Published online Dec 27, 2024. doi: 10.4254/wjh.v16.i12.1468
Revised: August 31, 2024
Accepted: September 19, 2024
Published online: December 27, 2024
Processing time: 206 Days and 0.4 Hours
Genetic and epigenetic alterations are related to metabolic dysfunction-associated steatotic liver disease (MASLD) pathogenesis.
To evaluate micro (mi)RNAs and lipophagy markers in an experimental model of metabolic dysfunction-associated steatohepatitis (MASH).
Adult male Sprague Dawley rats were randomized into two groups: Control group (n = 10) fed a standard diet; and intervention group (n = 10) fed a high-fat-choline-deficient diet for 16 weeks. Molecular evaluation of li
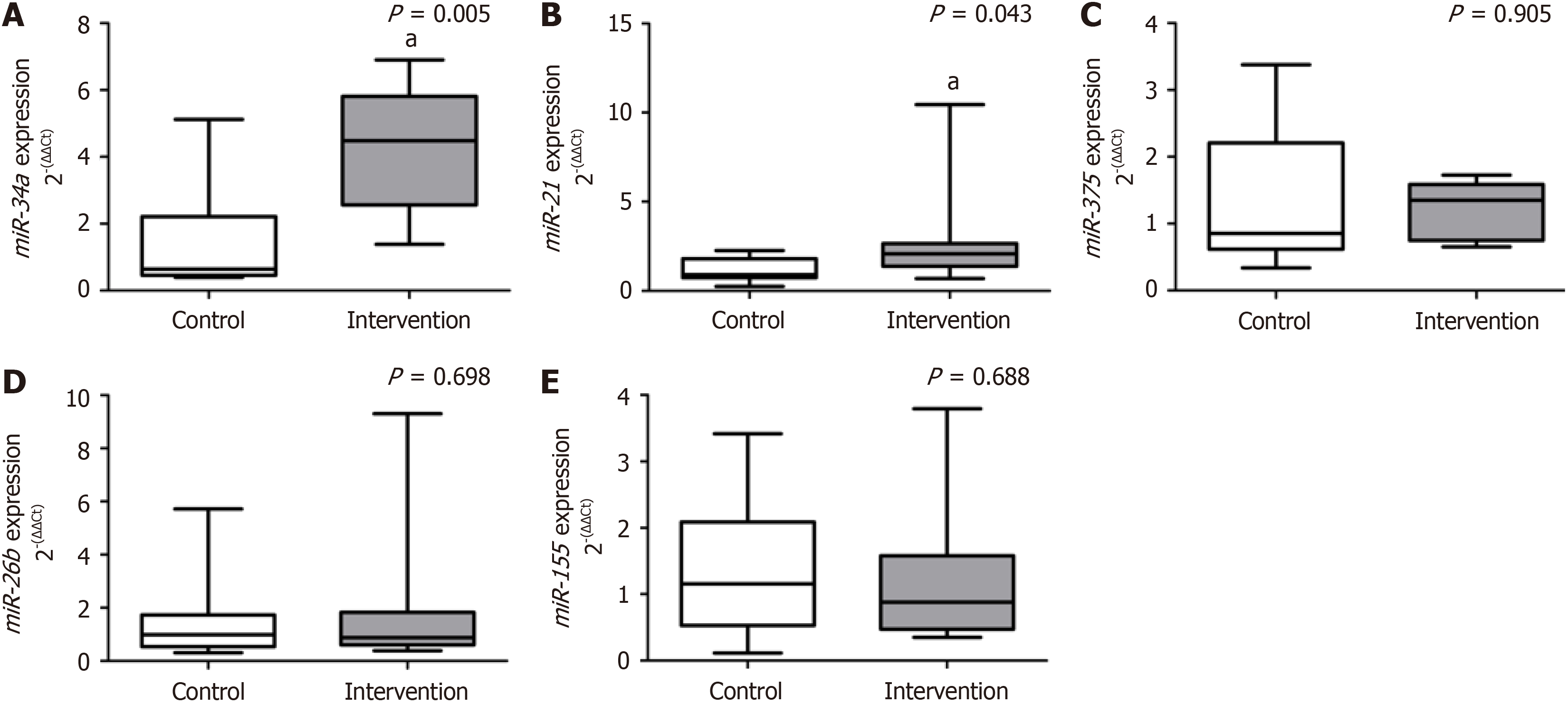
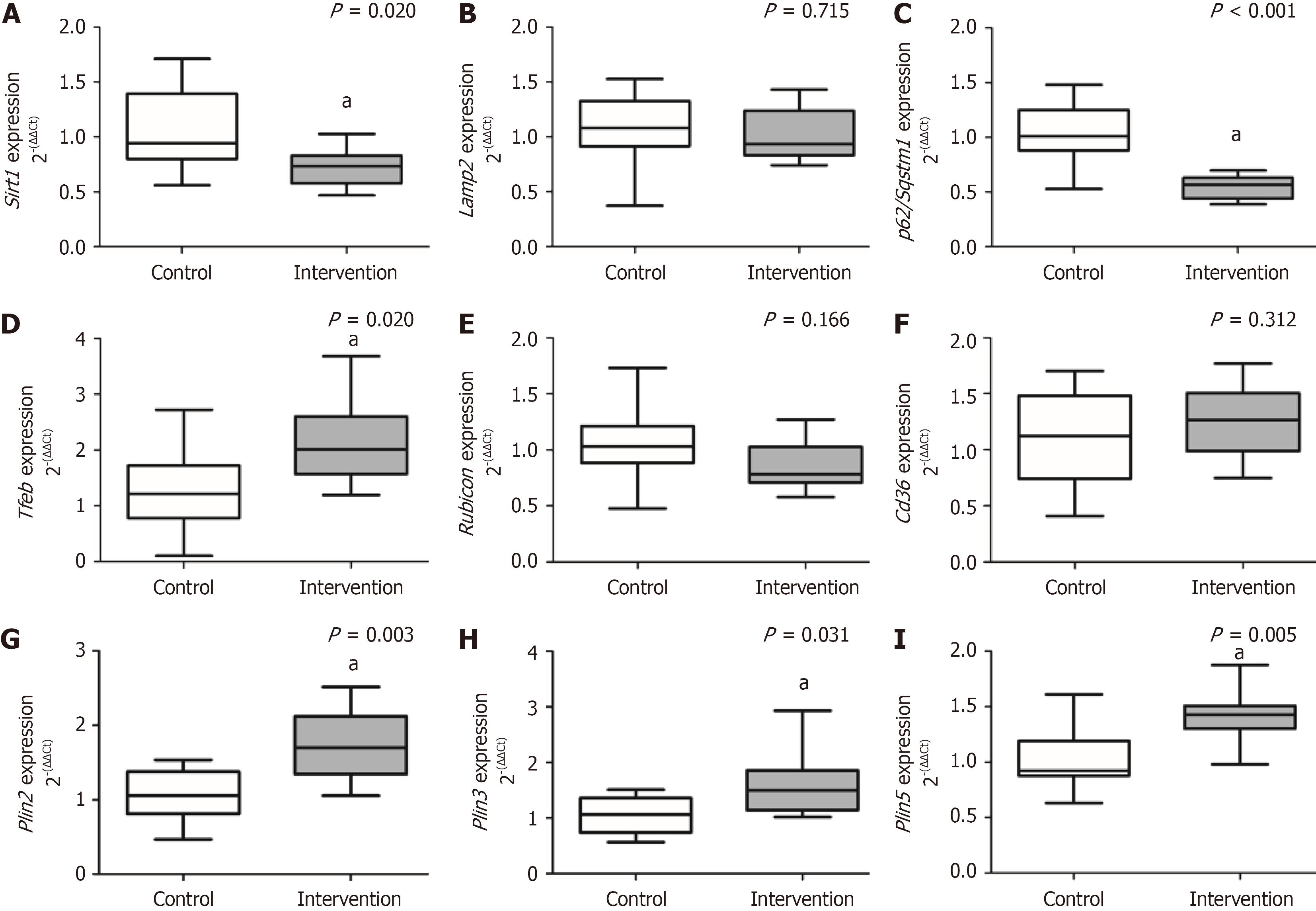
Animals in the intervention group developed MASH and showed a significant decrease in sirtuin-1 (P = 0.020) and p62/sequestosome-1 (P < 0.001); the opposite was reported for transcription factor-EB (P = 0.020), Plin2 (P = 0.003), Plin3 (P = 0.031), and Plin5 (P = 0.005) compared to the control group. There was no significant difference between groups for lysosome-associated membrane proteins-2 (P = 0.715), rubicon (P = 0.166), and Cd36 (P = 0.312). The intervention group showed a significant increase in miR-34a (P = 0.005) and miR-21 (P = 0.043) compared to the control. There was no significant difference between groups for miR-375 (P = 0.905), miR-26b (P = 0.698), and miR-155 (P = 0.688).
