Published online Apr 27, 2025. doi: 10.4254/wjh.v17.i4.105660
Revised: March 20, 2025
Accepted: April 1, 2025
Published online: April 27, 2025
Processing time: 81 Days and 1.7 Hours
Liver cirrhosis (LC) affect millions of people worldwide. The pathogenesis of cirrhosis involves complex interactions between immune responses and gut microbiota. Recent studies have highlighted the role of the interleukin-36 (IL-36) subfamily in inflammation and immune regulation. However, the relationship between serum IL-36 subfamily levels and gut microbiota in cirrhosis patients remains unclear. This study aimed to explore the clinical significance of serum IL-36 subfamily levels and their association with gut microbiota in cirrhosis patients.
To explore the clinical significance of serum IL-36 subfamily levels and their relationship with gut microbiota among cirrhosis patients.
Sixty-one cirrhosis patients were enrolled from Lihuili Hospital of Ningbo University from May 2022 to November 2023 as the LC group and 29 healthy volunteers as the healthy control (HC) group. The serum expressions of IL-36α, IL-36β, IL-36γ, IL-36Ra, and IL-38 were measured through ELISA, while 16S rRNA gene sequencing was employed to rate microbial community in human fecal samples.
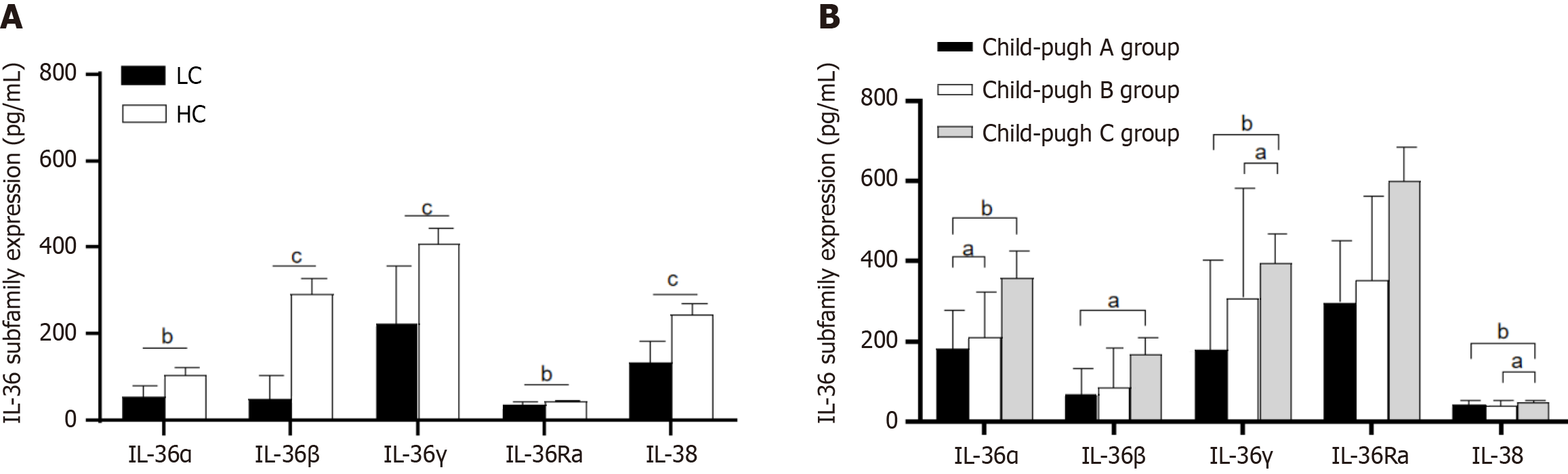
The serum levels of IL-36α, IL-36γ, IL-36Ra, and IL-38 in the LC group remarkably exceeded those in the HC group (P < 0.05). IL-36α, IL-36γ, and IL-38 were related positively to the Child-Pugh score (P < 0.05) and prominently exceeded those in the Child-Pugh C group (P < 0.05). The absolute abundance of harmful bacteria (Bacteroides, Bifidobacterium, Faecalibacterium) remarkably rose, while the beneficial bacteria (Firmicutes, Bacteroides, Escherichia-Shigella) notably decreased in the LC group (P < 0.05). IL-36α, IL-36γ, and IL-38 related positively to Lactobacillus
IL-36γ and IL-38 show promise as potential biomarkers for LC progression, but further validation is required.
Core Tip: This study investigated the clinical relevance of serum interleukin-36 (IL-36) subfamily levels and their correlation with gut microbiota in 61 patients with liver cirrhosis (LC) and 29 healthy controls. We found that IL-36α, IL-36γ, IL-36Ra, and IL-38 Levels were significantly higher in cirrhosis patients and strongly correlated with disease progression. These cytokines may serve as novel predictive markers for LC. Our findings highlight the potential of IL-36 subfamily members as diagnostic biomarkers, contributing valuable insights to the field.
- Citation: Pan YZ, Chen WT, Jin HR, Liu Z, Gu YY, Wang XR, Wang J, Lin JJ, Zhou Y, Xu LM. Correlation between the interleukin-36 subfamily and gut microbiota in patients with liver cirrhosis: Implications for gut-liver axis imbalance. World J Hepatol 2025; 17(4): 105660
- URL: https://www.wjgnet.com/1948-5182/full/v17/i4/105660.htm
- DOI: https://dx.doi.org/10.4254/wjh.v17.i4.105660
Liver diseases affect millions of people worldwide, with conditions, like hepatitis B and C[1], nonalcoholic fatty[2], alcoholic[2], and autoimmune liver diseases[3] potentially leading to the development of cirrhosis. Globally, over 1.06 million people live with liver cirrhosis (LC)[4], which accounted for > 1.32 million deaths[5], often due to complications such as gastrointestinal bleeding, sepsis, hepatic encephalopathy (HE), and spontaneous bacterial peritonitis and so on[6].
