Golding et al[11] stated that IFN-α, promotes proliferation of FoxP3Low/NegIFN-γPos activated Teffs while simultaneously suppresses the development of FoxP3HIIFN-γNeg activated Tregs. These data coincided with our findings in group II in relation to the other groups.
CD4 gene product is a membrane glycoprotein of T lymphocytes that mediates initiation and augments early phase of T-cell activation[20-22]. Findings of the present study demonstrated significant elevation of CD4 gene in all HCV groups in comparison to control group suggesting increase in activated T cells function. More significant elevation of CD4 was observed in HCV after treatment and in HCV with auto-antibodies groups.
CD25 is a type I transmembrane protein present on activated T cells, activated B cells and in memory CD8 T cells[23-27]. Our results demonstrated significant elevation of CD25 in all HCV studied groups with more significant elevation after Peg/Riba therapy. Similar findings were reported by Caetano et al[28] in chronic HCV patients during Peg/Riba treatment who presented an amplified CD8 T-cell responses specific to HCV and more increase Teffs.
Moorman et al[12] showed that many inhibitory signaling pathways were up-regulated during chronic HCV infection, resulting in expansion of Tregs and contraction of Teffs. Thus, this inhibitory pathway may not only regulate proliferation and differentiation of naïve T cells, but also control responses of Teffs, memory cells, and expansion of Tregs[29].
These facts coincided with our results that showed significant elevation of Tregs specific genes (CD69, FoxP3, and granzyme 2) and significant elevation of Teffs specific genes (TNFRSF4, INF-γ) and CD4, CD25 genes in both groups of HCV whether naïve or after treatment in comparison to healthy controls.
Granzyme B encodes a protein that is essential in induction of cell-mediated immune response for the faster initiation of target cell apoptosis by cytolytic T lymphocytes[30]. Tregs possess granzyme B, enabling them to induce apoptosis in effector T-cells[31,32]. Our results demonstrated that granzyme B was found to be significantly elevated in naïve HCV patients with significant decrease after Peg/Riba therapy. More significant decrease in its levels was observed in HCV patients with autoantibodies favoring autoimmune response in those patients.
CD69 expression was studied by Colbeck et al[33] and they stated that Tregs expressing CD69(+) are more proliferative and more suppressive than their CD69(-) counterparts. This finding explains our results that showed significant lower CD69 expression in HCV patients with autoantibodies suggesting inhibition of Tregs activity favoring autoimmune environment.
IFN-γ is a member of the type II class of interferons[34]. Longhi et al[31] stated that CD8+ T cells when cultured on their own secrete much higher levels of IFN-γ in patients with autoimmune hepatitis when compared to normal subjects and have a high proliferation rate. This finding coincided with our results that demonstrated significant higher levels of IFN-γ in patients with autoantibodies than healthy controls.
The protein encoded by the TNFRSF4 gene belongs to tumor necrosis factor - receptor superfamily that has essential roles in CD4+ T cell response[35,36].
Our findings showed that, in HCV patients with ANA +ve, TNFRSF4 and IFN-γ (T effector gene) were significantly higher as compared to HCV naïve and healthy control groups. This means that T effector cells dominate over T regulatory cells favoring autoimmunity and initiating pathological immune response with production of autoantibodies. Similar findings were reported by González-Amaro et al[37].
Also, our results agreed with similar findings reported by Longhi et al[31] who stated that regulatory T cells are decreased numerically and impaired functionally in autoimmune hepatitis. Similar to our results, several studies reported that HCV infection induces a dramatic increase in Tregs, which contributes to the immune response failure during HCV infection[38].
In conclusion, chronic HCV patients exhibited significant higher levels of both Teffs and Tregs in comparison to healthy controls. Moreover, elevated levels of Treg cells in patients with chronic HCV dampen both the CD4+ and CD8+ autologous T cell immune response. IFN-α and ribavirin therapy suppress proliferation of Tregs and do not restore the Teffs/Tregs imbalance. More significant suppression of Tregs was observed in HCV patients with autoantibodies favoring pathological autoimmune response.
