Published online May 27, 2024. doi: 10.4254/wjh.v16.i5.716
Revised: February 14, 2024
Accepted: April 9, 2024
Published online: May 27, 2024
Processing time: 144 Days and 0.1 Hours
Liver cancer, primarily hepatocellular carcinoma, remains a global health challenge with rising incidence and limited therapeutic options. Genetic factors play a pivotal role in the development and progression of liver cancer. This state-of-the-art paper provides a comprehensive review of the current landscape of genetic screening strategies for liver cancer. We discuss the genetic underpinnings of liver cancer, emphasizing the critical role of risk-associated genetic variants, somatic mutations, and epigenetic alterations. We also explore the intricate interplay between environmental factors and genetics, highlighting how genetic screening can aid in risk stratification and early detection via using liquid biopsy, and advancements in high-throughput sequencing technologies. By synthesizing the latest research findings, we aim to provide a comprehensive overview of the state-of-the-art genetic screening methods for liver cancer, shedding light on their potential to revolutionize early detection, risk assessment, and targeted therapies in the fight against this devastating disease.
Core Tip: Unraveling the intricate genetic underpinnings of hepatocellular carcinoma (HCC) is paramount for understanding its development and progression. In line with this, genetic screening could be a powerful tool for patient risk stratification, spotlighting risk-associated genetic variants, somatic mutations, and epigenetic alterations contributing to HCC. Moreover, embracing next-generation sequencing and exploring genetic biomarkers, including circulating tumor DNA, opens new frontiers for effective risk assessment and early detection of liver cancer.
- Citation: Peruhova M, Banova-Chakarova S, Miteva DG, Velikova T. Genetic screening of liver cancer: State of the art. World J Hepatol 2024; 16(5): 716-730
- URL: https://www.wjgnet.com/1948-5182/full/v16/i5/716.htm
- DOI: https://dx.doi.org/10.4254/wjh.v16.i5.716
Primary liver cancer, with hepatocellular carcinoma (HCC) as its predominant form (75% of all cases), remains a for
Globally, primary liver cancer has the highest incidence in Asia and Africa (> 8.4 cases per 100000 person-years). However, HCC incidence declined in many Asian countries but increased in India, America, Oceania and Europe[2].
Due to the intricate interplay of environmental factors contributing to liver carcinogenesis, it becomes increasingly apparent that genetic predisposition plays a pivotal role in shaping individual susceptibility[3]. The significance of genetic screening in the landscape of liver cancer cannot be overstated. Early detection has long been recognized as a linc
However, in the field of genetic screening for liver cancer, questions and challenges emerge. Are the currently available screening methodologies robust enough? What are the limitations and pitfalls of the existing approaches, and how can we overcome them? How do we integrate genetic screening into broader liver cancer prevention and management strategies?
This review critically examines the current state-of-the-art in genetic screening for liver cancer. Beyond providing an overview of prevalence and emphasizing the urgency of early detection, we scrutinize the rationale behind existing gene
It is of great importance for the overall survival of patients with HCC, to be detected at early stages of disease. For example, it has been determined that HCC detection before stage IV, reduces cancer-related deaths by ≥ 15% within 5 years[4]. Unfortunately, the screening tests that are used in clinical practice [ultrasound and serum level of alpha fetoprotein (AFP)] have low sensitivity and specificity[5].
The effectiveness of HCC surveillance depends on whether the screening tests in clinical practice can detect HCC at an early, which is a treatable stage. The recommended screening tests (ultrasound and AFP) are widely available and low cost, but the sensitivity and specificity of these tests are suboptimal. In a noncirrhotic liver, the sensitivity of ultrasound for detecting small HCC lesions is estimated to be only 60%[6].
A new perspective diagnostic tool, called multi-cancer early detection (MCED) has the potential to achieve early cancer detection by using signals for cancers from cell-free DNA (cfDNA) or other circulating analytes in the blood shed by tumors. It has to be underlined that the results from clinical studies of these tests have shown promising results concer
Recently published studies, demonstrated significant sensitivity for early detection of HCC using MCED assays. These methods use cfDNA mutation-based and circulating tumor DNA (ctDNA) methylation-based indicators, both in isolation and in conjunction with cancer-associated serum protein levels[9]. For example, this method is crucial for patients with non-alcoholic fatty liver disease (NAFLD) who may not meet the criteria for routine surveillance due to cost-effectiveness. Another interesting fact that has to be pointed out is that circulating tumor cells (CTCs) are not sufficiently sensitive for early detection of HCC, but can be used as a feasible tool in the surveillance of patients after liver resection, to detect tumor recurrence or tumor progression[10].
We also question their efficacy and explore the gaps in our current understanding, hypothesizing about untapped potentials and areas that demand further exploration. The objectives of this paper are twofold: To comprehensively survey the existing genetic screening landscape for liver cancer and to spur a critical dialogue that propels future research in directions that enhance our ability to combat this formidable adversary. By addressing these essential aspects, we aim to not only present the current state of affairs but also to stimulate further inquiry and innovation in the field of liver cancer genetics.
We conducted an extensive literature review to compile relevant studies on genetic screening for liver cancer. The search was performed in the major databases, including PubMed, Scopus, and Medline. The following combination of MESH and free-text terms was utilized to ensure a comprehensive search: ("hepatocellular carcinoma" OR "liver cancer") AND ("genetic screening" OR "genetic testing" OR "genomic profiling"); ("risk-associated genetic variants" OR "genetic risk factors" OR "genetic predisposition") AND ("liver neoplasms" OR "hepatocellular carcinoma"); ("somatic mutations" OR "tumor mutations" OR "genetic alterations") AND ("liver cancer" OR "hepatocellular carcinoma"); ("epigenetic alterations" OR "DNA methylation" OR "histone modification") AND ("genetic screening" OR "liver cancer"); ("genetic biomarkers" OR "biomolecular markers" OR "genetic signatures") AND ("hepatocellular carcinoma" OR "liver neoplasms"); ("circulating tumor DNA" OR "liquid biopsy" OR "ctDNA") AND ("genetic screening" OR "liver cancer"); ("next-generation sequencing" OR "NGS" OR "genomic sequencing") AND ("hepatocellular carcinoma" OR "liver cancer").
The retrieved papers were selected based on their relevance to the genetic landscape of liver cancer, emphasizing risk factors, somatic mutations, epigenetic modifications, genetic biomarkers, and advanced sequencing techniques. The search strategy aimed to encompass a broad spectrum of genetic aspects associated with liver cancer, ensuring a comprehensive and up-to-date review.
The most common reasons associated with the development of HCC are hepatitis B (HBV) and hepatitis C (HCV) infection. Other risk factors that are attributed to the evolution of HCC are male gender, heavy alcohol drinking, older age, and some monogenic liver diseases such as hemochromatosis, alpha1-antitrypsin deficiency, and porphyria cutanea tarda[11]. It is essential to underline that transmission of HBV and HCV among family members, in association with other environmental risk factors, could be part of the familial aggregation of liver cancer. For example, in Eastern Asia, the percentage of familial clustering of HCC is significantly higher due to a higher incidence of HBV infection[12,13]. It has to be underlined that family history is a substantial risk factor associated with the development of HCC, even in cases without infections with HBV and HCV[14,15].
A study by Turati et al[16] investigates the role of the family history of HCC in the non-Asian population[16]. The authors established that a family history of liver cancer is a risk factor, independently from the presence of HBV and HCV. It was estimated that families with a positive family history have 2-to 3-fold in their HCC risk. Moreover, in cases with a positive family history of liver cancer and hepatitis B/C, it was estimated that these subjects have 70-fold elevated HCC risk compared to those without a family history and hepatitis[16].
It has to be underlined that the mechanism of development of HCC is based on the accumulation of epigenetic and genetic alterations in hepatocytes. The environmental factors most indirectly, lead to chronic inflammation of liver cells, generating liver disease. For example, different toxins and viruses have the potential directly to induce genomic alteration in cancer-driver genes in hepatocytes. For example, aflatoxin B1, alcohol consumption, or smoking could induce DNA damage, and HBV can directly activate oncogenes in liver cells through viral insertion mutagenesis[17,18].
It has to be pointed out that the development of severe chronic liver disease or liver cancer is strongly associated with individual background and gene polymorphisms, which can modulate the risk of HCC[19].
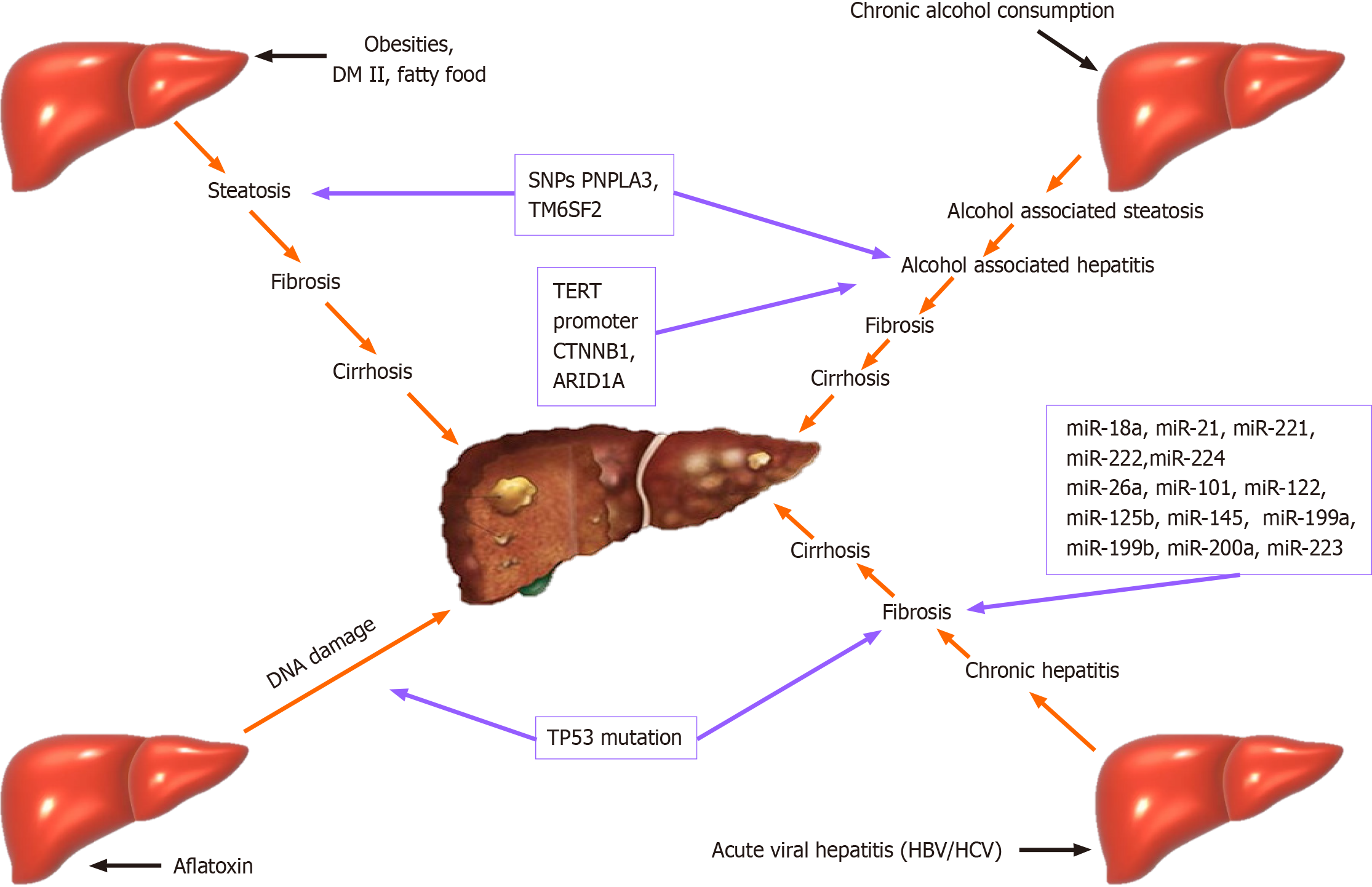
For example, it has been established that genetic polymorphisms in several genes (PNPLA3, TM6SF2, and HSD17B13) that encode proteins involved in lipid metabolism, are strongly connected with modulating the severity of non-alcoholic steatohepatitis (NASH) and alcohol-related chronic liver diseases. Another interesting fact is that these gene polymorphisms also modulate the risk of HCC. associated with either one of these risk factors. Genetic polymorphisms in several genes (PNPLA3, TM6SF2, and HSD17B13) that encode proteins involved in lipid metabolism modulate the severity of NASH and alcohol-related chronic liver diseases. These gene polymorphisms also modulate the risk of HCC[19].
