Published online Dec 27, 2025. doi: 10.4254/wjh.v17.i12.113660
Revised: October 22, 2025
Accepted: November 21, 2025
Published online: December 27, 2025
Processing time: 117 Days and 5.3 Hours
Xietu Hemu prescription (XHP), a Chinese patent formula, is optimized based on the theory of “phlegm-dampness” and has been clinically validated to effectively combat metabolic dysfunction-associated steatotic liver disease (MASLD). It not
To elucidate the mechanisms by which XHP inhibits adipocyte differentiation and maintains lipid metabolism homeostasis.
The therapeutic efficacy of XHP in metabolic-related disorders was analyzed using HepG2 cells and 3T3-L1 cells, along with transcriptomics to assess gene expression alterations during white adipogenesis. The primary metabolites of XHP were identified through ultra-performance liquid chromatography, and metabolic pathways were examined via serum metabolomics. Network analysis was employed to predict therapeutic targets. The accumulation of lipid droplets and the expression of associated proteins were confirmed using oil red O staining and Western blotting, respectively. Molecular docking was utilized to identify core targets and signaling pathways, which were substantiated through immunofluorescence and siRNA interference.
XHP-containing serum (XHPS) significantly inhibited the transformation of normal HepG2 cells into fatty liver cells. Concurrently, the treatment suppressed the differentiation of 3T3-L1 cells, reduced lipid droplet accumulation and total cholesterol/triglyceride levels, and downregulated the expression of PPARγ, C/EBPα, and FABP4. Through transcriptomics and network pharmacological intersectionality analyses, 24 core targets were identified, predominantly enriched in the AMPK signaling pathway. Molecular docking validated the strong binding affinity of XHP metabolites to targets such as leptin (-11.3 kcal/mol) and ADIPOQ (-9.4 kcal/mol). ELISA results indicated that XHPS augmented leptin autocrine secretion, thereby activating the AMPK signaling pathway (P < 0.05). Con
XHP effectively inhibits adipogenesis and enhances lipid metabolism homeostasis through the LEP/AM
Core Tip: Xietu Hemu prescription, a traditional Chinese medicine formula, ameliorates metabolic dysfunction-associated steatotic liver disease-related obesity by inhibiting adipogenesis. Integrating serum metabolomics, transcriptomics, and network analysis, this study identified the LEP/AMPK/PPARγ signaling axis as the central mechanism. The formula promotes leptin autocrine secretion, activates AMPK phosphorylation, and subsequently downregulates PPARγ and SREBPs, thereby reducing lipid accumulation and improving metabolic homeostasis, offering a novel multi-target therapeutic strategy.
- Citation: Cheng Z, Lu YF, He YX, Wei W, Xie YX, Lv TS, Wei Y, Lou Y, Yu JY, Zhou XQ. Integrated serum metabolomics reveal molecular mechanism of Xietu Hemu prescription on metabolic dysfunction-associated steatotic liver disease-related obesity. World J Hepatol 2025; 17(12): 113660
- URL: https://www.wjgnet.com/1948-5182/full/v17/i12/113660.htm
- DOI: https://dx.doi.org/10.4254/wjh.v17.i12.113660
The development of metabolic dysfunction-associated steatotic liver disease (MASLD) is fundamentally linked to disruptions in lipid metabolism. When the body is unable to store surplus lipids, they are released into the bloodstream and subsequently accumulate in hepatocytes. This accumulation can lead to lipotoxicity, which damages hepatocytes and triggers the activation of hepatic stellate cells, resulting in hepatic fibrosis[1]. Addressing obesity to mitigate lipotoxicity is recognized as one of the most effective strategies for managing MASLD. Obesity, a chronic global metabolic disorder, has escalated to epidemic proportions over recent decades, posing a significant public health challenge. According to a 2021 report by the World Health Organization, the global population of obese individuals has surpassed 650 million, with figures continuing to rise. In recent decades, shifts in lifestyle and dietary habits have contributed to a sharp increase in obesity rates across both developing and developed nations, imposing a substantial economic burden on healthcare systems[2].
Obesity and MASLD exhibit shared pathophysiological mechanisms, including imbalances in energy metabolism, insulin resistance, chronic inflammation, and dysregulation of lipid metabolism[3-5]. Research indicates that between 70% and 90% of obese individuals develop fatty liver disease, compared to 20%-30% in the general population, with the severity of the disease intensifying as obesity progresses[6,7]. This high rate of comorbidity underscores a complex pathophysiological connection between obesity and MASLD. Weight loss remains the most effective therapeutic intervention, with a reduction of 5%-10% in body weight significantly decreasing hepatic fat accumulation[8]. Therefore, managing obesity is a primary approach in the treatment of MASLD.
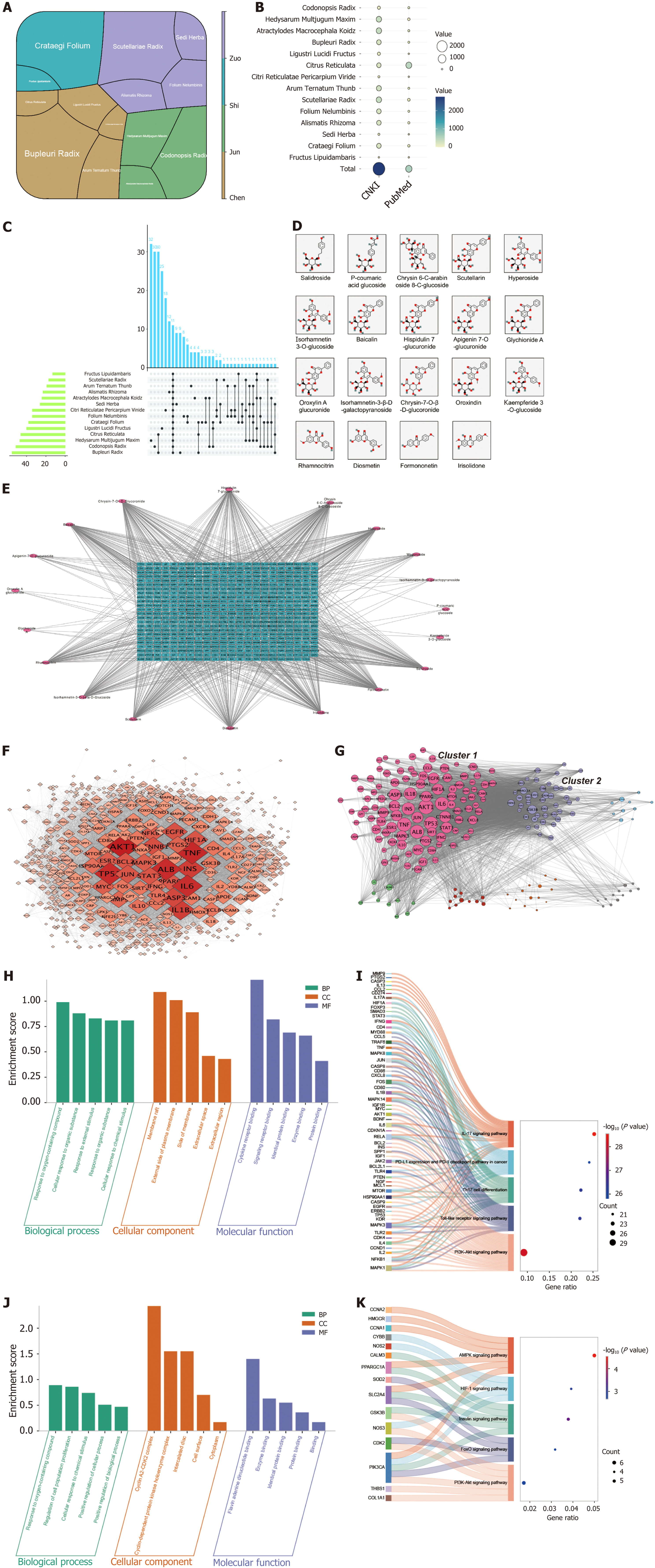
The Xietu Hemu prescription (XHP), a patented herbal formula (Chinese patent No. ZL 2022 1 0250245.4, authorized in 2023), was developed by the Department of Endocrinology at Jiangsu Provincial Hospital of Chinese Medicine to address obesity and MASLD. Early clinical research on XHP has shown its potential in reducing the fatty liver index, MASLD fibrosis score, body mass index, and visceral fat area[9]. The composition of XHP includes 14 botanical drugs, with Codonopsis Radix, Hedysarum Multijugum Maxim, and Atractylodes Macrocephala Koidz as the principal metabolites. The additional 11 botanical drugs are Bupleuri Radix, Ligustri Lucidi Fructus, Citrus Reticulata, Citri Reticulatae Pericarpium Viride, Arum Ternatum Thunb, Scutellariae Radix, Folium Nelumbinis, Alismatis Rhizoma, Sedi Herba, Crataegi Folium, and Fructus Liquidambaris.
Currently, the United States Food and Drug Administration (FDA) has not approved any pharmacological agents that are highly effective for treating obesity. Most Western medications that are clinically used for obesity demonstrate limited efficacy or present unacceptable side effects. In contrast, prescriptions from traditional Chinese medicine (TCM), which adopt a multi-target, cocktail-like therapeutic approach, have attracted increasing interest. According to TCM theory, the primary pathogenesis of obesity is attributed to the accumulation of “phlegm and dampness”. XHP was developed specifically to target this underlying syndrome. It combines the spleen-fortifying and dampness-resolving properties of Shenling Baizhu San, as originally documented in the Tai Ping Hui Min He Ji Ju Fang during the Song dynasty, with the liver-soothing and qi-regulating effects of Chaihu Shugan San from the Jing Yue Quan Shu of the Ming dynasty. XHP is tailored to address the combined pathological pattern of “phlegm-dampness accumulation and liver stagnation with spleen deficiency”, which typifies the TCM syndrome commonly observed in obese patients with MASLD. The the
The following primary antibodies were acquired: Anti-C/EBPα (8178), anti-FABP4 (3544), anti-PPARγ (2443), anti-mTOR (2983), and anti-p-mTOR (5536) from CST (United States); anti-GAPDH (10494-1-AP) from Proteintech (Wuhan, China); anti-AMPK (ab32047) from Abcam (United States); and anti-SREBP1/2 (sc-365513/sc-13552; for cytoplasmic protein detection) from Santa Cruz (United States). Other primary antibodies included anti-p-AMPK (AP0116), anti-leptin (A1300), anti-LEPR (A19076), anti-Lamin B1 (A11495), and anti-SREBP1/2 (A15586/A13049; for nuclear protein detection) obtained from Abclonal (Wuhan, China). Secondary antibodies (AS003/AS014) were sourced from Abclonal (Wuhan, China). The ECL developer (BL520) was supplied by Biosharp (Hefei, China), and the protease inhibitor (P001) was from New Cell & Molecular Biotech Co., Ltd (Suzhou, China). The specific reagents for 3T3-L1 preadipocytes, including medium (ZM0089) and serum-free cell freezing solution (CSP042), were procured from Zhongqiao Xinzhou Biotechnology Co., Ltd (Shanghai, China). Bovine insulin (I8040) and Dexamethasone (D4902) were available from Solarbio (Beijing, China) and Sigma (Germany), respectively. 3-Isobutyl-1-methylxanthine (I5879) and oil red O powder (O0625) were also supplied by Sigma (Germany). Additional reagents included fetal bovine serum (FBS), trypsin, and high glucose DMEM from Gibco (United States); CCK8 reagent (HY-K0301) and GW9662 (HY-16578) from MCE (United States); and Silibinin (CAS No. 22888-70-6, B21185) from Yuanye Bio-Technology Co., Ltd (Shanghai, China). Palmitic acid (PA) and oleic acid (OA) were procured from Kunchuang Technology Development Co., Ltd (Xi’an, China), along with other consumables and reagents from Servicebio (Wuhan, China).