Animals with MASH presented expression changes in markers related to lysosomal stress and autophagy as well as in miRNAs related to inflammation and fibrogenesis, processes that promote MASLD progression.
Core Tip: The model of metabolic dysfunction-associated steatohepatitis (MASH) can evaluate pathophysiological mechanisms and therapeutic targets for metabolic dysfunction-associated steatotic liver disease (MASLD). Animals with MASH showed increased expression of miR-34a and miR-21, which contribute to the inflammatory process and fibrogenesis of the disease. Additionally, the lipophagy process was reduced due to increased perilipin gene expression and decreased sirtuin-1. Lysosomal stress and autophagic activity increased in MASH through increased expression of transcription factor-EB and decreased levels of p62/sequestosome-1. Alterations in the expression of these genes are related to the inflammatory process and fibrogenesis, which promote the progression of MASLD.
- Citation: Schütz F, Longo L, Keingeski MB, Filippi-Chiela E, Uribe-Cruz C, Álvares-da-Silva MR. Lipophagy and epigenetic alterations are related to metabolic dysfunction-associated steatotic liver disease progression in an experimental model. World J Hepatol 2024; 16(12): 1468-1479
- URL: https://www.wjgnet.com/1948-5182/full/v16/i12/1468.htm
- DOI: https://dx.doi.org/10.4254/wjh.v16.i12.1468
Metabolic dysfunction-associated steatotic liver disease (MASLD) is a multifactorial pathology involving genetic, metabo
Although there has been progress in understanding the underlying factors related to the pathogenesis of the disease, its mechanisms are not fully elucidated[5,6]. In recent years, epigenetics, which explores the mechanisms by which phenotypes are altered through non-DNA sequence variation, has become the target of studies[7]. Also, micro (mi)RNAs[7] and dysregulated autophagy[8] can influence progression. Several miRNAs, such as miR-34a, miR-375, miR-26b, miR-21, and miR-155, are involved in the modulation of autophagy and can influence the expression or suppression of genes responsible for worsening liver damage through de novo lipogenesis, inflammatory processes, proapoptotic mechanisms, and cell death, leading to the progression of liver injury to hepatocellular carcinoma[9-12]. However, the relationship between miRNAs and autophagy in MASLD remains unclear[12].
Lipophagy, a specific form of autophagy in which lipid droplets are first engulfed by autophagosomes and next degraded in lysosomes, specifically aims to eliminate excess lipids in the liver and maintain lipid homeostasis in cells[5,8]. It is known that the blockage of lipophagy promotes the accumulation of hepatocellular lipids, and its activation causes the clearance of hepatocellular lipid droplets[8]. However, these mechanisms are not fully understood, and promoting lipophagy may be an effective strategy in preventing or delaying MASLD progression[5,8,13].
There is currently no fully approved therapy for MASH by any regulatory agency. However, several clinical protocols are being implemented directing the treatment to specific liver targets, such as hepatocyte dysregulation and inflammation pathways associated with fibrosis in addition to extrahepatic targets, which include the gut-liver signaling axis[1,14]. Therefore, experimental models play a fundamental role in the evaluation of the pathophysiological mechanisms related with the evolution of MASLD and in the evaluation of possible therapeutic targets[15,16]. Therefore, this study aimed to evaluate epigenetic alterations and the mechanisms of autophagy involved in the lipophagy process in an experimental model of MASH mimicking metabolic abnormalities found in humans.
Twenty adult (60-day-old) male Sprague Dawley rats weighing 280-350 g were included in this MASLD experimental study. Rats were maintained on a standard 12-h light-dark cycle in a temperature-controlled environment (22 ± 2 °C). The animals were housed in polypropylene cages with sawdust-lined flooring. The Institutional Ethics Committee approved all experiments and procedures for the use of animals (protocol number: 2022-0117). The procedures for the use of scientific animals were conducted in accordance with the Guide for the Care and Use of Laboratory Animals (8th ed, 2011) and law No. 11.794 (Brazil, 2008).
After becoming accustomed to the laboratory, the animals were randomized by weight into two groups, as described in a previous publication[15]: A control group (n = 10) received standard diet (Nuvilab CR-1; Quimtia SA, Colombo, Brazil); and an intervention group (n = 10) received a high-fat and choline-deficient (HFCD) diet, consisting of 31.5% total fat and enriched with 54.0% trans fatty acids (Rhoster Ltda, Sao Paulo, Brazil) for MASH induction. Both groups received water and food ad libitum during the experiment (Figure 1). The rats were subjected to anesthesia with isoflurane and eutha
The serum circulating miRNAs were extracted through miRNeasy serum/plasma kit (Qiagen, Hilden, Germany), and the procedures were described elsewhere[17]. The sequences and codes of the assessed miRNAs are described in Supplementary Table 1. Values were calculated by the formula 2-(ΔΔCt).
The total RNA was extracted from fragments of liver tissue using the RNeasy mini kit (Qiagen) following the manu
Sample size estimation was performed using the WINPEPI 11.20 program (Brixton Health, Israel) based on a published study by our research group that demonstrated the development of liver damage in an experimental model of MASH[15]. Normality was verified for all variables using Shapiro-Wilk test and histograms. Student t and Mann-Whitney U tests were performed. Quantitative variables were expressed as mean ± standard deviation or median and interquartile ranges (25th-75th). Pearson correlation coefficient was performed with a moderate (0.3 < r < 0.6), strong (0.6 < r < 0.9), or very strong (0.9 < r < 1.0) correlation were adopted. We also performed a correlation analysis between all variables to integrate all gene expression data. P ≤ 0.05 was considered statistically significant. Data were analyzed using the Statistical Package for Social Sciences (SPSS version 28.0; IBM Corp., Armonk, NY, United States).
