Published online Jan 27, 2023. doi: 10.4254/wjh.v15.i1.107
Peer-review started: September 21, 2022
First decision: October 12, 2022
Revised: October 21, 2022
Accepted: November 14, 2022
Article in press: November 14, 2022
Published online: January 27, 2023
Processing time: 117 Days and 3.7 Hours
Hepatitis C virus is known for its oncogenic potential, especially in hepatocellular carcinoma and non-Hodgkin lymphoma. Several studies have shown that chronic hepatitis C (CHC) has an increased risk of the development of colorectal cancer (CRC).
To analyze this positive relationship and develop an artificial intelligence (AI)-based tool using machine learning (ML) algorithms to stratify these patient popu
To develop the AI automated calculator, we applied ML to train models to predict the probability and the number of adenomas detected on colonoscopy. Data sets were split into 70:30 ratios for training and internal validation. The Scikit-learn standard scaler was used to scale values of continuous variables. Colonoscopy fin
Of 415 patients, 206 had colonoscopy results. On internal validation, the Bernoulli naive Bayes model predicted the probability of adenoma detection with the highest accuracy of 56%, precision of 55%, recall of 55%, and F1 measure of 54%. Support vector regressor predicted the number of adenomas with the least mean absolute error of 0.905.
Our AI-based tool can help providers stratify patients with CHC for early referral for screening colonoscopy. Along with providing a numerical percentage, the calculator can also comment on the number of adenomatous polyps a gastroenterologist can expect, prompting a higher adenoma detection rate.
Core Tip: Hepatitis C is associated with a wide array of extra-hepatic manifestations. In this study, we evaluated the incidence of colorectal adenomas and adenoma detection rates in hepatitis C patients. We developed an artificial intelligence-based tool to guide physicians in the detection and diagnosis of pre-malignant and malignant colorectal pathologies in these patient populations.
- Citation: Singh Y, Gogtay M, Yekula A, Soni A, Mishra AK, Tripathi K, Abraham G. Detection of colorectal adenomas using artificial intelligence models in patients with chronic hepatitis C. World J Hepatol 2023; 15(1): 107-115
- URL: https://www.wjgnet.com/1948-5182/full/v15/i1/107.htm
- DOI: https://dx.doi.org/10.4254/wjh.v15.i1.107
Hepatitis C is the most common blood-borne infection worldwide despite being gravely under-diagnosed[1,2]. Data from the National Health and Nutrition Examination Survey 2013-2016 and four other high-risk populations; homeless, incarcerated, active-duty military, and nursing homes estimates that approximately 4.1 million persons in the United States (approximately 1.7% of the population) were hepatitis C virus (HCV) antibody-positive indicating past exposure and 2.4 million persons (approximately 1% of the population) were HCV RNA positive indicating an active infection[3].
Although HCV is thought of as a primary disease of the liver, it has a wide array of extrahepatic manifestations, including skin, blood, lymphatic and intestinal pathologies[4,5]. The United States Chronic Hepatitis Cohort Study showed an increased incidence of rectal cancer with increased mortality in chronic HCV, but not colon cancers[4-6]. Colorectal cancer (CRC) is the third most commonly diagnosed cancer and the second most common cause of cancer-related death worldwide[3,7].
The current gold standard for diagnosing CRC is colonoscopy. The United States Preventive Services Task Force, American College of Gastroenterology, and American Cancer Society recommend screening for CRC starting at age 45 years in average-risk individuals[8-10]. There are no specific screening guidelines for CRC in patients with hepatitis C, despite there being an increased association.
Artificial intelligence (AI) is the new silk road. It is a technique that allows machines to store large amounts of data from various sources, process them accurately, reason, and even simulate human intelligence to provide a plan for clinical treatment. AI has augmented medical research on an enormous scale in recent years and continues to do so. It has significantly helped reduce the workload of clinicians and healthcare staff[11].
There are multiple facets of AI used in gastroenterology such as convolutional neural network (CNN), deep learning (DL), machine learning (ML), and computer-aided design (CAD)[12]. ML is a subset of AI, DL is a subset of ML and neural networks make up the backbone of DL algorithms. Multiple AI-assisted systems such as EndoBRAIN CAD by Kudo et al[13] and a CNN-based auxiliary model by Ding et al[14] have been used to detect and diagnose bowel pathologies including colonic adenomas and neoplasms with higher sensitivity and specificity as compared to trained endoscopic experts[13,14].