The 14 botanical drugs comprising XHP were sourced from the Department of Traditional Chinese Medicine at Jiangsu Provincial Hospital of Chinese Medicine. The quantities and respective percentages of these botanical drugs used in XHP are detailed in Table 1. These drugs were combined in precise proportions in a ceramic pot, submerged in an adequate volume of water, and subjected to a sequential process of softening for 0.5 hours, followed by heating and decoction for 1 hour. This decoction process was repeated for an additional hour with replenished water. Subsequently, the mixture was filtered to remove solid residues and the excess liquid was evaporated using a rotary evaporator to achieve the desired concentration of XHP for administration to rats. The prepared XHP was then stored at -80 °C, and warmed to 37 °C for use after thawing.
| TCM name | Latin names | Plant part | Occupied percent (%) | Dosage for adults (g/day/60 kg) | Dosage for rat (g/day/kg) |
| Dangshen | Codonopsis Radix | Root | 7.246 | 10 | 1.050 |
| Huangqi | Hedysarum Multijugum Maxim | Root | 7.246 | 10 | 1.050 |
| Baizhu | Atractylodes Macrocephala Koidz | Stolon | 7.246 | 10 | 1.050 |
| Chaihu | Bupleuri Radix | Root | 4.348 | 6 | 0.630 |
| Nüzhenzi | Ligustri Lucidi Fructus | Fruit | 10.870 | 15 | 1.575 |
| Chenpi | Citrus Reticulata | Pericarp | 4.348 | 6 | 0.630 |
| Qingpi | Citri Reticulatae Pericarpium Viride | Pericarp | 7.246 | 10 | 1.050 |
| Banxia | Arum Ternatum Thunb | Tuber | 7.246 | 10 | 1.050 |
| Huangqin | Scutellariae Radix | Root | 4.348 | 6 | 0.630 |
| Heye | Folium Nelumbinis | Leaf | 7.246 | 10 | 1.050 |
| Zexie | Alismatis Rhizoma | Tuber | 7.246 | 10 | 1.050 |
| Chuipencao | Sedi Herba | All | 10.870 | 15 | 1.575 |
| Shanzha | Crataegi Folium | Fruit | 7.246 | 10 | 1.050 |
| Lulutong | Fructus Lipuidambaris | Fruit | 7.246 | 10 | 1.050 |
| Total | - | - | 100.000 | 138 | 14.490 |
Male Sprague-Dawley rats, each weighing approximately 200 g (± 10 g), were procured from Jiangsu Qinglongshan Biotechnology Co., Ltd., for the purpose of drug serum preparation. Following the clinical guidelines provided by the Department of Endocrinology at Jiangsu Provincial Hospital of Chinese Medicine, an adult weighing 60 kg requires a daily intake of 138 g of XHP, equivalent to 2.3 g/kg. Accordingly, the dosage for rats was set at 14.49 g/kg, approximately 6.3 times the clinical dose[10]. The rats were administered 10 mL/kg/day of either drinking water or XHP for seven days to harvest the serum, subsequently designated as vehicle serum (VehS) or XHP-containing serum (XHPS). This serum was then inactivated, filtered, and stored in aliquots for further use in cellular experiments.
Ultra-performance liquid chromatography (UPLC)-quadrupole time-of-flight tandem mass spectrometry (QTOF/MS/MS) represents a sophisticated analytical method that combines UPLC with QTOF/MS/MS. This technology is primarily utilized for the separation, identification, and structural elucidation of complex biochemical samples.
Herbal prescription extraction: A small aliquot of the concentrated XHP was desiccated at 45 °C. Following this, 0.1 g of the solid extract was solubilized in 10 mL of 70% methanol and subjected to ultrasonic extraction for 30 minutes, a process that was repeated twice. The resultant supernatants were amalgamated, and 1 mL of this solution was then evaporated to dryness under a stream of nitrogen gas. The residual matter was reconstituted in 1 mL of ultrapure water and sub
Serum sample extraction: Pooled serum samples were prepared by combining 30 μL aliquots from each subject in both the XHP-treated group (n = 3, labeled XHPS) and the control group (n = 3, labeled VehS). To precipitate proteins, 0.9 mL of ice-cold methanol was added to each pooled sample, followed by vigorous vortexing and sonication in an ice bath for 10 minutes. The samples were then incubated at 4 °C for 60 minutes and centrifuged at 13000 rpm for 15 minutes. A 400 μL aliquot of the supernatant from each sample was evaporated to dryness under nitrogen. The dried residues were reconstituted in 300 μL of 70% methanol, centrifuged again at 13000 rpm for 15 minutes, and the refined supernatant was subsequently injected into the liquid chromatography quadrupole time-of-flight mass spectrometry system.
Chromatographic separation was effected on a Waters Acquity HSS T3 column (2.1 mm × 150 mm, 1.8 μm) maintained at 35 °C. The mobile phase comprised water with 0.1% formic acid and acetonitrile, delivered at a flow rate of 0.3 mL/minute. The volume of each injection was 2 μL, with the elution gradient specified in Supplementary Table 1. Mass spectrometric analysis was executed using a SCIEX Exion LC system coupled to an X500B Q-TOF mass spectrometer (AB Sciex, Foster City, CA, United States) equipped with an electrospray ionization source. Data acquisition occurred in both positive and negative ion modes across a scan range of m/z 100-1250. Detailed parameters for these analyses are provided in Supplementary Table 2.
HepG2 cells are frequently used to establish lipid metabolism-related fatty liver diseases, including MASLD and MASH, while 3T3-L1 cells are commonly employed for studies on adipogenesis and obesity. The objectives and methods of the experiment on HepG2 cells and 3T3-L1 cells referred to published researches[11,12].
HepG2 cells, generously provided by Servicebio, were cultured to approximately 80% confluence. Subsequently, these cells underwent a starvation period of 12 hours in DMEM before being subjected to a 24-hour modeling phase with PA (0.5 mmol/L) or PA + OA (PA: 0.25 mmol/L, OA: 0.5 mmol/L) in 5% VehS. Following this, cells were treated with varying concentrations of XHPS or VehS for 24 hours, after which lipid accumulation was assessed using oil red O staining.
The 3T3-L1 preadipocytes, provided by Professor Guang Chen’s laboratory at Tongji Medical College, Huazhong University of Science and Technology, were used within ten passages for all differentiation experiments. Initially, these preadipocytes were maintained in a growth medium supplemented with newborn calf serum. To promote dense cell-to-cell contact, cells were seeded in culture vessels until confluent, followed by an additional two-day culture period. This stage was designated as day 0, marking the transition to a quiescent state. The differentiation induction phase com
For viability assays, cells were plated in 96-well plates and exposed to different ratios of drug serum. After 72 hours, cell viability was evaluated using the Cell Counting Kit-8 assay, following the manufacturer’s protocol. Absorbance was measured at 450 nm using a PerkinElmer EnSpire microplate reader.
Following differentiation induction, 3T3-L1 preadipocytes cultured in 24-well plates were prepared for staining. The medium was carefully aspirated, and cells were washed three times with phosphate-buffered saline (PBS). Cells were then fixed in 10% neutral buffered formalin for 60 minutes. The oil red O working solution was prepared by diluting solution O0625 with deionized water in a 3:2 ratio, followed by filtration and a 10-minute room temperature incubation to eliminate impurities. Approximately 1.5 mL of this working solution was added to each well of a 6-well plate, and the cells were stained for 30-60 minutes. Post-staining, the solution was discarded, and residual dye was removed by a brief rinse with 60% isopropanol, lasting no longer than 10 seconds per well. Cells were washed 2-3 times with PBS and counterstained with hematoxylin for one minute. After a final wash with PBS, the stained cells were visualized and documented using an Olympus CKX41 inverted microscope.
The study utilized 3T3-L1 cells across three distinct passages, each comprising control and model groups. By the sixth day of the modeling process, mature lipid droplets were prominently visible under microscopic examination. The cells were initially rinsed gently with PBS, followed by the addition of Trizol reagent to lyse the cells. The lysates were immediately harvested and preserved at -80 °C. For the assessment of sample quality, polyadenylated mRNA was enriched using magnetic beads coated with oligo (dT). The isolated RNA was fragmented in a fragmentation buffer, and first-strand cDNA synthesis was carried out using random N6 primers. This step was followed by the synthesis of double-stranded DNA, ligation of sticky ends, and adapters to the 3’ ends, along with phosphorylation of the 5’ ends. Polymerase chain reaction (PCR) amplification was then performed, and the PCR products were denatured into single-stranded DNA by heating. Circularization of the single-stranded DNA was achieved using a bridging primer, resulting in the formation of a single-stranded circular DNA library. The libraries underwent stringent quality control measures, and only those that met the quality criteria were selected for sequencing. The analysis of gene expression included evaluating the stability of gene expression and identifying differentially expressed genes (DEGs). Functional enrichment analyses, such as those involving Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, were conducted to elucidate potential mechanisms inferred from the data.
For assays targeting nuclear proteins, samples were prepared utilizing the Nuclear and Cytoplasmic Protein Extraction Kit from KeyGEN (KGB5302), adhering strictly to the manufacturer’s protocol. For assays not concerning nucleoproteins, the following procedures were employed: Protein lysis buffer was fortified with a 1/100 volume of both protease and phosphatase inhibitors to prepare the protein lysis mixture. Following a PBS wash, 100 μL of the lysis mixture was applied to each well to ensure coverage of the cell monolayer. Cells were then dislodged and lysed using a cell sonicator, and the mixture was agitated gently at low temperature for 30 minutes. Post-centrifugation at low temperature, the supernatant was collected, and a quarter volume of protein loading buffer was added. The samples were then heated in boiling water for 10 minutes and were either immediately subjected to electrophoresis or stored. Protein concentration was quantified using a BCA assay kit (Servicebio, G2006) with a microplate reader (PerkinElmer, EnSpire). A standard load of 30 μg of protein per lane was applied to an SDS-PAGE gel. Following electrophoresis, proteins were transferred to a PVDF membrane. The membrane was blocked and subsequently incubated overnight at 4 °C with primary antibodies. The following day, after washing with Tris buffered saline Tween, the membrane was incubated with secondary antibodies at room temperature for 1 hour. Post-washing, the membrane underwent chemiluminescent detection using General Electric’s ImageQuant LAS 4000, and the acquired images were analyzed with ImageJ software.
The Mouse Leptin ELISA Kit (RK00380) was procured from Abclonal (Wuhan, China). The assay was conducted in accordance with the manufacturer’s protocol, utilizing duplicate wells for each sample. Results were quantified using the established standard curve.
Cells were seeded onto plates with pre-positioned coverslips and cultured until the 3T3-L1 cells adhered. Following treatment with the drug, cells were rinsed with ice-cold PBS and fixed with 4% paraformaldehyde for 15 minutes. Subsequent washes with PBS were performed before cells were permeabilized with 0.3% Triton X-100 for 10 minutes and washed again. Non-specific binding was minimized by incubating the cells with 2% bovine serum albumin at room temperature for one hour. After the removal of the blocking solution, cells were incubated with primary antibodies overnight at 4 °C. The next day, the cells were washed with PBST and incubated with fluorophore-conjugated secondary antibodies or DAPI staining solution for one hour at room temperature in the dark. After additional washes with PBST, the coverslips were mounted on glass slides using an anti-fade mounting medium. Fluorescent images were obtained using an Olympus CKX41 fluorescence microscope.