In previous studies, we demonstrated that animals in the intervention group developed MASH after ingestion of HFCD for 16 weeks[15,18]. In this experimental model, there was a substantial deposition of body fat, changes in miRNAs levels, receptors, mediators, and inflammatory cytokines that contributed to the development and progression of liver damage, in addition to increasing the risk of developing cardiovascular disease. Regarding the histopathological evaluation, we demonstrated that these animals developed MASH with inflammation, ballooning, and intense deposition of collagen fibers in the hepatic tissue[15,18]. The same animal tissues were used in the present study.
The results obtained from the gene expression of circulating miRNAs are reported in Figure 2. We reported a significant increase in miR-34a (P = 0.005) and miR-21 (P = 0.043) gene expression in the intervention group compared to the control group. We did not observe a significant difference between the experimental groups for the gene expression of miR-375 (P = 0.905), miR-26b (P = 0.698), and miR-155 (P = 0.688).
The results obtained for the expression of genes involved in hepatic lipophagy are demonstrated in Figure 3. There was a significant decrease in gene expression of Sirt1 (P = 0.020) and p62/Sqstm1 (P < 0.001) in the intervention group compared to the control group. The intervention group showed a significant increase in the gene expression of Tfeb (P = 0.020), Plin2 (P = 0.003), Plin3 (P = 0.031), and Plin5 (P = 0.005) compared to the control group. We did not observe a significant difference between the experimental groups for the expression of Lamp2 (P = 0.715), rubicon (P = 0.166), and Cd36 (P = 0.312).
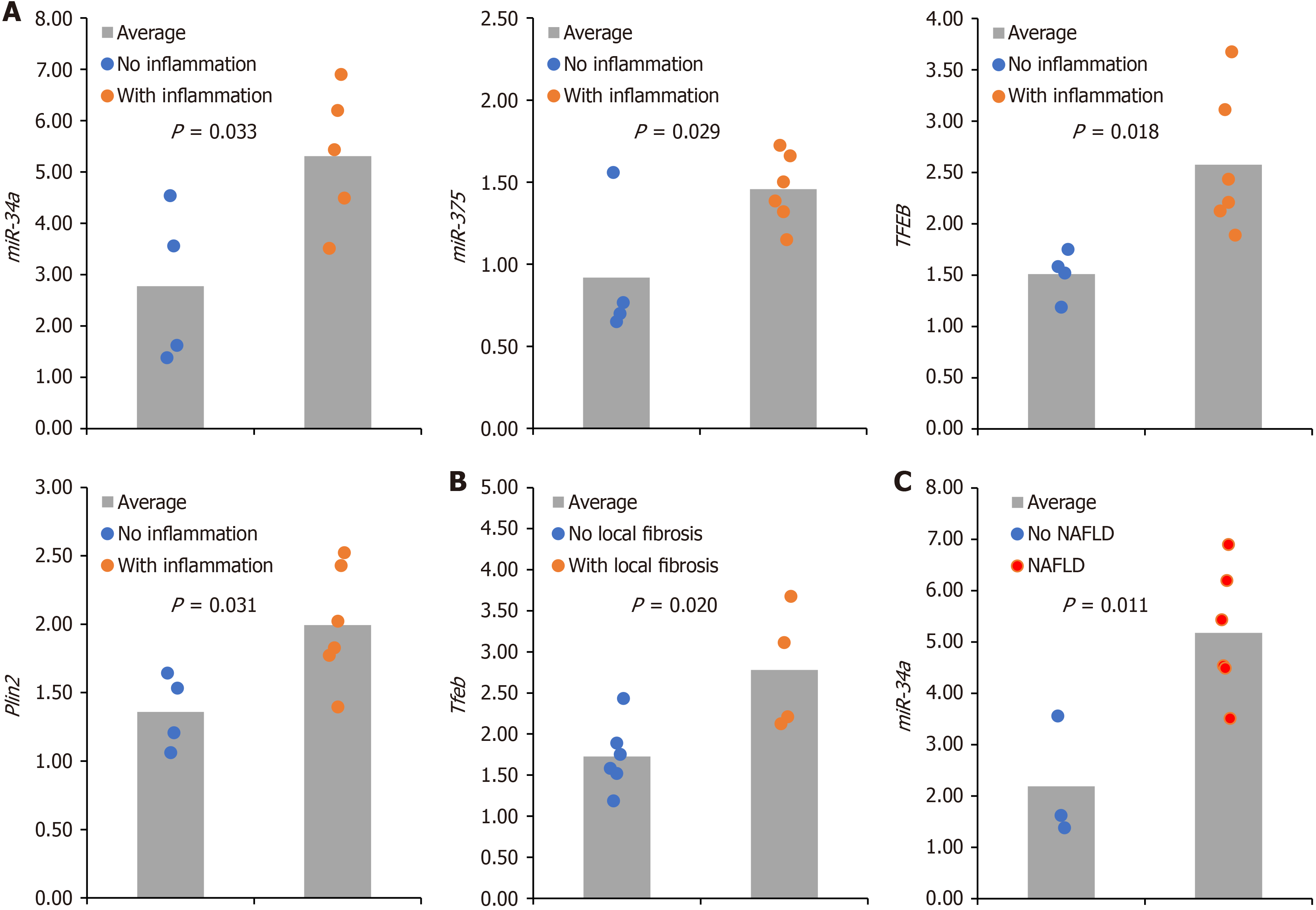
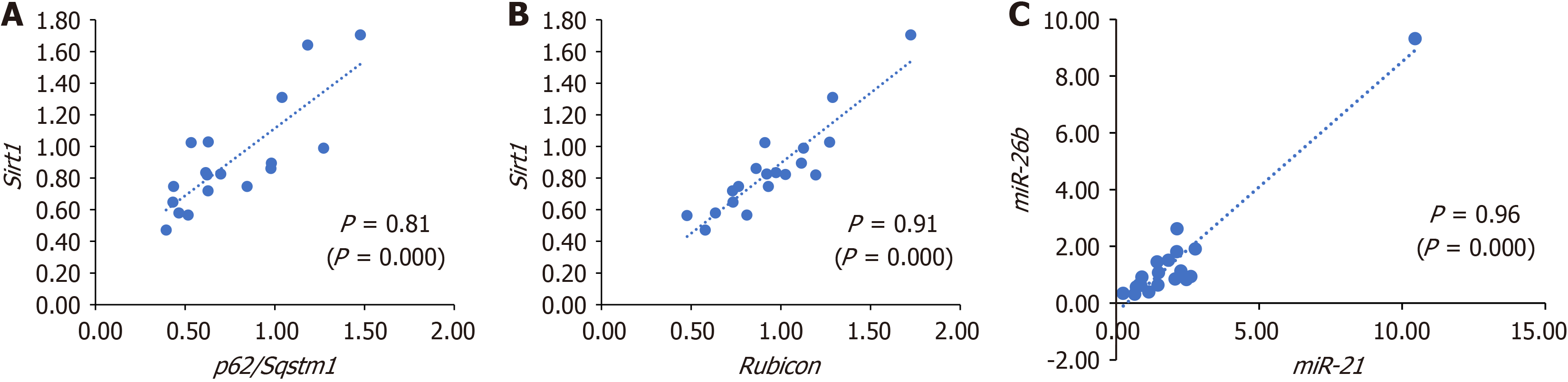
We then evaluated the relationship between histopathological and molecular markers. We divided the intervention group according to each histopathological variable into ‘present’ or ‘absent’ (e.g., with hepatic inflammation vs no hepatic inflammation; with hepatic fibrosis vs no hepatic fibrosis). After that, we compared the averages of genes and miRNA expression between groups. The results are demonstrated in Figure 4. We observed higher levels of miR-34a (P = 0.033), miR-375 (P = 0.029), Tfeb (P = 0.018), and Plin2 (P = 0.033) in animals with inflammation in tissue. We also found higher levels of Tfeb (P = 0.020) in the group of animals with fibrosis and higher levels of miR-34a (P = 0.011) in animals with MASLD compared to the group without MASLD. Finally, looking for potential cross-regulation between genes and miRNAs, we did a correlation analysis between circulating miRNAs and gene expression. The results are shown in Figure 5. We found a statistically significant correlation between Sirt1 and p62/Sqstm1 (P = 0.81; P = 0.000), between Sirt1 and rubicon (P = 0.91; P = 0.000), and between miR-26b and miR-21 (P = 0.96; P = 0.000). All other correlations are shown in the Table 1. Altogether, these integrative analyses suggest potential relations between histopathological and molecular markers in MASLD. Furthermore, correlation analysis suggests potential crosstalk and interplay between genes controlling autophagy, lipophagy, and lysosomal function.