It was established that genetic polymorphisms in WNT3A/WNT9A or in TERT have the potential to modulate the risk of HCC without impacting the process of development of chronic liver disease[20].
In recent years, technological advances have helped elucidate different genetic alterations in genes and signaling pathways that underlie HCC. Understanding these alterations, such as different types of mutations, polymorphisms, genomic instability, and even target genes involved in HCC progression, will contribute to early diagnosis and development of workable target therapies for HCC.
It has been established that multiple genetic changes accumulate slowly in some genetic loci during the early stages of the hepatocarcinogenesis process. With the progressive development of HCC, the accumulation of these changes accelerates[21]. Research has shown that the affected genes in HCC are significant numbers and may be associated with a predisposition, faster progression, worse prognosis, and different signaling pathways[22-25].
All genomic methodologies and technologies that have been used in the past decade, such as genome-wide association studies, microarray analysis, flow cytometry, array comparative genome hybridization, the random amplified polymorphic DNA, Omics profiling, next-generation sequencing (NGS), etc. have improved our understanding of the biology and genetic of HCC.
In 2016, a study revealed the genetic heterogeneity of HCC[26]. It summarizes the genetic changes involved in the progression of HCC. The authors discussed the role of genomic/chromosomal instability and possible associations with clinical and pathological characteristics of HCC. They also summarized single nucleotide polymorphisms (SNPs) associated with HCC susceptibility and risk, frequently recurring somatic mutations and various signaling pathways involved in HCC[26].
Over the years, there is different evidence that mutations in TERT, CTNNB1, TP53, AXIN1, ARID1A, ARID2, NFE2L2, KEAP1, RPS6KA3 and many other genes are associated with HCC[27-32]. Evidence also suggests an association between SNPs and susceptibility to HCC[33-36]. Research has identified various signaling pathways, such as Wnt/β-catenin signaling and PI3K-AKT-mTOR pathway, involved in the HCC development[26,37-40].
All these data show that molecular mechanisms and genetic alterations must be understood well, i.e., the entire mutational profile in HCC, to develop preventive strategies and quality treatment for HCC patients.
Multiple factors are involved in the occurrence of HCC. In developing HCC, the etiological factors and the underlying genetic landscape are essential. Most frequently, HCC develops based on cirrhosis-approximately 90%. Chronic viral hepatitis B and C are found in more than 70% of the cases of primary liver carcinoma. The other risk factors for HCC are NASH, chronic alcohol consumption, and aflatoxin[41]. Each one activates a different oncogenic pathway and has a specific genetic sign.
A plethora of studies recently described genetic alterations that appear in transformed liver tissues[42].
miRNAs (miRs) are small, single-stranded, non-coding RNAs (18-24 nucleotides) that post-transcriptionally regulate the expression of various oncogenes and tumor suppressor genes[43]. For instance, it has been established that many miRs are dysregulated during the process of transformation from HBV-infected liver to HCC. Some are shown in the table below[41,44] (Table 1).
| Upregulated | Downregulated |
| miR-18a | miR-26a |
| miR-21 | miR-101 |
| miR-221 | miR-122 |
| miR-222 | miR-125b |
| miR-224 | miR-145 |
| miR-199a | |
| miR-199b | |
| miR-200a | |
| miR-223 |
Numerous studies investigate different miR panels for early diagnosis of HCC in patients with cirrhosis. Ganesan and Kulik[45] found that a combination of some serologic markers and miR-16 in patients with cirrhosis can help in early diagnosis of liver cancer in 92.4% of cases[45].
The occurrence of HCC is associated with various genetic changes. One of the most common is TERT-alternation. The involvement of several other molecular pathways in hepatocarcinogenesis has also been proven. Schulze et al[46] divided genetic alterations in HCC into eleven categories, including telomere maintenance, Wnt/β-catenin signaling, p53/cell cycle, oxidative stress, epigenetic regulation, PI3K-Akt-mTOR, MAPK, and hepatic differentiation[46]. One of the most often aberrated in HCC are the genes involved in the signaling of Wnt/β-catenin and p53/cell cycle. Based on this fact, it was established that the heterogeneity of HCC is due to several oncogenic pathways being involved in the process of carcinogenesis and not a single mutation[47].
Some specific mutations are found in HBV-related HCC, like-TP53 mutation. The same mutation is found in patients with liver cancer who are exposed to aflatoxin B1[48]. In patients with alcohol liver disease, mutation of TERT promoter C is catenin beta-1, and AT-rich interaction domain 1 is associated with the appearance of HCC[48,49] (Figure 1).
On the other hand, a loss-of-function of some genes like 17-beta-hydroxysteroid dehydrogenase 13 (HSD17B13 rs72613567) was considered protective for HCC in patients with alcoholic liver disease (ALD)[48].
Epigenetic modifications have emerged as pivotal contributors to the intricate landscape of HCC development and pro
These modifications, encompassing DNA methylation, histone modifications, and non-coding RNA-mediated regulation, profoundly influence gene expression patterns, cell signaling, and tumor microenvironment interactions. In this section, we focus on the multifaceted role of epigenetic modifications in driving HCC progression, unraveling the intricate regulatory networks that orchestrate carcinogenic processes. By exploring the molecular nuances of epigenetic dysregulation, we aim to deepen insights into HCC pathogenesis and open new avenues for targeted therapeutic inter
Hypomethylation in HCC stimulated protooncogenes, such as c-jun, c-myc, etc., and increased genomic instability via influencing mitotic recombinations, eventually promoting carcinogenesis. Furthermore, Calvisi et al[53] also demon
Hypermethylation, associated with WNT/β-catenin signaling activation, APC inactivation, p16INK4A activation, RASSF1A and NORE1A activation, Mismatch repair system genes (hMLH1, hMSH2, and hMSH3) inactivation, CTF1, PDK4, FZD8, ZNF334, MAD2L1, CCNB1, CDC20, CCND1, AR, ESR1, p53 and MAPK signaling regulation, also is linked to HCC[50].
Future research may allow the estimation of the prognostic and diagnostic value of integrated bioinformatic data from HCC cells and tissues. Bai et al[54] described methylation sites typical for HCC but not cholangiocarcinoma[54]. As Guo et al[55] showed, mTOR signaling could also be involved in the progression of HCC via epigenetic regulations[55].
An integrated bioinformatic analysis may also help to identify diagnostic biomarkers to differentiate between different types of liver cancer.
The alteration of chromatin structure, which allows or prevents transcription factors from accessing gene regulatory sites, may also contribute to cancer pathogenesis. Histone deacetylase (HDAC) 1 (HDAC1) and HDAC2 are associated with the development of HCC, but they do so in different ways. HDAC1 expression was found to be correlated with tumors that were moderately and poorly differentiated[56], while HDAC2 expression was found to be an independent negative survival predictor[57]. HDAC2 is involved in the epigenetic regulation of cell cycle, apoptosis, and differentiation, and it was discovered that HCC patients frequently had elevated levels of HDAC2[58]. Other histone modifications associated with HCC are upregulated HDAC8, HDAC5, HDAC9, and downregulated HDAC3, HDAC5.
It has been discovered that several miRs, including let-7, miR-34a, miR-221, miR-222, and miR-122, miR-369, miR-3174, miR-383, miR-361-5p, miR-186, are implicated in the pathogenesis of liver cancer[59,60]. However, synthetic inhibitors, including antisense oligonucleotides or AntimiRs, can target the dysfunction of particular miRs[61].
Furthermore, epigenetic regulation may affect the mechanisms of resistance to systemic therapy in HCC. Oura et al[62] showed some of the mechanisms of resistance to drugs, such as sorafenib, regorafenib, and lenvatinib, associated with epigenetic modifications and tumor microenvironment regulation[62].
According to a growing body of evidence, long non-coding RNAs (lncRNAs) are implicated in the pathophysiology of HCC[63]. HBV, one of the main etiologic factors for HCC, can alter the host's epigenome through various methods. Of the viral proteins, Hepatitis B viral protein has been identified as a key epigenetic regulator[64].
Both ALD and NAFLD are common etiologic factors for HCC. Published data suggest that specific epigenetic modifications may be linked to these etiologic factors for HCC. For example, miR-21, miR-34a, miR-182 upregulation and miR-122 downregulation have been linked to NAFLD[65].
Most preclinical studies showed that epigenetic remodeling is potentially reversible by drugs, and the list of available epigenetic modifiers and inhibitors is continuously expanding[59]. Consequently, in the era of precision medicine, unraveling the epigenetic modifications occurring in HCC may offer new insights for screening potential therapeutic targets and studying personalized intervention strategies for managing this tumor. The optimization of scheduling strategies is a very relevant issue when considering combination therapies with conventional chemotherapeutics, other targeted agents, or immune checkpoint inhibitors. This is because the simultaneous, sequential, or alternate administration of epigenetic drugs is necessary to maximize efficacy, potentiate synergists, and overcome resistances[66].