Studies indicate that patients with LC have severe intestinal microorganism dysbiosis, tightly connected to the disease progression[7-9]. Damage of gut tight junction proteins increases intestinal mucosal permeability, allowing endotoxins from the intestine to enter the liver through the portal vein system[10]. Gut microbiota dysbiosis increases lipopolysaccharide levels, which activates the NF-κB and TLR4 signaling pathways, aggravating inflammatory cytokine release[11-13]. Excessive pro-inflammatory cytokines can exacerbate liver damage and fibrosis, while a relative increase in anti-inflammatory signals may relieve inflammation and potentially slow the progression to end-stage liver disease[14-16]. Therefore, the equilibrium between anti-inflammatory and pro-inflammatory mediators is critical.
Research indicates that the interleukin-36 (IL-36) subfamily is beneficial to the etiology of diverse inflammatory disorders. As a new-found member of the IL-1 family, the IL-36 subfamily contains three receptor agonists (IL-36α, IL-36β, and IL-36γ) as well as two receptor antagonists (IL-36Ra and IL-38)[17]. Three receptor agonists attach to IL-36 receptors and activate NF-κB, promoting inflammatory cytokines secretion. IL-36Ra inhibits the activation of these pathways[18,19]. Additionally, IL-38 and IL-36Ra share 43% sequence homology. Indeed, IL-38 exhibits similar anti-inflammatory efficacy among bodies[20]. IL-36 family cytokines have been affected various inflammations, like psoriatic arthritis, lupus erythematosus, and inflammatory bowel disease (IBD)[21-24]. In recent years, studies have gradually revealed the complex relationship between the IL-36 subfamily and the intestinal microbiota[25]. For example, IL-36γ is upregulated in the biopsy samples of the colonic mucosa of patients with IBD, especially ulcerative colitis (UC). It can induce colonic epithelial cells and fibroblasts to produce pro-inflammatory cytokines, thereby exacerbating intestinal inflammation[21,26]. However, the expression and significance of the IL-36 subfamily among cirrhosis patients are still unknown.
Our research investigated the relation IL-36 subfamily and intestinal microbiota dysbiosis while illustrating the function and importance of IL-36 subfamily in LC progression.
Sixty-one cirrhosis patients were enrolled from Lihuili Hospital of Ningbo University as the LC group between May 2022 and November 2023, and 29 healthy volunteers served as the healthy control (HC) group. The inclusion criteria were listed below: (1) Age from 18 to 65 years old; (2) Cirrhosis of various etiologies reaching the diagnostic standards suggested by the "Chinese Guidelines for the Management of LC," which include histological, endoscopic, or radiological evidence[27]; and (3) Understanding and signing an informed consent. The exclusion criteria were listed below: (1) Diagnosed or suspected cases of malignant tumors; (2) Use of antibiotics, hormones, immunosuppressants, or probiotics within 2 weeks before enrollment; (3) Coexistence of autoimmune diseases, like rheumatoid arthritis, psoriasis, etc.; (4) Other health conditions, like infection, fever, diabetes mellitus, coronary heart disease, and chronic obstructive pulmonary disease; (5) Pregnant or lactating women; and (6) Other conditions deemed unsuitable for participation by the researchers. Blood and fecal samples were gathered from the research participants. This study followed the principles of the Helsinki Declaration and receive approvals from the ethics committee of the hospital, with ethics approval number KY2022PJ064.
Sex, age, etiology of LC, and findings from abdominal ultrasound, magnetic resonance imaging, etc., were recorded. The collected clinical data contained alanine aminotransferase, albumin (Alb), alkaline phosphatase (ALP), aspartate aminotransferase (AST), creatinine, platelet (PLT), hemoglobin (HB), gamma-glutamyltransferase (γ-GGT), total bilirubin (TBIL), white blood cell (WBC), direct bilirubin, and indirect bilirubin levels.
Frozen serum samples were preserved at -80 °C. ELISA kits (Wuhan Huamei Biotech Co. Ltd, China) were utilized to gauge the expressions of three receptor agonists and two receptor antagonists following the manufacturer’s specifications.
Fecal samples were gathered and preserved at -80°C. Absolute quantification sequencing of the gut microbiota 16S rRNA gene was conducted by Shanghai Tianhao Biotechnology Co. Ltd. The primer used was as follows: Target region 16S V3V4 Primer F = Illumina adapter sequence 1 + CCTACGGGNGGCWGCAG, Primer r = Illumina adapter sequence 2 + GACTACHVGGGTATCTAATCC.
Following the library quality was evaluated, a paired-end sequencing strategy of Illumina 2 × 250 bp was used to sequence the libraries on the Illumina NovaSeq 6000 platform. Subsequently, Usearch (v10) software was used for data trimming and clustering analysis, sorting reads by abundance from highest to lowest, followed by clustering at 97% similarity, with the intention of attaining operational taxonomic units (OTUs). Each OTU was taxonomically classified, and the abundance of each species was annotated at various taxonomic levels to generate a species abundance table for in-depth analysis.
Statistical analyses were implemented through SPSS Statistics 21.0 (v 21.0). Continuous variables are denoted as mean ± SD, while categorical variables are denoted as medians (interquartile ranges). For continuous variables that conformed to a normal distribution, independent samples t-tests were used, while Fisher's exact probability assays or non-parametric assays were employed for continuous variables that did not conform to a Gaussian distribution and for categorical variables. For pairwise relationship analysis, Pearson's test was applied to Gaussian-distributed data, with Spearman's correlation analysis used for non-Gaussian-distributed data, with the correlation coefficient represented by the r value. For comparisons among multiple groups, one-way analysis of variance (ANOVA) was applied to Gaussian-distributed data, while the Kruskal-Wallis assay was utilized for data without conforming to a normal distribution. As to contrasts between two groups, the Wilcoxon rank-sum assay was employed. A P value threshold of 0.05 was adopted to measure significance.