| Variables | mRNAs | Gene expression of hepatic lipophagy | ||||||||||||
| miR-375 | miR-21 | miR-26b | miR-155 | p62/Sqstm1 | Rubicon | Cd36 | Tfeb | Plin2 | Plin3 | Plin5 | Lamp2 | Sirt1 | ||
| miRNAs | miR-34a | 0.245 | 0.573a | 0.303 | -0.105 | -0.389 | -0.002 | 0.480a | 0.470a | 0.455 | 0.159 | 0.317 | 0.075 | -0.397 |
| miR-375 | 0.111 | 0.220 | 0.196 | 0.040 | 0.086 | -0.091 | -0.008 | -0.107 | -0.087 | -0.246 | -0.444 | -0.144 | ||
| miR-21 | 0.9622,b | 0.579b | -0.389 | -0.273 | -0.035 | 0.227 | 0.099 | -0.173 | 0.501a | -0.040 | -0.376 | |||
| miR-26b | 0.7091,b | -0.071 | -0.14 | -0.072 | 0.023 | -0.157 | -0.290 | 0.366 | -0.099 | -0.245 | ||||
| miR-155 | 0.177 | 0.141 | -0.007 | -0.209 | -0.318 | -0.004 | 0.098 | -0.347 | 0.002 | |||||
| Gene expression of hepatic lipophagy | p62/Sqstm1 | 0.7761,b | 0.313 | -0.165 | -0.283 | -0.250 | -0.334 | 0.065 | 0.8061,b | |||||
| Rubicon | 0.488a | 0.357 | 0.043 | -0.107 | -0.194 | 0.104 | 0.9092,b | |||||||
| Cd36 | 0.6241,b | 0.488a | 0.553a | 0.289 | 0.460a | 0.339 | ||||||||
| Tfeb | 0.7881,b | 0.266 | 0.6791,b | 0.428 | 0.275 | |||||||||
| Plin2 | 0.466 | 0.6551,b | 0.199 | -0.024 | ||||||||||
| Plin3 | 0.378 | 0.138 | -0.142 | |||||||||||
| Plin5 | 0.256 | -0.066 | ||||||||||||
| Lamp2 | 0.192 | |||||||||||||
In this study, conducted in a nutritional experimental model, we reported a significant decrease in the gene expression of Sirt1 and p62/Sqstm1 in animals with MASH compared to controls. It is known that p62/SQSTM1 serves as a necessary receptor for selective macroautophagy, and its reduction indicates an impairment of the autophagic flux[19,20]. Fur
It is important to contextualize the animal model used to develop this study, as demonstrated in previous manuscripts[15,18]. In the studies using the same model, we showed that after 16 weeks of unrestricted access to an HFCD diet, the model resulted in significant fat accumulation in both the body and liver and activation of miRNAs, receptors, mediators, and inflammatory cytokines[15]. Additionally, we observed alterations in gut microbiota composition accompanied by an increase in the gene expression of markers related to intestinal permeability[15,18]. This model induced histopathological changes akin to those seen in human MASH, including significant collagen fiber deposition in liver tissue[15]. Fur
MiRNAs are extremely stable and act in inflammatory mechanisms and hepatic fibrogenesis culminating in the progression of MASLD[24]. In our experiment, we demonstrated a significant increase in miR-34a gene expression in animals with MASH compared with healthy controls. Also, we observed higher levels of miR-34a in animals from the intervention group showing hepatic inflammation. Consistent with this result, increased miR-34a gene expression is associated with more advanced stages of MASLD, and therefore its mechanism of action is an attractive target for future intervention in addition to its function as a biomarker for diagnosing and monitoring the progression of MASLD[25-27].
Furthermore, in our study, we revealed a decrease in Sirt1 gene expression in animals with MASH compared with the no intervention group. It was already demonstrated that adipose triglyceride lipase promotes lipophagy via SIRT1[28]. Also consistent with our finding, a previous study reported that the increase in miR-34 family expression contributed to the suppression of SIRT1 and peroxisome proliferator-activated receptor-α expression, both regulators of the storage pathways of lipid metabolism that contribute to the exacerbation of hepatic steatosis[29].
The intervention group also showed an increase in miR-21 expression, which was induced by unsaturated fatty acids and acts on lipid accumulation, inflammation, and liver fibrosis[30,31]. In a previous study conducted in the same experimental model, we reported hepatic and systemic inflammation and an increased risk of developing cardiovascular disease in animals with MASH. These findings were consistent with what was observed in this study regarding the mechanism of action of miR-21[15,18]. Moreover, experimental studies report that knockdown or deletion of miR-21 in mice fed a high-fat diet can reduce lipogenesis and hepatic steatosis by modulating the expression of transcription factors involved in fatty acid uptake, de novo lipogenesis, gluconeogenesis, and glucose production[32,33]. However, expression of miR-21 as an early diagnostic biomarker for MASLD provides mixed results, and further studies are needed before we can translate bedside observations to the clinical setting[30].
In this study, when comparing the experimental groups, no significant difference was found in the gene expression of miR-375, miR-26b, and miR-155. Nevertheless, it is worth noting that we observed higher levels of miR-375 in rats from the intervention group, which also showed hepatic inflammation. Interestingly, we found a strong correlation between miR-26b and miR-21. This correlation was already reported before in a very different model that analyzed the miRNAs in human saliva in response to stress[34].