To inhibit LEPR expression in 3T3-L1 cells, siRNA targeting LEPR was utilized, prepared by Nanjing Tsingke Biotechnology Co., Ltd. The sequence number is listed in Supplementary Table 3. Cells were seeded in 6-well plates and left for 24 hours until the cell density reached 70%-80%. For transfection, 125 μL of serum-free Opti-MEM and 100 pmol of siRNA or vehicle were added to a sterile centrifuge tube, while another tube received 125 μL of Opti-MEM and 6 μL of Lipo3.0 transfection reagent (Abbkine, BMU111-CN). Both mixtures were combined and incubated for 15 minutes to form the transfection reagent/siRNA complex. The cells were then cultured in this 250 μL mixture supplemented with 1.5 mL of complete medium for 24 hours. Post-transfection, the efficacy of the siRNA was assessed, and differentiation experiments of 3T3-L1 cells were initiated.
Data collection: Target information for each metabolite in XHPS was sourced from the TCM systems pharmacology[13] database and the PubChem metabolite database[14]. Additional targets were predicted using PharmMapper (https://www.lilab-ecust.cn/pharmmapper/index.html), and this information was integrated. The clinical drug database DrugBank[15] was queried using the keyword “obesity” to retrieve entries related to a body mass index greater than 30 kg/m2, including FDA-approved drugs for obesity and their corresponding targets. Disease-related targets were obtained from the disease gene databases DisGeNET[16] and GeneCards[17].
Data transformation: To eliminate invalid data and ensure the accuracy of cross-species gene ID conversions, the String database[18] was employed. Upon selecting the relevant species, the active metabolites of XHP and the associated disease targets for obesity, along with their corresponding clinical drug targets, were uploaded to the String platform. This step facilitated the conversion of all data into standardized gene symbols, thereby ensuring consistency and accuracy for subsequent analyses.
Protein-protein interaction network construction: The protein-protein interaction (PPI) network was constructed using Cytoscape 3.6.0 software[19]. Initially, a table delineating the one-to-one correspondence between XHP-predicted metabolites and their corresponding targets was imported into Cytoscape to establish the metabolite-target network. The PPI network pertinent to XHP’s role in obesity treatment was developed by integrating XHP-putative targets, obesity-related targets, and DEGs derived from the 3T3-L1 adipocyte differentiation experiments. Analysis of the PPI network was performed via the String platform prior to its visualization in Cytoscape 3.6.0. Subsequently, GO and KEGG pathway enrichment analyses were conducted.
Graphical visualization: Graphical visualizations including box plots, principal coordinate analysis (PCoA) plots, heatmaps, volcano plots, Venn diagrams, bubble charts, and chord diagrams were generated using the Bioinformatics platform (https://www.bioinformatics.com.cn). Mechanism summary diagrams were produced and exported using FigDraw (https://www.figdraw.com).
Key proteins and metabolites’ molecular structures were retrieved from the Protein Data Bank (PDB)[20] and PubChem databases, respectively. These structures were optimized with Autodock and Chem3D software, then converted to the “pdbqt” format. Water molecules were removed, and polar hydrogen atoms were appended to the protein structures. The docking box was delineated based on data from the source literature. All structures were subsequently imported into Autodock Vina 1.2.2[21] for molecular docking validation, setting a docking score criterion of ≤ -5.0 to signify successful docking; lower scores indicated greater reliability of the results. The docking outcomes were analyzed using PyMOL software, which facilitated the selection of optimal docking poses based on the ligand-binding regions of the original protein structures. The molecular docking diagrams were subsequently exported. Network analysis and RNA sequencing (RNA-seq) data screening were employed to predict potential targets of XHP for obesity treatment, selecting combinations with the highest predicted efficacy for docking. Protein structures included in this study comprised leptin (PDB ID: 1AX8), PPARG (PDB ID: 2PRG), ADIPOQ (PDB ID: 5 LX9), IGF1 (PDB ID: 6FF3), LIPE (hormone-sensitive type, PDB ID: 8ZVQ), FASN (PDB ID: 7MHD), and SLC2A4 (PDB ID: 7WSM). Settings for the docking center and active pocket were based on structural data or literature references from the corresponding PDB IDs.
Statistical analyses were conducted using SPSS software, version 27.0. Data are presented as the mean ± SD. Comparisons among three or more groups were performed using one-way analysis of variance (ANOVA), while pairwise comparisons were executed using Dunnett’s t-test. A P value of less than 0.05 was considered statistically significant.
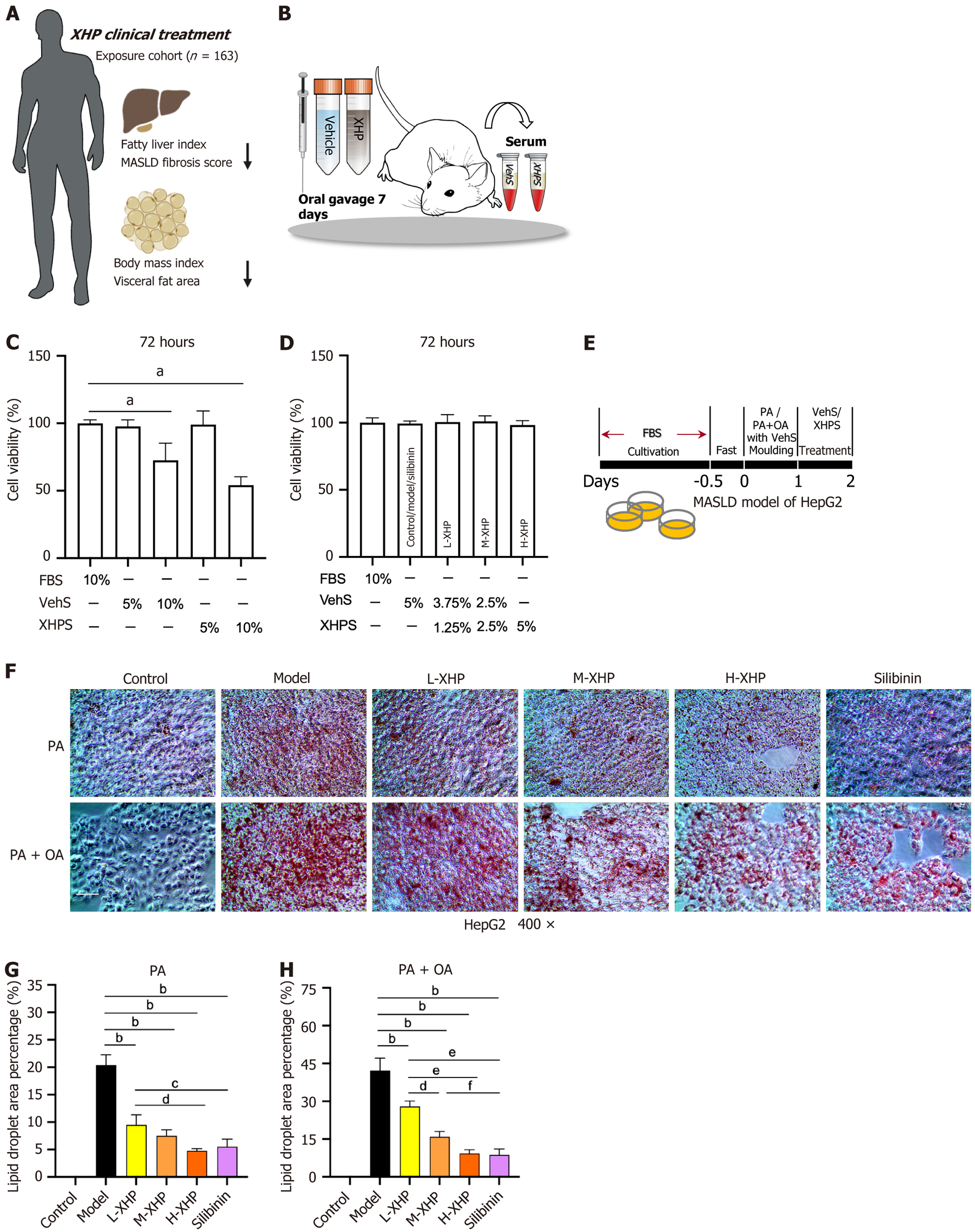
In a preceding clinical study involving 163 patients diagnosed with MASLD, treatment with XHP led to improvements in metabolic homeostasis. This was evidenced by reductions in the fatty liver index, MASLD fibrosis score, body mass index, and visceral fat area (Figure 1A).
To corroborate the lipid-regulatory effects of XHP in vitro, serum containing the drug was utilized. XHPS and VehS were derived through transabdominal aortic sampling from rats subjected to continuous gavage for one week (Figure 1B). Given the complexity of rat serum compared to FBS, initial adjustments in the concentrations of XHPS and VehS provided to the cells were necessary. HepG2 cells were cultured with varying concentrations of XHPS and VehS for a duration of 72 hours to ascertain an optimal comparison with FBS. The outcomes of the CCK8 assay indicated that concentrations of 5% XHPS and VehS were optimal (Figure 1C). Further experiments to assess the impact of different XHPS concentrations on the differentiation process revealed that all tested doses were deemed safe (Figure 1D). Consequently, for the entirety of the cellular experiments, concentrations of 5% XHPS, 2.5% XHPS + 2.5% VehS, and 1.25% XHPS + 3.75% VehS were employed as high, medium, and low doses of drug-serum, respectively, and were designated as the high dose of XHP (H-XHP) group, medium dose of XHP (M-XHP) group, and low dose of XHP (L-XHP) group. The control, model, and silibinin groups were incubated with 5% VehS throughout the incubation period. The experimental protocol is depicted in Figure 1E.
After a 24-hour induction with PA or PA + OA, a subsequent 24-hour intervention with XHPS mitigated lipid accumulation in HepG2 cells, as evidenced by oil red O staining (Figure 1F-H). Under both PA and PA + OA induction protocols, the model group exhibited the largest lipid droplet area within comparable fields of view relative to other groups. In contrast, lipid droplet areas progressively diminished in the L-XHP, M-XHP, and H-XHP groups, all of which remained significantly smaller than those observed in the model group. This suggests that XHPS treatment alleviated hepatic lipotoxicity in a dose-dependent manner. Silibinin, a standard clinical intervention for MASLD, did not display a statistically significant difference in efficacy when administered at 0.1 mmol/L compared to H-XHPS therapy.
Given the clinical and experimental efficacy of XHP in treating MASLD with noted reductions in lipid accumulation (related to obesity), this study aims to systematically analyze its pharmacological effects.