The LC group comprised 51 males and 10 females, showing an average age of 53.89 ± 9.38 years, while the HC group had 23 males and 6 females, showing a mean age of 49.21 ± 8.49 years. No remarkable disparities emerged in age (P = 0.278) or sex (P = 0.623) between both groups. In the LC group, patients were classified according to etiology: 29 cases (47.54%) were attributed to hepatitis B-related cirrhosis, 20 (32.79%) to alcohol-related cirrhosis, and 12 (19.67%) to other causes of cirrhosis. Furthermore, based on Child-Pugh scores, patients with cirrhosis were stratified into Grade A (n = 20), Grade B (n = 24), and Grade C (n = 17) categories.
The LC group had considerably higher levels of ALP, AST, and TBIL and lower levels of WBC, HB, PLT, and Alb than the HC group (P < 0.05; Table 1).
| Variables | HC (n = 29) | LC (n = 61) | χ²/t | P value |
| Sex (male) | 23.00 (79.30%) | 50.00 (82.00%) | 0.248 | 0.763 |
| Age (years) | 51.65 ± 8.34 | 53.89 ± 9.38 | -1.091 | 0.278 |
| WBC (109/L) | 5.58 ± 1.21 | 4.60 ± 2.48 | 2.024 | 0.046 |
| HB (g/L) | 146.22 ± 35.68 | 110.82 ± 29.99 | 4.918 | < 0.001 |
| PLT (109/L) | 230.10 ± 54.88 | 109.62 ± 73.07 | 7.877 | < 0.001 |
| Alb (g/L) | 47.09 ± 4.74 | 32.87 ± 6.92 | 9.990 | < 0.001 |
| ALT (U/L) | 20.59 ± 6.92 | 43.64 ± 6.92 | -1.803 | 0.075 |
| AST (U/L) | 23.00 ± 9.39 | 58.62 ± 60.48 | -3.145 | 0.002 |
| ALP (U/L) | 70.28 ± 15.33 | 117.16 ± 52.84 | -4.673 | < 0.001 |
| γ-GGT (U/L) | 23.17 ± 12.72 | 123.61 ± 333.17 | -1.618 | 0.109 |
| TBIL (umol/L) | 10.81 ± 3.52 | 65.82 ± 93.82 | -3.147 | 0.002 |
| Cr (umol/L) | 71.25 ± 8.79 | 71.93 ± 28.85 | -0.149 | 0.882 |
Figure 1 exhibits that the serum levels of IL-36α, IL-36γ, IL-36Ra, and IL-38 in the LC group remarkably exceeded those in the HC group (P < 0.05). However, remarkably disparity emerged in serum IL-36β levels between both groups (P > 0.05).
The serum IL-36 subfamily expressions were compared among cirrhosis patients based on etiology, through which there were no statistically remarkable disparities occurred in serum IL-36α, IL-36β, IL-36γ, IL-36Ra, and IL-38 expression levels among patients with cirrhosis caused by hepatitis B, alcohol and other etiologies (P > 0.05).
Figure 2 presents that the serum expressions of IL-36α were prominently elevated in the B group in contrast with the Child-Pugh A group (P = 0.037). Additionally, the serum expressions of IL-36α (P = 0.007), IL-36β (P = 0.01), IL-36γ (P = 0.001), and IL-38 (P = 0.002) were prominently up-regulated in the C group relative to the A group. The serum expression levels of IL-36γ (P = 0.015) and IL-38 (P = 0.02) were excessively high relative to the C group to the B group.
IL-36α significantly related positively to PLT, Alb, AST, ALP, γ-GGT, TBil, and Child-Pugh scores (P < 0.05) (Table 2). IL-36γ was related positively to HB, PLT, Alb, AST, ALP, TBil, and Child-Pugh scores (P < 0.05). IL-36Ra was related positively to HB, PLT, and TBil (P < 0.05). IL-38 was related positively to Alb, AST, ALP, TBil, and Child-Pugh scores (P < 0.05).
