Published online Sep 27, 2021. doi: 10.4254/wjh.v13.i9.1069
Peer-review started: February 22, 2021
First decision: May 3, 2021
Revised: May 12, 2021
Accepted: August 10, 2021
Article in press: August 10, 2021
Published online: September 27, 2021
Processing time: 211 Days and 9.6 Hours
The hepatitis C virus has a high mutation capacity that leads to the emergence of resistance-associated substitutions (RAS). However, the consequence of resistance selection during new direct-acting antiviral drug (DAA) treatment is not nece
Core Tip: The presence of resistance-associated substitutions (RAS) to hepatitis C virus (HCV) treatment is a frequent event. Direct-acting antiviral (DAA) treatment repre
- Citation: Ridruejo E, Pereson MJ, Flichman DM, Di Lello FA. Hepatitis C virus treatment failure: Clinical utility for testing resistance-associated substitutions. World J Hepatol 2021; 13(9): 1069-1078
- URL: https://www.wjgnet.com/1948-5182/full/v13/i9/1069.htm
- DOI: https://dx.doi.org/10.4254/wjh.v13.i9.1069
For years, the only available treatment for chronic hepatitis C virus (HCV) infection was pegylated interferon and its combination with ribavirin (PEG-IFN/RBV) therapy. However, the sustained viral response (SVR) to treatment of infected patients was limited, varying between 42% and 46% for HCV genotype 1, about 60% for HCV genotype 4, and 76% to 80% for HCV genotype 2 or 3[1-5]. The outcomes were troublesome in patients coinfected with human immunodeficiency virus /HCV, whose SVR rates were even lower[6-9]. Fortunately, treatment against HCV infection has improved significantly in the last decade, changing from a nonspecific immunomodulatory therapy with multiple and severe side effects, such as PEG-IFN/RBV, to specific viral target options such as direct-acting antiviral (DAA) drugs against NS3, NS5A, and NS5B proteins. Thus, since the development of the latest generation of DAA drugs, the SVR is achieved in 95% to 99% of treated patients[10]. Although this scenario is very encouraging, the 1% to 5% of patients who do not achieve SVR are the pitfall of DAA therapy. Therefore, the current complex challenge is to rescue patients who fail to one or more DAA schemes.
Response to treatment with PEG-IFN/RBV was associated with viral variants and single nucleotide polymorphisms[11-17]. The introduction of DAA drugs implied a higher specific and targeted pressure on the virus, which favor the selection of resistance-associated substitutions (RAS) to different antiviral agents. In this context, virological failure was associated with RAS that may be present either from the beginning (baseline RAS) of treatment or acquired during it[18].
Naturally, HCV produces approximately 1012 viral particles per day[19]. In addition, the viral replication complex lacks proofreading activity, resulting in a large amount of viral variants in each infected individual. Although, in theory, all possible mutants can be produced in just 1 day, not all of them are able to remain in the population. That is because some viral genome regions have constraints and most mutations generate variants that impair viral fitness and, therefore, do not proliferate. As a result, a large mutant spectrum known as quasispecies is generated[20]. The quasispecies, that represent the lowest level of viral diversity, drives virus adaptability and constitute the greatest challenge to treatment resistance[20].