ML is one of the subsets of AI where algorithms are trained for specific tasks. This is being in
The use of AI has shown an increase in both polyp detection rate and adenoma detection rate (ADR) which are the primary colonoscopy quality indicators. Each 1% increase in ADR leads to an approximately 3% decrease in the future risk of cancer[15]. As timely intervention in the form of screening colonoscopy can help detect pre-neoplastic or early stages of CRC[15], it is imperative to develop a tool that can help clinicians distinguish which patients with chronic hepatitis C (CHC) infection require early referral for colonoscopy. Rustagi et al[16] in a case-control study showed a significantly higher number of adenomas detected on screening colonoscopy in the CHC group. In addition, building an ML model which can help predict the expected number of adenomas can reduce missed adenomas, decreasing subsequent cancer and the overall burden of CRC on healthcare. This is the aim of our research in developing an AI/ML-based tool.
This observational study with cross-sectional data collection and analysis was conducted at a com
Several ML models were trained and tested, and the model that could predict percentage probability with the highest accuracy was saved for the deployment stage.
The dataset used to train and test the ML algorithms was collected manually and stored in a comma-separated file format. The dataset contained several attribute vectors from 415 patients [i.e., sex, age, body mass index (BMI), obesity class, oral contraceptive use, significant alcohol use, hypothyroidism, intravenous drug use, diabetes mellitus, human immunodeficiency virus, concomitant statin use, controlled attenuation parameter (CAP) grade, HCV status, genotype, aspartate aminotransferase, alanine aminotransferase, platelet count, hemoglobin A1c, and triglycerides]. Some patients had missing data, which were replaced by zeros for training and testing. The task was to predict the percentage probability of adenoma occurrence in colonoscopy, followed by calculating the number of polyps. For this purpose, two ML models, i.e., classification and regression models, were trained, and the best models were saved from both ML categories.
The dataset was loaded onto a pandas DataFrame. The output label was colorectal adenomatous polyps. Numerical feature vectors were replaced with categorical variables. Categorical attributes such as HCV genotype and gene polymorphisms underwent label encoding. The variables with numerical values were scaled using the scikit-learn StandardScaler function to scale down to the desired range of 0-1. The complete dataset was split into a 70:30 ratio, in which 70% of the total dataset was used for training. The remaining 30% of the entire dataset was used to test internal validity and select the ML model with the highest predictive value.
Several ML algorithms were trained to predict hepatic steatosis and CAP grades using the above-listed vectors. These models included: Support vector classifier, random forest, Bernoulli naïve Bayes (BNB), Gradient boosting classifier (GBC), logistic regression, and stochastic gradient descent classifier. All models were trained using 70% of the dataset. After training, each model was tested using the remaining 30% of the dataset The model with the highest testing accuracy, the GBC model, was chosen for external validity testing.
The GBC model is a set of ML algorithms that additively combines multiple weaker ML models to produce a final predictive model. Our model assigned a binary classification to datasets and used multiple regressions along several decision trees to refine its attempts to predict the steatosis classification correctly. The model graded each attempt on a loss function which evaluates the extent to which the previous tree was inaccurate.