| Variables | IL-36α | IL-36γ | IL-36Ra | IL-38 | ||||
| R value | P value | R value | P value | R value | P value | R value | P value | |
| WBC (109/L) | 0.005 | 0.963 | 0.047 | 0.661 | -0.111 | 0.298 | 0.048 | 0.655 |
| HB (g/L) | -0.296 | 0.005 | -0.245 | 0.020 | -0.240 | 0.023 | -0.186 | 0.079 |
| PLT (109/L) | -0.345 | 0.001 | -0.275 | 0.009 | -0.255 | 0.015 | -0.153 | 0.151 |
| Alb (g/L) | 0.525 | < 0.001 | -0.437 | < 0.001 | -0.185 | 0.082 | -0.346 | 0.001 |
| ALT (U/L) | 0.180 | 0.090 | -0.187 | 0.077 | -0.020 | 0.852 | 0.165 | 0.119 |
| AST (U/L) | 0.395 | < 0.001 | 0.584 | < 0.001 | 0.172 | 0.104 | 0.495 | < 0.001 |
| ALP (U/L) | 0.482 | < 0.001 | 0.354 | < 0.001 | 0.136 | 0.202 | 0.406 | < 0.001 |
| γ-GGT (U/L) | 0.437 | < 0.001 | -0.023 | 0.828 | 0.036 | 0.736 | -0.043 | 0.689 |
| TBIL (umol/L) | 0.471 | < 0.001 | 0.559 | < 0.001 | 0.228 | 0.031 | 0.540 | < 0.001 |
| Cr (umol/L) | -0.080 | 0.451 | -0.024 | 0.823 | -0.187 | 0.078 | 0.003 | 0.981 |
| Child-pug score | 0.418 | 0.001 | 0.387 | 0.002 | 0.203 | 0.117 | 0.380 | 0.003 |
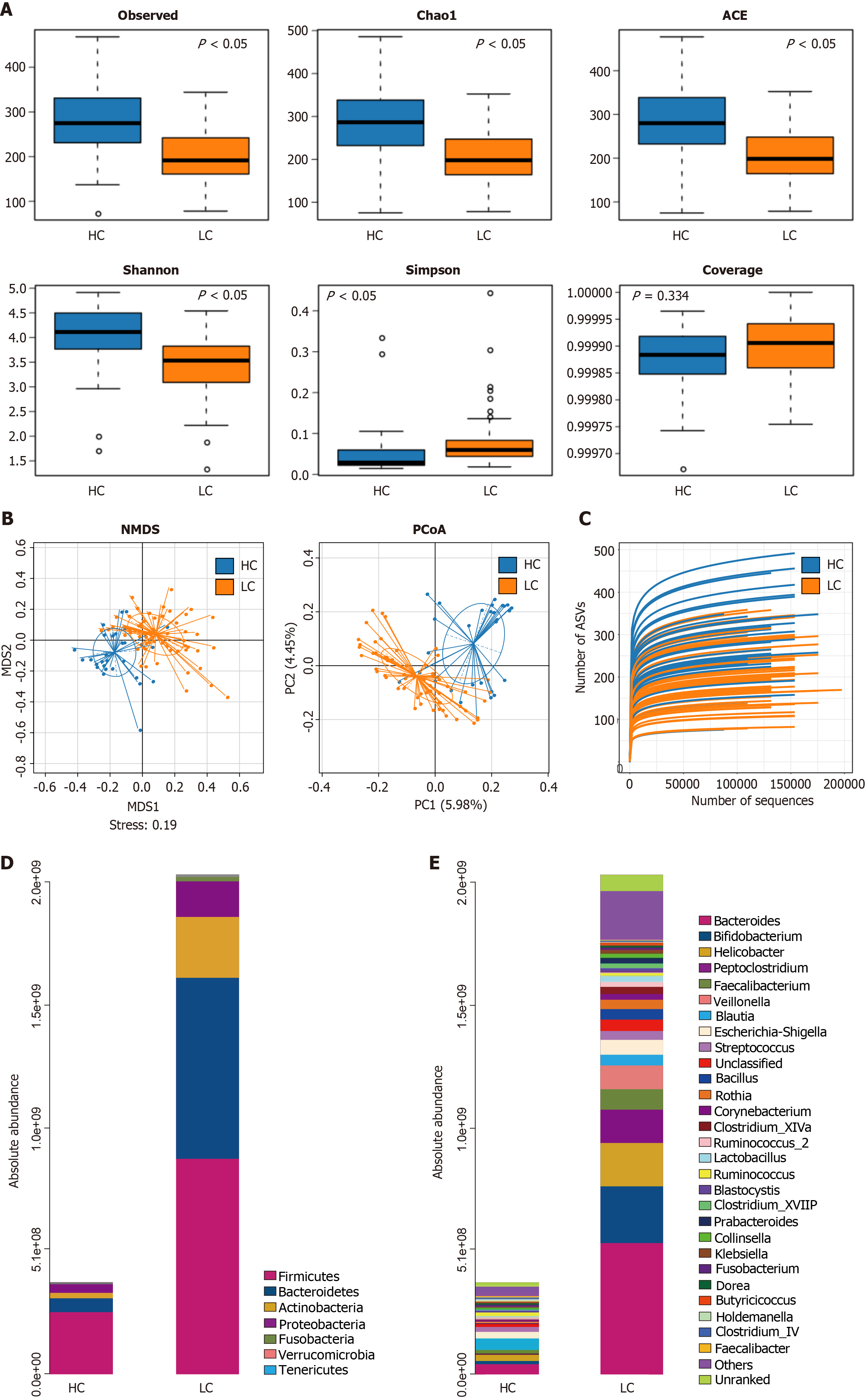
Figure 2A shows that the alpha diversity in intestinal microorganisms of cirrhosis patients exhibits a decrease in Observe, Chado1, ACE compared with the HC group (P < 0.05), and the Simpson index rose in the LC group (P < 0.05). Additionally, the gut microbiota coverage in the LC and HC groups approached 1, with no remarkable disparity between both groups (P > 0.05), indicating adequate sequencing depth in both groups. As shown in Figure 2B, principal coordinates analysis (PCoA) and non-metric multidimensional scaling analysis exhibited remarkable disparities in beta multiformity of gut microbes between both groups (P < 0.05). These results suggest a marked difference in beta diversity of gut microbiota between both groups.
As shown in Figure 2C, the rarefaction curves were observed for evaluating the sequencing depth of 90 samples. The results indicated that the rarefaction curves tended to flatten, suggesting that the sampling in this study was sufficient and the sequencing depth adequately covered most of the species present in the samples, thereby reflecting the majority of gut microbiota information.
