Published online Nov 27, 2013. doi: 10.4254/wjh.v5.i11.642
Revised: September 28, 2013
Accepted: October 17, 2013
Published online: November 27, 2013
Processing time: 106 Days and 22.5 Hours
AIM: To investigate the evidence of homogeneous phenomenon on CYP3A5*3 MDR1-3435 and CYP3A4*18 of the liver graft after living donor liver transplantation (LDLT).
METHODS: We identified the proportional change of the CYP3A5*3, MDR1-3435 and CYP3A4*18 from the peripheral blood mononuclear cell of 41 pairs recipient/donor with different genotype polymorphisms and 119 liver graft biopsy samples used with the pyrosequencing technique after LDLT. Polymerase chain reaction/ligase detection reaction assay and restriction fragment length polymorphism was employed for genotyping the CYP3A5*3 and CYP3A4*18 single nucleotide polymorphisms (SNPs). All of the recipients and donors expressed with the similar SNP genotype of CYP3A5*3, MDR1-3435 or CYP3A4*18 were excluded.
RESULTS: The final genetic polymorphisms of the liver graft biopsy samples of CYP3A5*3, MDR1-3435 and CYP3A4*18 was predominated depends on the donor with restriction fragment length polymorphism and seems to be less related to the recipient. The proportional changes of G to A alleles of the 119 samples of CYP3A5*3 (included A > A/G, A/G > A, A/G > G, G > A, G > A/G and A > G), C to T alleles of the 108 samples of MDR1-3435 (included C > C/T, C/T > C, C/T > T, T > C/T and T > C), and T to C alleles of 15 samples of CYP3A4*18 (included T/C > T and T > C/T) were significant different between the recipients and the liver graft biopsy samples (P < 0.0001) and less difference when compared with the donors in the pyrosequencing analysis after LDLT.
CONCLUSION: The CYP3A5*3, MDR1-3435 and CYP3A4*18 of the recipient could be modified by the donor so-called homogenous phenomenon when the recipient’s blood drained into the liver graft.
Core tip: The most innovative concept is that pyrosequencing can deeply clarify the proportional change of the A and G alleles in CYP3A5*3, C and T alleles in MDR1-3435, and T and C alleles in CYP3A4*1 when the different genotype of single nucleotide polymorphism after living donor liver transplantation. The biogenetic characteristic of the recipient could be modified by a donor you want to change the genetic characteristic. For further confirmation, homogeneous phenomenon was the truly occurred in the cytochrome P450 system when the recipients and donors with different genotype of the single nucleotide polymorphism.
- Citation: Chiu KW, Nakano T, Chen KD, Hsu LW, Lai CY, Chiu HC, Huang CY, Cheng YF, Goto S, Chen CL. Homogeneous phenomenon of the graft when using different genotype characteristic of recipients/donors in living donor liver transplantation. World J Hepatol 2013; 5(11): 642-648
- URL: https://www.wjgnet.com/1948-5182/full/v5/i11/642.htm
- DOI: https://dx.doi.org/10.4254/wjh.v5.i11.642
The cytochrome P450 is a very important system for the drugs metabolism in the liver. Therefore, the metabolic characteristic of this system is considerable affected when the recipient received a liver graft with different genotype of the drug metabolic isoenzymes after liver transplantation. In our previous study, it was very interesting that the genotypes of the CYP2C19 in the peripheral blood mononuclear cell does not change even the recipient received a liver graft with individually differences genotype of the CYP2C19 after living donor liver transplantation (LDLT)[1]. Nevertheless, the fast drug metabolic characteristic of the liver graft seems to be presenting a more frequency of the abnormal liver function after LDLT[2]. The different genotype of the drug metabolic isoenzyme represented with an uncertain phenomenon when the new liver graft continuously circulated with the original recipient peripheral blood[3]. Recently, we successfully identified the homogeneous phenomenon of the single nucleotide polymorphism (SNP) from the liver graft biopsy specimen using by the pyrosequencing method[4]. Herein, we would like to expose all of the polymorphisms of CYP3A5*3, MDR1-3435 and CYP3A4*18 because of the importance for the immunosuppressive agent metabolism in the liver transplantation setting.