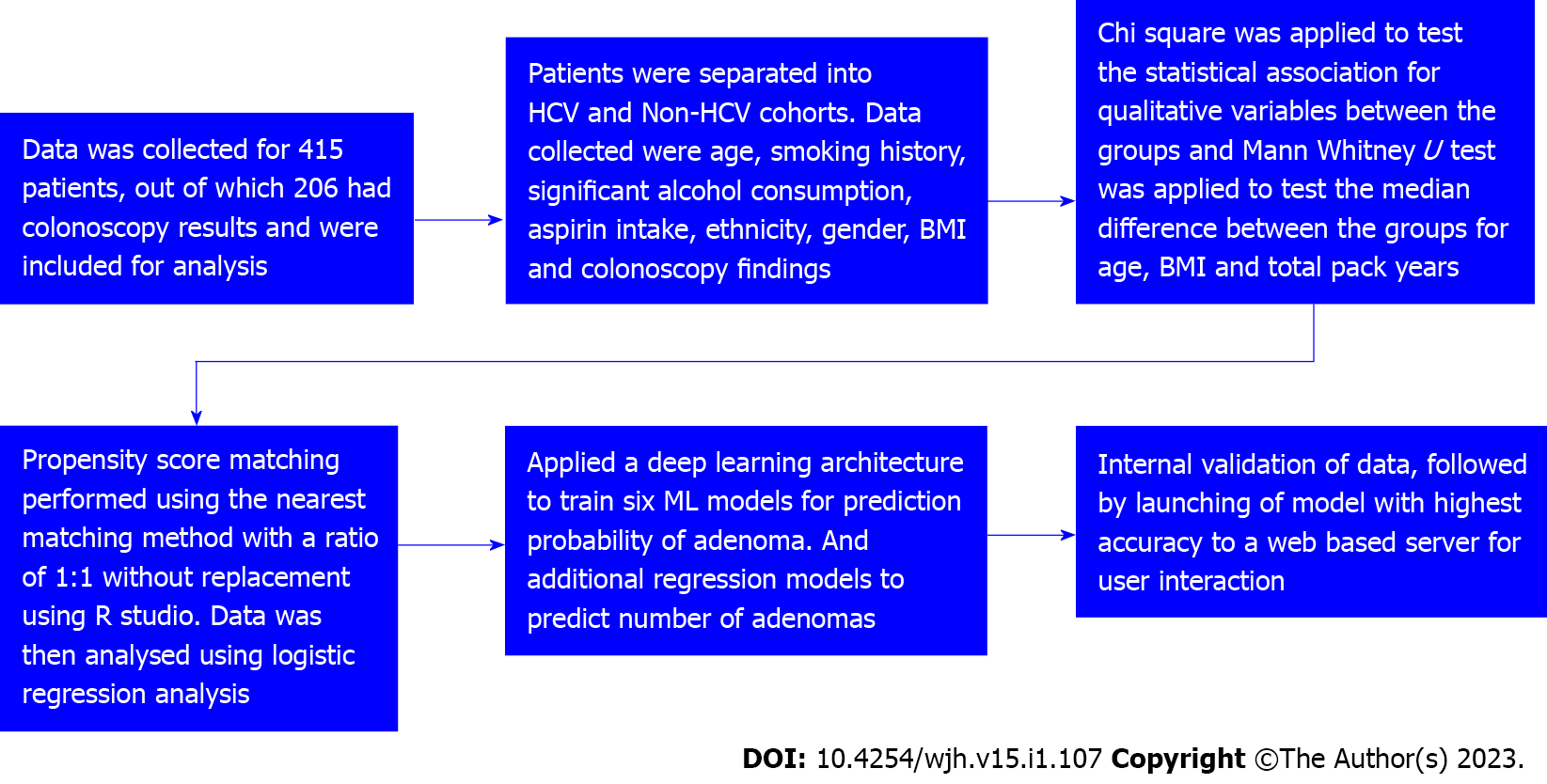
Patients were divided into cases (HCV) and controls (non-HCV). Data was tabulated as described in Table 1. Figure 1 is a strobe diagram with an overview of the methodology and results. The performance of different ML algorithms for training and testing to detect colorectal adenomatous polyps is shown in Table 2.
| Variables | Case (n = 109) | Control (n = 97) | OR | 95%CI | P value |
| Age (mean ± SD in yr) | 62.73 ± 9.146 | 60.20 ± 7.062 | 0.02a | ||
| Age median (yr), n (%) | 49 (50.5) | 58 (53.2) | 0.54 | 0.31-0.95 | 0.03a |
| Gender, n (%) - female | 48 (44) | 46 (47.4) | 0.87 | 0.50-1.51 | 0.62 |
| BMI, median (IQR) | 28 (7) | 32 (7) | 0.001a | ||
| Family history of hepatitis, n (%) | 10 (9.2) | 6 (6.2) | 1.53 | 0.55-4.38 | 0.42 |
| Aspirin use, n (%) | 22 (20.2) | 35 (36.1) | 0.44 | 0.24-0.83 | 0.01a |
| Smoking, n (%) | 2.31 | 1.31-4.07 | 0.004a | ||
| No | 51 (46.8) | 65 (67) | |||
| Yes | 38 (34.9) | 15 (15.5) | |||
| Former | 20 (18.3) | 17 (17.5) | |||
| Total pack years, median (IQR) | 25 (21) | 25 (11) | 0.75 | ||
| Alcohol use, n (%) | 12 (11) | 24 (24.7) | 0.37 | 0.17-0.80 | 0.01a |
| DM, n (%) | 27 (24.8) | 31 (32) | 0.87 | 0.50-1.51 | 0.25a |
| HIV, n (%) | 3 (2.8) | 1 (1) | 2.71 | 0.27-26.56 | 0.62a |