The gut microbiota is primarily composed of the Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria. As shown in Figure 2D and E, at the level of phylum, the absolute abundance of Bacteroidetes in the LC group markedly surpassed that in the HC group (P < 0.05), and no statistically substantial difference emerged between both groups for Firmicutes, Actinobacteria, and Proteobacteria (P > 0.05). At the level of genus, the absolute abundance of Bacteroides Veillonella and Fusobacterium in the LC group prominently surpassed that in the HC group. In contrast, the absolute abundance of Lactobacillus, Parabacteroides, Bifidobacterium, Collinsella, and Gemmiger in the LC group remarkably came short of that in the HC group (P < 0.05). No statistically marked disparities emerged between both groups for the genera Blautia, Lachnospira, Faecalibacterium, Escherichia-Shigella, Prevotella, Streptococcus, Rothia, Allobaculum, Akkermansia, Ruminococcus_2, Clostridium_XVIII, Klebsiella, Dorea, Butyricicoccus, Holdemanella, and Clostridium_IV (P > 0.05).
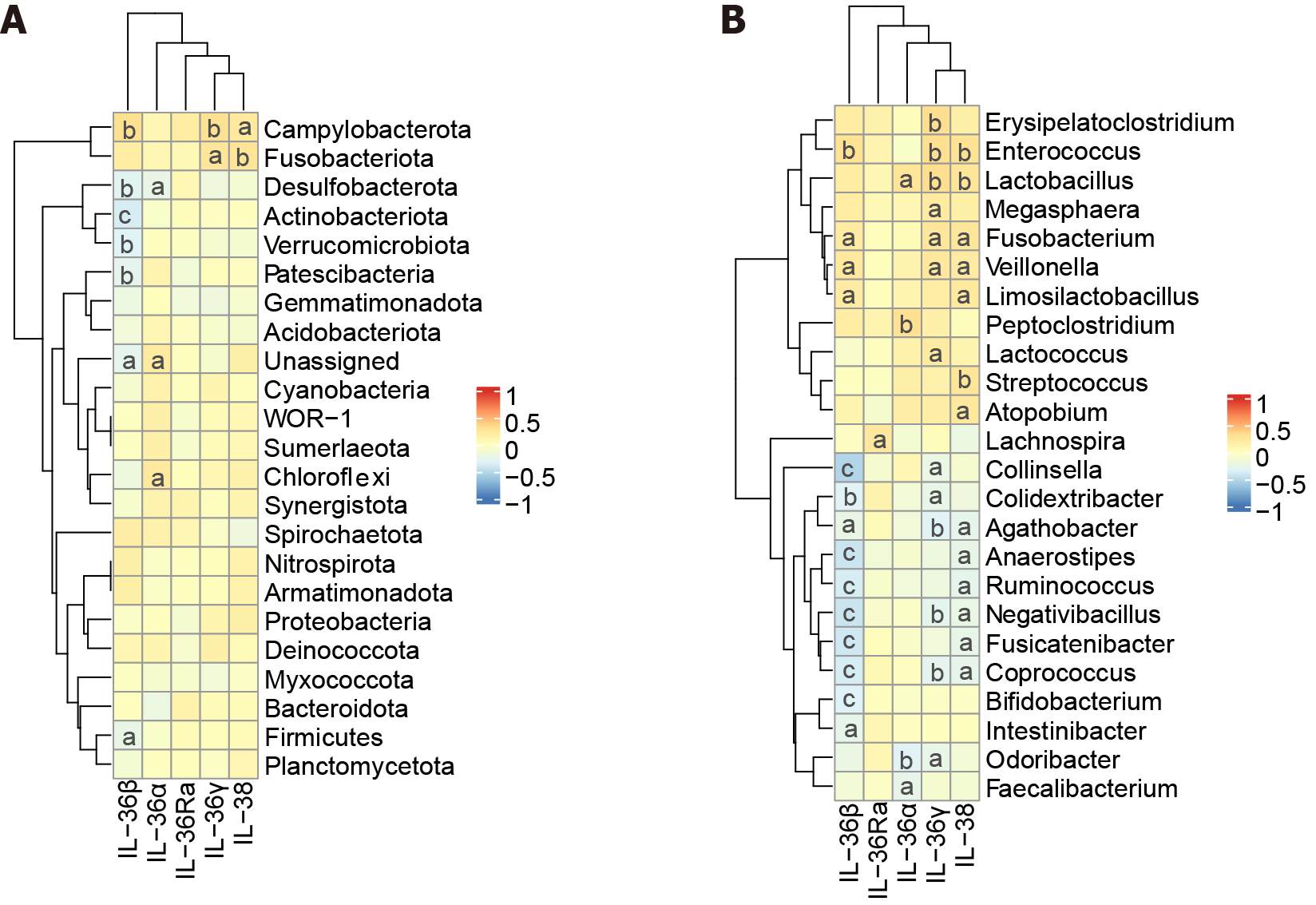
As shown in Figure 3A, at the phylum level, IL-36γ positively related to Fusobacteriota and Campylobacterota (P < 0.05). IL-36β negatively related to Desulfobacterota, Actinobacteriota, Verrucomicrobiota and Firmicutes and positively correlated with Campylobacterota (P < 0.05).
Figure 3B exhibits that, at the level of genus, IL-36Ra positively correlated with Lachnospira (P < 0.05). IL-36γ related positively to Fusobacterium, Lactobacillus, Veillonella, Enterococcus, and Erysipelatoclostridium (P < 0.05) and related negatively to Coprococcus, Collinsella, Negativibacillus (P < 0.05). IL-38 related positively to Fusobacterium, Veillonella, Streptococcus, Atopobium, Lactobacillus, and Enterococcus (P < 0.05) and related negatively to Anaerostipes, and Fusicatenibacter (P < 0.05). IL-36α related positively to Peptoclostridium and Lactobacillus (P < 0.05) and related negatively to Faecalibacterium (P < 0.05). IL-36β related positively to Enterococcus, Veillonella, and Fusobacterium (P < 0.05) and related negatively to Bifidobacterium and Fusicatenibacter (P < 0.05).