Hepatitis C virus (HCV) is accompanied with different autoimmune manifestations, and could be a stimulator for the autoimmune reactions causing production of autoantibodies. More recently, Acay et al stated that the auto-antibodies in chronic HCV infection are highly incident. The authors stated that high percentages of patients with chronic hepatitis C had anti-mitochondrial antibodies (AMA), anti-smooth muscle antibodies (ASMA), anti-nuclear antibodies (ANA), thyroid antibody and anti-liver kidney microsomal antibodies (anti-LKM-1). The old HCV therapeutic protocol was pegylated interferon (IFN) and ribavirin. IFN-α leads to down regulation of CD4+ FoxP3hiIFN-γneg activated T regulatory cells (aTregs) while at the same time induces induction of of CD4+ FoxP3low/negIFN-γpos T-activated cells (aTeffs). Together, these observations support the fact that sustained elevation of IFN-α reverses Tregs/Teffs balance towards Teffs activation, generation of auto antibody and development of autoimmunity. The objective of the present study is to evaluate the extent of Teffs/Tregs imbalance in chronic HCV and its association with old standard of care as well as the presence of ANA.
IFN-α/ribavirin old therapeutic protocol enhances CD4 effector (Teffs) functions by inhibiting Tregs activation. The old protocol reverses Tregs/Teffs balance towards Teffs activation, generation of auto antibody and development of autoimmunity.
New direct acting antiviral drugs do not induce Tregs/Teffs imbalance, whereas the old standard of care IFN-α and ribavirin induce Tregs/Teffs imbalance. Replacing the old therapeutic protocol by the new direct acting antiviral drugs is mandatory because beside its efficacy, the new direct acting antiviral drugs do not induce autoimmunity.
Chronic HCV patients exhibited significant higher levels of both Teffs and Tregs in comparison to healthy controls. Moreover, elevated levels of Treg cells in patients with chronic HCV dampen both the CD4+ and CD8+ autologous T cell immune response. IFN-α and ribavirin therapy suppress proliferation of Tregs and do not restore the Teffs/Tregs imbalance. More significant suppression of Tregs was observed in HCV patients with autoantibodies favoring pathological autoimmune response. Repalcing the old therapeutic protocol; IFN-α and ribavirin by the new direct acting antiviral drugs is mandatory and is also essential avoid Teffs/Tregs imbalance.
Tregs also known as suppressor T cells, are a subpopulation of T cells that down-regulates or suppress induction, proliferation and activation of effector T cells. Tregs also maintain tolerance to self-antigens and prevent autoimmunity. Teffs includes various T cell types that actively respond to antigenic stimuli, such as co-stimulation. This includes helper T cells, cytotoxic or killer T cells, and potentially other T cell types as memory cells. HCV-induced autoantibodies involves: AMA, ASMA, ANA, thyroid antibody and anti-LKM-1. Forkhead box P3 is a member of forkhead/winged-helix transcriptional regulators which master the development and function of Tregs. CD4 gene product is a membrane glycoprotein of T lymphocytes that mediates initiation and augments early phase of T-cell activation. Granzyme B is a protein that enables Tregs to induce apoptosis in effector T-cells. TNFRSF4 gene belongs to tumor necrosis factor -receptor superfamily that plays essential roles in CD4+ T cell response.
The manuscript presents the gene expression of immune molecules in peripheral blood mononuclear cell by reverse transcription polymerase chain reaction. And to investigate the immune changes induced by HCV, as well as after PR therapy. This paper may be interest to the readers of the journal and provide some knowledge for clinicians and researchers.