Based on the liver graft biopsy samples with the different genotype between recipient and donor, 119 liver graft biopsy specimens in 41 cases of CYP3A5*3, 108 liver graft biopsies in 41 cases of MDR1-3435 and 15 liver graft biopsies in 3 cases of 3A4*18 (38 recipients/donors had the similar polymorphism genotype for CYP3A4*18) were enrolled for pyrosequencing analysis prospectively. All of the recipients and donors expressed with the similar SNP genotype of CYP3A5*3, MDR1-3435 or CYP3A4*18 were excluded in this study. We performed 119 liver biopsies in 41 recipients because of the evidence of clinical investigation of abnormal liver function after LDLT as our previous reported[2,5]. They were performed once in 14 recipients, twice in 7, 3 times in 9, 4 times in 1, 5 times in 6, 6 times in 1, 7 times in 2, and 10 times in 1. Base of the haplotypes of the SNP from the polymerase chain reaction (PCR)-restriction fragment length polymorphism (RFLP), there were 10 cases in A > A/G, 3 cases in A/G > A, 12 cases in A/G > G, 2 cases in G > A, 13 cases in G > A/G and 1 case in A > G on the polymorphism genotypes of CYP3A5*3; 14 cases in C > C/T, 16 cases in C/T > C, 3 cases in C/T > T, 7cases in T > C/T and 1case in T > C on the polymorphism genotypes of MDR1-3435; and only 2 cases in T/C > T and 1 case in T > T/C on the polymorphism genotypes of CYP3A4*18 due to 92.7% (38/41) recipients/donors were the similar CYP3A4*18 genotype SNP.
Genomic DNA was isolated from the 0.5 mL EDTA-treated whole blood and liver biopsy specimens using the QIAamp DNA mini kit (Qiagen) in accordance with the manufacture’s instruction.
PCR/ligase detection reaction assay (LDR) and RFLP was employed for genotyping the CYP3A5*3 and CYP3A4*18 SNPs. A PCR assay for the CYP3A5*3 was using forward primer (5′-CATGACTTAGTAGACAGAT GAC-3′) and reverse primer (5′-GGTCCAAACAGGGAAGAAATA-3') was performed in a 25 μL of reaction volume; and for genotyping the CYP3A4*18, a PCR assay was using forward primer (5′-CACCCTGATGTCCAGCAGAAA CT-3′) and reverse primer (5′-AATAGAAAGCAGATGAACCAGAGCC-3′). The Probe for the CYP3A5*3-A was using (5′-TTTTTTTTTTTTTTTTTTTT TGTGGTCCAAACAGGGAAGAGATAT-3′) and for the CYP3A5*3-G was using (5′-TTTTTTTTTTTTTTTTTTTTTTTGTGGTCCAAACAGGGAAGAGATAC-3′). The Probe for the CYP3A4*18-G was using (5′-TTTTTTTTTTTTTTT TTTTTTTTTACCTCCTCCCTCCTTCTCCATGTAC-3′) and for the CYP3A4*18-A (5′-TTTTTTTTTTTTTTTTTTTTTTACCTCCTCCCTCCTTCTCC ATGTAT-3′. The PCR conditions consisted of a denaturation step at 95 °C for 15 min, followed by 35 cycles of 94 °C for 30 s, 65 °C for 1 min, and 72 °C for 1 min, followed by a final extension step at 72 °C for 7 min. The specific amplified fragments were used in an LDR assay to identify the mutations associated with CYP3A5*3 and. CYP3A4*18. The LDR assay was performed as follows: 10 μL of the reaction mix contained 1 μL of 1 × ligase reaction buffer (New England Biolabs, United States), 1 μL of probes (12.5 pmol/μL each), 0.05 μL (2 U) of thermostable Taq DNA ligase (New England Biolabs), and 1 μL of PCR product. The ligation reaction was performed with a GeneAmp PCR System 9600 (Perkin Elmer, United States) as follows: 15 min at 95 °C, followed by 35 cycles of 30 s at 94 °C and 2 min at 60 °C. PCR-RFLP was performed to genotype intron 3 (6986 A > G) variant alleles in the CYP 3A5*3 gene and exon 10 (878 T > C) variant alleles in the CYP3A4*18 gene, with slight modifications. PCR products were digested with