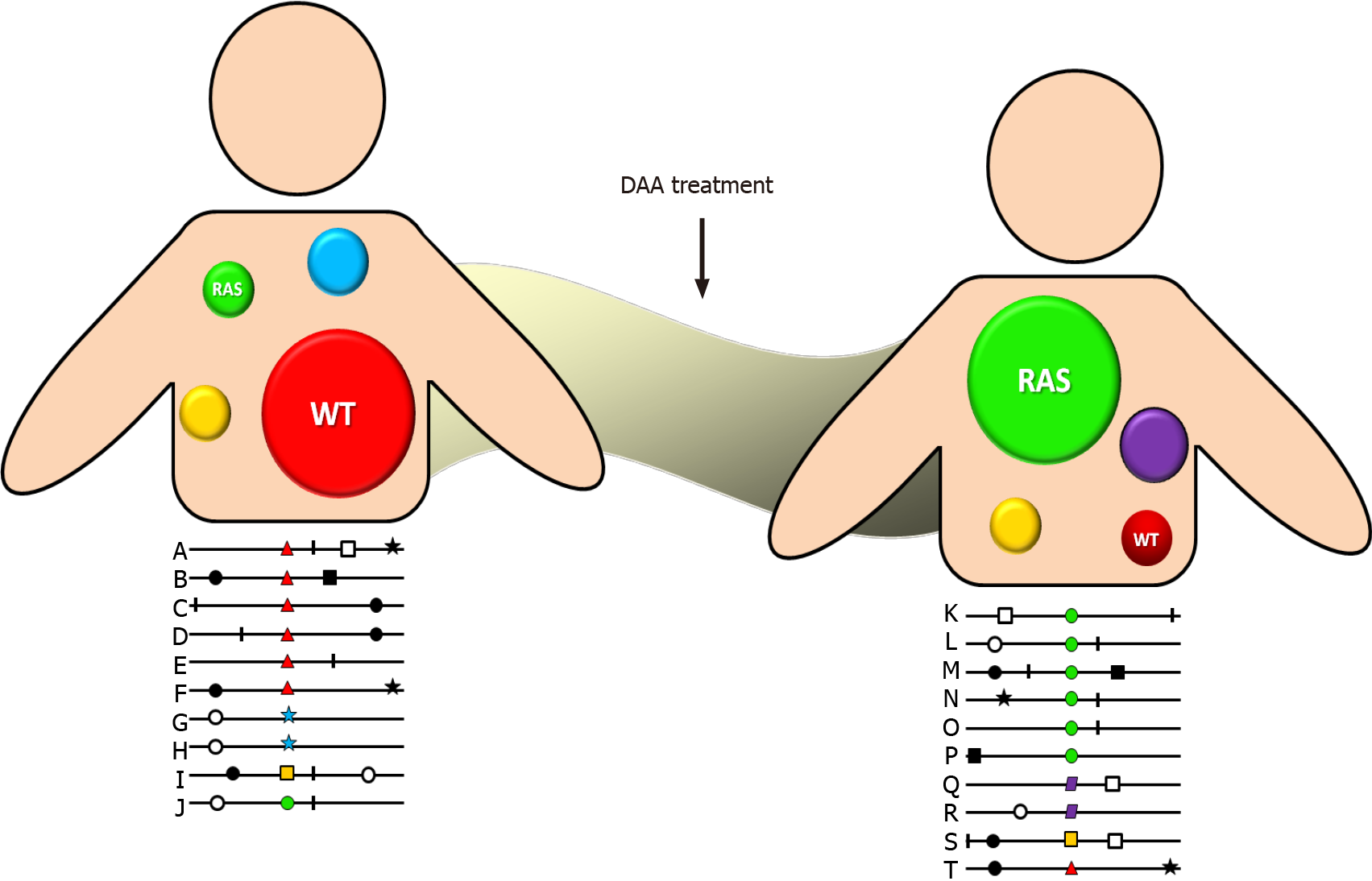
DAA drug administration inhibits wild-type HCV variants allowing the selection of reduced susceptibility variants, which present a better fitness to this environment. Although initially they do so inefficiently, over time they develop compensatory amino acid substitutions that have a higher fitness and increase the frequency of resistant variants in the quasispecies spectrum (Figure 1). Additionally, each antiviral drug has a different genetic barrier that is characterized by a threshold above which DAA resistance develops. The threshold is determined by several factors including the number of required nucleotide mutations, the level of resistance, and the viral variant fitness. Therefore, even when a viral variant with a RAS emerges, it does not mean that it is sufficient to lead to therapeutic failure. In that way, therapeutic outcome will depend on a finely poised and complex balance between the DAA genetic barrier and viral-resistant variant fitness. Consequently, a highly resistant strain with a low replication capacity will be clinically less relevant than a less resistant one that replicates more efficiently. Fortunately, more powerful DAA drugs with greater genetic barriers have been developed in the last few years[21].