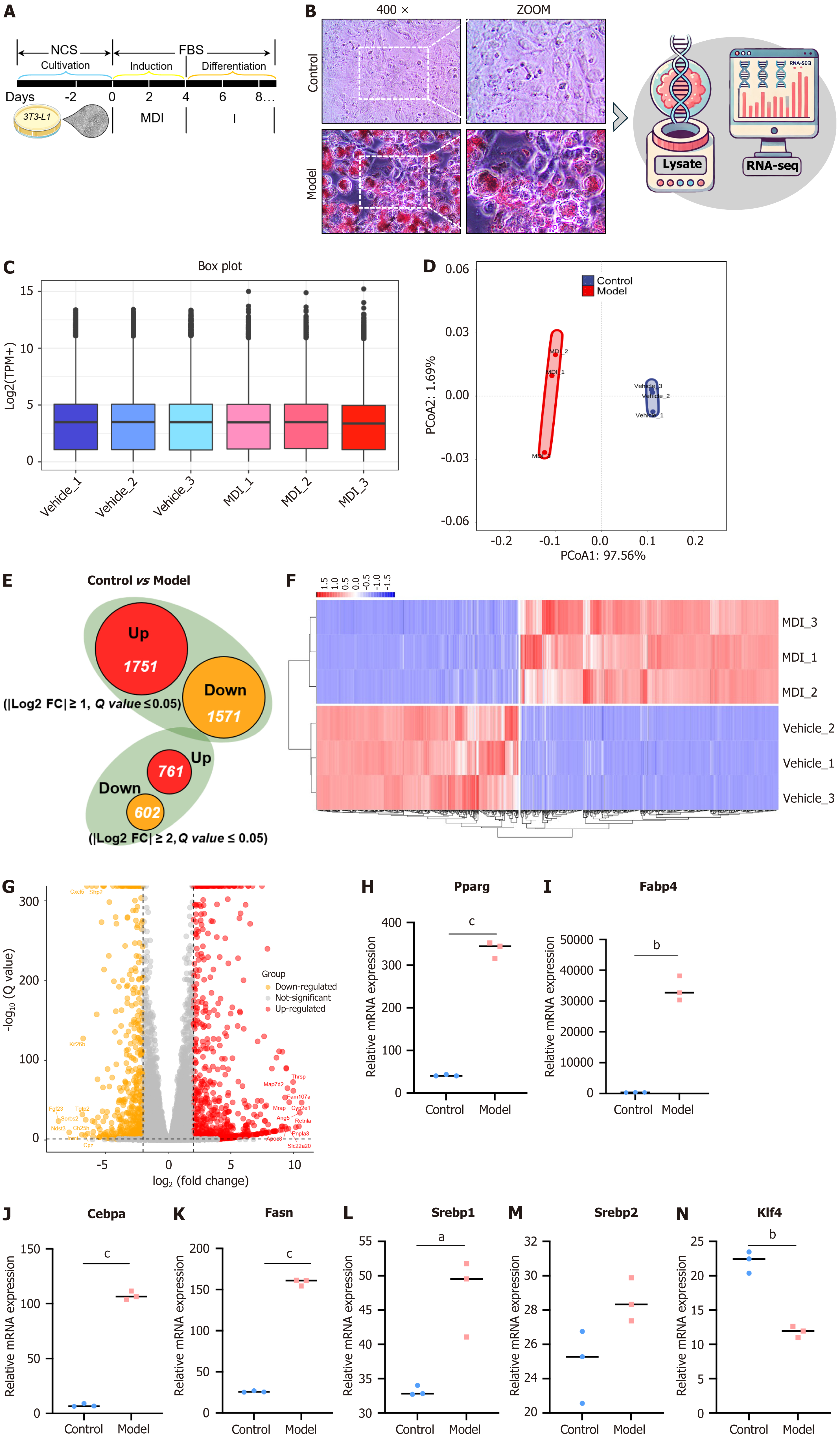
The process by which 3T3-L1 cells differentiate into mature white adipocytes is depicted in Figure 2A. This study aimed to elucidate the changes in gene expression accompanying the differentiation of 3T3-L1 cells. For this purpose, RNA-seq analysis was conducted on both control and model groups. Following four days of induction with 3-isobutyl-1-methylxanthine, dexamethasone, and insulin (MDI) and a subsequent two days of insulin treatment, significant lipid droplet accumulation was observed in the model group relative to the control, as indicated by oil red O staining. This accumulation suggests successful differentiation into an adipose-like phenotype (Figure 2B). Consequently, RNA-seq analysis was employed to examine the differences in gene expression between the two groups. Figure 2C illustrates that gene expression distribution was remarkably consistent across samples from both groups, indicating stable expression levels across all samples. PCoA employing Bray’s distance algorithm revealed significant mRNA expression differences between the control and model groups, with similar expression patterns within each group (Figure 2D).
The differential expression analysis identified 1751 genes with increased expression and 1571 genes with decreased expression, using |Log2FC| ≥ 1 and a q value ≤ 0.05 as the inclusion criteria. However, when the threshold for |Log2FC| was set to a minimum of 2, to place a greater emphasis on the multiplicity of differentials, the number of DEGs was reduced to 761 and 602, respectively (Figure 2E). These genes are considered highly differential and warrant further analysis. Figure 2F and G present a heatmap and a volcano plot of differential gene expression based on the aforementioned screening criteria. Among the top 10 differentially up/down-regulated genes, Thrsp, Mrap, Apoe3, Cyp2e1, Retnla, Pnpla3, Ch25h, Sorbs2, and Sfrp2, all of which have been reported to relate directly or indirectly to lipid metabolism, were notably ranked. In Figure 2H-N, key genes implicated in lipid metabolism were further extracted from the RNA-seq raw data, and their comparative transcripts per million values in each sample group were displayed. The results indicated that the model group exhibited significantly higher expression of key adipogenic and lipid storage transcription factors, such as Pparg, Cebpa, Fabp4, Fasn, and Srebp1/2, consistent with the oil red O staining results. Additionally, Klf4, which enhances key adipogenic transcription factors, was expressed at lower levels in the model group, potentially indicating alterations from the dysregulated lipid regulatory system observed in severely obese individuals. The observed changes in Klf4 mRNA levels may be attributable to a negative feedback mechanism regulating lipid droplet synthesis within the intracellular compartment during 3T3-L1 cell differentiation.
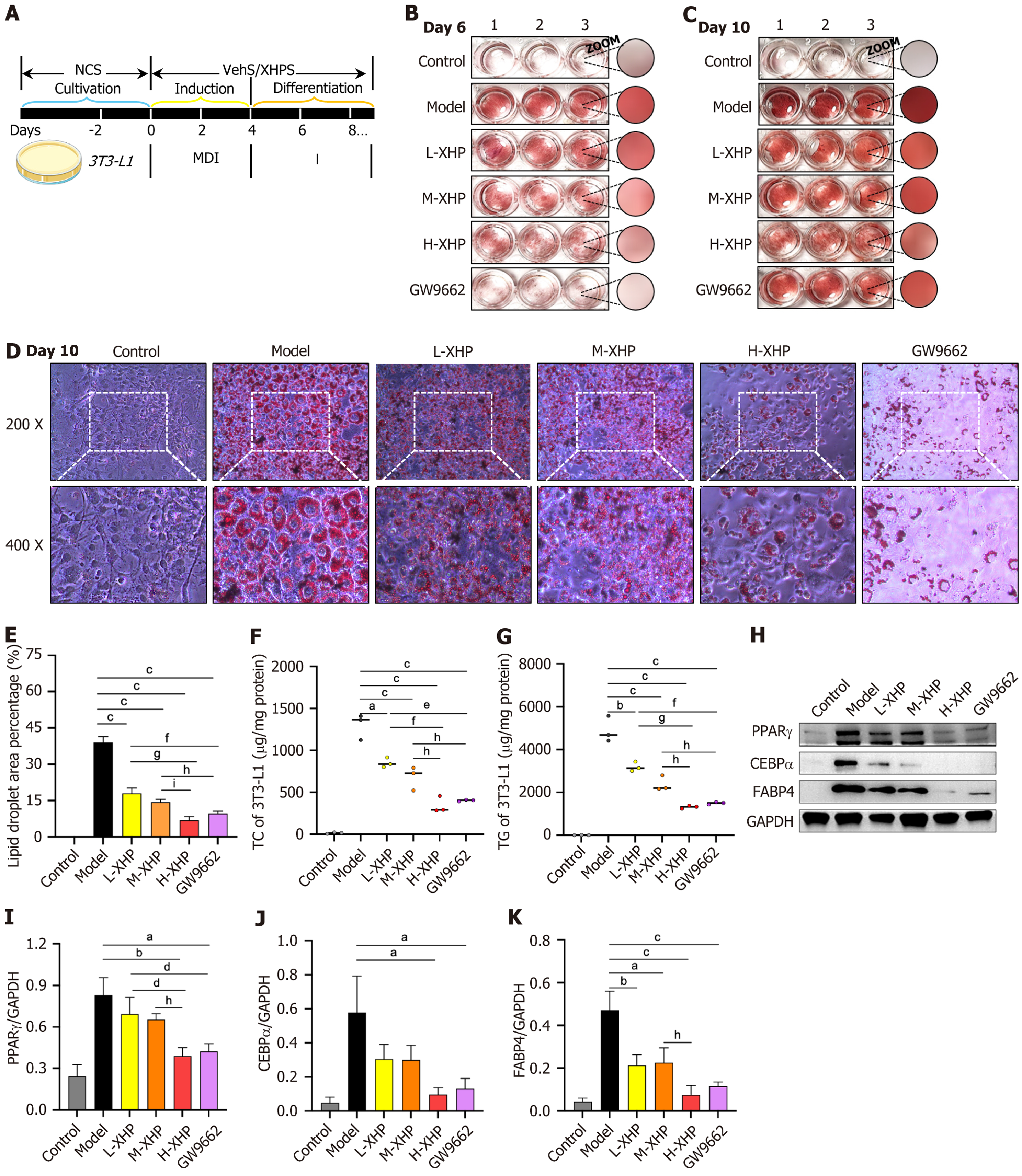
The protocol for the differentiation of 3T3-L1 cells treated with XHPS is depicted in Figure 3A. Following a 6-10 days treatment period, the 3T3-L1 cells were stained with oil red O, enabling the visualization of differences in lipid accumulation among various groups (Figure 3B and C). On day 6, XHPS exhibited a dose-dependent inhibition of lipid droplet formation, which, although less potent than GW9662, a PPARγ inhibitor, still demonstrated significant efficacy. As the experimental period progressed, initially small lipid droplets coalesced into larger formations. Notably, by day 10, XHPS continued to effectively inhibit the maturation of 3T3-L1 cells, with minimal differences observed between the H-XHP group and the GW9662 treatment. Given the multi-targeted nature of XHPS as a cocktail therapy, the inhibition observed in the H-XHP group is particularly noteworthy in terms of drug safety and efficacy conversion rates. Microscopic analysis on day 10 further clarified the lipogenic inhibitory effects of XHPS, with statistically significant differences noted (Figure 3D and E). Consequently, all subsequent experiments were conducted with samples harvested following a 10-day differentiation cycle. Corresponding trends were observed in the intracellular levels of total cholesterol (TC) and triglyceride (TG), which paralleled the findings from the oil red O staining (Figure 3F and G). Immunoblotting analysis indi
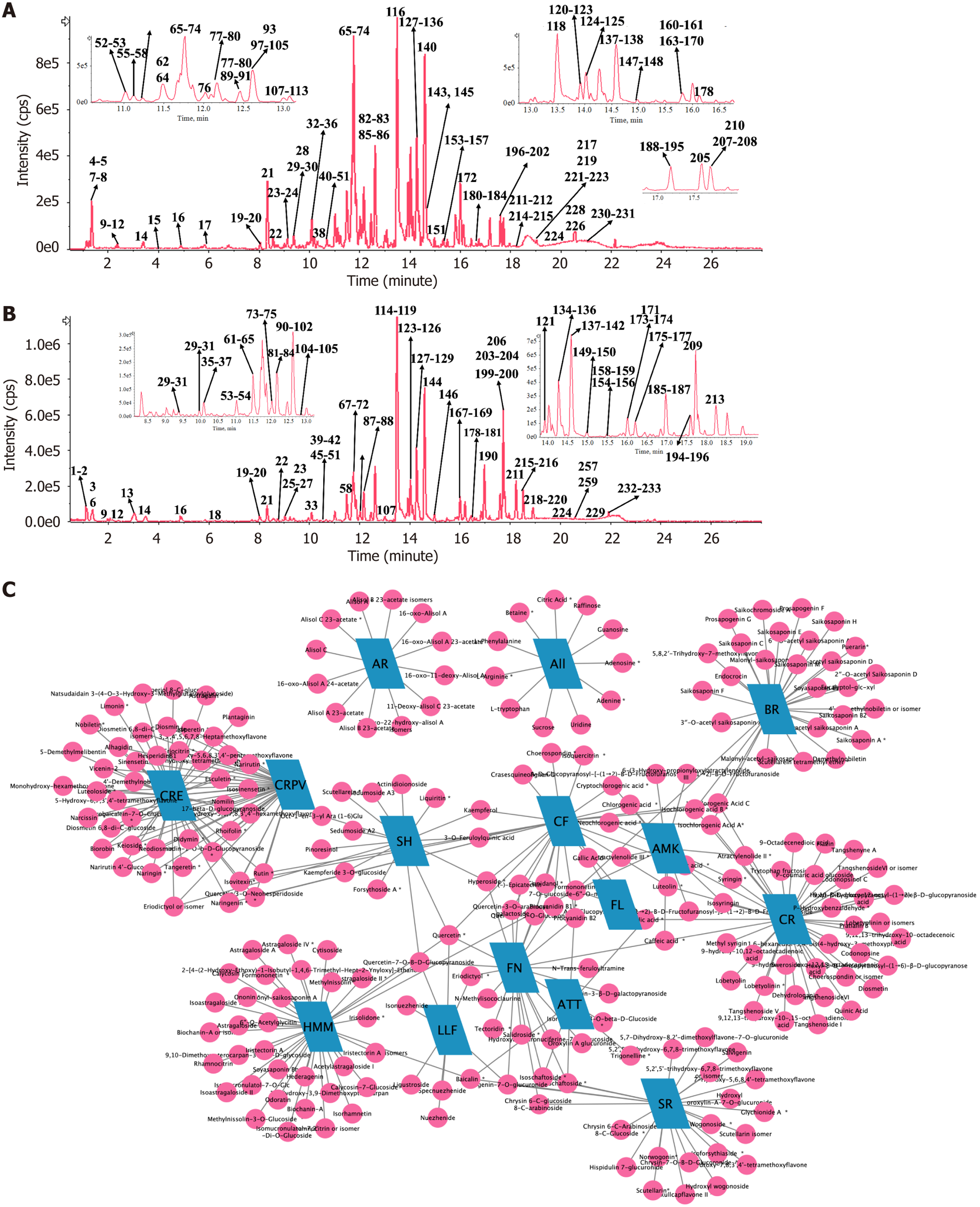
Through the application of UPLC-QTOF/MS/MS analytical techniques, our study successfully identified 233 chemical metabolites in XHP. The metabolites comprise 113 flavonoids, 20 phenylpropanoids, 5 polyacetylenes, 23 terpenoids, and 26 triterpenoid saponins, in addition to a diverse array of other metabolite classes including alkaloids, amino acids, organic acids, and iridoids. Figure 4 displays the base peak chromatogram of XHP. The structural features and fra