On the other hand, we have also discovered that epigenetic regulatory circuits are incredibly complicated, and we have only begun exploring the tip of the iceberg. Therefore, to fully comprehend the pathophysiological implications of the epigenetic machinery, all of its dimensions must be fully unlocked. Considering the sheer size of this task, artificial intelligence tools seem necessary for the molecular analyses of relevant experimental models and clinical samples[67].
One of the challenges in early detecting HCC is the lack of highly sensitive and specific screening techniques. Over the years, ultrasound, computed tomography, and magnetic resonance imaging techniques have significantly improved their detection sensitivity, but their safety and cost disadvantages remain.
The future of early HCC detection probably lies in high-throughput tests that allow specific detection of HCC. These tests are promising tools for diagnosis, prognostication, and patients' selection for personalized therapy in HCC. One of the most important discoveries in the field lies in the use of circulating liquid biomarkers ("liquid biopsy") [68]. Liquid biopsy involves molecular diagnostics of nucleic acids (DNA/RNA) derived from tumors or extracellular vesicles (EVs) in the blood. The method uses blood/plasma, ascitic fluid or urine. These biological samples contain except standard HCC biomarkers [AFP, AFP with a high lectin affinity (AFP-L3), Des-γ-Carboxy Prothrombin (DCP), Glypican-3 (GPC3), osteopontin (OPN), Dickkopf Wnt signaling pathway inhibitor 1] and CTCs, tumor cfDNA, EVs or tumor-educated platelets that can be used for biomarkers[69,70]. They can be taken consecutively at different periods and give robust information about the development in progression. In recent years, liquid biopsy technologies have significantly advanced and become more precise, especially with the introduction of the NGS technique. NGS technology is up and coming, but multiple factors still hinder efforts to develop personalized therapy.
cfDNA are small degraded nucleic acids (under 200 bp) from damaged cells. They are of malignant origin and circulate in the blood, called ctDNA. Their analysis allows for early diagnosis by conducting liquid biopsy in real-time. Many studies suggest they are suitable tools that accurately represent the HCC tumor development and genetic profile[71-75].
Over the years, the methylation pattern of cfDNA in liquid biopsy of HCC has been investigated because methylation patterns are known to be unique for each cell type and remain stable in some pathological conditions[76]. Methylation changes in such conditions are known to occur early in tumor development[77], and therefore, guidelines have been proposed for more accurate analysis of methylated circulating DNA in liquid biopsies[78].
Guo et al[72] recently demonstrated that epigenetic variants carried by ctDNA can be biomarkers for early HCC detection[72]. They used enzymatic methylation sequencing and identified over 200 CpGs with significant methylation differences between HCC and non-HCC samples. А study revealed that HCC could be distinguished from normal controls by quantitative analysis of multiple methylated genes in plasma[79,80]. By adding miR data to the methylation pattern of some of these genes, a predictive model for the diagnosis of HCC in patients with low AFP values was obtained. Simultaneous detection of tumor somatic mutations and quantification of cfDNA can also be used for early diagnosis of HCC. Research shows that using high-depth targeted massive sequencing of ctDNA could reveal tumor somatic genetic changes[81,82]. Droplet Digital PCR can detect ctDNA in HCC patients by targeting hot mutations[83].
Indeed, those early studies that focused on detecting tumor-specific mutations by deep sequencing, which resulted from inter- and/or intratumor heterogeneity, reduced the sensitivity of these methods. Therefore, an alternative approach is to investigate unique tumor DNA methylation profiles, as they are known to be a hallmark of cancer[84]. A pan-cancer study also strongly supported the fact that methylation profiles are more precise and sensitive cancer classifiers of cfDNA than detected tumor-specific mutations[85]. One of the most successful screening biomarkers to be used was SEPT9. To diagnose HCC, Oussalah et al[86] show the precision of PCR-based analysis of methylation of SEPT9 promoter in circulating cfDNA (mSEPT9)[86]. The mSEPT9 test showed high diagnostic accuracy and may be a promising epigenetic biomarker for HCC diagnosis.
In their study, Xu et al[87] identified an HCC-specific DNA methylation marker panel by comparing the methylation profiles of HCC tumor tissue and normal leukocytes[87]. They showed that plasma ctDNA and tumor DNA were highly correlated, although they did not consider cfDNA from other tissues found in the blood at different levels. This diagnostic prediction model showed high specificity and sensitivity, but if there is DNA contamination, it can potentially yield false positives. To address this challenge, Cheishvili et al[88] used over 12000 methylation profiles in The Cancer Genome Atlas (https://portal.gdc.cancer.gov/) and the Gene Expression Omnibus data repository collections (https://www.ncbi.nlm.nih.gov/geo/info/submission.html)[88]. They determined the DNA methylation sites, which are consistently unmethylated in all blood DNA and tissue samples. Thus, they identified four CpGs positions that were highly meth
One way of intercellular communication is through small EVs. They are "cargo" vesicles that consist of proteins, DNA, RNA, miR and lipids. They are released from healthy, malignant, inflamed or apoptotic cells[89,90]. Liver cells are of different types and are capable of producing EVs. They can be found in serum, plasma, urine and other biological fluids. They are protected from degradation, making them suitable liquid biomarkers for diagnosing liver diseases[91-93]. In their study, Manea et al[69] have detailed the studies in the literature that reveal the role of EVs in the early diagnosis of HCC[69].
NGS technology is used for DNA and RNA sequencing and variant/mutation detection. With it, hundreds and thou
The accuracy of NGS generally depends on the length of coverage, and whole-genome sequencing (WGS) has a cove
Deep sequencing and WERS reveal the heterogeneity of HCC. Deep sequencing performed revealed frequent substitutions (C>T/G>A and T>C/A>G) that may explain the changes in the methylation profile observed in HCC[97]. A comparative study of whole exomes in advanced HCC cases compared with exomes from primary tumors revealed а lot of mutated genes[99]. Since mutations are found in primary tumors, they appear acquired during HCC progression.
The whole-genome array comparative genomic hybridization technique is widely used in screening for copy number changes[100]. This technique has been used in HCC research and has shown that there are chromosomal losses and gains in HCC. More than 30 recurrently altered regions have been associated with HCC[30,101,102]. Although many alterations have been identified, it is still challenging to associate these genes with altered regions and specific HCC phenotypes.
Applying NGS technologies in HCC does not bring immediate patient benefits but rapidly changes our understanding of HCC. Advances in NGS technologies and liquid biopsy techniques will undoubtedly lead to the possibility of earlier diagnosis, prognosis, and a more personalized approach to HCC treatment. Searching for specific substances/biomarkers secreted from the surface of circulating HCC cells would help HCC be diagnosed in the early stage and reduce mortality.
In the relentless battle against liver cancer, early detection stands as a paramount strategy for improving patient out
Additionally, biomarkers, such as ctDNA, offer a non-invasive window into the molecular intricacies of liver cancer, which include the current landscape of genetic and biomarker-based approaches, aiming to unravel their diagnostic prowess and illuminate the promising avenues for enhancing the timely detection of liver cancer[104]. Since developing a treatment plan depends on knowing each patient's exact cause and stage, it is widely acknowledged that early diagnosis can significantly increase the chance of a patient's survival by implementing curative treatment. Accordingly, methods for diagnosing precision medication should be developed for early identification of HCC[104].
Early diagnosis is critical to the prognosis of HCC patients. Serum AFP is currently the most widely used biomarker for HCC diagnosis, but its sensitivity and specificity are about 50%[105].
Other biomarkers for early detection of liver cancer are AFP and its isoform AFP-L3, GPC3, DCP, OPN, golgi protein-73, etc.[41]. Genetic markers for HCC are miRs as miR-224, miR-766, miR-23, miR-10b, miR-106b, and miR-181 etc. Cancer antigen 19-9, carcinoembryonic antigen, matrix metalloproteinase 7, CYFRA 21-1-a fragment of cytokeratin 19, interleukin 6, S100 calcium-binding protein A6, cfDNA, cell-free RNA and lncRNAs, etc. are also liver cancer biomarkers[41,104].
It is important to note that liquid biopsy has been developed and implemented in clinical practice over the past few decades, and it primarily detects ctDNA, CTCs, exosomes, and circulating tumor RNA in bodily fluids such as plasma, urine, and cerebrospinal fluid. ctDNA is the most widely applied genetic biomarker derived from tumor tissue. It carries somatic mutations, single-nucleotide variants, DNA methylations, viral sequences, and physical characteristics linked to carcinogenesis[104].
We discuss the advantages and disadvantages, of genetic and other screening technologies for HCC in Table 2.
| Advantages | Disadvantages | |
| AFP testing | Widely used; relatively cost-effective | Limited sensitivity, especially in early-stage HCC; prone to false positives/negatives |
| Genomic profiling (next-Generation sequencing, whole-genome sequencing) | High sensitivity for detecting genetic alterations; provides comprehensive genomic information | Costly; complex data analysis; may identify variants of uncertain significance |
| Liquid biopsy (circulating tumor DNA) | Non-invasive; potential for early detection; provides real-time monitoring | Limited sensitivity in early stages; technical challenges; standardization concerns |
| Imaging techniques (MRI, CT, ultrasound) | Commonly used; assesses tumor characteristics and location | Limited sensitivity for small lesions; exposure to radiation (CT); may not detect molecular changes |
| Molecular biomarkers (miRNA, methylation patterns) | High specificity; potential for early detection | Variable sensitivity; limited standardization; assay complexity |
| Circulating tumor cells analysis | May reflect metastatic potential; potential for real-time monitoring | Rare in early stages; technical challenges; standardization issues |
The choice of screening technology depends on the specific clinical context, resource availability, and the desired balance between sensitivity and specificity. A multimodal approach combining different technologies may offer a more comprehensive strategy for HCC screening.
Liver cancer, predominantly HCC, poses a formidable global health challenge, necessitating innovative approaches for early detection and risk stratification. Here, we present some intricacies of genetic screening, elucidating the challenges and opportunities that shape its current landscape[106].