Ssp I for CYP3A5*3 and with Hpa II for CYP3A4*18. The products were separated by agarose gel electrophoresis and analyzed by an ABI PRISM 377 DNA sequencer[6]. Genotyping was performed using an independent external contractor (Biowing Applied Biotechnology Co. Ltd., China). Genomic DNA was isolated from whole blood using the UltraPure™ Genomic DNA Isolation Kit (Shanghai SBS Genetech Technology Co., China). PCR-RFLP was performed to genotype exon 26 (C3435T) variant alleles in the MDR1 gene, with slight modifications. A PCR assay was using forward primer (5′-TGCTGGTCCTGAAGTTGATCTGTGAAC-3′) and reverse primer (5′-ACATTAGGCAGTGACTCGATGAAGGCA-3′). The PCR conditions consisted of a denaturation step at 95 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 54 °C to 59 °C for 50 s, and elongation at 72 °C for 1 min, followed by a final extension at 72 °C for 10 min. PCR products were digested with Sau3A I (C3435T) and analyzed by electrophoretic separation on agarose gels, followed by direct visualization over an ultraviolet transilluminator after ethidium bromide staining[7,8].
DNA amplification: One of the known primers of CYP3A5*3, MDR1-3435 and CYP3A4*18 was used for amplification of DNA for PCR analysis was biotinylated, respectively. Primers for pyrosequencing of CYP3A5*3, MDR1-3435 and CYP3A4*18 were designed with PyroMark Assay Design Software 2.0. In the PCR assay (PyroMark PCR Kit-Qiagen), we used a forward primer and a reverse primer that was biotinylated at the 5′- end of the CYP3A5*3, MDR1-3435 and CYP3A4*18 respectively. The assay was performed in a 25-μL reaction volume. The PCR conditions consisted of initial denaturation at 95 °C for 15 min, followed by 45 cycles of denaturation at 94 °C for 30 s, annealing at 60 °C for 30 s, extension at 72 °C for 30 s, and a final extension step at 72 °C for 10 min. The PCR products were separated on 2% agarose gels.
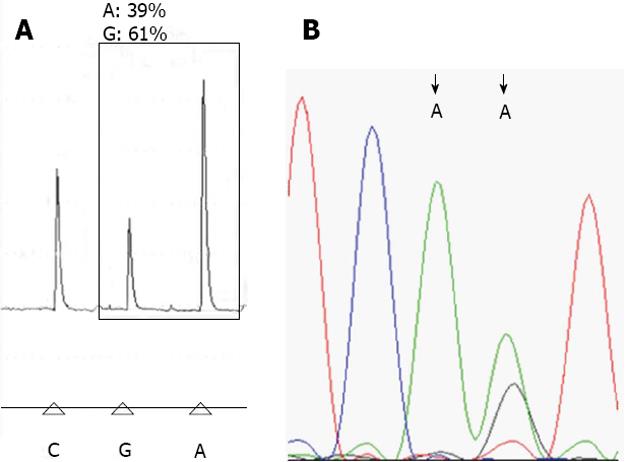
Pyrosequencing analysis: Biotinylated PCR products were immobilized on streptavidin-coated Sepharose beads (Streptavidin Sepharose High Performance, GE Healthcare). All of the streptavidin-coated Sepharose beads (2 μL per sample) were mixed with binding buffer (40 μL per sample) in a tube. High-purity water was then added to a total volume of 80 μL per well, including the PCR product (20 μL). This immobilization mix was incubated for 10 min at 25 °C with continuous mixing (1400 rpm) on a shaking device, and the sequencing primer was then diluted to 0.3 μmol/L in annealing buffer. Next, 25 μL of the solution was transferred to each well of a PyroMark Q24 Plate. After immobilization, the liquid was removed by aspirating the beads with a Vacuum Prep Tool and the beads were treated for approximately 5 s with 75% ethanol, 5 s with denaturation buffer, and 5 s with washing buffer. The PyroMark Q24 Plate containing the samples was heated at 80 °C for 2 min using a PyroMark Q24 Plate Holder and a heating block. The plate was then removed from the plate holder and the samples were allowed to cool to room temperature (15-25 °C) for at least 5 min, and the reagents, including enzyme and substrate mixtures, and nucleotides were added to the cartridge (PyroMark Q24, Qiagen)[4,9]. The samples were analyzed using a PyroMark Q24 system (Qiagen) according to standard protocols. The order of nucleotide dispensation was chosen based on suggestions provided by the PyroMark Assay Design Software 2.0 (Figure 1A).