Codonopsis Radix: This botanical drug yielded 41 metabolites across various chemical classes such as phenylpropanoids (e.g., Tangshenoside I), polyacetylenes (e.g., Lobetyolinin), and alkaloids (e.g., Codonopsine), which serve as characteristic markers. Hedysarum Multijugum Maxim: Analysis revealed 35 metabolites, primarily flavonoids and triterpenoid saponins, with Astragaloside IV highlighted as a diagnostic marker. Atractylodes Macrocephala Koidz: The analysis identified 13 metabolites, predominantly polysaccharides and sesquiterpenes (e.g., atractylenolides), indicative of this botanical drug. Citrus Reticulata and Citri Reticulatae Pericarpium Viride: These related samples contained 44 metabolites, predominantly flavonoids with a minor presence of limonoids (e.g., limonin). Bupleuri Radix: The composition analysis indicated 29 metabolites, with saikosaponins (e.g., saikosaponin A) as the primary markers. Scutellariae Radix: This sample was characterized by 24 flavonoid metabolites, particularly flavonoid glucuronides (e.g., baicalin, wogonoside). Ligustrum Lucidum Fructus: The analysis yielded 8 metabolites, notably iridoids (e.g., specnuezhenide) as characteristic markers. Folium Nelumbinis: This sample contained 23 metabolites, including both flavonoids and alkaloids. Alismatis Rhizoma: Identified 12 triterpenic acids, serving as characteristic metabolites. Additional botanical drugs: Fructus Liquidambaris and Arum Ternatum Thunb predominantly contained primary metabolites such as amino acids and nucleosides, lacking distinctive markers. Sedi Herba revealed 15 metabolites primarily comprising flavonoids and monoterpene glycosides. Crataegi Folium yielded 27 metabolites, dominated by flavonoids and phenylpropanoids.
The literature review reveals a substantial association between XHP and obesity: XHP comprises 14 botanical drugs. In TCM formulations, the compatibility of botanical drugs is essential. Within XHP, Codonopsis Radix, Hedysarum Multijugum Maxim, and Atractylodes macrocephala Koidz function as the “monarch drugs” (Jun), forming the core of the formula. The remaining nine botanical drugs are categorized as “minister” (Chen), “assistant” (Zuo), and “guide” (Shi) drugs, which hierarchically support the monarch drugs to augment therapeutic efficacy. Figure 5A illustrates the proportions of the botanical drugs in XHP, with color coding denoting their roles in the TCM formulation. A search of CNKI (https://www.cnki.net) and PubMed (https://pubmed.ncbi.nlm.nih.gov) regarding the association of botanical drugs in XHP with obesity shows that all are implicated in obesity treatment to varying extents. Figure 5B and Supplementary Table 4 display the number of literature references related to obesity for these botanical drugs, while Figure 5C depicts the frequency of occurrence of XHP metabolites in each herb.
Serum metabolites identification of XHP: Based on in vitro UPLC analysis, the extraction of blood entry metabolites was performed. The analysis detected a total of 23 metabolites in rat plasma, with flavonoids being the most absorbed type of metabolites in vivo. The specific distribution of their prototype metabolites is presented in Supplementary Table 5. Nineteen of these metabolites are listed in PubChem and are included in subsequent network pharmacological analyses, and Figure 5D illustrates their structural composition.
Network pharmacological analyses of XHPS: A total of 527 putative targets associated with 19 XHPS metabolites were identified and analyzed using various modular approaches. The relationship between the 19 XHPS metabolites and the 527 putative targets is outlined in Figure 5E. Following the construction of the PPI network and the decomposition using the MCODE model, GO and KEGG pathway enrichment analyses were conducted. The PPI network identifies AKT1, TNF, and ALB as potential hub-targets of XHPS (Figure 5F). The PPI network is subsequently divided into seven clusters (Figure 5G, each containing more than ten nodes). Functional analyses (Figure 5H-K) suggest that cluster 1 may represent the immunological module, involving pathways such as the “IL-17 signaling pathway” and “Th17 cell differentiation”. Meanwhile, cluster 2 appears to be the lipid metabolism module, involving pathways like the “AMPK signaling pathway”, “Insulin signaling pathway”, and “PI3K-AKT signaling pathway”. Collectively, these network pharmacological analyses of XHPS confirm a significant correlation with obesity.
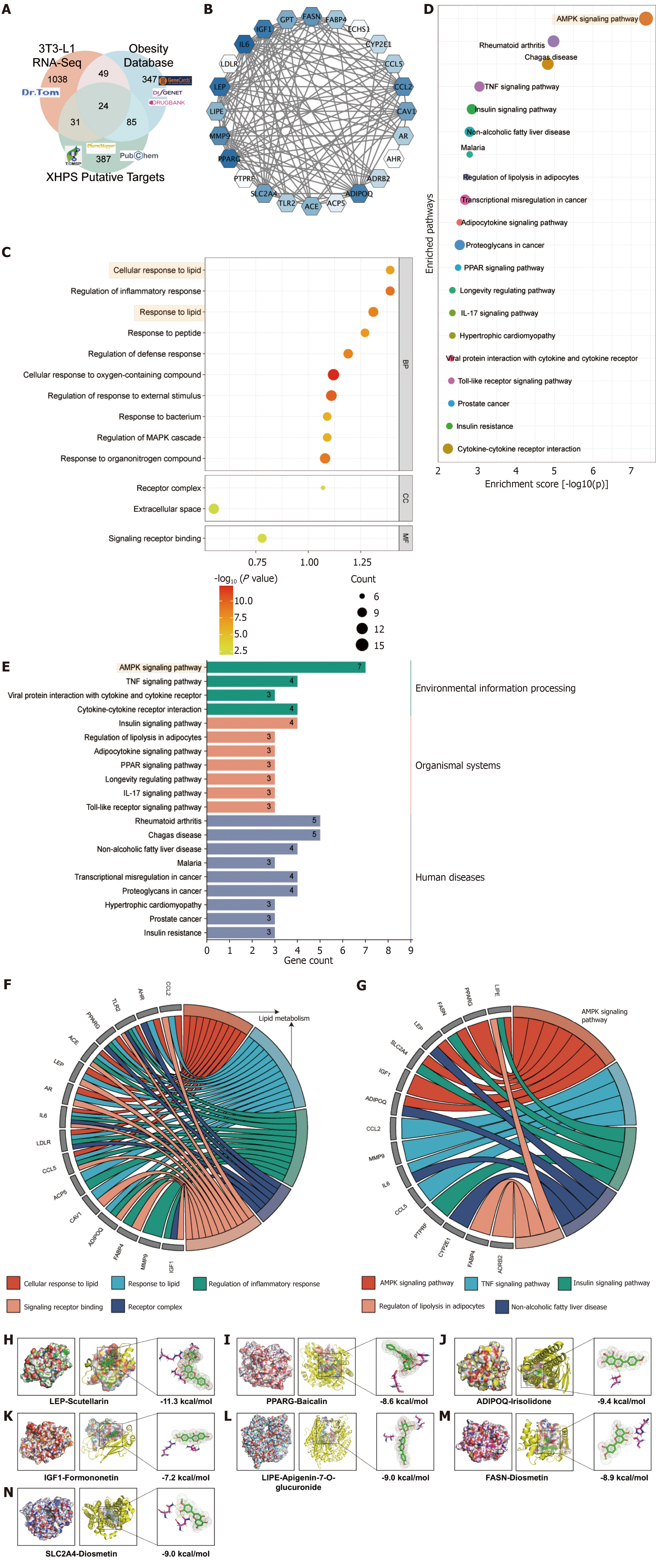
Data integration identifies 24 core targets: To further elucidate the signaling pathways by which XHP regulates lipid metabolism, databases related to disease targets, including DisGeNET, GeneCards, and DrugBank, were employed to assemble a collection of obesity-related genes. By intersecting these obesity-related genes with DEGs exhibiting a |Log2FC| of at least 2 from 3T3-L1 cell RNA-seq data and putative targets of XHP, 24 genes were pinpointed as poten
Big data analysis predicts the AMPK signaling pathway as the central mechanism of XHP treatment: Functional enri
To further investigate the direct and indirect targets of XHP, the top 20 KEGG pathways were visualized, emphasizing the pathway categories and their associated core genes. The AMPK signaling pathway was noted for containing the highest number of core genes (Figure 6E). Figure 6F and G delineate the relationships among all top GO biological process terms, KEGG pathways, and core genes. Notably, the AMPK signaling pathway implicates key genes such as PPARG, FASN, IGF1, SLC2A4 (also known as GLUT4), LEP, LIPE, and ADIPOQ, all of which require further validation to elucidate their interactions with XHP.