| Adenomatous polyps present | 58 (53.2) | 33 (34) | 2.20 | 1.25-3.87 | 0.006a |
| Bowel preparation (%) | 0.14 | ||||
| Good | 90 (82.6) | 88 (90.7) | |||
| Fair | 11(10.1) | 7 (7.2) | |||
| Poor | 8 (7.3) | 2 (2.1) |
| ML model | Test accuracy (%) | Precision (%) | Recall (%) | F1 score (%) |
| Support vector classifier | 52 | 51 | 51 | 51 |
| Random forest | 53 | 53 | 53 | 53 |
| Bernoulli naïve Bayes | 56 | 55 | 55 | 54 |
| Gradient boosting | 50 | 49 | 49 | 48 |
| Logistic regression | 50 | 48 | 48 | 47 |
| Deep neural networks | 53 | 53 | 53 | 51 |
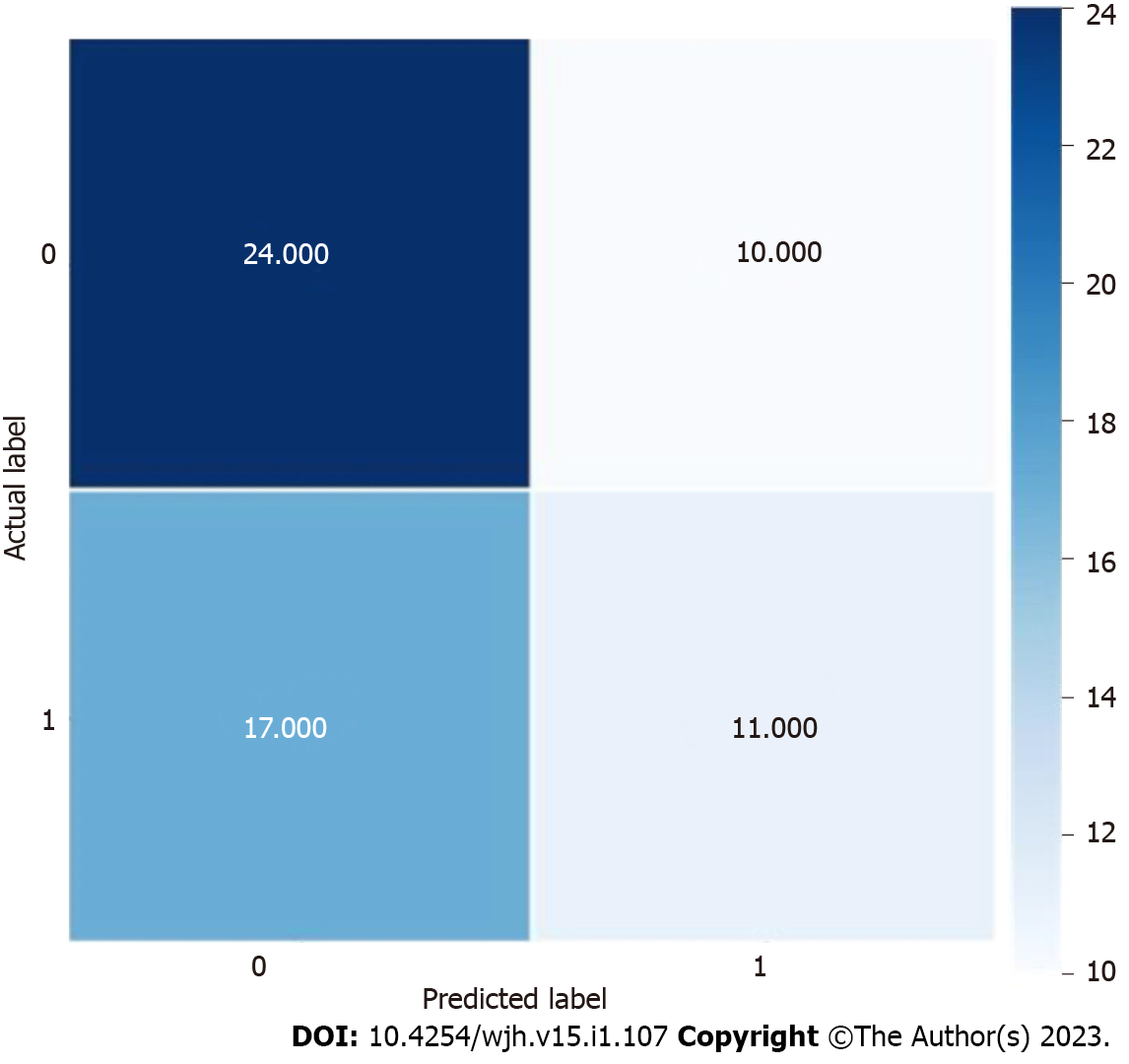
Using the colonoscopy results determined by the pathology of the biopsied polyp as the output, we applied a DL architecture using the above variables to train and test several ML models using a 415-patient dataset. As shown in Table 2, our results demonstrated that the BNB model had the highest testing accuracy (56%), precision (55%), recall (55%), and F1 score (54%). The distribution of actual and predicted labels during internal testing can be seen in Figure 2. As depicted in Table 3, our results demonstrated that the support vector regressor model had the lowest mean absolute error - 0.905, indicating it was the most suitable ML model to calculate an approximate number of polyps.