This research was conducted in accordance with the Declaration of Helsinki (2000) of World Medical Association and institutional standards and was granted ethical approval by the institute review board from Chang Gung Memorial hospital (No: 100-2953C). Written informed consent for participation in the study was obtained from participants or from a parent or guardian in case of minor participants. All of the participants had been provided and obtained the written informed consent to participate in this study and the ethics committees had approved all of the consent procedure.
Statistical analyses were performed using SPSS software (version 12.0; SPSS, Chicago, IL, United States). Comparisons of parameters of the haplotypes of CYP3A5*3, MDR1-3435 and CYP3A4*18 between the donors and recipients were performed using the Student’s t-test with 2SD. P values less than 0.05 were considered statistically significant.
We performed 119 liver graft biopsies as part of clinical investigations after LDLT. For the SNP of CYP3A5*3, A allele was the wide type and G allele was mutant variant. The detail proportional change of A and G alleles of the CYP3A5*3 in the pyrosequencing between the recipients, donors and liver graft biopsy samples were showed on Table 1. The proportional change of A and G alleles of the CYP3A5*3 SNP in the pyrosequencing with RFLP was significant different between the liver graft biopsy sample and the peripheral blood mononuclear cells of the recipients in A > A/G (P < 0.0001), A/G > A (P < 0.001), A/G > G (P < 0.0001), G > A (P < 0.0001), G > A/G (P < 0.0001), but less difference to those of the donors (Table 1). For the SNP of MDR1-3435, C allele was the wide type and T was mutant variant. The detail proportional change of C and T alleles of the MDR1-3435 SNP in the pyrosequencing between the recipients, donors and liver graft biopsy samples were showed on Table 2. This phenomenon with proportional changes of C and T alleles in the MDR1-3435 was also significant different between the liver graft biopsy samples and the recipients in C > C/T (P < 0.0001), C/T > C (P < 0.0001), C/T > T (P < 0.001) and T > C/T (P < 0.0001), and less difference to those of the donors (Table 2). For the SNP of CYP3A4*18, T allele was the wide type and C allele was mutant variant. The detail proportional change of T and C alleles of the CYP3A5*3 SNP in the pyrosequencing between the recipients, donors and liver graft biopsy samples were showed on Table 3. Of the 41 pairs of recipient/donor, there were only 3 pairs (7.3%, 3/41) recipient/donor with different genotype and performed 15 liver graft biopsy samples for further analysis. There was also significant difference of the proportion changes of T and C alleles between the liver graft biopsy samples and the peripheral blood mononuclear cell of the recipient (P < 0.0001) but less difference to those of the peripheral blood mononuclear cell of the donor (P = 0.033) (Table 3).