Molecular docking suggests LEP as a candidate core target of XHP on the AMPK signaling pathway: To evaluate the binding affinity and adaptability between candidate targets and their corresponding metabolites in XHP, molecular docking simulations were conducted using Autodock Vina 1.2.2. Based on bioinformatics enrichment analysis, it is postulated that XHP inhibits the differentiation and maturation of white adipose tissue via the AMPK signaling pathway. Consequently, all predicted targets of XHP within this pathway were selected for docking with their corresponding metabolites in the formula. PPARG, identified as the hub target, was associated with the highest number of corresponding metabolites (six), whereas FASN, IGF1, SLC2A4 (also known as GLUT4), LEP, LIPE, and ADIPOQ were each associated with one, one, one, two, two, and one metabolite(s), respectively. A docking score threshold of ≤ -5.0 was employed to define successful docking, resulting in the identification of 14 successful docking pairs. These pairs demonstrated binding through visible hydrogen bonds and robust electrostatic interactions. The detailed docking scores are presented in Supplementary Table 6. Figure 6H-N and Supplementary Table 7 illustrate seven representative docking pairs with highly stable binding conformations. Notably, the pairs “scutellarin to LEP” and “diosmetin to ADIPOQ” exhibited binding energies of -11.3 kcal/mol and -10.5 kcal/mol, respectively, signifying the most stable interactions. These findings suggest that XHP may modulate the AMPK signaling pathway cascade through LEP, potentially sup
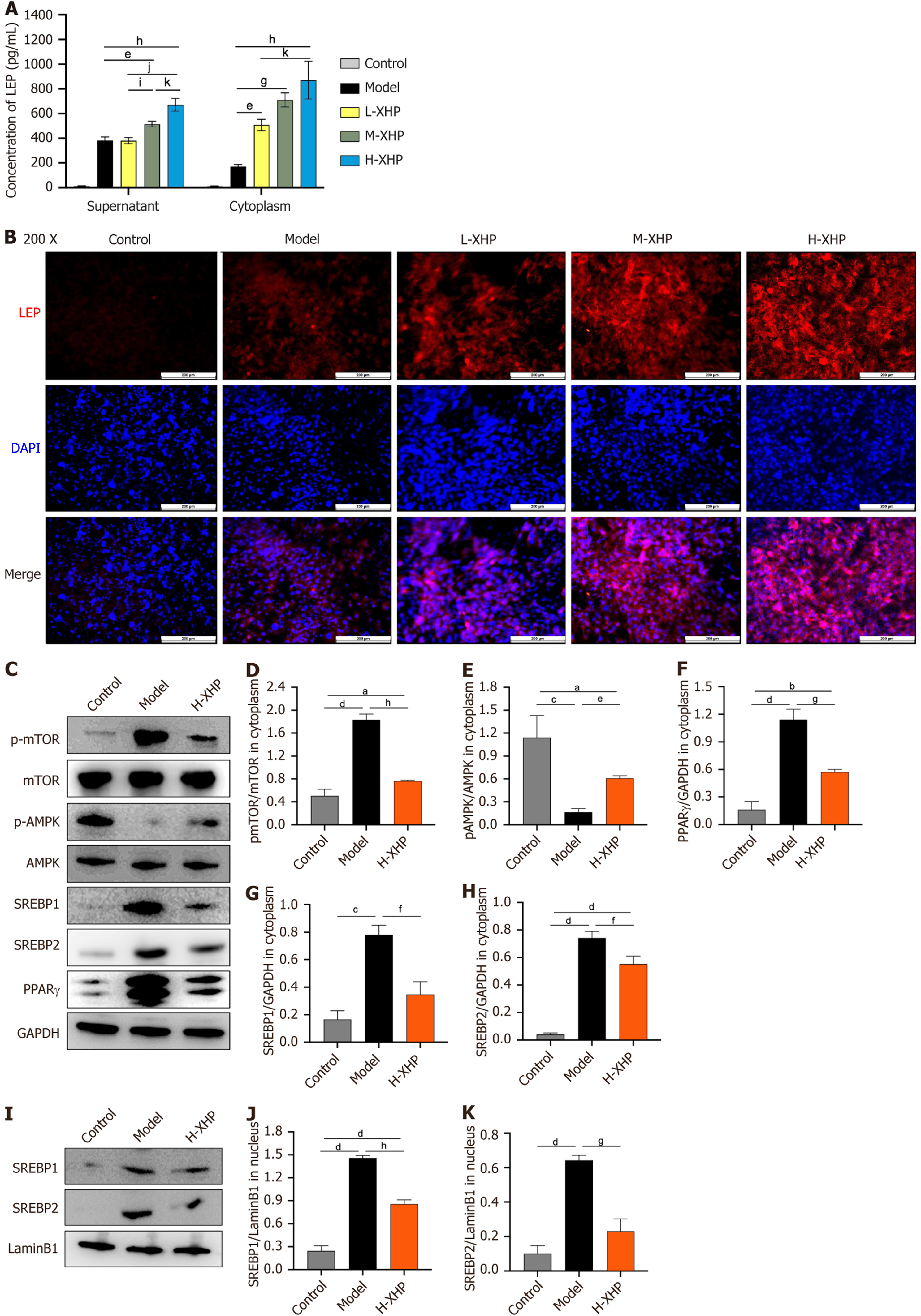
XHP activates AMPK phosphorylation by promoting LEP release during 3T3-L1 lipogenesis: Based on Figure 3, which demonstrated that XHP effectively inhibited lipogenesis and the synthesis of TC and TG in 3T3-L1 cells on day 10, the experimental conditions described in this section were replicated for mechanism validation. An ELISA assay was initially conducted to measure LEP levels. Since the 3T3-L1 cells in the control group were in an undifferentiated state, leptin secretion was virtually undetectable. Figure 7A indicates that XHP dose-dependently increased leptin secretion compared to the model group, with a significant rise in both intracellular and supernatant LEP levels, and a statistically significant increase in these levels in the H-XHP group compared to the L-XHP group. LEP-associated immunofluorescence staining of 3T3-L1 cell specimens revealed that XHP dose-dependently stimulated LEP protein expression (Figure 7B). Cyto
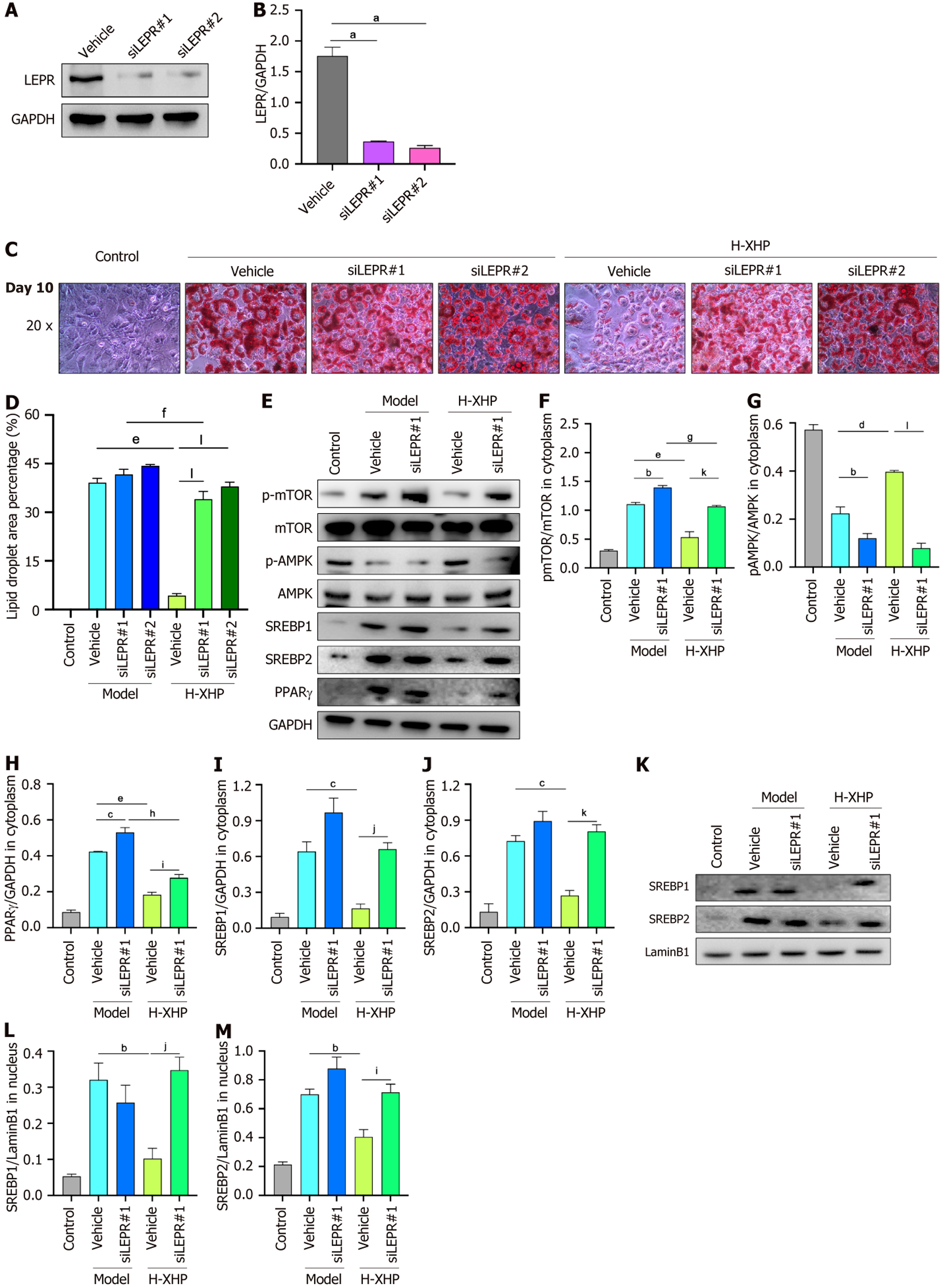
Inhibitory effect of H-XHP on lipogenesis in 3T3-L1 cells disappears after disruption of LEP-LEPR binding accom
Figure 8C and D shows that, in the absence of H-XHP (the three model groups), the size of lipid droplets in equivalent fields of view was marginally and not significantly different in the siLEPR1 and siLEPR2 cell lines compared to the vehicle control lines. Conversely, in the H-XHP-treated cells (the three H-XHP groups), lipid droplets were substantially reduced in the vehicle control lines compared to those in siLEPR1 and siLEPR2, suggesting that LEP-LEPR interaction plays a crucial role in H-XHP’s therapeutic efficacy. Subsequent immunoblotting of control, vehicle, and siLEPR1 cell lines revealed significant alterations: A decrease in the ratio of p-AMPK to total AMPK, an increase in the ratio of p-mTOR to total mTOR, and in PPARγ levels in the model-siLEPR1 group compared to the model-vehicle group. While SREBP1/2 increased slightly, this difference was not statistically significant. However, these changes were more pronounced in the H-XHP-Vehicle group compared to the H-XHP-siLEPR1 group, indicating that leptin secretion crucially influences lipidogenic differentiation in 3T3-L1 cells, and that the therapeutic effect of H-XHP is mediated through the interaction of LEP with LEPR, as shown in Figure 8E-J. Further immunoblotting of nuclear proteins showed that the expression trends of SREBP1/2 in the nucleus mirrored those of their cytoplasmic counterparts (Figure 8K-M), suggesting that the inhibitory action of H-XHP on the synthesis of nuclear SREBP1/2 precursors is also mediated by its impact on leptin secretion.
The 3T3-L1 cell line, recognized for its established use and widespread acceptance as an in vitro model for adipocyte differentiation, replicates a transcriptional profile highly akin to that of human adipose tissue. This model can be reliably induced to differentiate into white adipocytes, rendering it invaluable for phenotypic and molecular studies of adi
In our investigations, MDI-induced differentiation of 3T3-L1 cells manifested pronounced adipogenic characteristics, as evidenced by morphological alterations and the upregulated expression of lipid metabolism markers such as PPARγ, C/EBPα, FABP4, and SREBP1. These observations affirm the efficacy and reliability of the 3T3-L1 model for assessing the anti-obesity effects of XHP in vitro. Transcriptomic analysis of 3T3-L1 cells identified distinctive gene expression profiles between the control and model groups subsequently. The ten most highly expressed genes in the model group included Thrsp, Map7d2, Fam107a, Mrap, Ang5, Cyp2e1, Retnla, Pnpla3, Apoe3, and Slc22a20, while the control group exhibited elevated expression of genes such as Cxcl5, Sfrp2, Kif26b, Fgf23, Ndst3, Tgtp2, Sorbs2, Ch25h, Cpz, and Inmt. Given that the cells of the control group remained undifferentiated, their gene expression profile may not accurately represent the adipose phenotype of lean individuals. Conversely, the differentiated state of the model group provides a more representative basis for modeling obese phenotypes, thus rendering the exploration of highly expressed genes in the model group particularly pertinent. Several of these DEGs have been previously associated with obesity and lipid metabolism, meriting further exploration and discussion.