LC is a severity global health challenge[1]. Recent research has found the important function of intestinal microbes in the development of LC. Research has shown that as high as 50%-70% of cirrhosis patients suffer from HE, which tightly correlates with intestinal microbe imbalance[28,29]. In this study, LC patients possessed much lower alpha diversity indicators (Chao1, observed OTUs, and PD Whole-tree; P < 0.05). The beta diversity of intestinal microbes among LC patients was prominently different from healthy controls (PCoA analysis, P < 0.05), indicating that patients with LC have gut microbiota imbalance. We found that the absolute abundance of harmful bacteria such as Veillonella, Bacteroides and Fusobacterium in the LC group were prominently more elevated than in the HC group (P < 0.05). The absolute abundance of beneficial bacteria, like Lactobacillus and Bifidobacterium in the LC group markedly came short of that in the HC group (P < 0.05). Previous researches have indicated that intestinal microorganism imbalance is capable of exacerbating hepatic inflammation and activating inflammatory pathways, which tightly correlates with the emergence of complications regarding LC[28,30,31]. Hence, investigations on the variations in inflammatory factors among cirrhosis patients contributes to the precision assessment and prediction of the progression of the disease.
Recent research has indicated the function of IL-36 subfamily in both pro-inflammatory and anti-inflammatory reactions across a variety of diseases[32]. For instance, it has been demonstrated that IL-36α is an essential player in the progression of hepatocellular carcinoma (HCC), suggesting that it might become a prospective therapeutic target and a predictive biomarker to treat HCC[33]. In addition, IL-38 Lessens the level of inflammatory factors while impeding inflammations among collagen-induced arthritis mice[34]. However, there have been no reports on the relationship between IL-36 subfamily and LC in literature to date. Our study manifested the serum expressions of pro-inflammatory IL-36α and IL-36γ, along with anti-inflammatory IL-36Ra and IL-38 in the LC group surpassed those in the HC group (P < 0.05). This suggests that in LC, the pro-inflammatory and anti-inflammatory cytokines of IL-36 are compensatorily increased. While pro-inflammatory signaling pathways are activated, the body simultaneously initiates protective mechanisms, increasing anti-inflammatory cytokines. Additionally, we found the expressions of IL-36α, IL-36γ, and IL-38 tightly related positively to the Child-Pugh score (P < 0.05), suggesting their close relationship to cirrhosis progression.
It is still unclear whether IL-36 subfamily engages in the gut-liver axis imbalance in LC. Research has illustrated that IL-36 subfamily participates in adjusting gut microbiota. In conditions with gut microbiota dysbiosis, such as IBD, the level of IL-36 cytokines is altered[35]. Studies have shown that the expression of IL-36γ is regulated by the gut microbiota. For example, after dextran sulfate sodium (DSS) induction, the expression of IL-36γ in the colon tissue of germ-free mice was significantly lower than that in wild - type mice, indicating that the gut microbiota may promote the production of IL-36γ through some mechanisms[26]. In addition, Bacteroides can inhibit the colonization of Klebsiella pneumoniae through the IL-36 signaling pathway, thus maintaining the balance of the intestinal microecology. In IL-36R knockout (IL-36R-/-) mice, the gut microbiota was significantly altered after DSS induction, and the expression of the antibacterial peptide Lcn2 was significantly decreased[36]. These results suggest that IL-36γ not only participates in the intestinal inflammatory response but also affects the balance of the intestinal microecology by regulating the composition of the gut microbiota. As an antagonist of the IL-36 receptor, IL-38 has anti-inflammatory effects and can inhibit the IL-36-mediated inflammatory response. Studies have shown that the expression level of IL-38 is increased in patients with IBD, and it may alleviate disease symptoms by inhibiting the intestinal inflammatory response[37]. In addition, IL-38 may exert its anti-inflammatory effect by regulating the composition of the gut microbiota.
Our study found that pro-inflammatory factor IL-36γ and anti-inflammatory factor IL-38 were the most meaningful indicators, which positively correlated with harmful bacteria Veillonella and Fusobacterium (P < 0.05). It is found that pro-inflammatory factor IL-36γ is elevated in IBD and promotes wound healing and intestinal inflammations, contributing to gut inflammatory disorders, like UC and IBD[35]. Actually, IL-38 is a vital player in IBD, it is elevated among IBD patients and it suppresses intestinal inflammation. IL-38 is an anti-inflammatory cytokine that antagonizes the IL-36R, thereby reducing inflammatory responses[25]. These discoveries illuminate that IL-36γ and IL-38 might be novel warning indicators for LC. IL-36 subfamily are the key players in regulating inflammatory pathways and maintaining intestinal microbe balance.
In summary, IL-36γ and IL-38 are linked to gut microbiota imbalances in patients with cirrhosis. Targeted regulation of gut microbiota and inhibition of IL-36 subfamily inflammatory cytokines may provide a theoretical basis for microbiota-based therapies to slow LC progression and improve patient outcomes. Our research still has several restrictions. Initially, we should increase the sample size. Second, emphasizing the demand for further animal experiments to elucidate the regulatory mechanisms linking the IL-36 subfamily with liver damage and gut microbiota dysbiosis in LC.