In preclinical and in real-life studies, the reported prevalence rate of baseline RAS is around 5% to 40%, raising concern of the effect on reducing SVR[22-28]. Eventually, the adverse impact of baseline RAS could be minimized by extending treatment duration or optimizing DAA regimens. However, that is not always clinically possible, as a considerable proportion of treatment failures are caused by RAS acquired during it[29,30]. Table 1 shows the most relevant RAS reported for the currently most used DAA drugs.
| Drug family | Drug | Licensed for genotype | RAS |
| NS3 inhibitors | Glecaprevir (GLE) | 1, 2, 3, 4, 5, 6 | 36M, 56H, 156G/V, 168A/K/L/R |
| Grazoprevir (GZR) | 1, 4 | 36A/L/M, 56H/F, 155G/K/L/Q/T/S, 156T/V, 168any | |
| Paritaprevir (PTV) | 1, 4 | 36A/M, 43L, 155C/K/Q/H, 156T/V, 168any | |
| Voxilaprevir (VOX) | 1, 2, 3, 4, 5, 6 | 36A/G/L/M, 41K/R/S/V, 43L/S7V, 54S, 55A/I, 56H/F, 80K/L, 122D/G/N, 155G/K/N/K/T/W, 156L/S/T/V, 168any | |
| NS5A inhibitors | Daclatasvir (DCV) | 1, 3, 4 | 24H, 28A/M/S/T, 30D/E/G/H/K/N/Q/R/S/T, 31I/F/M/V, 32L/del, 58A/D/N/S, 62L, 93C/H/N/R/S/W |
| Elbasvir (EBR) | 1, 3, 4 | 28G/T, 30G/H/K/R/V/Y, 31F/M/V, 58D, 93C/H/N | |
| Ledipasvir (LDV) | 1, 3, 4, 5, 6 | 24N/G, 28A/M/T, 30E/G/H/K/N/R/S/T/Y, 31I/M/V, 32L/del, 38F, 58D, 92K/T, 93C/H/N/R/S/T/W | |
| Ombitasvir (OBV) | 1, 4 | 28M/S/T/V, 30E/Q/R/Y, 31I/F/V, 32del, 58D, 92T, 93C/H/N/S | |
| Pibrentasvir (PIB) | 1, 2, 3, 4, 5, 6 | 24R, 28G/K/S, 30K/R, 31I/M, 32del, 58C/D, 93H/N | |
| Velpatasvir (VEL | 1, 2, 3, 4, 5, 6 | 28V, 30E/H/K, 31M/V, 32L, 93H/N/R/S/W | |
| NS5B nucleoside analogs inhibitors | Dasabuvir (DSV) | 1 | 316Y, 368T, 395G, 411S, 414T, 444K, 445F, 448C/H, 451S, 553T/V, 554S, 556G/N/R, 557R,558R, 559G/N, 561H, 565F |
| NS5B non-nucleoside analogs inhibitors | Sofosbuvir (SOF) | 1, 2, 3, 4, 5, 6 | 159F, 282R/T, 289L, 320I/V, 321A |
Unfortunately, the lack of a large market of standardized commercial assays for RAS determination has led to developing in-house RAS assays, which has created a great disparity the techniques that are used, the determined RAS, and their interpretation. Two main techniques for RAS detection have been applied. One is direct sequencing (Sanger) with sensitivity that allows detecting viral species present in between 15% and 25% within quasispecies, and the second is next generation sequencing (NGS), which allows the detection of variants present in less than 1%[31,32]. NGS is thus a more sensitive technique, but it is also much more expensive. It is therefore very likely that direct sequencing will continue to be the technique of choice because of its cost/benefit in the context of the high SVR rates of currently used DAA regimens.