It is worth remembering that the gene expression of miR-375, miR-26b, and miR-155 has been associated with inflammatory cell recruitment, modulation of autophagy, and tumorigenesis by acting on regulators of cell differentiation in hepatocellular carcinoma[12,35,36]. In this study, gene expression of these miRNAs was examined in animals with initial liver injury, i.e. without cirrhosis and/or hepatocellular carcinoma, which may explain the lack of significant differences between experimental groups. Finally, it is known that the variability and heterogeneity in the expression of these miRNAs is dependent on the liver disease and its developmental stage, which requires further studies to elucidate these mechanisms.
Impairments in hepatic autophagy play a critical role in the evolution of MASLD by promoting the storage of triglycerides in lipid droplets[37]. Autophagy refers to the degradation of nonspecific cytoplasmic proteins as an adaptive response to extracellular and intracellular stress, i.e. a mechanism regulating cell death[37,38]. The autophagy process is regulated by several markers, including p62/SQSTM1 and Beclin-1, which are differentially expressed depending on the organ and the developmental stage of the lesion[39]. Our experiment showed a significant decrease in p62/Sqstm1 gene expression in animals with MASH compared with healthy controls. The lack of autophagy leads to the accumulation of p62, a process detrimental to liver cells that can contribute to the progression of MASLD and liver fibrosis[19,38]. In addition, deletion of p62/SQSTM1 has been shown to promote hyperphagia-induced obesity due to abnormal leptin signaling as well as insulin resistance, impaired glucose tolerance, and hepatic steatosis[40].
However, Sahani et al[41] reported that the expression level of p62/Sqstm1 does not always inversely correlate with autophagic activity, as this process is influenced by transcriptional alterations, amino acid availability from lysosomes, and autophagic degradation[41]. In addition, we speculate that posttranscriptional regulation could explain what is really happening. Accordingly, the promoter may be more active and transcribe more, but there may be miRNAs regulating the levels of miRNA of p62/SQSTM1.
Some studies show the regulation of p62/SQSTM1 levels by miR-372 and miR-100[42,43]. The decrease in p62/Sqstm1 gene expression in animals with MASH could be responsible for the accumulation of lipophagosomes since SQSTM1 is described as important for lipophagy, at least in an in vitro model with ethanol[44]. We also observed a strong correlation between the expression of p62/Sqstm1 and Sirt1. It was already demonstrated that Sirt1 is upregulated to deacetylate and stabilize p62/Sqstm1 in hepatocellular carcinoma[45]. It is also worth highlighting that even though we did not find a significant difference between the experimental groups for the expression of rubicon, we found a strong correlation between the expression of rubicon and Sirt1. It is known that rubicon is a negative regulator of autophagosome-lysosome fusion, and an elevation of RUBICON protein in samples taken from patients with nonalcoholic fatty liver disease (MASLD former name) was reported[46,47].
At the molecular level, autophagy is essentially controlled by autophagy-related genes (ATGs) and adaptors, especially p62/SQSTM1. These genes are responsible for mediating autophagosome formation and component uptake into the organelle[48,49]. It is worth mentioning that there are studies demonstrating that polymorphisms in ATG genes, speci
In the autophagy process, TFEB has been identified as a promoter of ATGs and consequently as a key regulator of lipid metabolism and lysosomal function[8,52,53]. Here we show an increase in Tfeb gene expression in animals with MASH. Also, we observed a significant increase in Tfeb gene expression when comparing the presence or absence of inflammation and the presence or absence of fibrosis in the intervention group. Nutritional status (starvation and/or caloric restriction) and lysosomal stress can promote transcriptional regulation of TFEB, which plays a key role in hepatic lipid metabolism[8,52,54]. Thus, it is possible to speculate that after ingestion of HFCD for 16 weeks, MASH animals showed an increase in Tfeb expression through lysosomal stress. In this sense, TFEB agonists, such as some clinically approved drugs (e.g., digoxin or the marine-derived natural product (e.g., ikarugamycin), or other drugs that mirror the cellular effects of hunger and/or caloric restriction are promising therapeutic options for common metabolic disorders such as obesity and fatty liver as it is a key regulator of lipid metabolism and lysosomal biogenesis and function[54].
Hepatocytes store triacylglycerides (TAG) in lipid droplets due to an overload of free fatty acids[55,56]. Access to the surface of lipid droplets is controlled by PLINs that regulate TAG storage and hydrolysis through lipolysis and lipophagy[55,56]. In our study, animals with MASH showed a significant increase in gene expression of Plin2, Plin3, and Plin5 compared with healthy animals. In the PLIN protein family, PLIN2 is the most abundant member in steatosis of mice and humans, and its expression is strongly correlated with lesion severity, which are results that the data obtained in our study corroborate[55-57].
Experimental models with PLIN2 deficiency have invariably shown a decrease in hepatic TAG, leading to reduced liver inflammation and fibrosis, which supports its role as a target for MASH therapy[56-58]. Degradation of PLIN2 and PLIN3 by chaperone-mediated autophagy (CMA) is a prerequisite for lipolysis to occur[59,60]. Increased expression of Plin2 and Plin3 and/or inactivity of CMA promotes decreased lipophagy and intracellular lipid accumulation, as corroborated by our study[59,60]. In this study high levels of Plin2 gene expression were observed in animals from the intervention group showing hepatic inflammation.
Another perilipin, PLIN3 has been shown to be required for the recruitment of proteins involved with lipophagy, and stimulation of this mechanism may contribute to liver protection and thus provide a therapeutic strategy for MASLD[59]. PLIN5 is expressed in highly oxidative tissues and is an early response to excessive lipid uptake in hepatocytes[53,61]. It is key in the regulation of hepatic lipophagy and fatty acid oxidation by SIRT1. PLIN5 translocates to the nucleus and promotes transcriptional regulation of SIRT1, which contributes to the increase in expression of downstream target genes that control mitochondrial biogenesis, oxidative metabolism, and lipophagy[55,62]. Data that corroborate the results obtained in this study reported not only a decrease in the expression of p62/Sqstm1 in animals with MASH but also an increase in lipophagy flux[62].
In this study, no significant difference was observed between groups for Lamp2a gene expression; however, the results reported in the literature on the activity of this marker in CMA are controversial[63,64]. LAMP2A supports the CMA process, which occurs at a late stage of the autophagy process, and therefore we can justify the results obtained in this study because the animals showed liver damage at an early stage[63].
Autophagic degradation of lipids or lipophagy was first reported in 2009[65]. The mechanisms associated with this process are complex, but the general picture of how abnormal lipophagy causes MASLD has not been elucidated[66,67]. Moreover, several epigenetic and lipophagy markers show potential therapeutic effects in nutritional models of MASH[6,67]. In this study, we evaluated the markers based on gene expression only, which should be considered a limitation. However, we found that animals fed a MASH-inducing diet exhibited an increased expression of miR-34a and miR-21, which contribute to the inflammatory process and fibrogenesis of the disease[25,26,30].