Identification of at-risk populations and the need for targeted screening is critical. Unraveling the genetic under
Nevertheless, ethical considerations in genetic screening for liver cancer have become essential. As genetic screening becomes more sophisticated, ethical considerations loom larger. Balancing the potential benefits of early detection with protecting individuals' privacy and autonomy requires careful navigation. Ensuring informed consent, addressing poten
We also must think about integrating genetic data into personalized medicine approaches. Genetic insights pave the way for personalized medicine in liver cancer. Tailoring interventions based on an individual's genetic profile holds immense promise for treatment efficacy. There is a need to explore the integration of genetic data into the broader land
However, there are also some limitations and areas for future research in liver cancer genetics. While genetic screening presents a transformative tool, acknowledging its limitations is crucial. Challenges like the complexity of gene-enviro
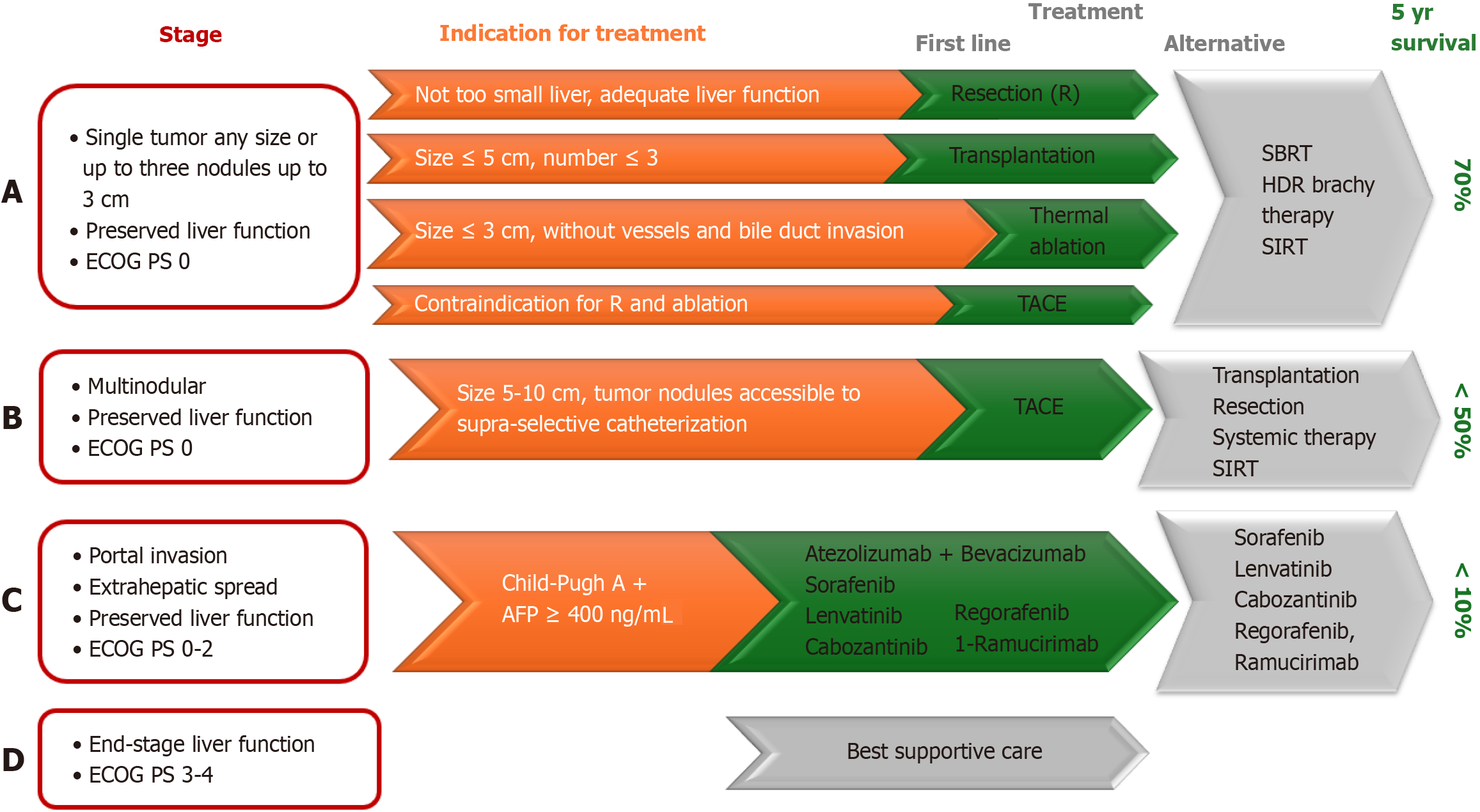
There are numerous staging systems for primary liver carcinoma, such as TNM, Okuda, (cancer of the liver Italian program), and CLIP system. Barcelona Clinic's liver cancer staging system is most commonly used for liver carcinoma because it reports not only the characteristics of cancer–its number, size and spread of cancer, but also the liver function and general health of the patient. It is recommended by American Association for the Study of Liver Diseases and the European Association for the Study of the Liver[110]. This staging system was updated for the last time in 2022 (Figure 2).
The earlier HCC is diagnosed, the better the survival rate–more than 70% of early-stage patients survive 5 years, while in advanced stages, only 20%[111].
Curative methods for HCC carcinoma are resection and liver transplantation (LT). Results from LT are very good with 1-year and 5-year survival, 90% and 70%, respectively. Patients on the transplant list have to wait. For this, local ablative methods such as transarterial chemoembolization (TACE) are applied. Guo et al[112] investigated which gene could predict prognosis after TACE (transarterial chemoembolization). Increased expressions of (PKM2) pyruvate kinase and peptide arginine deiminase IV indicate TACE resistance and poor prognosis[112]. Conversely, high expression of chromobox homolog 4 is associated with longer overall survival after TACE and higher sensitivity to doxorubicin[112].
Patients with elevated expression of some genes, like SERPINE1 have better drug sensitivity to lenvatinib[113].
In line with this, emerging therapies targeting specific genetic alterations in liver cancer are promising. The rapidly evolving landscape of liver cancer therapies is increasingly shaped by a nuanced understanding of genetic alterations driving HCC. This includes therapeutic advancements, exploring innovative strategies directly targeting specific genetic abnormalities, etc. From targeted molecular therapies to gene-based interventions, the arsenal against liver cancer is expanding, ushering in a new era where precision medicine meets the unique genetic signatures of individual tumors[114]. A critical aspect is also the patient education and awareness regarding genetic screening benefits. Empowering patients through education and fostering awareness about the benefits of genetic screening form a pivotal component of effective liver cancer management. The latter requires proactive patient engagement, unraveling the potential of genetic screening in facilitating early detection and personalized treatment plans. Patient education emerges as a linchpin as the medical landscape becomes more personalized, ensuring informed decisions and active participation in their healthcare journey[115,116].
Advancements in technology and their potential impact on genetic screening: The horizon of genetic screening for liver cancer is poised for transformation through technological leaps: Cutting-edge advancements, from enhanced NGS techniques to integrating artificial intelligence and machine learning. These innovations promise heightened precision, sensitivity, and cost-effectiveness in detecting genetic alterations associated with HCC, fostering a more accessible and comprehensive approach to population-wide screening[117].
Integrating multi-omics data for a comprehensive understanding of liver cancer genetics is the future of genetic scre
Collaborative efforts and research initiatives to enhance genetic screening practices are a must. The journey towards effective genetic screening necessitates collaboration across disciplines and institutions. Highlighting collaborative research initiatives, it is mandatory to include collective efforts underway to standardize protocols, share data, and establish large-scale genetic databases. These endeavors foster a unified approach, ensuring the amalgamation of diverse datasets for more robust genetic screening practices[119].
Potential breakthroughs and innovations in liver cancer prevention through genetic insights hold the key to revolutionary breakthroughs in liver cancer prevention: the potential for targeted interventions, risk stratification, and personalized prevention strategies. As genetic screening evolves, identifying high-risk individuals and guiding preventive measures becomes increasingly pivotal. It offers hope for a future where liver cancer can be intercepted at its earliest, most treatable stages[120].
It is well known that the HCC is characterized by significant clinical and molecular variability. Over the past 20 years, it has been published a lot of data, related to the most common molecular alterations in HCC, but unfortunately, this knowledge has not led to better prognostic evaluation or treatment of this cancer.
To open the door for new therapeutic approaches, every effort should be made to establish a connection between therapeutic response and the molecular subtypes of HCC. It is anticipated that the introduction of NGS technology will contribute significantly to clinical oncology paving the way for novel therapeutic strategies.
The molecular characterization of genetic alterations in HCC will facilitate the creation of prognostic biomarkers that can be employed in routine clinical practice. These biomarkers can be a useful tool in predicting prognosis and therapeutic response. The patient stratification for appropriate adjuvant and palliative treatments (including non-targeted therapies), early diagnosis, and surveillance of patients in the risk group, all depend on these markers (found in tumor tissue, blood, urine, etc.)[110].
In conclusion, this review has provided a comprehensive exploration of the current state-of-the-art in genetic screening for liver cancer, emphasizing its pivotal role in early detection and personalized prevention. The intricate interplay of risk-associated genetic variants, somatic mutations, and epigenetic alterations has been dissected to underscore the nuanced understanding of HCC development and progression. Key takeaways include emerging technologies' transformative potential, multi-omics data integration, and the imperative for collaborative efforts to refine screening practices. As we navigate these frontiers, it is evident that genetic insights will reshape the landscape of liver cancer prevention and management. This prompts a resounding call to action for continued advancements in liver cancer genetics, urging researchers, clinicians, and policymakers to forge ahead in a unified effort. Through sustained innov
Personalized medicine promises that it will allow us to treat HCC on an individual basis by determining the patient's genetic and epigenetic background and then conceiving a specific course of treatment that takes into account both the patient's best outcome and lowest risk. Future developments hold out hope for early HCC detection, which is the most important obstacle in achieving good treatment results in these patients.