| 1. | Jiménez-Sousa MA, Fernández-Rodríguez A, Guzmán-Fulgencio M, García-Álvarez M, Resino S. Meta-analysis: implications of interleukin-28B polymorphisms in spontaneous and treatment-related clearance for patients with hepatitis C. BMC Med. 2013;11:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 73] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 2. | AASLD/IDSA HCV Guidance Panel. Hepatitis C guidance: AASLD-IDSA recommendations for testing, managing, and treating adults infected with hepatitis C virus. Hepatology. 2015;62:932-954. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 912] [Cited by in RCA: 997] [Article Influence: 90.6] [Reference Citation Analysis (0)] |
| 3. | Mohamoud YA, Mumtaz GR, Riome S, Miller D, Abu-Raddad LJ. The epidemiology of hepatitis C virus in Egypt: a systematic review and data synthesis. BMC Infect Dis. 2013;13:288. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 234] [Cited by in RCA: 253] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 4. | McClory S, Hughes T, Freud AG, Briercheck EL, Martin C, Trimboli AJ, Yu J, Zhang X, Leone G, Nuovo G. Evidence for a stepwise program of extrathymic T cell development within the human tonsil. J Clin Invest. 2012;122:1403-1415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 73] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 5. | Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. New York: Garland Science 2002; 1367. Lymphocytes and the Cellular Basis of Adaptive Immunity Available from: http://www.ncbi.nlm.nih.gov/books/NBK26921/. |
| 6. | Vantourout P, Hayday A. Six-of-the-best: unique contributions of γδ T cells to immunology. Nat Rev Immunol. 2013;13:88-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 989] [Cited by in RCA: 999] [Article Influence: 76.8] [Reference Citation Analysis (0)] |
| 7. | Jadali Z, Alavian SM. Autoimmune diseases co-existing with hepatitis C virus infection. Iran J Allergy Asthma Immunol. 2010;9:191-206. [PubMed] |
| 8. | Himoto T, Masaki T. Extrahepatic manifestations and autoantibodies in patients with hepatitis C virus infection. Clin Dev Immunol. 2012;2012:871401. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 46] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 9. | Acay A, Demir K, Asik G, Tunay H, Acarturk G. Assessment of the Frequency of Autoantibodies in Chronic Viral Hepatitis. Pak J Med Sci. 2015;31:150-154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 10. | National Institutes of Health. National Institutes of Health Consensus Development Conference Statement: Management of hepatitis C: 2002--June 10-12, 2002. Hepatology. 2002;36:S3-S20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 227] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 11. | Golding A, Rosen A, Petri M, Akhter E, Andrade F. Interferon-alpha regulates the dynamic balance between human activated regulatory and effector T cells: implications for antiviral and autoimmune responses. Immunology. 2010;131:107-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 38] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 12. | Moorman JP, Wang JM, Zhang Y, Ji XJ, Ma CJ, Wu XY, Jia ZS, Wang KS, Yao ZQ. Tim-3 pathway controls regulatory and effector T cell balance during hepatitis C virus infection. J Immunol. 2012;189:755-766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 84] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 13. | Zhang L, Zhao Y. The regulation of Foxp3 expression in regulatory CD4(+)CD25(+)T cells: multiple pathways on the road. J Cell Physiol. 2007;211:590-597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 119] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 14. | Marson A, Kretschmer K, Frampton GM, Jacobsen ES, Polansky JK, MacIsaac KD, Levine SS, Fraenkel E, von Boehmer H, Young RA. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature. 2007;445:931-935. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 579] [Cited by in RCA: 590] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 15. | Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057-1061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6077] [Cited by in RCA: 6497] [Article Influence: 282.5] [Reference Citation Analysis (0)] |
| 16. | Walker CM. Adaptive immunity to the hepatitis C virus. Adv Virus Res. 2010;78:43-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 58] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 17. | Belkaid Y, Chen W. Regulatory ripples. Nat Immunol. 2010;11:1077-1078. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 18. | Zhai N, Chi X, Li T, Song H, Li H, Jin X, Crispe IN, Su L, Niu J, Tu Z. Hepatitis C virus core protein triggers expansion and activation of CD4(+)CD25(+) regulatory T cells in chronic hepatitis C patients. Cell Mol Immunol. 2015;12:743-749. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 35] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 19. | Langhans B, Nischalke HD, Arndt S, Braunschweiger I, Nattermann J, Sauerbruch T, Spengler U. Ribavirin exerts differential effects on functions of Cd4+ Th1, Th2, and regulatory T cell clones in hepatitis C. PLoS One. 2012;7:e42094. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 20. | Geer LY, Marchler-Bauer A, Geer RC, Han L, He J, He S, Liu C, Shi W, Bryant SH. The NCBI BioSystems database. Nucleic Acids Res. 2010;38:D492-D496. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 556] [Cited by in RCA: 592] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 21. | Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2010;38:D5-D16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 351] [Cited by in RCA: 347] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 22. | Soldevila B, Alonso N, Martínez-Arconada MJ, Morillas RM, Planas R, Sanmartí AM, Martínez-Cáceres EM. A prospective study of T- and B-lymphocyte subpopulations, CD81 expression levels on B cells and regulatory CD4(+) CD25(+) CD127(low/-) FoxP3(+) T cells in patients with chronic HCV infection during pegylated interferon-alpha2a plus ribavirin treatment. J Viral Hepat. 2011;18:384-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 23. | Starbeck-Miller GR, Xue HH, Harty JT. IL-12 and type I interferon prolong the division of activated CD8 T cells by maintaining high-affinity IL-2 signaling in vivo. J Exp Med. 2014;211:105-120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 130] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 24. | Janeway CA, Travers P, Walport M, Shlomchik MJ. Immunobiology: The Immune System in Health and Disease. New York: Garland Science 2001; Available from: http://www.ncbi.nlm.nih.gov/books/NBK10757/. |
| 25. | Triplett TA, Curti BD, Bonafede PR, Miller WL, Walker EB, Weinberg AD. Defining a functionally distinct subset of human memory CD4+ T cells that are CD25POS and FOXP3NEG. Eur J Immunol. 2012;42:1893-1905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 38] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 26. | Wherry EJ, Ahmed R. Memory CD8 T-cell differentiation during viral infection. J Virol. 2004;78:5535-5545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 632] [Cited by in RCA: 711] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 27. | Burchill MA, Golden-Mason L, Wind-Rotolo M, Rosen HR. Memory re-differentiation and reduced lymphocyte activation in chronic HCV-infected patients receiving direct-acting antivirals. J Viral Hepat. 2015;22:983-991. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 91] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 28. | Caetano J, Martinho A, Paiva A, Pais B, Valente C, Luxo C. Differences in hepatitis C virus (HCV)-specific CD8 T-cell phenotype during pegylated alpha interferon and ribavirin treatment are related to response to antiviral therapy in patients chronically infected with HCV. J Virol. 2008;82:7567-7577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 29. | Mengshol JA, Golden-Mason L, Arikawa T, Smith M, Niki T, McWilliams R, Randall JA, McMahan R, Zimmerman MA, Rangachari M. A crucial role for Kupffer cell-derived galectin-9 in regulation of T cell immunity in hepatitis C infection. PLoS One. 2010;5:e9504. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 161] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 30. | Dahl CA, Bach FH, Chan W, Huebner K, Russo G, Croce CM, Herfurth T, Cairns JS. Isolation of a cDNA clone encoding a novel form of granzyme B from human NK cells and mapping to chromosome 14. Hum Genet. 1990;84:465-470. [PubMed] |
| 31. | Longhi MS, Hussain MJ, Mitry RR, Arora SK, Mieli-Vergani G, Vergani D, Ma Y. Functional study of CD4+CD25+ regulatory T cells in health and autoimmune hepatitis. J Immunol. 2006;176:4484-4491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 242] [Cited by in RCA: 245] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 32. | Azzi J, Skartsis N, Mounayar M, Magee CN, Batal I, Ting C, Moore R, Riella LV, Ohori S, Abdoli R. Serine protease inhibitor 6 plays a critical role in protecting murine granzyme B-producing regulatory T cells. J Immunol. 2013;191:2319-2327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 33. | Colbeck EJ, Hindley JP, Smart K, Jones E, Bloom A, Bridgeman H, McPherson RC, Turner DG, Ladell K, Price DA. Eliminating roles for T-bet and IL-2 but revealing superior activation and proliferation as mechanisms underpinning dominance of regulatory T cells in tumors. Oncotarget. 2015;6:24649-24659. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 34. | Gray PW, Goeddel DV. Structure of the human immune interferon gene. Nature. 1982;298:859-863. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 356] [Cited by in RCA: 367] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 35. | Latza U, Dürkop H, Schnittger S, Ringeling J, Eitelbach F, Hummel M, Fonatsch C, Stein H. The human OX40 homolog: cDNA structure, expression and chromosomal assignment of the ACT35 antigen. Eur J Immunol. 1994;24:677-683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 96] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 36. | Keoshkerian E, Helbig K, Beard M, Zaunders J, Seddiki N, Kelleher A, Hampartzoumian T, Zekry A, Lloyd AR. A novel assay for detection of hepatitis C virus-specific effector CD4(+) T cells via co-expression of CD25 and CD134. J Immunol Methods. 2012;375:148-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 37. | González-Amaro R, Cortés JR, Sánchez-Madrid F, Martín P. Is CD69 an effective brake to control inflammatory diseases? Trends Mol Med. 2013;19:625-632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 150] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 38. | Hashempoor T, Bamdad T, Merat S, Janzamin E, Nemati L, Jabbari H, Sharifi AH, Zamini H. Expansion of CD4+ CD25+ FoxP3+ regulatory T cells in chronic hepatitis C virus infection. Iran J Immunol. 2010;7:177-185. [PubMed] |
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Egypt
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
P- Reviewer: Baddour N, Barili F, Ji FP S- Editor: Gong XM L- Editor: A E- Editor: Li D