We would like to express our sincere gratitude to all the contributors involved in this study. Special thanks go to the patients and their families who participated in this research, as their contributions were essential to our findings.
| 1. | Wu XN, Xue F, Zhang N, Zhang W, Hou JJ, Lv Y, Xiang JX, Zhang XF. Global burden of liver cirrhosis and other chronic liver diseases caused by specific etiologies from 1990 to 2019. BMC Public Health. 2024;24:363. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 68] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 2. | Tapper EB, Parikh ND. Diagnosis and Management of Cirrhosis and Its Complications: A Review. JAMA. 2023;329:1589-1602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 223] [Article Influence: 74.3] [Reference Citation Analysis (33)] |
| 3. | Liu YB, Chen MK. Epidemiology of liver cirrhosis and associated complications: Current knowledge and future directions. World J Gastroenterol. 2022;28:5910-5930. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 89] [Cited by in RCA: 103] [Article Influence: 25.8] [Reference Citation Analysis (21)] |
| 4. | Xiao J, Wang F, Wong NK, He J, Zhang R, Sun R, Xu Y, Liu Y, Li W, Koike K, He W, You H, Miao Y, Liu X, Meng M, Gao B, Wang H, Li C. Global liver disease burdens and research trends: Analysis from a Chinese perspective. J Hepatol. 2019;71:212-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 423] [Article Influence: 60.4] [Reference Citation Analysis (1)] |
| 5. | GBD 2017 Cirrhosis Collaborators. The global, regional, and national burden of cirrhosis by cause in 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol. 2020;5:245-266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1184] [Cited by in RCA: 1121] [Article Influence: 186.8] [Reference Citation Analysis (5)] |
| 6. | Gülcicegi DE, Goeser T, Kasper P. Prognostic assessment of liver cirrhosis and its complications: current concepts and future perspectives. Front Med (Lausanne). 2023;10:1268102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 21] [Reference Citation Analysis (0)] |
| 7. | Lee S, Arefaine B, Begum N, Stamouli M, Witherden E, Mohamad M, Harzandi A, Zamalloa A, Cai H, Williams R, Curtis MA, Edwards LA, Chokshi S, Mardinoglu A, Proctor G, Moyes DL, McPhail MJ, Shawcross DL, Uhlen M, Shoaie S, Patel VC. Oral-gut microbiome interactions in advanced cirrhosis: characterisation of pathogenic enterotypes and salivatypes, virulence factors and antimicrobial resistance. J Hepatol. 2024;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 17] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 8. | Guan H, Zhang X, Kuang M, Yu J. The gut-liver axis in immune remodeling of hepatic cirrhosis. Front Immunol. 2022;13:946628. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 37] [Reference Citation Analysis (0)] |
| 9. | Li O, Xu H, Kim D, Yang F, Bao Z. Roles of Human Gut Microbiota in Liver Cirrhosis Risk: A Two-Sample Mendelian Randomization Study. J Nutr. 2024;154:143-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 10. | Di Vincenzo F, Del Gaudio A, Petito V, Lopetuso LR, Scaldaferri F. Gut microbiota, intestinal permeability, and systemic inflammation: a narrative review. Intern Emerg Med. 2024;19:275-293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 626] [Article Influence: 313.0] [Reference Citation Analysis (0)] |
| 11. | An L, Wirth U, Koch D, Schirren M, Drefs M, Koliogiannis D, Nieß H, Andrassy J, Guba M, Bazhin AV, Werner J, Kühn F. The Role of Gut-Derived Lipopolysaccharides and the Intestinal Barrier in Fatty Liver Diseases. J Gastrointest Surg. 2022;26:671-683. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 175] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 12. | Ma N, Ma D, Liu X, Zhao L, Ma L, Ma D, Dong S. Bisphenol P exposure in C57BL/6 mice caused gut microbiota dysbiosis and induced intestinal barrier disruption via LPS/TLR4/NF-κB signaling pathway. Environ Int. 2023;175:107949. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 34] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 13. | Zhang X, Yuan S, Zhang X, Zhang T, Meng C, Zhuang K, Dang S. ANGPTL4 regulates CD163 expression and Kuppfer cell polarization induced cirrhosis via TLR4/NF-κB pathway. Exp Cell Res. 2021;405:112706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 14. | Shi S, Zhou Y, Zhang H, Zhu Y, Jiang P, Xie C, Feng T, Zeng Y, He H, Luo Y, Chen J. The Causal Relationship between Inflammatory Cytokines and Liver Cirrhosis in European Descent: A Bidirectional Two-Sample Mendelian Randomization Study and the First Conclusions. Biomedicines. 2024;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Reference Citation Analysis (0)] |
| 15. | Karatayli E, Hall RA, Weber SN, Dooley S, Lammert F. Effect of alcohol on the interleukin 6-mediated inflammatory response in a new mouse model of acute-on-chronic liver injury. Biochim Biophys Acta Mol Basis Dis. 2019;1865:298-307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 16. | Xiang X, Feng D, Hwang S, Ren T, Wang X, Trojnar E, Matyas C, Mo R, Shang D, He Y, Seo W, Shah VH, Pacher P, Xie Q, Gao B. Interleukin-22 ameliorates acute-on-chronic liver failure by reprogramming impaired regeneration pathways in mice. J Hepatol. 2020;72:736-745. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 153] [Article Influence: 25.5] [Reference Citation Analysis (1)] |