| ML model | Mean absolute error |
| Linear regression | 1.072 |
| LGBM regressor | 1.106 |
| XGBoost regressor | 1.273 |
| ElasticNet | 0.941 |
| Gradient boosting regressor | 1.139 |
| Support vector regressor | 0.905 |
A flask-based web app was developed using the model with the highest accuracy for the application phase. A flash application programming interface (API) was used to deploy the trained ML model as a web application. The web-based API was then deployed into a web server using Heroku, a cloud application platform (https://adenomadetection.herokuapp.com/).
The initial analyses of the variables have demonstrated that median age of 62 years, with higher BMI, smoking, alcohol use, and concomitant aspirin use are related to adenoma detection with statistical significance among the data collected from patients with CHC who underwent colonoscopies. Using these initial datasets, DL architecture was used to train six ML models, which were prepared and validated using a 70:30 ratio of the dataset. Of the multiple models, the BNB model predicted the probability of adenoma detection with the highest accuracy, precision, and recall. Flask API was used to deploy the ML model, and Heroku web API was used to develop the AI tool. The model training and performance are shown in Tables 2-4.
| ML model | Performance |
| Support vector classifier | Good |
| Random forest | Good |
| Bernoulli naïve Bayes | Optimal |
| Gradient boosting | Inadequate |
| Logistic regression | Inadequate |
| Deep neural networks | Good |
The AI tool may be useful in the clinical setting to triage patients with hepatitis C, who have not received a formal CRC screening to stratify them into high-risk and low-risk groups. This can guide the gastroenterologist during the colonoscopy to help increase the ADRs. This knowledge of the increased risk of CRC incidence in hepatitis C and isolating patients that are high risk can prompt physicians to start CRC screening early and more frequently in these specific subsets.
Although multiple review articles have been published, there are very few clinical studies. To our knowledge, this is one of two retrospective analyses performed comparing CRC in hepatitis C and non-hepatitis C individuals. One of the main limiting factors was that the study was performed in the patient population that visited the primary care clinic at a community hospital in a single city in the northeast United States; it is limited by the geographical and socioeconomic aspects of the patient population. Analysis and validation were carried out on this limited sample size. Another drawback was that the study was primarily a retrospective analysis. Timing of the colonoscopy and the patient having active hepatitis infection might not always be documented, and timing was not specific in many patients, which limited the sample size that was analyzed.
AI application to healthcare has been increasingly pursued in the past decade, changing how we deliver advanced patient care[17,18]. AI and ML ensure the automation of many time-consuming activities and provide better insight into patient data that are not evident to providers[17]. Although the hype behind AI is promising, various barriers prevent the real-world application of these new AI systems. AI has helped in increased interoperability of extensive data, reasonable data-driven decisions involving evidence-based medicine, and hence a higher quality of care.
CRC is a commonly diagnosed malignancy in men and women with increasing mortality[19]. Many epidemiological studies have shown a positive association of CHC infection with extra-hepatic malignancies, especially gastrointestinal malignancies[16,19]. Our study focused on this positive association between CHC and CRC and on using AI and ML models to further ease the diagnosis in these patient populations.