| 1. | McGlynn KA, Petrick JL, El-Serag HB. Epidemiology of Hepatocellular Carcinoma. Hepatology. 2021;73 Suppl 1:4-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 820] [Cited by in RCA: 1503] [Article Influence: 300.6] [Reference Citation Analysis (2)] |
| 2. | Valery PC, Laversanne M, Clark PJ, Petrick JL, McGlynn KA, Bray F. Projections of primary liver cancer to 2030 in 30 countries worldwide. Hepatology. 2018;67:600-611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 253] [Article Influence: 31.6] [Reference Citation Analysis (1)] |
| 3. | Dragani TA. Risk of HCC: genetic heterogeneity and complex genetics. J Hepatol. 2010;52:252-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 167] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 4. | Clarke CA, Hubbell E, Kurian AW, Colditz GA, Hartman AR, Gomez SL. Projected Reductions in Absolute Cancer-Related Deaths from Diagnosing Cancers Before Metastasis, 2006-2015. Cancer Epidemiol Biomarkers Prev. 2020;29:895-902. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 5. | Ding J, Wen Z. Survival improvement and prognosis for hepatocellular carcinoma: analysis of the SEER database. BMC Cancer. 2021;21:1157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 86] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 6. | Lok AS, Sterling RK, Everhart JE, Wright EC, Hoefs JC, Di Bisceglie AM, Morgan TR, Kim HY, Lee WM, Bonkovsky HL, Dienstag JL; HALT-C Trial Group. Des-gamma-carboxy prothrombin and alpha-fetoprotein as biomarkers for the early detection of hepatocellular carcinoma. Gastroenterology. 2010;138:493-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 376] [Cited by in RCA: 457] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 7. | Owens DK, Davidson KW, Krist AH, Barry MJ, Cabana M, Caughey AB, Donahue K, Doubeni CA, Epling JW Jr, Kubik M, Ogedegbe G, Pbert L, Silverstein M, Simon MA, Tseng CW, Wong JB; US Preventive Services Task Force. Screening for Hepatitis C Virus Infection in Adolescents and Adults: US Preventive Services Task Force Recommendation Statement. JAMA. 2020;323:970-975. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 224] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 8. | Su YH, Kim AK, Jain S. Liquid biopsies for hepatocellular carcinoma. Transl Res. 2018;201:84-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 9. | Klein EA, Richards D, Cohn A, Tummala M, Lapham R, Cosgrove D, Chung G, Clement J, Gao J, Hunkapiller N, Jamshidi A, Kurtzman KN, Seiden MV, Swanton C, Liu MC. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann Oncol. 2021;32:1167-1177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 630] [Article Influence: 126.0] [Reference Citation Analysis (0)] |
| 10. | Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, Douville C, Javed AA, Wong F, Mattox A, Hruban RH, Wolfgang CL, Goggins MG, Dal Molin M, Wang TL, Roden R, Klein AP, Ptak J, Dobbyn L, Schaefer J, Silliman N, Popoli M, Vogelstein JT, Browne JD, Schoen RE, Brand RE, Tie J, Gibbs P, Wong HL, Mansfield AS, Jen J, Hanash SM, Falconi M, Allen PJ, Zhou S, Bettegowda C, Diaz LA Jr, Tomasetti C, Kinzler KW, Vogelstein B, Lennon AM, Papadopoulos N. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926-930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1752] [Cited by in RCA: 2031] [Article Influence: 253.9] [Reference Citation Analysis (0)] |
| 11. | Stuver S, Trichopoulos D. Cancer of the Liver and Biliary Tract'. In: Textbook of Cancer Epidemiology. New York: 2008: 308-332. Available from: https://doi.org/10.1093/acprof:oso/9780195311174.003.0012. |
| 12. | Yu MW, Chang HC, Liaw YF, Lin SM, Lee SD, Liu CJ, Chen PJ, Hsiao TJ, Lee PH, Chen CJ. Familial risk of hepatocellular carcinoma among chronic hepatitis B carriers and their relatives. J Natl Cancer Inst. 2000;92:1159-1164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 160] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 13. | Chen CH, Huang GT, Lee HS, Yang PM, Chen DS, Sheu JC. Clinical impact of screening first-degree relatives of patients with hepatocellular carcinoma. J Clin Gastroenterol. 1998;27:236-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 14. | Evans AA, Chen G, Ross EA, Shen FM, Lin WY, London WT. Eight-year follow-up of the 90,000-person Haimen City cohort: I. Hepatocellular carcinoma mortality, risk factors, and gender differences. Cancer Epidemiol Biomarkers Prev. 2002;11:369-376. [PubMed] |
| 15. | Hassan MM, Spitz MR, Thomas MB, Curley SA, Patt YZ, Vauthey JN, Glover KY, Kaseb A, Lozano RD, El-Deeb AS, Nguyen NT, Wei SH, Chan W, Abbruzzese JL, Li D. The association of family history of liver cancer with hepatocellular carcinoma: a case-control study in the United States. J Hepatol. 2009;50:334-341. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 69] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 16. | Turati F, Edefonti V, Talamini R, Ferraroni M, Malvezzi M, Bravi F, Franceschi S, Montella M, Polesel J, Zucchetto A, La Vecchia C, Negri E, Decarli A. Family history of liver cancer and hepatocellular carcinoma. Hepatology. 2012;55:1416-1425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 90] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 17. | Ng SWK, Rouhani FJ, Brunner SF, Brzozowska N, Aitken SJ, Yang M, Abascal F, Moore L, Nikitopoulou E, Chappell L, Leongamornlert D, Ivovic A, Robinson P, Butler T, Sanders MA, Williams N, Coorens THH, Teague J, Raine K, Butler AP, Hooks Y, Wilson B, Birtchnell N, Naylor H, Davies SE, Stratton MR, Martincorena I, Rahbari R, Frezza C, Hoare M, Campbell PJ. Convergent somatic mutations in metabolism genes in chronic liver disease. Nature. 2021;598:473-478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 131] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 18. | Schulze K, Imbeaud S, Letouzé E, Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C, Shinde J, Soysouvanh F, Calatayud AL, Pinyol R, Pelletier L, Balabaud C, Laurent A, Blanc JF, Mazzaferro V, Calvo F, Villanueva A, Nault JC, Bioulac-Sage P, Stratton MR, Llovet JM, Zucman-Rossi J. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47:505-511. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1006] [Cited by in RCA: 1409] [Article Influence: 128.1] [Reference Citation Analysis (0)] |
| 19. | Trépo E, Caruso S, Yang J, Imbeaud S, Couchy G, Bayard Q, Letouzé E, Ganne-Carrié N, Moreno C, Oussalah A, Féray C, Blanc JF, Clément B, Hillon P, Boursier J, Paradis V, Calderaro J, Gnemmi V, Nault JC, Guéant JL, Devière J, Archambeaud I, Vitellius C, Turlin B, Bronowicki JP, Gustot T, Sutton A; GENTHEP Consortium, Ziol M, Nahon P, Zucman-Rossi J. Common genetic variation in alcohol-related hepatocellular carcinoma: a case-control genome-wide association study. Lancet Oncol. 2022;23:161-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 66] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 20. | Nault JC, Ningarhari M, Rebouissou S, Zucman-Rossi J. The role of telomeres and telomerase in cirrhosis and liver cancer. Nat Rev Gastroenterol Hepatol. 2019;16:544-558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 174] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 21. | Takai A, Dang HT, Wang XW. Identification of drivers from cancer genome diversity in hepatocellular carcinoma. Int J Mol Sci. 2014;15:11142-11160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 46] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 22. | Nishida N, Goel A. Genetic and epigenetic signatures in human hepatocellular carcinoma: a systematic review. Curr Genomics. 2011;12:130-137. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 153] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 23. | Miteva D, Peshevska-Sekulovska M, Snegarova V, Peruhova M, Vasilev GH, Vasilev GV, Sekulovski M, Lazova S, Gulinac M, Tomov L, et al. Microbiome and Genetic Factors in the Pathogenesis of Liver Diseases. Gastroenterol Insights. 2023;14:575-597. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 24. | Teufel A, Staib F, Kanzler S, Weinmann A, Schulze-Bergkamen H, Galle PR. Genetics of hepatocellular carcinoma. World J Gastroenterol. 2007;13:2271-2282. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 94] [Cited by in RCA: 97] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 25. | Müller M, Bird TG, Nault JC. The landscape of gene mutations in cirrhosis and hepatocellular carcinoma. J Hepatol. 2020;72:990-1002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 128] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 26. | Niu ZS, Niu XJ, Wang WH. Genetic alterations in hepatocellular carcinoma: An update. World J Gastroenterol. 2016;22:9069-9095. [PubMed] [DOI] [Full Text] |
| 27. | Khemlina G, Ikeda S, Kurzrock R. The biology of Hepatocellular carcinoma: implications for genomic and immune therapies. Mol Cancer. 2017;16:149. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 191] [Cited by in RCA: 331] [Article Influence: 36.8] [Reference Citation Analysis (1)] |
| 28. | Kan Z, Zheng H, Liu X, Li S, Barber TD, Gong Z, Gao H, Hao K, Willard MD, Xu J, Hauptschein R, Rejto PA, Fernandez J, Wang G, Zhang Q, Wang B, Chen R, Wang J, Lee NP, Zhou W, Lin Z, Peng Z, Yi K, Chen S, Li L, Fan X, Yang J, Ye R, Ju J, Wang K, Estrella H, Deng S, Wei P, Qiu M, Wulur IH, Liu J, Ehsani ME, Zhang C, Loboda A, Sung WK, Aggarwal A, Poon RT, Fan ST, Hardwick J, Reinhard C, Dai H, Li Y, Luk JM, Mao M. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res. 2013;23:1422-1433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 355] [Cited by in RCA: 419] [Article Influence: 32.2] [Reference Citation Analysis (0)] |
| 29. | Kawai-Kitahata F, Asahina Y, Tanaka S, Kakinuma S, Murakawa M, Nitta S, Watanabe T, Otani S, Taniguchi M, Goto F, Nagata H, Kaneko S, Tasaka-Fujita M, Nishimura-Sakurai Y, Azuma S, Itsui Y, Nakagawa M, Tanabe M, Takano S, Fukasawa M, Sakamoto M, Maekawa S, Enomoto N, Watanabe M. Comprehensive analyses of mutations and hepatitis B virus integration in hepatocellular carcinoma with clinicopathological features. J Gastroenterol. 2016;51:473-486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 92] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 30. | Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6:674-687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1461] [Cited by in RCA: 1576] [Article Influence: 78.8] [Reference Citation Analysis (0)] |