| CYP3A5*3intron36986A > GA (wide type)Ssp I1 | PS | Recipient | Donor | Graft(liver biopsy) | P value | ||
| R vs G | D vs G | ||||||
| A > A/G, n = 10 | A | 86.23 ± 7.67 | 24.78 ± 3.90 | n = 35 | 35.57 ± 7.52 | < 0.0001 | >0.05 |
| G | 13.77 ± 7.67 | 75.23 ± 3.90 | 64.43 ± 7.52 | ||||
| A/G > A, n = 3 | A | 21.97 ± 1.32 | 73.13 ± 19.2 | n = 5 | 68.92 ± 11.75 | < 0.001 | >0.05 |
| G | 78.03 ± 1.32 | 26.87 ± 19.2 | 31.08 ± 11.75 | ||||
| A/G > G, n = 12 | A | 21.73 ± 5.81 | 6.44 ± 3.38 | n = 33 | 9.95 ± 5.68 | < 0.0001 | >0.05 |
| G | 78.27 ± 5.81 | 93.56 ± 3.38 | 90.05 ± 5.68 | ||||
| G > A, n = 2 | A | 3.06 ± 0.27 | 94.08 ± 5.87 | n = 10 | 47.29 ± 9.44 | < 0.0001 | < 0.0001 |
| G | 96.94 ± 0.27 | 5.92 ± 5.87 | 52.71 ± 9.44 | ||||
| G > A/G, n = 13 | A | 4.55 ± 1.88 | 22.97 ± 3.84 | n = 31 | 18.75 ± 6.43 | < 0.0001 | = 0.033 |
| G | 95.45 ± 1.88 | 77.03 ± 3.84 | 81.25 ± 6.43 | ||||
| A > G, n = 1 | A | 84.58 | 2.26 | n = 5 | 11.60 ± 5.26 | - | - |
| G | 15.42 | 97.74 | 88.39 ± 5.26 | ||||
| Total, n = 41 | n = 119 | ||||||
| MDR1-3435Exon263435C>TC (wide type)Sau3A I1 | PS | Recipient | Donor | Graft (liver biopsy) | P value | ||
| R vs G | D vs G | ||||||
| C > C/T, n = 14 | C | 93.10 ± 2.71 | 46.38 ± 1.94 | n = 40 | 56.97 ± 5.17 | < 0.0001 | < 0.0001 |
| T | 6.90 ± 2.71 | 53.62 ± 1.94 | 43.03 ± 5.17 | ||||
| C/T > C, n = 16 | C | 46.36 ± 1.75 | 92.94 ± 2.15 | n = 31 | 81.96 ± 5.42 | < 0.0001 | < 0.0001 |
| T | 53.64 ± 1.75 | 7.06 ± 2.15 | 18.04 ± 5.42 | ||||
| C/T > T, n = 3 | C | 46.35 ± 2.77 | 2.88 ± 1.49 | n = 5 | 12.38 ± 6.46 | < 0.001 | > 0.05 |
| T | 53.65 ± 2.77 | 97.12 ± 1.49 | 87.62 ± 6.46 | ||||
| T > C/T, n = 7 | C | 2.33 ± 0.48 | 47.52 ± 1.00 | n = 27 | 34.38 ± 6.21 | < 0.0001 | > 0.05 |
| T | 97.67 ± 0.48 | 52.47 ± 1.00 | 65.62 ± 6.21 | ||||
| T > C, n = 1 | C | 4.19 | 94.19 | n = 5 | 63.93 ± 5.25 | - | - |
| T | 95.81 | 5.81 | 36.07 ± 5.25 | ||||
| Total, n = 41 | n = 108 | ||||||
| CYP3A4*18Exon10878T>CT (wide type)Hpa II1 | PS | Recipient | Donor | Graft (liver biopsy) | P value | ||
| R vs G | D vs G | ||||||
| T/C > T, n = 2 | T | 58.59 ± 3.08 | 99.02 ± 1.39 | n = 12 | 91.86 ± 4.21 | < 0.0001 | 0.033 |
| C | 41.42 ± 3.08 | 0.99 ± 1.39 | 8.14 ± 4.21 | ||||
| T > T/C, n = 1 | T | 100 | 55.92 | n = 3 | 65.23 ± 3.31 | - | - |
| C | 0 | 44.08 | 34.77 ± 3.31 | ||||
| Total, n = 3 | n = 15 | ||||||
Anti-rejection agents such as tacrolimus usually target CYP3A5, MDR-1 and CYP3A4, which are the major metabolic isoenzymes of cytochrome P450. In the present study, all of the alleles A and G in CYP3A5*3 SNP, alleles C and T in MDR1-3435 SNP and alleles T and C in CYP3A4*18 SNP of the liver graft biopsy samples were expressed significant proportional change when compared with the recipients after LDLT. The most innovative finding suggested that the characteristic activity of the drug metabolism in the cytochrome P450 should be modified by the donor with individual characteristic. In our previous study showed that the CYP2C19 has 3 variant genotypes which have been classified as homozygous extensive metabolizers (HomEM), heterozygous extensive metabolizers (HetEM), and poor metabolizers in characteristic drug metabolism in cytochrome P450. But the donor graft does not affect CYP2C19 genotype expressed on the peripheral blood in recipient with LDLT[1]. Interestingly, the liver graft still presented the original CYP2C19 genotype characteristics of the donor when liver graft biopsy for a part of the clinical investigation at that moment. This clinical phenomenon could not be demonstrated used with Western blotting analysis[10] but it represented from the traditional sequencing so-called homogeneous phenomenon[3,4]. In the present study, pyrosequencing goes forward to identify the proportional change