The process of lipophagy was also reduced in the face of increased PLIN gene expression and decreased Sirt1. Lysosomal stress and autophagic activity increased, as evidenced by an increase in Tfeb and a decrease in p62/Sqstm1 expression, respectively. Further evaluation by single-cell sequencing, proteomics, and next-generation sequencing will allow characterization of the mechanistic details of lipophagy and epigenetic perturbations in metabolic dysfunction-associated fatty liver disease progression to fully exploit its potential as a target for chemoprevention of lesions[67].
We demonstrated in this animal model that p62/SQSTM1 and Sirt1 decreased and Plin2, Plin3, and Plin5 increased in MASLD in comparison to controls, suggesting that both autophagy and lipophagy play a role in the disease development. In addition, our study highlighted the differential expression of miRNAs, such as the increased expression of miR-34a and miR-21, which are associated with hepatic inflammation and lipid metabolism. This observation further supports their potential role as biomarkers and therapeutic targets in MASLD progression. The mechanisms underlying the pathophysiology of MASLD caused by epigenetic processes have not yet been elucidated. Therefore, further preclinical studies are essential to better explain the MASLD metabolic pathway.
| 1. | Friedman SL, Neuschwander-Tetri BA, Rinella M, Sanyal AJ. Mechanisms of NAFLD development and therapeutic strategies. Nat Med. 2018;24:908-922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1376] [Cited by in RCA: 3204] [Article Influence: 400.5] [Reference Citation Analysis (2)] |
| 2. | Huang DQ, El-Serag HB, Loomba R. Global epidemiology of NAFLD-related HCC: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2021;18:223-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 840] [Cited by in RCA: 1368] [Article Influence: 273.6] [Reference Citation Analysis (0)] |
| 3. | Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5322] [Cited by in RCA: 7942] [Article Influence: 794.2] [Reference Citation Analysis (8)] |
| 4. | Xian YX, Weng JP, Xu F. MAFLD vs. NAFLD: shared features and potential changes in epidemiology, pathophysiology, diagnosis, and pharmacotherapy. Chin Med J (Engl). 2020;134:8-19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 78] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 5. | Nassir F. NAFLD: Mechanisms, Treatments, and Biomarkers. Biomolecules. 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 267] [Article Influence: 66.8] [Reference Citation Analysis (0)] |
| 6. | Xu X, Poulsen KL, Wu L, Liu S, Miyata T, Song Q, Wei Q, Zhao C, Lin C, Yang J. Targeted therapeutics and novel signaling pathways in non-alcohol-associated fatty liver/steatohepatitis (NAFL/NASH). Signal Transduct Target Ther. 2022;7:287. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 180] [Cited by in RCA: 254] [Article Influence: 63.5] [Reference Citation Analysis (0)] |
| 7. | Juanola O, Martínez-López S, Francés R, Gómez-Hurtado I. Non-Alcoholic Fatty Liver Disease: Metabolic, Genetic, Epigenetic and Environmental Risk Factors. Int J Environ Res Public Health. 2021;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 203] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 8. | Martinez-Lopez N, Singh R. Autophagy and Lipid Droplets in the Liver. Annu Rev Nutr. 2015;35:215-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 279] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 9. | Huang PS, Liao CJ, Huang YH, Yeh CT, Chen CY, Tang HC, Chang CC, Lin KH. Functional and Clinical Significance of Dysregulated microRNAs in Liver Cancer. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 10. | Rodrigues PM, Afonso MB, Simão AL, Islam T, Gaspar MM, O'Rourke CJ, Lewinska M, Andersen JB, Arretxe E, Alonso C, Santos-Laso Á, Izquierdo-Sanchez L, Jimenez-Agüero R, Eizaguirre E, Bujanda L, Pareja MJ, Prip-Buus C, Banales JM, Rodrigues CMP, Castro RE. miR-21-5p promotes NASH-related hepatocarcinogenesis. Liver Int. 2023;43:2256-2274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 28] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 11. | Xie D, Yuan P, Wang D, Jin H, Chen H. Expression and prognostic significance of miR-375 and miR-221 in liver cancer. Oncol Lett. 2017;14:2305-2309. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 12. | Jin F, Wang Y, Li M, Zhu Y, Liang H, Wang C, Wang F, Zhang CY, Zen K, Li L. MiR-26 enhances chemosensitivity and promotes apoptosis of hepatocellular carcinoma cells through inhibiting autophagy. Cell Death Dis. 2017;8:e2540. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 178] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 13. | Gao Y, Zhang W, Zeng LQ, Bai H, Li J, Zhou J, Zhou GY, Fang CW, Wang F, Qin XJ. Exercise and dietary intervention ameliorate high-fat diet-induced NAFLD and liver aging by inducing lipophagy. Redox Biol. 2020;36:101635. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 203] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 14. | Trauner M, Fuchs CD. Novel therapeutic targets for cholestatic and fatty liver disease. Gut. 2022;71:194-209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 149] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 15. | Longo L, Tonin Ferrari J, Rampelotto PH, Hirata Dellavia G, Pasqualotto A, P Oliveira C, Thadeu Schmidt Cerski C, Reverbel da Silveira T, Uribe-Cruz C, Álvares-da-Silva MR. Gut Dysbiosis and Increased Intestinal Permeability Drive microRNAs, NLRP-3 Inflammasome and Liver Fibrosis in a Nutritional Model of Non-Alcoholic Steatohepatitis in Adult Male Sprague Dawley Rats. Clin Exp Gastroenterol. 2020;13:351-368. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 16. | Santhekadur PK, Kumar DP, Sanyal AJ. Preclinical models of non-alcoholic fatty liver disease. J Hepatol. 2018;68:230-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 290] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 17. | Michalczuk MT, Longo L, Keingeski MB, Basso BS, Guerreiro GTS, Ferrari JT, Vargas JE, Oliveira CP, Uribe-Cruz C, Cerski CTS, Filippi-Chiela E, Álvares-da-Silva MR. Rifaximin on epigenetics and autophagy in animal model of hepatocellular carcinoma secondary to metabolic-dysfunction associated steatotic liver disease. World J Hepatol. 2024;16:75-90. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (3)] |