| 31. | Li S, Mao M. Next generation sequencing reveals genetic landscape of hepatocellular carcinomas. Cancer Lett. 2013;340:247-253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 52] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 32. | Hirotsu Y, Zheng TH, Amemiya K, Mochizuki H, Guleng B, Omata M. Targeted and exome sequencing identified somatic mutations in hepatocellular carcinoma. Hepatol Res. 2016;46:1145-1151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 33. | Wang B, Yeh CB, Lein MY, Su CM, Yang SF, Liu YF, Tang CH. Effects of HMGB1 Polymorphisms on the Susceptibility and Progression of Hepatocellular Carcinoma. Int J Med Sci. 2016;13:304-309. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 34. | Qu LS, Jin F, Guo YM, Liu TT, Xue RY, Huang XW, Xu M, Chen TY, Ni ZP, Shen XZ. Nine susceptibility loci for hepatitis B virus-related hepatocellular carcinoma identified by a pilot two-stage genome-wide association study. Oncol Lett. 2016;11:624-632. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 35. | Miki D, Ochi H, Hayes CN, Abe H, Yoshima T, Aikata H, Ikeda K, Kumada H, Toyota J, Morizono T, Tsunoda T, Kubo M, Nakamura Y, Kamatani N, Chayama K. Variation in the DEPDC5 locus is associated with progression to hepatocellular carcinoma in chronic hepatitis C virus carriers. Nat Genet. 2011;43:797-800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 140] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 36. | Kumar V, Kato N, Urabe Y, Takahashi A, Muroyama R, Hosono N, Otsuka M, Tateishi R, Omata M, Nakagawa H, Koike K, Kamatani N, Kubo M, Nakamura Y, Matsuda K. Genome-wide association study identifies a susceptibility locus for HCV-induced hepatocellular carcinoma. Nat Genet. 2011;43:455-458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 296] [Cited by in RCA: 295] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 37. | Lachenmayer A, Alsinet C, Savic R, Cabellos L, Toffanin S, Hoshida Y, Villanueva A, Minguez B, Newell P, Tsai HW, Barretina J, Thung S, Ward SC, Bruix J, Mazzaferro V, Schwartz M, Friedman SL, Llovet JM. Wnt-pathway activation in two molecular classes of hepatocellular carcinoma and experimental modulation by sorafenib. Clin Cancer Res. 2012;18:4997-5007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 259] [Article Influence: 18.5] [Reference Citation Analysis (1)] |
| 38. | Yang S, Luo C, Gu Q, Xu Q, Wang G, Sun H, Qian Z, Tan Y, Qin Y, Shen Y, Xu X, Chen SH, Chan CC, Wang H, Mao M, Fang DD. Activating JAK1 mutation may predict the sensitivity of JAK-STAT inhibition in hepatocellular carcinoma. Oncotarget. 2016;7:5461-5469. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 44] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 39. | Wands JR, Kim M. WNT/β-catenin signaling and hepatocellular carcinoma. Hepatology. 2014;60:452-454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 56] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 40. | Kudo M. Signaling pathway/molecular targets and new targeted agents under development in hepatocellular carcinoma. World J Gastroenterol. 2012;18:6005-6017. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 37] [Cited by in RCA: 36] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 41. | Baj J, Bryliński Ł, Woliński F, Granat M, Kostelecka K, Duda P, Flieger J, Teresiński G, Buszewicz G, Furtak-Niczyporuk M, Portincasa P. Biomarkers and Genetic Markers of Hepatocellular Carcinoma and Cholangiocarcinoma-What Do We Already Know. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 42. | Cancer Genome Atlas Research Network. Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell. 2017;169:1327-1341.e23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1578] [Cited by in RCA: 1823] [Article Influence: 202.6] [Reference Citation Analysis (1)] |
| 43. | Peruhova M, Peshevska-Sekulovska M, Krastev B, Panayotova G, Georgieva V, Konakchieva R, Nikolaev G, Velikova TV. What could microRNA expression tell us more about colorectal serrated pathway carcinogenesis? World J Gastroenterol. 2020;26:6556-6571. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 15] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 44. | Chidambaranathan-Reghupaty S, Fisher PB, Sarkar D. Hepatocellular carcinoma (HCC): Epidemiology, etiology and molecular classification. Adv Cancer Res. 2021;149:1-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 330] [Cited by in RCA: 639] [Article Influence: 106.5] [Reference Citation Analysis (0)] |
| 45. | Ganesan P, Kulik LM. Hepatocellular Carcinoma: New Developments. Clin Liver Dis. 2023;27:85-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 296] [Article Influence: 98.7] [Reference Citation Analysis (1)] |
| 46. | Schulze K, Nault JC, Villanueva A. Genetic profiling of hepatocellular carcinoma using next-generation sequencing. J Hepatol. 2016;65:1031-1042. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 232] [Article Influence: 23.2] [Reference Citation Analysis (1)] |
| 47. | Shibata T, Aburatani H. Exploration of liver cancer genomes. Nat Rev Gastroenterol Hepatol. 2014;11:340-349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 156] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 48. | Caruso S, O'Brien DR, Cleary SP, Roberts LR, Zucman-Rossi J. Genetics of Hepatocellular Carcinoma: Approaches to Explore Molecular Diversity. Hepatology. 2021;73 Suppl 1:14-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 65] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 49. | Zhang Y, Jiang HH, Wang ZY, Zhai B, Lin MB. Alcohol dehydrogenase 4 is a TP53-associated gene signature for the prediction of prognosis in hepatocellular carcinoma. Oncol Lett. 2023;25:3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 50. | Wolinska E, Skrzypczak M. Epigenetic Changes Affecting the Development of Hepatocellular Carcinoma. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 51. | Tao S, Liang S, Zeng T, Yin D. Epigenetic modification-related mechanisms of hepatocellular carcinoma resistance to immune checkpoint inhibition. Front Immunol. 2022;13:1043667. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 26] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 52. | Wahid B, Ali A, Rafique S, Idrees M. New Insights into the Epigenetics of Hepatocellular Carcinoma. Biomed Res Int. 2017;2017:1609575. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 74] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 53. | Calvisi DF, Ladu S, Gorden A, Farina M, Lee JS, Conner EA, Schroeder I, Factor VM, Thorgeirsson SS. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J Clin Invest. 2007;117:2713-2722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 293] [Cited by in RCA: 323] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 54. | Bai Y, Tong W, Xie F, Zhu L, Wu H, Shi R, Wang L, Yang L, Liu Z, Miao F, Zhao Q, Zhang Y. DNA methylation biomarkers for diagnosis of primary liver cancer and distinguishing hepatocellular carcinoma from intrahepatic cholangiocarcinoma. Aging (Albany NY). 2021;13:17592-17606. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 55. | Guo M, Li N, Zheng J, Wang W, Wu Y, Han X, Guo J, Chen W, Bai Z, Bai W, Wu J. Epigenetic Regulation of Hepatocellular Carcinoma Progression through the mTOR Signaling Pathway. Can J Gastroenterol Hepatol. 2021;2021:5596712. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 56. | Ler SY, Leung CH, Khin LW, Lu GD, Salto-Tellez M, Hartman M, Iau PT, Yap CT, Hooi SC. HDAC1 and HDAC2 independently predict mortality in hepatocellular carcinoma by a competing risk regression model in a Southeast Asian population. Oncol Rep. 2015;34:2238-2250. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 76] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 57. | Quint K, Agaimy A, Di Fazio P, Montalbano R, Steindorf C, Jung R, Hellerbrand C, Hartmann A, Sitter H, Neureiter D, Ocker M. Clinical significance of histone deacetylases 1, 2, 3, and 7: HDAC2 is an independent predictor of survival in HCC. Virchows Arch. 2011;459:129-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 105] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 58. | Lee YH, Seo D, Choi KJ, Andersen JB, Won MA, Kitade M, Gómez-Quiroz LE, Judge AD, Marquardt JU, Raggi C, Conner EA, MacLachlan I, Factor VM, Thorgeirsson SS. Antitumor effects in hepatocarcinoma of isoform-selective inhibition of HDAC2. Cancer Res. 2014;74:4752-4761. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 79] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 59. | Braghini MR, Lo Re O, Romito I, Fernandez-Barrena MG, Barbaro B, Pomella S, Rota R, Vinciguerra M, Avila MA, Alisi A. Epigenetic remodelling in human hepatocellular carcinoma. J Exp Clin Cancer Res. 2022;41:107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 57] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 60. | Syyam A, Akbar HR, Jilkova ZM, Afzal S. Role of Genetic and Epigenetic Modifications in the Progression of Hepatocellular Carcinoma in Chronic HCV Patients. Livers. 2023;3:82-92. [RCA] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 61. | Xu X, Peng Q, Jiang X, Tan S, Yang Y, Yang W, Han Y, Chen Y, Oyang L, Lin J, Xia L, Peng M, Wu N, Tang Y, Li J, Liao Q, Zhou Y. Metabolic reprogramming and epigenetic modifications in cancer: from the impacts and mechanisms to the treatment potential. Exp Mol Med. 2023;55:1357-1370. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 156] [Article Influence: 52.0] [Reference Citation Analysis (0)] |
| 62. | Oura K, Morishita A, Hamaya S, Fujita K, Masaki T. The Roles of Epigenetic Regulation and the Tumor Microenvironment in the Mechanism of Resistance to Systemic Therapy in Hepatocellular Carcinoma. Int J Mol Sci. 2023;24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 33] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 63. | Lanzafame M, Bianco G, Terracciano LM, Ng CKY, Piscuoglio S. The Role of Long Non-Coding RNAs in Hepatocarcinogenesis. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 72] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 64. | Park IY, Sohn BH, Yu E, Suh DJ, Chung YH, Lee JH, Surzycki SJ, Lee YI. Aberrant epigenetic modifications in hepatocarcinogenesis induced by hepatitis B virus X protein. Gastroenterology. 2007;132:1476-1494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 249] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 65. | Eslam M, Valenti L, Romeo S. Genetics and epigenetics of NAFLD and NASH: Clinical impact. J Hepatol. 2018;68:268-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 732] [Cited by in RCA: 726] [Article Influence: 90.8] [Reference Citation Analysis (0)] |
| 66. | Morel D, Jeffery D, Aspeslagh S, Almouzni G, Postel-Vinay S. Combining epigenetic drugs with other therapies for solid tumours - past lessons and future promise. Nat Rev Clin Oncol. 2020;17:91-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 322] [Article Influence: 46.0] [Reference Citation Analysis (0)] |