AI is integrated into our everyday life to such an extent that we barely remember what life was before it. This includes face recognition to unlock our phones, self-driven cars, chatbots for almost every business, and even ML-based financial fraud detection. AI-driven models are increasingly utilized to screen, diagnose and monitor multiple clinical conditions. Many AI algorithms have been used in CRC detection. Hu et al[20] performed ML simulations using S-Kohonen, Backpropagation and SVM neural networks, showing the S-Kohonen method’s effectiveness for colon cancer classification[20].
Zhang et al[21] derived a cost-effective and sensitive method for detecting BRAF mutations in CRC using a counter propagation artificial neural network to distinguish mutant BRAF vs wild type[20,21]. Many such ML models are increasingly being utilized in precision oncology to precisely guide diagnosis and management decisions in CRC patients[19-21].
AI tools will potentially transform our practice by leveraging massive amounts of data to personalize care to the right patient, in the right amount, at the right time. These novel tools assist physicians in the detection and early diagnosis of pre-malignant and malignant lesions in general and high-risk populations.
Our AI tool can be further modified based on the treatment of hepatitis C with the new direct-acting antivirals, and how treated and cured hepatitis C alters the incidence of CRC in these groups. Long-term prospective studies, including a subgroup analysis between patients cured of hepatitis C, who had a relapse of the disease, and who refused or were untreated and how it affected CRC detection, would help guide diagnostics. Further validation with randomized controlled trials and multicenter part
Several studies have shown that chronic hepatitis C virus (HCV) increases the risk of developing colorectal cancer (CRC). We conducted a study to analyze this positive relationship. We developed an artificial intelligence (AI) based tool using machine learning (ML) algorithms to stratify these patient populations into risk groups for CRC/adenoma detection.
We acknowledge the increased applications of AI with ML in medicine. Gastroenterology and hepatology have immense potential for AI integration. Hence, to develop an AI automated calculator, we applied ML to train models to predict the probability and the number of adenomas detected on colonoscopy.
Our objective was the create a readily available AI tool in the form of a calculator that healthcare providers throughout the globe can access to predict the prevalence of adenoma/CRC.
We used colonoscopy findings as the gold standard and applied a deep learning architecture to train ML models for prediction. The institutional review board approved the study.
Data on 415 patients were collected. We discovered a higher incidence of adenoma/CRC in patients with chronic HCV in the untreated patient population. On internal validation, the Bernoulli naive Bayes ML model showed the highest predictive accuracy and recall for adenoma detection rates.
Our AI-based tool shows an association between HCV and colorectal adenomas. This tool can help providers stratify their patients at increased risk of CRC and prompt early referral for colonoscopy.
In the future, we would like to see this calculator being used in clinical practice as a preventative measure to increase early diagnosis of high-risk adenomas/CRC.
| 1. | Lauer GM, Walker BD. Hepatitis C virus infection. N Engl J Med. 2001;345:41-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2042] [Cited by in RCA: 2026] [Article Influence: 81.0] [Reference Citation Analysis (0)] |
| 2. | Denniston MM, Klevens RM, McQuillan GM, Jiles RB. Awareness of infection, knowledge of hepatitis C, and medical follow-up among individuals testing positive for hepatitis C: National Health and Nutrition Examination Survey 2001-2008. Hepatology. 2012;55:1652-1661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 270] [Cited by in RCA: 305] [Article Influence: 21.8] [Reference Citation Analysis (1)] |
| 3. | Hofmeister MG, Rosenthal EM, Barker LK, Rosenberg ES, Barranco MA, Hall EW, Edlin BR, Mermin J, Ward JW, Ryerson AB. Estimating Prevalence of Hepatitis C Virus Infection in the United States, 2013-2016. Hepatology. 2019;69:1020-1031. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 364] [Cited by in RCA: 387] [Article Influence: 55.3] [Reference Citation Analysis (0)] |