only requires a standard biotinylation of one of the PCR primers. It should be easier and more accurate to demonstration the SNP allele when compared with the traditional sequence (Figure 1). The bias of our study was the lack of microdisection of the hepatocyte[11-13] or isolation of the hepatocyte[14,15] from the graft, because we believed that the genetic characteristic of the graft hepatocyte is belongs to the donor originally. The peripheral blood of the recipient continuous goes through into the graft but it may not be contribute the genetic characteristic of the donor graft. During liver graft biopsy process, both of the graft hepatocyte and contaminated peripheral blood from the recipient would be mixed together. Hence, the liver graft biopsy sample is still presented with the original SNP genotype characteristic from the donor but the proportional change of the allele should be contributed with the recipient. It is so-called homogeneous phenomenon and expressed on the polymorphisms of CYP3A5*3, MDR1-3435 and CYP3A4*18. In the early stage within 1 mo after LDLT, this proportional change of the alleles was not stabilized. After a longitudinal followed up liver graft biopsy, the proportional change of the alleles was going to constant more than 1 mo after LDLT[4]. Followed up liver graft biopsy sampling more than 5 times per recipient was 24.4% (10/41) in CYP3A5*3, 17.1% (7/41) in MDR1-3435 and 66.7% (2/3) in CYP3A4*18. It was also suggested that the acute rejection is easily occurred within one month after liver transplantation[2,5] and the anti-rejection agent concentration in the serum was higher in one month later after LDLT[4].
As we know, the effect of MDR1-3435, CYP3A5*3, and CYP3A4*18 SNPs on cyclosporine A pharmacokinetics is very important[16-22]. In our study, the variant polymorphisms of CYP3A5*3 and MDR1-3435 had much differences between the recipients/donors which represented on the liver graft biopsy after LDLT. In contrast, the wide type and mutant variant was only 7.3% (3/41) differences in the polymorphisms of CYP3A4*18 needed liver graft biopsy after LDLT. This results suggested that the metabolic activity of the CYP3A4*18 was relative stabilized at the early stage after LDLT when compared with the CYP3A5*3 and MDRR1-3435, and corresponded that the CYP3A4*18B genotype affects cyclosporine A pharmacokinetics during the first month following surgery in Chinese renal transplant recipients. Patients with CYP3A4*18B alleles may require higher doses of cyclosporine A to reach the target levels[23-25].
In conclusion, pyrosequencing technique could be successful establishment of homogeneous phenomenon of the CYP3A5*3, MDR1-3435 and CYP3A4*18. The genetic polymorphisms characteristic of these isoenzymes of the recipient could be modified by the donor representing a biogenetic change when the peripheral blood drained into the new liver graft.
Homogenous phenomenon of graft liver CYP2C19 genotypes including homozygous extensive metabolizers, heterozygous extensive metabolizers, and poor metabolizers after living donor liver transplantation.
It is very useful and promising information about immunosuppression acted major role in bringing of organ transplantation in these days.
This clinical study is important to deeply clarify the biogenetic characteristic of the cytochrome P450 system when the recipients and donors with different genotype of the single nucleotide polymorphism.
Pyrosequencing technique could be successful establishment of homogeneous phenomenon of the CYP3A5*3, MDR1-3435 and CYP3A4*18.
Homogenous phenomenon: The graft liver from the donor mixed with the recipient circulating blood might be the result of present biogenetic phenomenon of genotype homogenous characteristics.