| 67. | Fernández-Barrena MG, Arechederra M, Colyn L, Berasain C, Avila MA. Epigenetics in hepatocellular carcinoma development and therapy: The tip of the iceberg. JHEP Rep. 2020;2:100167. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 83] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 68. | Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32:579-586. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1413] [Cited by in RCA: 1731] [Article Influence: 144.3] [Reference Citation Analysis (0)] |
| 69. | Manea I, Iacob R, Iacob S, Cerban R, Dima S, Oniscu G, Popescu I, Gheorghe L. Liquid biopsy for early detection of hepatocellular carcinoma. Front Med (Lausanne). 2023;10:1218705. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 19] [Reference Citation Analysis (0)] |
| 70. | Adeniji N, Dhanasekaran R. Current and Emerging Tools for Hepatocellular Carcinoma Surveillance. Hepatol Commun. 2021;5:1972-1986. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 71. | Nguyen VC, Nguyen TH, Phan TH, Tran TT, Pham TTT, Ho TD, Nguyen HHT, Duong ML, Nguyen CM, Nguyen QB, Bach HT, Kim VV, Pham TA, Nguyen BT, Nguyen TNV, Huynh LAK, Tran VU, Tran TTT, Nguyen TD, Phu DTB, Phan BHH, Nguyen QT, Truong DK, Do TT, Nguyen HN, Phan MD, Giang H, Tran LS. Fragment length profiles of cancer mutations enhance detection of circulating tumor DNA in patients with early-stage hepatocellular carcinoma. BMC Cancer. 2023;23:233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 22] [Reference Citation Analysis (0)] |
| 72. | Guo P, Zheng H, Li Y, Xiao Y, Zheng J, Zhu X, Xu H, He Z, Zhang Q, Chen J, Qiu M, Jiang M, Liu P, Chen H. Hepatocellular carcinoma detection via targeted enzymatic methyl sequencing of plasma cell-free DNA. Clin Epigenetics. 2023;15:2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 73. | Phan TH, Chi Nguyen VT, Thi Pham TT, Nguyen VC, Ho TD, Quynh Pham TM, Tran TH, Nguyen TD, Khang Le ND, Nguyen TH, Duong ML, Bach HT, Kim VV, Pham TA, Nguyen BT, Vo Nguyen TN, Bieu Phu DT, Huu Phan BH, Nguyen DS, Truong DK, Do TT, Giang H, Nguyen HN, Phan MD, Tran LS. Circulating DNA methylation profile improves the accuracy of serum biomarkers for the detection of nonmetastatic hepatocellular carcinoma. Future Oncol. 2022;18:4399-4413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 74. | Wang P, Song Q, Ren J, Zhang W, Wang Y, Zhou L, Wang D, Chen K, Jiang L, Zhang B, Chen W, Qu C, Zhao H, Jiao Y. Simultaneous analysis of mutations and methylations in circulating cell-free DNA for hepatocellular carcinoma detection. Sci Transl Med. 2022;14:eabp8704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 49] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 75. | Chen L, Abou-Alfa GK, Zheng B, Liu JF, Bai J, Du LT, Qian YS, Fan R, Liu XL, Wu L, Hou JL, Wang HY; PreCar Team. Genome-scale profiling of circulating cell-free DNA signatures for early detection of hepatocellular carcinoma in cirrhotic patients. Cell Res. 2021;31:589-592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 81] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 76. | Bergman Y, Cedar H. DNA methylation dynamics in health and disease. Nat Struct Mol Biol. 2013;20:274-281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 402] [Cited by in RCA: 442] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 77. | Okajima W, Komatsu S, Ichikawa D, Miyamae M, Ohashi T, Imamura T, Kiuchi J, Nishibeppu K, Arita T, Konishi H, Shiozaki A, Morimura R, Ikoma H, Okamoto K, Otsuji E. Liquid biopsy in patients with hepatocellular carcinoma: Circulating tumor cells and cell-free nucleic acids. World J Gastroenterol. 2017;23:5650-5668. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 71] [Cited by in RCA: 78] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 78. | Kerachian MA, Azghandi M, Mozaffari-Jovin S, Thierry AR. Guidelines for pre-analytical conditions for assessing the methylation of circulating cell-free DNA. Clin Epigenetics. 2021;13:193. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 79. | Huang ZH, Hu Y, Hua D, Wu YY, Song MX, Cheng ZH. Quantitative analysis of multiple methylated genes in plasma for the diagnosis and prognosis of hepatocellular carcinoma. Exp Mol Pathol. 2011;91:702-707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 92] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 80. | Lu CY, Chen SY, Peng HL, Kan PY, Chang WC, Yen CJ. Cell-free methylation markers with diagnostic and prognostic potential in hepatocellular carcinoma. Oncotarget. 2017;8:6406-6418. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 51] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 81. | Ye Q, Ling S, Zheng S, Xu X. Liquid biopsy in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. Mol Cancer. 2019;18:114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 290] [Article Influence: 41.4] [Reference Citation Analysis (0)] |
| 82. | De Mattos-Arruda L, Weigelt B, Cortes J, Won HH, Ng CKY, Nuciforo P, Bidard FC, Aura C, Saura C, Peg V, Piscuoglio S, Oliveira M, Smolders Y, Patel P, Norton L, Tabernero J, Berger MF, Seoane J, Reis-Filho JS. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann Oncol. 2014;25:1729-1735. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 298] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 83. | Huang A, Zhang X, Zhou SL, Cao Y, Huang XW, Fan J, Yang XR, Zhou J. Detecting Circulating Tumor DNA in Hepatocellular Carcinoma Patients Using Droplet Digital PCR Is Feasible and Reflects Intratumoral Heterogeneity. J Cancer. 2016;7:1907-1914. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 87] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 84. | Gai W, Sun K. Epigenetic Biomarkers in Cell-Free DNA and Applications in Liquid Biopsy. Genes (Basel). 2019;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 95] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 85. | Liu MC, Oxnard GR, Klein EA, Swanton C, Seiden MV; CCGA Consortium. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann Oncol. 2020;31:745-759. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 707] [Cited by in RCA: 993] [Article Influence: 165.5] [Reference Citation Analysis (0)] |
| 86. | Oussalah A, Rischer S, Bensenane M, Conroy G, Filhine-Tresarrieu P, Debard R, Forest-Tramoy D, Josse T, Reinicke D, Garcia M, Luc A, Baumann C, Ayav A, Laurent V, Hollenbach M, Ripoll C, Guéant-Rodriguez RM, Namour F, Zipprich A, Fleischhacker M, Bronowicki JP, Guéant JL. Plasma mSEPT9: A Novel Circulating Cell-free DNA-Based Epigenetic Biomarker to Diagnose Hepatocellular Carcinoma. EBioMedicine. 2018;30:138-147. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 127] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 87. | Xu RH, Wei W, Krawczyk M, Wang W, Luo H, Flagg K, Yi S, Shi W, Quan Q, Li K, Zheng L, Zhang H, Caughey BA, Zhao Q, Hou J, Zhang R, Xu Y, Cai H, Li G, Hou R, Zhong Z, Lin D, Fu X, Zhu J, Duan Y, Yu M, Ying B, Zhang W, Wang J, Zhang E, Zhang C, Li O, Guo R, Carter H, Zhu JK, Hao X, Zhang K. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater. 2017;16:1155-1161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 478] [Cited by in RCA: 657] [Article Influence: 73.0] [Reference Citation Analysis (0)] |
| 88. | Cheishvili D, Wong C, Karim MM, Kibria MG, Jahan N, Das PC, Yousuf MAK, Islam MA, Das DC, Noor-E-Alam SM, Szyf M, Alam S, Khan WA, Al Mahtab M. A high-throughput test enables specific detection of hepatocellular carcinoma. Nat Commun. 2023;14:3306. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 22] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 89. | Thietart S, Rautou PE. Extracellular vesicles as biomarkers in liver diseases: A clinician's point of view. J Hepatol. 2020;73:1507-1525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 136] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 90. | Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, Ayre DC, Bach JM, Bachurski D, Baharvand H, Balaj L, Baldacchino S, Bauer NN, Baxter AA, Bebawy M, Beckham C, Bedina Zavec A, Benmoussa A, Berardi AC, Bergese P, Bielska E, Blenkiron C, Bobis-Wozowicz S, Boilard E, Boireau W, Bongiovanni A, Borràs FE, Bosch S, Boulanger CM, Breakefield X, Breglio AM, Brennan MÁ, Brigstock DR, Brisson A, Broekman ML, Bromberg JF, Bryl-Górecka P, Buch S, Buck AH, Burger D, Busatto S, Buschmann D, Bussolati B, Buzás EI, Byrd JB, Camussi G, Carter DR, Caruso S, Chamley LW, Chang YT, Chen C, Chen S, Cheng L, Chin AR, Clayton A, Clerici SP, Cocks A, Cocucci E, Coffey RJ, Cordeiro-da-Silva A, Couch Y, Coumans FA, Coyle B, Crescitelli R, Criado MF, D'Souza-Schorey C, Das S, Datta Chaudhuri A, de Candia P, De Santana EF, De Wever O, Del Portillo HA, Demaret T, Deville S, Devitt A, Dhondt B, Di Vizio D, Dieterich LC, Dolo V, Dominguez Rubio AP, Dominici M, Dourado MR, Driedonks TA, Duarte FV, Duncan HM, Eichenberger RM, Ekström K, El Andaloussi S, Elie-Caille C, Erdbrügger U, Falcón-Pérez JM, Fatima F, Fish JE, Flores-Bellver M, Försönits A, Frelet-Barrand A, Fricke F, Fuhrmann G, Gabrielsson S, Gámez-Valero A, Gardiner C, Gärtner K, Gaudin R, Gho YS, Giebel B, Gilbert C, Gimona M, Giusti I, Goberdhan DC, Görgens A, Gorski SM, Greening DW, Gross JC, Gualerzi A, Gupta GN, Gustafson D, Handberg A, Haraszti RA, Harrison P, Hegyesi H, Hendrix A, Hill AF, Hochberg FH, Hoffmann KF, Holder B, Holthofer H, Hosseinkhani B, Hu G, Huang Y, Huber V, Hunt S, Ibrahim AG, Ikezu T, Inal JM, Isin M, Ivanova A, Jackson HK, Jacobsen S, Jay SM, Jayachandran M, Jenster G, Jiang L, Johnson SM, Jones JC, Jong A, Jovanovic-Talisman T, Jung S, Kalluri R, Kano SI, Kaur S, Kawamura Y, Keller ET, Khamari D, Khomyakova E, Khvorova A, Kierulf P, Kim KP, Kislinger T, Klingeborn M, Klinke DJ 2nd, Kornek M, Kosanović MM, Kovács ÁF, Krämer-Albers EM, Krasemann S, Krause M, Kurochkin IV, Kusuma GD, Kuypers S, Laitinen S, Langevin SM, Languino LR, Lannigan J, Lässer C, Laurent LC, Lavieu G, Lázaro-Ibáñez E, Le Lay S, Lee MS, Lee YXF, Lemos DS, Lenassi M, Leszczynska A, Li IT, Liao K, Libregts SF, Ligeti E, Lim R, Lim SK, Linē A, Linnemannstöns K, Llorente A, Lombard CA, Lorenowicz MJ, Lörincz ÁM, Lötvall J, Lovett J, Lowry MC, Loyer X, Lu Q, Lukomska B, Lunavat TR, Maas SL, Malhi H, Marcilla A, Mariani J, Mariscal J, Martens-Uzunova ES, Martin-Jaular L, Martinez MC, Martins VR, Mathieu M, Mathivanan S, Maugeri M, McGinnis LK, McVey MJ, Meckes DG Jr, Meehan KL, Mertens I, Minciacchi VR, Möller A, Møller Jørgensen M, Morales-Kastresana A, Morhayim J, Mullier F, Muraca M, Musante L, Mussack V, Muth DC, Myburgh KH, Najrana T, Nawaz M, Nazarenko I, Nejsum P, Neri