| 4. | Gill K, Ghazinian H, Manch R, Gish R. Hepatitis C virus as a systemic disease: reaching beyond the liver. Hepatol Int. 2016;10:415-423. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 110] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 5. | Pol S, Vallet-Pichard A, Hermine O. Extrahepatic cancers and chronic HCV infection. Nat Rev Gastroenterol Hepatol. 2018;15:283-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 63] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 6. | Moorman AC, Gordon SC, Rupp LB, Spradling PR, Teshale EH, Lu M, Nerenz DR, Nakasato CC, Boscarino JA, Henkle EM, Oja-Tebbe NJ, Xing J, Ward JW, Holmberg SD; Chronic Hepatitis Cohort Study Investigators. Baseline characteristics and mortality among people in care for chronic viral hepatitis: the chronic hepatitis cohort study. Clin Infect Dis. 2013;56:40-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 138] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 7. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 56656] [Article Influence: 7082.0] [Reference Citation Analysis (134)] |
| 8. | Shaukat A, Kahi CJ, Burke CA, Rabeneck L, Sauer BG, Rex DK. ACG Clinical Guidelines: Colorectal Cancer Screening 2021. Am J Gastroenterol. 2021;116:458-479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 537] [Article Influence: 107.4] [Reference Citation Analysis (0)] |
| 9. | USPSTF guidelines for colorectal cancer prevention released. Inpharma Wkly. 2007;1579:3. [DOI] [Full Text] |
| 10. | Minsky BD. Ask The Experts: Unique management considerations for rectal cancer and colon cancer. Colorectal Canc. 2012;1:205-107. [RCA] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 11. | Kuo CY, Chiu HM. Application of artificial intelligence in gastroenterology: Potential role in clinical practice. J Gastroenterol Hepatol. 2021;36:267-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 12. | Chen HY, Ge P, Liu JY, Qu JL, Bao F, Xu CM, Chen HL, Shang D, Zhang GX. Artificial intelligence: Emerging player in the diagnosis and treatment of digestive disease. World J Gastroenterol. 2022;28:2152-2162. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 4] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 13. | Kudo SE, Misawa M, Mori Y, Hotta K, Ohtsuka K, Ikematsu H, Saito Y, Takeda K, Nakamura H, Ichimasa K, Ishigaki T, Toyoshima N, Kudo T, Hayashi T, Wakamura K, Baba T, Ishida F, Inoue H, Itoh H, Oda M, Mori K. Artificial Intelligence-assisted System Improves Endoscopic Identification of Colorectal Neoplasms. Clin Gastroenterol Hepatol. 2020;18:1874-1881.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 162] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 14. | Ding Z, Shi H, Zhang H, Meng L, Fan M, Han C, Zhang K, Ming F, Xie X, Liu H, Liu J, Lin R, Hou X. Gastroenterologist-Level Identification of Small-Bowel Diseases and Normal Variants by Capsule Endoscopy Using a Deep-Learning Model. Gastroenterology. 2019;157:1044-1054.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 221] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 15. | Corley DA, Levin TR, Doubeni CA. Adenoma detection rate and risk of colorectal cancer and death. N Engl J Med. 2014;370:2541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 73] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 16. | Rustagi T, Zarookian EI, Qasba O, Diez LF. Chronic hepatitis C as a risk factor for colorectal adenoma. Int J Colorectal Dis. 2014;29:75-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (1)] |
| 17. | Bees J. How Artificial Intelligence Use Is Expanding in Health Care. NEJM Catal. 2022;. [DOI] [Full Text] |
| 18. | Gordon W. Moving Past the Promise of AI to Real Uses in Health Care Delivery. NEJM Catal. 2022;. [DOI] [Full Text] |
| 19. | Mitsala A, Tsalikidis C, Pitiakoudis M, Simopoulos C, Tsaroucha AK. Artificial Intelligence in Colorectal Cancer Screening, Diagnosis and Treatment. A New Era. Curr Oncol. 2021;28:1581-1607. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 155] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 20. | Hu HP, Niu ZJ, Bai YP, Tan XH. Cancer classification based on gene expression using neural networks. Genet Mol Res. 2015;14:17605-17611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 21. | Zhang X, Yang Y, Wang Y, Fan Q. Detection of the BRAF V600E Mutation in Colorectal Cancer by NIR Spectroscopy in Conjunction with Counter Propagation Artificial Neural Network. Molecules. 2019;24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 22. | Singh Y, Gogtay M, Gurung S, Trivedi N, Abraham GM. Assessment of Predictive Factors of Hepatic Steatosis Diagnosed by Vibration Controlled Transient Elastography (VCTE) in Chronic Hepatitis C Virus-Infected Patients. JCHIMP. 2022;12. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (2)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Liu S, China; Mijwil MM, Iraq S-Editor: Wang JJ L-Editor: Webster JR P-Editor: Wang JJ