Through pyrosequencing technology, the genotype polymorphisms of CYP3A5*3 MDR1-3435 and CYP3A4*18 were analysised from the peripheral blood mononuclear cell of 41 pairs recipient/donor with different genotype polymorphisms and 119 liver graft biopsy samples. The genetic polymorphisms characteristic of the CYP3A5*3, MDR1-3435 and CYP3A4*18 of the recipient could be modified by the donor representing a biogenetic change so-called homogenous phenomenon when the peripheral blood drained into the new liver graft. This study innovatively found that the liver graft could have the evidence to handle the cytochrome P450 drug metabolizing system in the recipient.
| 1. | Chiu KW, Tai WC, Nakano T, Tseng HP, Cheng YF, Jawan B, Goto S, Chen CL. Donor graft does not affect the P450 2C19 genotype expressed in peripheral blood in recipients of living donor liver transplantation. Clin Transplant. 2010;24:830-834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 2. | Chiu KW, Nakano T, Hu TH, Tseng HP, Cheng YF, Jawan B, Eng HL, Goto S, Chen CL. Influence of CYP2C19 genotypes on graft pathological findings and postoperative liver function in recipients after living-donor liver transplantation. Ann Transplant. 2010;15:38-43. [PubMed] |
| 3. | Chiu KW, Nakano T, Hu TH, Tseng HP, Cheng YF, Jawan B, Eng HL, Goto S, Chen CL. Homogenous phenomenon of graft liver CYP2C19 genotypes after living donor liver transplantation. Eur J Clin Invest. 2012;42:352-356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 4. | Chiu KW, Nakano T, Chen KD, Lai CY, Hsu LW, Chiu HC, Huang CY, Cheng YF, Goto S, Chen CL. Pyrosequencing to identify homogeneous phenomenon when using recipients/donors with different CYP3A5*3 genotypes in living donor liver transplantation. PLoS One. 2013;8:e71314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 5. | Chiu KW, Chen YS, de Villa VH, Wang CC, Eng HL, Wang SH, Liu PP, Jawan B, Huang TL, Cheng YF. Characterization of liver enzymes on living related liver transplantation patients with acute rejection. Hepatogastroenterology. 2005;52:1825-1827. [PubMed] |
| 6. | Fukushima-Uesaka H, Saito Y, Watanabe H, Shiseki K, Saeki M, Nakamura T, Kurose K, Sai K, Komamura K, Ueno K. Haplotypes of CYP3A4 and their close linkage with CYP3A5 haplotypes in a Japanese population. Hum Mutat. 2004;23:100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 129] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 7. | Hu YF, Tu JH, Tan ZR, Liu ZQ, Zhou G, He J, Wang D, Zhou HH. Association of CYP3A4*18B polymorphisms with the pharmacokinetics of cyclosporine in healthy subjects. Xenobiotica. 2007;37:315-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 51] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 8. | Chiu KW, Hu TH, Nakano T, Chen KD, Lai CY, Hsu LW, Tseng HP, Chiu HC, Cheng YF, Goto S. Biological interactions of CYP2C19 genotypes with CYP3A4*18, CYP3A5*3, and MDR1-3435 in living donor liver transplantation recipients. Transplant Res. 2013;2:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 9. | Eriksson S, Berg LM, Wadelius M, Alderborn A. Cytochrome p450 genotyping by multiplexed real-time dna sequencing with pyrosequencing technology. Assay Drug Dev Technol. 2002;1:49-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 42] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 10. | Chiu KW, Nakano T, Tseng HP, Cheng YF, Jawan B, Eng HL, Goto S, Chen CL. Western blotting analysis for quantitative detection of CYP2C19 expression in liver tissues in the setting of living donor liver transplantation. Hepatogastroenterology. 2012;59:805-808. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 11. | Tretiakova M, Hart J. Laser microdissection for gene expression study of hepatocellular carcinomas arising in cirrhotic and non-cirrhotic livers. Methods Mol Biol. 2011;755:233-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 12. | Mustafa A, Cenayko C, Mitry RR, Quaglia A. Laser microdissection microscopy: application to cell culture. Methods Mol Biol. 2012;806:385-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 13. | Munshaw S, Hwang HS, Torbenson M, Quinn J, Hansen KD, Astemborski J, Mehta SH, Ray SC, Thomas DL, Balagopal A. Laser captured hepatocytes show association of butyrylcholinesterase gene loss and fibrosis progression in hepatitis C-infected drug users. Hepatology. 