Pnpla3 has been well-established as a genetic determinant of fatty liver disease and a heritable risk factor for MASLD and metabolic syndrome. Specific alleles of Pnpla3 are significantly associated with an increased risk of liver-related mortality[27]. Cyp2e1 is a cytochrome P450 enzyme implicated in the decomposition and browning of white fat, as well as in the pathogenesis of fatty liver disease[28,29]. Notably, a deficiency in Cyp2e1 activates the expression of hepatic peri-PPARα target genes, including FGF21, which are secreted by the liver to enhance fat browning and energy expenditure, thereby reducing obesity[28]. Ang5 and Apoe3 both play roles in the regulation of lipoproteins. Thrsp is highly expressed in conditions of obesity and non-alcoholic fatty liver disease. Studies employing Thrsp-deficient mice have highlighted the role of Thrsp in adipogenesis and obesity[30]. However, despite enhanced adipogenic differentiation, Thrsp expression was paradoxically downregulated in obese individuals, indicating a potentially complex role in obesity that merits further investigation[31]. In contrast, the genes Map7d2, Fam107a, Mrap, Retnla, and Slc22a20 have been less extensively studied in the context of obesity. These genes may represent novel targets for future research and potential therapeutic interventions. Together, these DEGs constitute the phenotypic basis of adipocyte differentiation and offer potential molecular targets for XHP-targeted interventions.
XHP, a multi-metabolite, multi-target TCM prescription, has demonstrated considerable therapeutic effects in a 3T3-L1 cell-based obesity model. In contrast, GW9662, a known PPARγ antagonist, also exhibited inhibitory effects on adi
LEP, a hormone secreted by adipocytes, regulates energy metabolism via hypothalamic centers. It has been de
In the obese state, tissue AMPK signaling pathway activity tends to decrease, with the inhibitory effect being particularly pronounced in models of high-fat diet-induced obesity[45,46]. Reduced AMPK activity significantly diminishes fatty acid oxidation, which in turn, contributes to the development of lipid accumulation and insulin resistance[47]. In the central nervous system, LEP exerts its appetite-suppressing effects by specifically inhibiting AMPK activity in the hypothalamus[48]. In peripheral tissues, LEP’s modulation of lipid metabolism is also closely linked with AMPK signaling. For instance, in hepatic tissues, LEP and AMPK collaboratively inhibit lipogenesis by synergistically sup
During the conclusive experiments for mechanism validation, XHP was observed to significantly enhance leptin secretion levels in both the cytoplasm and culture supernatants. Additionally, the si-LEPR interference assay substantiated that LEP-LEPR binding is essential for XHP’s efficacy, indicating that XHP may impede white adipose production by fostering LEP autocrine secretion and activating AMPK, thereby down-regulating the expression of PPARγ within the adipogenic pathway, including SREBP1/2. XHP modulates energy metabolism and fat deposition across individuals via the LEP and AMPK pathways. The role of XHP in inhibiting leukocyte adipogenesis through the LEP/AMPK/PPARγ pathway provides an experimental basis for the clinical advancement of XHP in the prevention and treatment of obesity and MASLD.
There are also some limitations to the research. Although this paper was based on experiments in vitro that found XHP promotes leptin secretion, investigations into whether XHP improves LEP sensitivity in obese individuals remain unexplored, marking a limitation that will be addressed in forthcoming in vivo experiments. Additionally, given that the LEP/AMPK/PPARγ pathway is implicated in the distribution of visceral and subcutaneous adipose tissue as well as in adipose browning, it is imperative that the anti-obesity effects of XHP be validated in vivo with a focus on these specific aspects. Furthermore, prior clinical studies have demonstrated that XHP improves key obesity-related parameters and established MASLD surrogates, with a trend toward enhanced insulin sensitivity (P = 0.06; n = 262). These XHP-mediated improvements warrant further validation in animal models to determine whether their changes are associated with the LEP/AMPK/PPARγ signaling pathway under XHP stimulation. Finally, given the established link between insulin sensitivity and type 2 diabetes mellitus (T2DM), the potential of XHP as a therapeutic intervention for obesity-related T2DM also merits further investigation.
In summary, this integrative multi-omics investigation definitively elucidates the molecular mechanism underpinning XHP’s efficacy against obesity, establishing the LEP/AMPK/PPARγ signaling axis as its central therapeutic pathway. We demonstrate that XHP orchestrates its anti-adipogenic effects not through broad metabolic suppression but by specifically augmenting autocrine leptin secretion, which serves as the critical initiator for AMPK phosphorylation. This activation instigates a downstream cascade that transcriptionally represses the master regulators of adipogenesis, PPARγ and SREBPs, thereby attenuating lipid droplet accumulation and restoring metabolic homeostasis. The unequivocal abolition of XHP’s therapeutic impact upon LEPR knockdown provides conclusive evidence of the pathway’s indispensability, positioning XHP as a sophisticated, multi-targeted therapeutic strategy that counteracts MASLD-related obesity by directly targeting the foundational process of lipotoxicity.
| 1. | Tacke F, Puengel T, Loomba R, Friedman SL. An integrated view of anti-inflammatory and antifibrotic targets for the treatment of NASH. J Hepatol. 2023;79:552-566. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 178] [Cited by in RCA: 224] [Article Influence: 74.7] [Reference Citation Analysis (0)] |
| 2. | Ng M, Fleming T, Robinson M, Thomson B, Graetz N, Margono C, Mullany EC, Biryukov S, Abbafati C, Abera SF, Abraham JP, Abu-Rmeileh NM, Achoki T, AlBuhairan FS, Alemu ZA, Alfonso R, Ali MK, Ali R, Guzman NA, Ammar W, Anwari P, Banerjee A, Barquera S, Basu S, Bennett DA, Bhutta Z, Blore J, Cabral N, Nonato IC, Chang JC, Chowdhury R, Courville KJ, Criqui MH, Cundiff DK, Dabhadkar KC, Dandona L, Davis A, Dayama A, Dharmaratne SD, Ding EL, Durrani AM, Esteghamati A, Farzadfar F, Fay DF, Feigin VL, Flaxman A, Forouzanfar MH, Goto A, Green MA, Gupta R, Hafezi-Nejad N, Hankey GJ, Harewood HC, Havmoeller R, Hay S, Hernandez L, Husseini A, Idrisov BT, Ikeda N, Islami F, Jahangir E, Jassal SK, Jee SH, Jeffreys M, Jonas JB, Kabagambe EK, Khalifa SE, Kengne AP, Khader YS, Khang YH, Kim D, Kimokoti RW, Kinge JM, Kokubo Y, Kosen S, Kwan G, Lai T, Leinsalu M, Li Y, Liang X, Liu S, Logroscino G, Lotufo PA, Lu Y, Ma J, Mainoo NK, Mensah GA, Merriman TR, Mokdad AH, Moschandreas J, Naghavi M, Naheed A, Nand D, Narayan KM, Nelson EL, Neuhouser ML, Nisar MI, Ohkubo T, Oti SO, Pedroza A, Prabhakaran D, Roy N, Sampson U, Seo H, Sepanlou SG, Shibuya K, Shiri R, Shiue I, Singh GM, Singh JA, Skirbekk V, Stapelberg NJ, Sturua L, Sykes BL, Tobias M, Tran BX, Trasande L, Toyoshima H, van de Vijver S, Vasankari TJ, Veerman JL, Velasquez-Melendez G, Vlassov VV, Vollset SE, Vos T, Wang C, Wang X, Weiderpass E, Werdecker A, Wright JL, Yang YC, Yatsuya H, Yoon J, Yoon SJ, Zhao Y, Zhou M, Zhu S, Lopez AD, Murray CJ, Gakidou E. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2014;384:766-781. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7951] [Cited by in RCA: 8179] [Article Influence: 681.6] [Reference Citation Analysis (0)] |
| 3. | Polyzos SA, Kountouras J, Mantzoros CS. Obesity and nonalcoholic fatty liver disease: From pathophysiology to therapeutics. Metabolism. 2019;92:82-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 453] [Cited by in RCA: 887] [Article Influence: 126.7] [Reference Citation Analysis (0)] |
| 4. | Gallardo-Montejano VI, Saxena G, Kusminski CM, Yang C, McAfee JL, Hahner L, Hoch K, Dubinsky W, Narkar VA, Bickel PE. Nuclear Perilipin 5 integrates lipid droplet lipolysis with PGC-1α/SIRT1-dependent transcriptional regulation of mitochondrial function. Nat Commun. 2016;7:12723. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 131] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 5. | Wei D, Sun Q, Li Y, Li C, Li X, Sun C. Leptin Reduces Plin5 m(6)A Methylation through FTO to Regulate Lipolysis in Piglets. Int J Mol Sci. 2021;22:10610. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 6. | Coia H, Ma N, He AR, Kallakury B, Berry DL, Permaul E, Makambi KH, Fu Y, Chung FL. Detection of a lipid peroxidation-induced DNA adduct across liver disease stages. Hepatobiliary Surg Nutr. 2018;7:85-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 7. | Younossi ZM, Felix S, Jeffers T, Younossi E, Nader F, Pham H, Afendy A, Cable R, Racila A, Younoszai Z, Lam BP, Golabi P, Henry L, Stepanova M. Performance of the Enhanced Liver Fibrosis Test to Estimate Advanced Fibrosis Among Patients With Nonalcoholic Fatty Liver Disease. JAMA Netw Open. 2021;4:e2123923. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 55] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 8. | Alferink LJM, Erler NS, de Knegt RJ, Janssen HLA, Metselaar HJ, Darwish Murad S, Kiefte-de Jong JC. Adherence to a plant-based, high-fibre dietary pattern is related to regression of non-alcoholic fatty liver disease in an elderly population. Eur J Epidemiol. 2020;35:1069-1085. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 9. | Xiang LL, Cao YT, Wang XX, Wang GX, Zhang YJ, Li RH, Qi F, Huai JX, Sun J, He XJ, Zhou XQ. Xietu Hemu Prescription Improves Metabolic Dysfunction-Associated Steatotic Liver Disease: A Real-World Cohort Study. J Multidiscip Healthc. 2025;18:4377-4389. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 10. | Liu X, Pan F, Sha C, Wang Z, Liu G, Wang H, Ling S, Huang K. Fuzi decoction ameliorates intervertebral disc degeneration through ferroptosis modulation by suppressing NF-κB pathway. Int Immunopharmacol. 2025;148:114155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 11. | Wang M, Yang B, Li X, Ma H, Ou X, Dawa Z, Liu F, Zhu C, Lin C. Brunodelphinine A alleviates non-alcoholic fatty liver disease by inhibiting oxidative stress and regulating lipid metabolism via NOX4/SIRT1/PPARs axis. Phytomedicine. 2025;147:157202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 12. | Yu HC, Park SH, Jo HJ, Sim H, Lee MR, Kim G, Park SY, Lee Y, Bae EJ, Park BH. PAK4 phosphorylates cyclin-dependent kinase 2 to promote the G(1)/S transition during adipogenesis. Exp Mol Med. 2025;57:2121-2132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 13. | Ru J, Li P, Wang J, Zhou W, Li B, Huang C, Li P, Guo Z, Tao W, Yang Y, Xu X, Li Y, Wang Y, Yang L. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1623] [Cited by in RCA: 3443] [Article Influence: 286.9] [Reference Citation Analysis (0)] |
| 14. | Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA, Wang J, Yu B, Zhang J, Bryant SH. PubChem Substance and Compound databases. Nucleic Acids Res. 2016;44:D1202-D1213. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3081] [Cited by in RCA: 3338] [Article Influence: 333.8] [Reference Citation Analysis (0)] |