| 18. | Longo L, Rampelotto PH, Filippi-Chiela E, de Souza VEG, Salvati F, Cerski CT, da Silveira TR, Oliveira CP, Uribe-Cruz C, Álvares-da-Silva MR. Gut dysbiosis and systemic inflammation promote cardiomyocyte abnormalities in an experimental model of steatohepatitis. World J Hepatol. 2021;13:2052-2070. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 19. | Carotti S, Aquilano K, Zalfa F, Ruggiero S, Valentini F, Zingariello M, Francesconi M, Perrone G, Alletto F, Antonelli-Incalzi R, Picardi A, Morini S, Lettieri-Barbato D, Vespasiani-Gentilucci U. Lipophagy Impairment Is Associated With Disease Progression in NAFLD. Front Physiol. 2020;11:850. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 90] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 20. | Shroff A, Nazarko TY. SQSTM1, lipid droplets and current state of their lipophagy affairs. Autophagy. 2023;19:720-723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 40] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 21. | Zhu Y, Hu S, Pan X, Gopoju R, Cassim Bawa FN, Yin L, Xu Y, Zhang Y. Hepatocyte Sirtuin 6 Protects against Atherosclerosis and Steatohepatitis by Regulating Lipid Homeostasis. Cells. 2023;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 22. | Choudhary NS, Duseja A. Genetic and epigenetic disease modifiers: non-alcoholic fatty liver disease (NAFLD) and alcoholic liver disease (ALD). Transl Gastroenterol Hepatol. 2021;6:2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 65] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 23. | Jonas W, Schürmann A. Genetic and epigenetic factors determining NAFLD risk. Mol Metab. 2021;50:101111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 118] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 24. | Di Mauro S, Scamporrino A, Filippello A, Di Pino A, Scicali R, Malaguarnera R, Purrello F, Piro S. Clinical and Molecular Biomarkers for Diagnosis and Staging of NAFLD. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 25. | Ezaz G, Trivedi HD, Connelly MA, Filozof C, Howard K, L Parrish M, Kim M, Herman MA, Nasser I, Afdhal NH, Jiang ZG, Lai M. Differential Associations of Circulating MicroRNAs With Pathogenic Factors in NAFLD. Hepatol Commun. 2020;4:670-680. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 26. | Zhang Z, Moon R, Thorne JL, Moore JB. NAFLD and vitamin D: Evidence for intersection of microRNA-regulated pathways. Nutr Res Rev. 2023;36:120-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 27. | Xin S, Zhan Q, Chen X, Xu J, Yu Y. Efficacy of serum miRNA test as a non-invasive method to diagnose nonalcoholic steatohepatitis: a systematic review and meta-analysis. BMC Gastroenterol. 2020;20:186. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 28. | Sathyanarayan A, Mashek MT, Mashek DG. ATGL Promotes Autophagy/Lipophagy via SIRT1 to Control Hepatic Lipid Droplet Catabolism. Cell Rep. 2017;19:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 292] [Article Influence: 32.4] [Reference Citation Analysis (0)] |
| 29. | Wang JM, Qiu Y, Yang Z, Kim H, Qian Q, Sun Q, Zhang C, Yin L, Fang D, Back SH, Kaufman RJ, Yang L, Zhang K. IRE1α prevents hepatic steatosis by processing and promoting the degradation of select microRNAs. Sci Signal. 2018;11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 98] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 30. | Zhang T, Yang Z, Kusumanchi P, Han S, Liangpunsakul S. Critical Role of microRNA-21 in the Pathogenesis of Liver Diseases. Front Med (Lausanne). 2020;7:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 104] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 31. | Rodrigues PM, Afonso MB, Simão AL, Carvalho CC, Trindade A, Duarte A, Borralho PM, Machado MV, Cortez-Pinto H, Rodrigues CM, Castro RE. miR-21 ablation and obeticholic acid ameliorate nonalcoholic steatohepatitis in mice. Cell Death Dis. 2017;8:e2748. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 80] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 32. | Wu H, Ng R, Chen X, Steer CJ, Song G. MicroRNA-21 is a potential link between non-alcoholic fatty liver disease and hepatocellular carcinoma via modulation of the HBP1-p53-Srebp1c pathway. Gut. 2016;65:1850-1860. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 144] [Cited by in RCA: 166] [Article Influence: 16.6] [Reference Citation Analysis (1)] |
| 33. | Calo N, Ramadori P, Sobolewski C, Romero Y, Maeder C, Fournier M, Rantakari P, Zhang FP, Poutanen M, Dufour JF, Humar B, Nef S, Foti M. Stress-activated miR-21/miR-21* in hepatocytes promotes lipid and glucose metabolic disorders associated with high-fat diet consumption. Gut. 2016;65:1871-1881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 106] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 34. | Wiegand C, Heusser P, Klinger C, Cysarz D, Büssing A, Ostermann T, Savelsbergh A. Stress-associated changes in salivary microRNAs can be detected in response to the Trier Social Stress Test: An exploratory study. Sci Rep. 2018;8:7112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 35. | Fu X, Dong B, Tian Y, Lefebvre P, Meng Z, Wang X, Pattou F, Han W, Wang X, Lou F, Jove R, Staels B, Moore DD, Huang W. MicroRNA-26a regulates insulin sensitivity and metabolism of glucose and lipids. J Clin Invest. 2015;125:2497-2509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 210] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 36. | Blaya D, Pose E, Coll M, Lozano JJ, Graupera I, Schierwagen R, Jansen C, Castro P, Fernandez S, Sidorova J, Vasa-Nicotera M, Solà E, Caballería J, Trebicka J, Ginès P, Sancho-Bru P. Profiling circulating microRNAs in patients with cirrhosis and acute-on-chronic liver failure. JHEP Rep. 2021;3:100233. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 37. | Zhang L, Zhang Z, Li C, Zhu T, Gao J, Zhou H, Zheng Y, Chang Q, Wang M, Wu J, Ran L, Wu Y, Miao H, Zou X, Liang B. S100A11 Promotes Liver Steatosis via FOXO1-Mediated Autophagy and Lipogenesis. Cell Mol Gastroenterol Hepatol. 2021;11:697-724. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 65] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 38. | Rusten TE, Stenmark H. p62, an autophagy hero or culprit? Nat Cell Biol. 2010;12:207-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 201] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 39. | Jiang Y, Chen D, Gong Q, Xu Q, Pan D, Lu F, Tang Q. Elucidation of SIRT-1/PGC-1α-associated mitochondrial dysfunction and autophagy in nonalcoholic fatty liver disease. Lipids Health Dis. 2021;20:40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 40. | Miura I, Komine S, Okada K, Wada S, Warabi E, Uchida F, Oh S, Suzuki H, Mizokami Y, Shoda J. Prevention of non-alcoholic steatohepatitis by long-term exercise via the induction of phenotypic changes in Kupffer cells of hyperphagic obese mice. Physiol Rep. 2021;9:e14859. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 41. | Sahani MH, Itakura E, Mizushima N. Expression of the autophagy substrate SQSTM1/p62 is restored during prolonged starvation depending on transcriptional upregulation and autophagy-derived amino acids. Autophagy. 2014;10:431-441. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 260] [Cited by in RCA: 316] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 42. | Feng L, Ma Y, Sun J, Shen Q, Liu L, Lu H, Wang F, Yue Y, Li J, Zhang S, Lin X, Chu J, Han W, Wang X, Jin H. YY1-MIR372-SQSTM1 regulatory axis in autophagy. Autophagy. 2014;10:1442-1453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 60] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 43. | Ge YY, Shi Q, Zheng ZY, Gong J, Zeng C, Yang J, Zhuang SM. MicroRNA-100 promotes the autophagy of hepatocellular carcinoma cells by inhibiting the expression of mTOR and IGF-1R. Oncotarget. 2014;5:6218-6228. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 72] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 44. | Wang L, Zhou J, Yan S, Lei G, Lee CH, Yin XM. Ethanol-triggered Lipophagy Requires SQSTM1 in AML12 Hepatic Cells. Sci Rep. 2017;7:12307. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 43] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 45. | Feng L, Chen M, Li Y, Li M, Hu S, Zhou B, Zhu L, Yu L, Zhou Q, Tan L, An H, Wang X, Jin H. Sirt1 deacetylates and stabilizes p62 to promote hepato-carcinogenesis. Cell Death Dis. 2021;12:405. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 46. | Qian H, Chao X, Williams J, Fulte S, Li T, Yang L, Ding WX. Autophagy in liver diseases: A review. Mol Aspects Med. 2021;82:100973. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 270] [Article Influence: 54.0] [Reference Citation Analysis (0)] |