| 17. | Wang X, Yi P, Liang Y. The Role of IL-36 in Infectious Diseases: Potential Target for COVID-19? Front Immunol. 2021;12:662266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 18. | Towne JE, Renshaw BR, Douangpanya J, Lipsky BP, Shen M, Gabel CA, Sims JE. Interleukin-36 (IL-36) ligands require processing for full agonist (IL-36α, IL-36β, and IL-36γ) or antagonist (IL-36Ra) activity. J Biol Chem. 2011;286:42594-42602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 247] [Cited by in RCA: 279] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 19. | He Q, Chen HX, Li W, Wu Y, Chen SJ, Yue Q, Xiao M, Li JW. IL-36 cytokine expression and its relationship with p38 MAPK and NF-κB pathways in psoriasis vulgaris skin lesions. J Huazhong Univ Sci Technolog Med Sci. 2013;33:594-599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 20. | Yuan X, Peng X, Li Y, Li M. Role of IL-38 and its related cytokines in inflammation. Mediators Inflamm. 2015;2015:807976. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 21. | Soubières AA, Poullis A. Emerging Biomarkers for the Diagnosis and Monitoring of Inflammatory Bowel Diseases. Inflamm Bowel Dis. 2016;22:2016-2022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (1)] |
| 22. | Blumberg H, Dinh H, Trueblood ES, Pretorius J, Kugler D, Weng N, Kanaly ST, Towne JE, Willis CR, Kuechle MK, Sims JE, Peschon JJ. Opposing activities of two novel members of the IL-1 ligand family regulate skin inflammation. J Exp Med. 2007;204:2603-2614. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 274] [Cited by in RCA: 284] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 23. | Frey S, Derer A, Messbacher ME, Baeten DL, Bugatti S, Montecucco C, Schett G, Hueber AJ. The novel cytokine interleukin-36α is expressed in psoriatic and rheumatoid arthritis synovium. Ann Rheum Dis. 2013;72:1569-1574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 143] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 24. | Friedrich M, Tillack C, Wollenberg A, Schauber J, Brand S. IL-36γ sustains a proinflammatory self-amplifying loop with IL-17C in anti-TNF-induced psoriasiform skin lesions of patients with Crohn's disease. Inflamm Bowel Dis. 2014;20:1891-1901. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 61] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 25. | Andoh A, Nishida A. Pro- and anti-inflammatory roles of interleukin (IL)-33, IL-36, and IL-38 in inflammatory bowel disease. J Gastroenterol. 2023;58:69-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 27] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 26. | Scheibe K, Kersten C, Schmied A, Vieth M, Primbs T, Carlé B, Knieling F, Claussen J, Klimowicz AC, Zheng J, Baum P, Meyer S, Schürmann S, Friedrich O, Waldner MJ, Rath T, Wirtz S, Kollias G, Ekici AB, Atreya R, Raymond EL, Mbow ML, Neurath MF, Neufert C. Inhibiting Interleukin 36 Receptor Signaling Reduces Fibrosis in Mice With Chronic Intestinal Inflammation. Gastroenterology. 2019;156:1082-1097.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 172] [Article Influence: 24.6] [Reference Citation Analysis (12)] |
| 27. | Chinese Society of Hepatology; Chinese Medical Association. [Chinese guidelines on the management of liver cirrhosis]. Zhonghua Gan Zang Bing Za Zhi. 2019;27:846-865. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 40] [Reference Citation Analysis (0)] |
| 28. | Liu Y, Chen Z, Li C, Sun T, Luo X, Jiang B, Liu M, Wang Q, Li T, Cao J, Li Y, Chen Y, Kuai L, Xiao F, Xu H, Cui H. Associations between changes in the gut microbiota and liver cirrhosis: a systematic review and meta-analysis. BMC Gastroenterol. 2025;25:16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 12] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 29. | He X, Hu M, Xu Y, Xia F, Tan Y, Wang Y, Xiang H, Wu H, Ji T, Xu Q, Wang L, Huang Z, Sun M, Wan Y, Cui P, Liang S, Pan Y, Xiao S, He Y, Song R, Yan J, Quan X, Wei Y, Hong C, Liao W, Li F, El-Omar E, Chen J, Qi X, Gao J, Zhou H. The gut-brain axis underlying hepatic encephalopathy in liver cirrhosis. Nat Med. 2025;31:627-638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 33] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 30. | Gong Y, Tingxi Z, Qing L, Guozhen Z, Bing T, Xiaoliang Y, Yan W, Wenjuan J, Yan X, Hui L, Xue H, Zebo Y. Elevated production of IL-36α in chronic hepatitis B virus-infected patients correlates with viral load. Microb Pathog. 2017;113:412-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 31. | Wang HJ, Jiang YF, Wang XR, Zhang ML, Gao PJ. Elevated serum interleukin-38 level at baseline predicts virological response in telbivudine-treated patients with chronic hepatitis B. World J Gastroenterol. 2016;22:4529-4537. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 29] [Cited by in RCA: 28] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 32. | Baker K, O'Donnell C, Bendix M, Keogh S, Byrne J, O'Riordain M, Neary P, Houston A, Brint E. IL-36 signalling enhances a pro-tumorigenic phenotype in colon cancer cells with cancer cell growth restricted by administration of the IL-36R antagonist. Oncogene. 2022;41:2672-2684. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 33. | Song Y, Chu H, Liu F, Guo W, Gao N, Chen C, Bao S. The Pro-Tumor Biological Function of IL-36α Plays an Important Role in the Tumor Microenvironment of HCC. Cancer Manag Res. 2023;15:895-904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 34. | Liang S, Chen L, Liang R, Ling J, Hou M, Gao S, Ou M, Yang M. Emerging Role of Interleukin-38 (IL-38) in the Development of Rheumatoid Arthritis. Rheumatol Ther. 2024;11:349-362. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 35. | Neurath MF. Strategies for targeting cytokines in inflammatory bowel disease. Nat Rev Immunol. 2024;24:559-576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 191] [Article Influence: 95.5] [Reference Citation Analysis (0)] |
| 36. | Sequeira RP, McDonald JAK, Marchesi JR, Clarke TB. Commensal Bacteroidetes protect against Klebsiella pneumoniae colonization and transmission through IL-36 signalling. Nat Microbiol. 2020;5:304-313. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 100] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 37. | Xie C, Yan W, Quan R, Chen C, Tu L, Hou X, Fu Y. Interleukin-38 is elevated in inflammatory bowel diseases and suppresses intestinal inflammation. Cytokine. 2020;127:154963. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/