| 15. | Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, Assempour N, Iynkkaran I, Liu Y, Maciejewski A, Gale N, Wilson A, Chin L, Cummings R, Le D, Pon A, Knox C, Wilson M. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074-D1082. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3266] [Cited by in RCA: 5635] [Article Influence: 805.0] [Reference Citation Analysis (0)] |
| 16. | Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, Ronzano F, Centeno E, Sanz F, Furlong LI. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020;48:D845-D855. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 328] [Cited by in RCA: 1204] [Article Influence: 200.7] [Reference Citation Analysis (0)] |
| 17. | Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, Kaplan S, Dahary D, Warshawsky D, Guan-Golan Y, Kohn A, Rappaport N, Safran M, Lancet D. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr Protoc Bioinformatics. 2016;54:1.30.1-1.30.33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1289] [Cited by in RCA: 3424] [Article Influence: 342.4] [Reference Citation Analysis (0)] |
| 18. | Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607-D613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10161] [Cited by in RCA: 12506] [Article Influence: 1786.6] [Reference Citation Analysis (1)] |
| 19. | Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498-2504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24663] [Cited by in RCA: 35925] [Article Influence: 1633.0] [Reference Citation Analysis (7)] |
| 20. | Burley SK, Berman HM, Bhikadiya C, Bi C, Chen L, Di Costanzo L, Christie C, Dalenberg K, Duarte JM, Dutta S, Feng Z, Ghosh S, Goodsell DS, Green RK, Guranovic V, Guzenko D, Hudson BP, Kalro T, Liang Y, Lowe R, Namkoong H, Peisach E, Periskova I, Prlic A, Randle C, Rose A, Rose P, Sala R, Sekharan M, Shao C, Tan L, Tao YP, Valasatava Y, Voigt M, Westbrook J, Woo J, Yang H, Young J, Zhuravleva M, Zardecki C. RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019;47:D464-D474. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 677] [Cited by in RCA: 858] [Article Influence: 143.0] [Reference Citation Analysis (0)] |
| 21. | Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455-461. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26714] [Cited by in RCA: 16490] [Article Influence: 1030.6] [Reference Citation Analysis (12)] |
| 22. | Wang J, Zhang T, Gu R, Ke Y, Zhang S, Su X, Pan X, He Q, Li G, Zhang Z, Zhang L, Li J, Wu W, Chen C. Development and Evaluation of Reconstructed Nanovesicles from Turmeric for Multifaceted Obesity Intervention. ACS Nano. 2024;18:23117-23135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 26] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 23. | Wan X, Wang L, Khan MA, Peng L, Sun X, Yi X, Wang Z, Chen K. NAT10-mediated N4-acetylcytidine modification in KLF9 mRNA promotes adipogenesis. Cell Death Differ. 2025;32:1613-1629. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 24. | Sun Z, Zhang S, Liang J, Li C, Yang X, Liu QS, Zhou Q, Shi J, Zhao B, Jiang G. Effects of multiple novel bisphenol S analogs on adipogenesis in 3T3-L1 cells. J Hazard Mater. 2025;489:137689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 25. | Sun W, Yu Z, Yang S, Jiang C, Kou Y, Xiao L, Tang S, Zhu T. A Transcriptomic Analysis Reveals Novel Patterns of Gene Expression During 3T3-L1 Adipocyte Differentiation. Front Mol Biosci. 2020;7:564339. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 26. | Briand N, Prado C, Mabilleau G, Lasnier F, Le Lièpvre X, Covington JD, Ravussin E, Le Lay S, Dugail I. Caveolin-1 expression and cavin stability regulate caveolae dynamics in adipocyte lipid store fluctuation. Diabetes. 2014;63:4032-4044. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 61] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 27. | Vilar-Gomez E, Gawrieh S, Vuppalanchi R, Kettler C, Pike F, Samala N, Chalasani N. PNPLA3 rs738409, environmental factors and liver-related mortality in the US population. J Hepatol. 2025;82:571-581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 28. | Zhang Y, Yan T, Wang T, Liu X, Hamada K, Sun D, Sun Y, Yang Y, Wang J, Takahashi S, Wang Q, Krausz KW, Jiang C, Xie C, Yang X, Gonzalez FJ. Crosstalk between CYP2E1 and PPARα substrates and agonists modulate adipose browning and obesity. Acta Pharm Sin B. 2022;12:2224-2238. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 33] [Reference Citation Analysis (0)] |
| 29. | Zhu Y, Wang Y, Hoshitsuki K, Yang D, Kokai L, Ma X, Xie W, Fernandez CA. Induction of Cyp2e1 contributes to asparaginase-induced hepatocyte sensitization to lipotoxicity. Acta Pharm Sin B. 2025;15:963-972. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 30. | Anderson GW, Zhu Q, Metkowski J, Stack MJ, Gopinath S, Mariash CN. The Thrsp null mouse (Thrsp(tm1cnm)) and diet-induced obesity. Mol Cell Endocrinol. 2009;302:99-107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 38] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 31. | Ortega FJ, Vazquez-Martin A, Moreno-Navarrete JM, Bassols J, Rodriguez-Hermosa J, Gironés J, Ricart W, Peral B, Tinahones FJ, Fruhbeck G, Menendez JA, Fernández-Real JM. Thyroid hormone responsive Spot 14 increases during differentiation of human adipocytes and its expression is down-regulated in obese subjects. Int J Obes (Lond). 2010;34:487-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 32. | Vella KR, Ramadoss P, Lam FS, Harris JC, Ye FD, Same PD, O'Neill NF, Maratos-Flier E, Hollenberg AN. NPY and MC4R signaling regulate thyroid hormone levels during fasting through both central and peripheral pathways. Cell Metab. 2011;14:780-790. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 87] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 33. | Haberman ER, Sarker G, Arús BA, Ziegler KA, Meunier S, Martínez-Sánchez N, Freibergerová E, Yilmaz-Özcan S, Fernández-González I, Zentai C, O'Brien CJO, Grainger DE, Sidarta-Oliveira D, Chakarov S, Raimondi A, Iannacone M, Engelhardt S, López M, Ginhoux F, Domingos AI. Immunomodulatory leptin receptor(+) sympathetic perineurial barrier cells protect against obesity by facilitating brown adipose tissue thermogenesis. Immunity. 2024;57:141-152.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 15] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 34. | Mullur R, Liu YY, Brent GA. Thyroid hormone regulation of metabolism. Physiol Rev. 2014;94:355-382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 965] [Cited by in RCA: 1522] [Article Influence: 126.8] [Reference Citation Analysis (0)] |
| 35. | Mollé N, Krichevsky S, Kermani P, Silver RT, Ritchie E, Scandura JM. Ruxolitinib can cause weight gain by blocking leptin signaling in the brain via JAK2/STAT3. Blood. 2020;135:1062-1066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 36. | Morton GJ, Blevins JE, Williams DL, Niswender KD, Gelling RW, Rhodes CJ, Baskin DG, Schwartz MW. Leptin action in the forebrain regulates the hindbrain response to satiety signals. J Clin Invest. 2005;115:703-710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 100] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 37. | Minokoshi Y, Kim YB, Peroni OD, Fryer LG, Müller C, Carling D, Kahn BB. Leptin stimulates fatty-acid oxidation by activating AMP-activated protein kinase. Nature. 2002;415:339-343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1554] [Cited by in RCA: 1480] [Article Influence: 61.7] [Reference Citation Analysis (0)] |
| 38. | Folgueira C, Beiroa D, González-Rellán MJ, Porteiro B, Milbank E, Castelao C, García-Palacios M, Casanueva FF, López M, Diéguez C, Seoane LM, Nogueiras R. Uroguanylin Improves Leptin Responsiveness in Diet-Induced Obese Mice. Nutrients. 2019;11:752. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 39. | Harrison L, Schriever SC, Feuchtinger A, Kyriakou E, Baumann P, Pfuhlmann K, Messias AC, Walch A, Tschöp MH, Pfluger PT. Fluorescent blood-brain barrier tracing shows intact leptin transport in obese mice. Int J Obes (Lond). 2019;43:1305-1318. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 72] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 40. | Howard JK, Cave BJ, Oksanen LJ, Tzameli I, Bjørbaek C, Flier JS. Enhanced leptin sensitivity and attenuation of diet-induced obesity in mice with haploinsufficiency of Socs3. Nat Med. 2004;10:734-738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 358] [Cited by in RCA: 372] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 41. | Münzberg H, Myers MG Jr. Molecular and anatomical determinants of central leptin resistance. Nat Neurosci. 2005;8:566-570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 371] [Cited by in RCA: 368] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 42. | Ardid-Ruiz A, Ibars M, Mena P, Del Rio D, Muguerza B, Bladé C, Arola L, Aragonès G, Suárez M. Potential Involvement of Peripheral Leptin/STAT3 Signaling in the Effects of Resveratrol and Its Metabolites on Reducing Body Fat Accumulation. Nutrients. 2018;10:1757. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 43. | Bourebaba L, Kornicka-Garbowska K, Al Naem M, Röcken M, Łyczko J, Marycz K. MSI-1436 improves EMS adipose derived progenitor stem cells in the course of adipogenic differentiation through modulation of ER stress, apoptosis, and oxidative stress. Stem Cell Res Ther. 2021;12:97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 44. | Jayashankar V, Selwan E, Hancock SE, Verlande A, Goodson MO, Eckenstein KH, Milinkeviciute G, Hoover BM, Chen B, Fleischman AG, Cramer KS, Hanessian S, Masri S, Turner N, Edinger AL. Drug-like sphingolipid SH-BC-893 opposes ceramide-induced mitochondrial fission and corrects diet-induced obesity. EMBO Mol Med. 2021;13:e13086. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 45. | Fabelo C, Hernandez J, Chang R, Seng S, Alicea N, Tian S, Conde K, Wagner EJ. Endocannabinoid Signaling at Hypothalamic Steroidogenic Factor-1/Proopiomelanocortin Synapses Is Sex- and Diet-Sensitive. Front Mol Neurosci. 2018;11:214. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 46. | Morrow NM, Burke AC, Samsoondar JP, Seigel KE, Wang A, Telford DE, Sutherland BG, O'Dwyer C, Steinberg GR, Fullerton MD, Huff MW. The citrus flavonoid nobiletin confers protection from metabolic dysregulation in high-fat-fed mice independent of AMPK. J Lipid Res. 2020;61:387-402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 47. | Herzig S, Shaw RJ. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat Rev Mol Cell Biol. 2018;19:121-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1378] [Cited by in RCA: 3051] [Article Influence: 339.0] [Reference Citation Analysis (0)] |
| 48. | Chen YC, Chien CY, Hsu CC, Lee CH, Chou YT, Shiah SG, Liu SY, Yen CY, Hsieh AC, Wabitsch M, Shieh YS. Obesity-associated leptin promotes chemoresistance in colorectal cancer through YAP-dependent AXL upregulation. Am J Cancer Res. 2021;11:4220-4240. [PubMed] |
| 49. | Gong Z, Han S, Li C, Meng T, Huo Y, Liu X, Huang Y, Yang L. Rhinacanthin C Ameliorates Insulin Resistance and Lipid Accumulation in NAFLD Mice via the AMPK/SIRT1 and SREBP-1c/FAS/ACC Signaling Pathways. Evid Based Complement Alternat Med. 2023;2023:6603522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 25] [Reference Citation Analysis (0)] |
| 50. | Ferré P, Phan F, Foufelle F. SREBP-1c and lipogenesis in the liver: an update1. Biochem J. 2021;478:3723-3739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 109] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/