| 47. | Tanaka S, Hikita H, Tatsumi T, Sakamori R, Nozaki Y, Sakane S, Shiode Y, Nakabori T, Saito Y, Hiramatsu N, Tabata K, Kawabata T, Hamasaki M, Eguchi H, Nagano H, Yoshimori T, Takehara T. Rubicon inhibits autophagy and accelerates hepatocyte apoptosis and lipid accumulation in nonalcoholic fatty liver disease in mice. Hepatology. 2016;64:1994-2014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 292] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 48. | Nishimura T, Tooze SA. Emerging roles of ATG proteins and membrane lipids in autophagosome formation. Cell Discov. 2020;6:32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 239] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 49. | Levine B, Kroemer G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell. 2019;176:11-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1024] [Cited by in RCA: 2233] [Article Influence: 319.0] [Reference Citation Analysis (0)] |
| 50. | Baselli GA, Jamialahmadi O, Pelusi S, Ciociola E, Malvestiti F, Saracino M, Santoro L, Cherubini A, Dongiovanni P, Maggioni M, Bianco C, Tavaglione F, Cespiati A, Mancina RM, D'Ambrosio R, Vaira V, Petta S, Miele L, Vespasiani-Gentilucci U, Federico A, Pihlajamaki J, Bugianesi E, Fracanzani AL, Reeves HL, Soardo G, Prati D, Romeo S, Valenti LV; EPIDEMIC Study Investigators. Rare ATG7 genetic variants predispose patients to severe fatty liver disease. J Hepatol. 2022;77:596-606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 75] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 51. | Lin YC, Chang PF, Lin HF, Liu K, Chang MH, Ni YH. Variants in the autophagy-related gene IRGM confer susceptibility to non-alcoholic fatty liver disease by modulating lipophagy. J Hepatol. 2016;65:1209-1216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 75] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 52. | Schulze RJ, Sathyanarayan A, Mashek DG. Breaking fat: The regulation and mechanisms of lipophagy. Biochim Biophys Acta Mol Cell Biol Lipids. 2017;1862:1178-1187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 175] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 53. | Klionsky DJ, Abdel-Aziz AK, Abdelfatah S, Abdellatif M, Abdoli A, Abel S, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy. 2021;17:1-382. |
| 54. | Wang C, Niederstrasser H, Douglas PM, Lin R, Jaramillo J, Li Y, Oswald NW, Zhou A, McMillan EA, Mendiratta S, Wang Z, Zhao T, Lin Z, Luo M, Huang G, Brekken RA, Posner BA, MacMillan JB, Gao J, White MA. Small-molecule TFEB pathway agonists that ameliorate metabolic syndrome in mice and extend C. elegans lifespan. Nat Commun. 2017;8:2270. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 126] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 55. | Griffin JD, Bejarano E, Wang XD, Greenberg AS. Integrated Action of Autophagy and Adipose Tissue Triglyceride Lipase Ameliorates Diet-Induced Hepatic Steatosis in Liver-Specific PLIN2 Knockout Mice. Cells. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 56. | Graffmann N, Ring S, Kawala MA, Wruck W, Ncube A, Trompeter HI, Adjaye J. Modeling Nonalcoholic Fatty Liver Disease with Human Pluripotent Stem Cell-Derived Immature Hepatocyte-Like Cells Reveals Activation of PLIN2 and Confirms Regulatory Functions of Peroxisome Proliferator-Activated Receptor Alpha. Stem Cells Dev. 2016;25:1119-1133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 87] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 57. | Najt CP, Senthivinayagam S, Aljazi MB, Fader KA, Olenic SD, Brock JR, Lydic TA, Jones AD, Atshaves BP. Liver-specific loss of Perilipin 2 alleviates diet-induced hepatic steatosis, inflammation, and fibrosis. Am J Physiol Gastrointest Liver Physiol. 2016;310:G726-G738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 114] [Article Influence: 11.4] [Reference Citation Analysis (1)] |
| 58. | McManaman JL, Bales ES, Orlicky DJ, Jackman M, MacLean PS, Cain S, Crunk AE, Mansur A, Graham CE, Bowman TA, Greenberg AS. Perilipin-2-null mice are protected against diet-induced obesity, adipose inflammation, and fatty liver disease. J Lipid Res. 2013;54:1346-1359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 194] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 59. | Garcia-Macia M, Santos-Ledo A, Leslie J, Paish HL, Collins AL, Scott RS, Watson A, Burgoyne RA, White S, French J, Hammond J, Borthwick LA, Mann J, Bolaños JP, Korolchuk VI, Oakley F, Mann DA. A Mammalian Target of Rapamycin-Perilipin 3 (mTORC1-Plin3) Pathway is essential to Activate Lipophagy and Protects Against Hepatosteatosis. Hepatology. 2021;74:3441-3459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 42] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 60. | Kaushik S, Cuervo AM. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat Cell Biol. 2015;17:759-770. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 493] [Cited by in RCA: 573] [Article Influence: 52.1] [Reference Citation Analysis (0)] |
| 61. | Asimakopoulou A, Engel KM, Gassler N, Bracht T, Sitek B, Buhl EM, Kalampoka S, Pinoé-Schmidt M, van Helden J, Schiller J, Weiskirchen R. Deletion of Perilipin 5 Protects Against Hepatic Injury in Nonalcoholic Fatty Liver Disease via Missing Inflammasome Activation. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 62. | Zhang E, Cui W, Lopresti M, Mashek MT, Najt CP, Hu H, Mashek DG. Hepatic PLIN5 signals via SIRT1 to promote autophagy and prevent inflammation during fasting. J Lipid Res. 2020;61:338-350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 63. | Das S, Seth RK, Kumar A, Kadiiska MB, Michelotti G, Diehl AM, Chatterjee S. Purinergic receptor X7 is a key modulator of metabolic oxidative stress-mediated autophagy and inflammation in experimental nonalcoholic steatohepatitis. Am J Physiol Gastrointest Liver Physiol. 2013;305:G950-G963. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 54] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 64. | Sharma S, Mells JE, Fu PP, Saxena NK, Anania FA. GLP-1 analogs reduce hepatocyte steatosis and improve survival by enhancing the unfolded protein response and promoting macroautophagy. PLoS One. 2011;6:e25269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 166] [Cited by in RCA: 224] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 65. | Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131-1135. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3239] [Cited by in RCA: 3239] [Article Influence: 190.5] [Reference Citation Analysis (0)] |
| 66. | Maan M, Peters JM, Dutta M, Patterson AD. Lipid metabolism and lipophagy in cancer. Biochem Biophys Res Commun. 2018;504:582-589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 198] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 67. | Li HY, Peng ZG. Targeting lipophagy as a potential therapeutic strategy for nonalcoholic fatty liver disease. Biochem Pharmacol. 2022;197:114933. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/licenses/by-nc/4.0/.