2012;56:544-554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 14. | Donato MT, Lahoz A, Jiménez N, Pérez G, Serralta A, Mir J, Castell JV, Gómez-Lechón MJ. Potential impact of steatosis on cytochrome P450 enzymes of human hepatocytes isolated from fatty liver grafts. Drug Metab Dispos. 2006;34:1556-1562. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 111] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 15. | Brückner S, Tautenhahn HM, Winkler S, Stock P, Jonas S, Dollinger M, Christ B. Isolation and hepatocyte differentiation of mesenchymal stem cells from porcine bone marrow--”surgical waste” as a novel MSC source. Transplant Proc. 2013;45:2056-2058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 16. | Kolars JC, Schmiedlin-Ren P, Schuetz JD, Fang C, Watkins PB. Identification of rifampin-inducible P450IIIA4 (CYP3A4) in human small bowel enterocytes. J Clin Invest. 1992;90:1871-1878. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 361] [Cited by in RCA: 354] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 17. | Hughes SJ, Morse MA, Weghorst CM, Kim H, Watkins PB, Guengerich FP, Orringer MB, Beer DG. Cytochromes P450 are expressed in proliferating cells in Barrett’s metaplasia. Neoplasia. 1999;1:145-153. [PubMed] |
| 18. | Yamazaki H, Shibata A, Suzuki M, Nakajima M, Shimada N, Guengerich FP, Yokoi T. Oxidation of troglitazone to a quinone-type metabolite catalyzed by cytochrome P-450 2C8 and P-450 3A4 in human liver microsomes. Drug Metab Dispos. 1999;27:1260-1266. [PubMed] |
| 19. | Hoffmeyer S, Burk O, von Richter O, Arnold HP, Brockmöller J, Johne A, Cascorbi I, Gerloff T, Roots I, Eichelbaum M. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci USA. 2000;97:3473-3478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 557] [Cited by in RCA: 894] [Article Influence: 34.4] [Reference Citation Analysis (0)] |
| 20. | Cascorbi I, Gerloff T, Johne A, Meisel C, Hoffmeyer S, Schwab M, Schaeffeler E, Eichelbaum M, Brinkmann U, Roots I. Frequency of single nucleotide polymorphisms in the P-glycoprotein drug transporter MDR1 gene in white subjects. Clin Pharmacol Ther. 2001;69:169-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 522] [Cited by in RCA: 532] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 21. | Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, Watkins PB, Daly A, Wrighton SA, Hall SD. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27:383-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1583] [Cited by in RCA: 1619] [Article Influence: 64.8] [Reference Citation Analysis (0)] |
| 22. | Gawrońska-Szklarz B, Siuda A, Kurzawski M, Bielicki D, Marlicz W, Droździk M. Effects of CYP2C19, MDR1, and interleukin 1-B gene variants on the eradication rate of Helicobacter pylori infection by triple therapy with pantoprazole, amoxicillin, and metronidazole. Eur J Clin Pharmacol. 2010;66:681-687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 23. | Qiu XY, Jiao Z, Zhang M, Zhong LJ, Liang HQ, Ma CL, Zhang L, Zhong MK. Association of MDR1, CYP3A4*18B, and CYP3A5*3 polymorphisms with cyclosporine pharmacokinetics in Chinese renal transplant recipients. Eur J Clin Pharmacol. 2008;64:1069-1084. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 24. | Crettol S, Venetz JP, Fontana M, Aubert JD, Pascual M, Eap CB. CYP3A7, CYP3A5, CYP3A4, and ABCB1 genetic polymorphisms, cyclosporine concentration, and dose requirement in transplant recipients. Ther Drug Monit. 2008;30:689-699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 68] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 25. | Li DY, Teng RC, Zhu HJ, Fang Y. CYP3A4/5 polymorphisms affect the blood level of cyclosporine and tacrolimus in Chinese renal transplant recipients. Int J Clin Pharmacol Ther. 2013;51:466-474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
P- Reviewers: Aydin U, Li JD, Liu B, Qin JM S- Editor: Gou SX L- Editor: A E- Editor: Liu XM