C, Neri T, Nieuwland R, Nimrichter L, Nolan JP, Nolte-'t Hoen EN, Noren Hooten N, O'Driscoll L, O'Grady T, O'Loghlen A, Ochiya T, Olivier M, Ortiz A, Ortiz LA, Osteikoetxea X, Østergaard O, Ostrowski M, Park J, Pegtel DM, Peinado H, Perut F, Pfaffl MW, Phinney DG, Pieters BC, Pink RC, Pisetsky DS, Pogge von Strandmann E, Polakovicova I, Poon IK, Powell BH, Prada I, Pulliam L, Quesenberry P, Radeghieri A, Raffai RL, Raimondo S, Rak J, Ramirez MI, Raposo G, Rayyan MS, Regev-Rudzki N, Ricklefs FL, Robbins PD, Roberts DD, Rodrigues SC, Rohde E, Rome S, Rouschop KM, Rughetti A, Russell AE, Saá P, Sahoo S, Salas-Huenuleo E, Sánchez C, Saugstad JA, Saul MJ, Schiffelers RM, Schneider R, Schøyen TH, Scott A, Shahaj E, Sharma S, Shatnyeva O, Shekari F, Shelke GV, Shetty AK, Shiba K, Siljander PR, Silva AM, Skowronek A, Snyder OL 2nd, Soares RP, Sódar BW, Soekmadji C, Sotillo J, Stahl PD, Stoorvogel W, Stott SL, Strasser EF, Swift S, Tahara H, Tewari M, Timms K, Tiwari S, Tixeira R, Tkach M, Toh WS, Tomasini R, Torrecilhas AC, Tosar JP, Toxavidis V, Urbanelli L, Vader P, van Balkom BW, van der Grein SG, Van Deun J, van Herwijnen MJ, Van Keuren-Jensen K, van Niel G, van Royen ME, van Wijnen AJ, Vasconcelos MH, Vechetti IJ Jr, Veit TD, Vella LJ, Velot É, Verweij FJ, Vestad B, Viñas JL, Visnovitz T, Vukman KV, Wahlgren J, Watson DC, Wauben MH, Weaver A, Webber JP, Weber V, Wehman AM, Weiss DJ, Welsh JA, Wendt S, Wheelock AM, Wiener Z, Witte L, Wolfram J, Xagorari A, Xander P, Xu J, Yan X, Yáñez-Mó M, Yin H, Yuana Y, Zappulli V, Zarubova J, Žėkas V, Zhang JY, Zhao Z, Zheng L, Zheutlin AR, Zickler AM, Zimmermann P, Zivkovic AM, Zocco D, Zuba-Surma EK. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6453] [Cited by in RCA: 8277] [Article Influence: 1034.6] [Reference Citation Analysis (1)] |
| 91. | Lu X, Li Y, Zhang X, Shi J, Feng H, Gao Y, Yu Z. Advances of multi-omics applications in hepatic precancerous lesions and hepatocellular carcinoma: The role of extracellular vesicles. Front Mol Biosci. 2023;10:1114594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 92. | Lapitz A, Arbelaiz A, Olaizola P, Aranburu A, Bujanda L, Perugorria MJ, Banales JM. Extracellular Vesicles in Hepatobiliary Malignancies. Front Immunol. 2018;9:2270. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 93. | Yang J, Li C, Zhang L, Wang X. Extracellular Vesicles as Carriers of Non-coding RNAs in Liver Diseases. Front Pharmacol. 2018;9:415. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 94. | Dominguez DA, Wang XW. Impact of Next-Generation Sequencing on Outcomes in Hepatocellular Carcinoma: How Precise Are We Really? J Hepatocell Carcinoma. 2020;7:33-37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 95. | Nakagawa H, Shibata T. Comprehensive genome sequencing of the liver cancer genome. Cancer Lett. 2013;340:234-240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 41] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 96. | Marquardt JU, Andersen JB, Thorgeirsson SS. Functional and genetic deconstruction of the cellular origin in liver cancer. Nat Rev Cancer. 2015;15:653-667. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 241] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 97. | Fujimoto A, Furuta M, Shiraishi Y, Gotoh K, Kawakami Y, Arihiro K, Nakamura T, Ueno M, Ariizumi S, Nguyen HH, Shigemizu D, Abe T, Boroevich KA, Nakano K, Sasaki A, Kitada R, Maejima K, Yamamoto Y, Tanaka H, Shibuya T, Shibata T, Ojima H, Shimada K, Hayami S, Shigekawa Y, Aikata H, Ohdan H, Marubashi S, Yamada T, Kubo M, Hirano S, Ishikawa O, Yamamoto M, Yamaue H, Chayama K, Miyano S, Tsunoda T, Nakagawa H. Whole-genome mutational landscape of liver cancers displaying biliary phenotype reveals hepatitis impact and molecular diversity. Nat Commun. 2015;6:6120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 175] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 98. | Ngalah Bidii Stephen, Dhatri Madduru, Pranathi Pappu, Urvashi Vijay, Prashanth Suravajhala, Obul Reddy Bandapalli. Hepatocarcinogenesis and the role of next-generation sequencing in liver cancer. In: Theranostics and Precision Medicine for the Management of Hepatocellular Carcinoma, Volume 2. 2022: 45-57. [DOI] [Full Text] |
| 99. | Stefanska B, Huang J, Bhattacharyya B, Suderman M, Hallett M, Han ZG, Szyf M. Definition of the landscape of promoter DNA hypomethylation in liver cancer. Cancer Res. 2011;71:5891-5903. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 144] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 100. | Kim TM, Yim SH, Shin SH, Xu HD, Jung YC, Park CK, Choi JY, Park WS, Kwon MS, Fiegler H, Carter NP, Rhyu MG, Chung YJ. Clinical implication of recurrent copy number alterations in hepatocellular carcinoma and putative oncogenes in recurrent gains on 1q. Int J Cancer. 2008;123:2808-2815. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 81] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 101. | Kang L, Liu X, Gong Z, Zheng H, Wang J, Li Y, Yang H, Hardwick J, Dai H, Poon RT, Lee NP, Mao M, Peng Z, Chen R. Genome-wide identification of RNA editing in hepatocellular carcinoma. Genomics. 2015;105:76-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 102. | Skidmore ZL, Kunisaki J, Lin Y, Cotto KC, Barnell EK, Hundal J, Krysiak K, Magrini V, Trani L, Walker JR, Fulton R, Brunt EM, Miller CA, Wilson RK, Mardis ER, Griffith M, Chapman W, Griffith OL. Genomic and transcriptomic somatic alterations of hepatocellular carcinoma in non-cirrhotic livers. Cancer Genet. 2022;264-265:90-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 103. | Duan J, Wu Y, Liu J, Zhang J, Fu Z, Feng T, Liu M, Han J, Li Z, Chen S. Genetic Biomarkers For Hepatocellular Carcinoma In The Era Of Precision Medicine. J Hepatocell Carcinoma. 2019;6:151-166. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 104. | Basthi Mohan P, Lochan R et Shetty S. Biomarker in Hepatocellular Carcinoma. Indian J Surg Oncol. 2024;. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 105. | Bruix J, Sherman M; American Association for the Study of Liver Diseases. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53:1020-1022. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5972] [Cited by in RCA: 6630] [Article Influence: 442.0] [Reference Citation Analysis (1)] |
| 106. | McMahon B, Cohen C, Brown RS Jr, El-Serag H, Ioannou GN, Lok AS, Roberts LR, Singal AG, Block T. Opportunities to address gaps in early detection and improve outcomes of liver cancer. JNCI Cancer Spectr. 2023;7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 29] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 107. | Lee YT, Fujiwara N, Yang JD, Hoshida Y. Risk stratification and early detection biomarkers for precision HCC screening. Hepatology. 2023;78:319-362. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 84] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 108. | Storm C, Agarwal R, Offit K. Ethical and legal implications of cancer genetic testing: do physicians have a duty to warn patients' relatives about possible genetic risks? J Oncol Pract. 2008;4:229-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 109. | Anwanwan D, Singh SK, Singh S, Saikam V, Singh R. Challenges in liver cancer and possible treatment approaches. Biochim Biophys Acta Rev Cancer. 2020;1873:188314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 762] [Cited by in RCA: 979] [Article Influence: 163.2] [Reference Citation Analysis (2)] |
| 110. | Jianyong L, Lunan Y, Wentao W, Yong Z, Bo L, Tianfu W, Minqing X, Jiaying Y. Barcelona clinic liver cancer stage B hepatocellular carcinoma: transarterial chemoembolization or hepatic resection? Medicine (Baltimore). 2014;93:e180. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 111. | Calderon-Martinez E, Landazuri-Navas S, Vilchez E, Cantu-Hernandez R, Mosquera-Moscoso J, Encalada S, Al Lami Z, Zevallos-Delgado C, Cinicola J. Prognostic Scores and Survival Rates by Etiology of Hepatocellular Carcinoma: A Review. J Clin Med Res. 2023;15:200-207. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 57] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 112. | Guo Y, Hu H, Xu S, Xia W, Li H. Useful genes for predicting the efficacy of transarterial chemoembolization in hepatocellular carcinoma. J Cancer Res Ther. 2022;18:1860-1866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 113. | Lin Z, Xu Q, Miao D, Yu F. An Inflammatory Response-Related Gene Signature Can Impact the Immune Status and Predict the Prognosis of Hepatocellular Carcinoma. Front Oncol. 2021;11:644416. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 76] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 114. | Chakraborty E, Sarkar D. Emerging Therapies for Hepatocellular Carcinoma (HCC). Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 239] [Article Influence: 59.8] [Reference Citation Analysis (0)] |
| 115. | Shaw J, Patidar KR, Reuter B, Hajezifar N, Dharel N, Wade JB, Bajaj JS. Focused Education Increases Hepatocellular Cancer Screening in Patients with Cirrhosis Regardless of Functional Health Literacy. Dig Dis Sci. 2021;66:2603-2609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 116. | Chin JJ, Tham HW. Knowledge, Awareness, and Perception of Genetic Testing for Hereditary Disorders Among Malaysians in Klang Valley. Front Genet. 2020;11:512582. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 117. | Yang X, Dai J, Yao S, An J, Wen G, Jin H, Zhang L, Zheng L, Chen X, Yi Z, Tuo B. APOBEC3B: Future direction of liver cancer research. Front Oncol. 2022;12:996115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 118. | Wang J, Miao Y, Li L, Wu Y, Ren Y, Cui Y, Cao H. Multi-omics data integration for hepatocellular carcinoma subtyping with multi-kernel learning. Front Genet. 2022;13:962870. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 119. | Khoury MJ, Feero WG, Chambers DA, Brody LC, Aziz N, Green RC, Janssens ACJW, Murray MF, Rodriguez LL, Rutter JL, Schully SD, Winn DM, Mensah GA. A collaborative translational research framework for evaluating and implementing the appropriate use of human genome sequencing to improve health. PLoS Med. 2018;15:e1002631. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 120. | Taniguchi H. Liver Cancer 2.0. Int J Mol Sci. 2023;24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 18] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Bulgaria
Peer-review report’s classification
Scientific Quality: Grade C, Grade C
Novelty: Grade B, Grade B
Creativity or Innovation: Grade B, Grade B
Scientific Significance: Grade B, Grade C
P-Reviewer: Huang B, China; Wang ZX, China S-Editor: Liu H L-Editor: A P-Editor: Zhao YQ