Since the implementation of DAA agent, the main question that has been asked is the extent to which the RAS frequency impacts the outcome of treatment. It has been reported that the presence of a low proportion of viral variants carrying RAS within the quasispecies of an infected patient would have a lesser impact on SVR rates. In fact, some studies have reported a 15% cutoff of the viral population harboring RAS from in which a drop in the virological response rate was observed. Ikeda et al[33] (2017) reported that the SVR rates to daclatasvir (DCV)/asunaprevir (ASV) in HCV-infected patients with Y93H ratios of < 1%, 1%–25%, 26%-75%, and > 76% were 99%, 100%, 71%, and 23%, respectively[33]. Similarly, using a 15% NS5A pretreatment cutoff of ledipasvir (LDV)-specific RASs, Zeuzem et al[23] (2017) reported significant differences in SVR rates in patients treated with sofosbuvir (SOF)/LDV[23]. Overall, it has been established that SVR decreases as the proportion of RAS in the quasispecies infecting a patient increases. The second question was whether there was a differential impact of RAS depending on whether the patients were treatment naïve or previously treated. That question will be discussed in more detail below.
The clinical impact of RAS depends particularly on both the HCV genotype/subtype and the administered DAA regimen, which varies in efficacy according to the type of RAS as well as the treatment experience and presence of cirrhosis.
In naïve patients, the prevalence of RAS that significantly affect the response to treatment is estimated to be approximately 5%. In that case, the SVR rates of patients with RAS would be 91%, while for patients without RAS it would be approximately 99%[23,34,35]. In summary, RAS assessment prior to the beginning of treatment is not recommended for naïve patients. In previously treated patients, the situation is more complex and refers to subjects who have failed to respond to treatment with a DAA compound. In that case, the presence of post-failure RAS is more than 75%, and SVR rates are more affected. In fact, it has been reported that SVR rates are between 75% and 85% in patients with RAS, while for patients without RAS they continue to be remarkably high (> 95%)[23,34,35].
Identifying the HCV genotype/subtype before starting therapy in naïve patients, in the pangenotypic treatment era, remains useful and may be necessary when drug availability or lack of affordability require genotype-specific treatment or optimal treatment regimens. In that sense, HCV genotyping and subtyping should be performed by nucleotide sequence analysis of some coding regions, generally the core, NS3, or the NS5B coding regions, which accurately discriminates HCV subtypes[36,37]. Furthermore, the use of the NS3 or NS5B regions to determine the viral genotype and subtype also allows the detection of the baseline RAS[36]. On the other hand, as HCV subtypes, including 1l, 3b, 3g, 4r, 6u, 6v, among others, harbor a high frequency of baseline RAS, knowing the HCV subtype before treatment in regions or countries where these subtypes are prevalent (i.e. China, South-East Asia, and sub-Saharan Africa) is strongly recommended in order to optimize treatment[38-41]. Indeed, infrequent subtypes harboring RAS that confer resistance to NS5A inhibitors should be considered for treatment with the fixed-dose combinations SOF/velpatasvir (VEL)/ voxilaprevir (VOX) for 12 wk.
HCV-1 is the most prevalent genotype worldwide (46.2%), and one third of the HCV-1 that infects patients belongs to subtype 1a[42]. Several studies have reported that DAA-naïve individuals infected with HCV-1a are more difficult to treat than those infected with HCV-1b[23,43-45]. In fact, it has been observed that in the presence of cirrhosis, high baseline viral load, or failure of previous treatment with PEG-IFN/RBV, the SVR rates of patients treated with elbasvir (EBR)/grazoprevir (GZR), or SOF/LDV were significantly lower for HCV-1a compare with HCV-1b infected in
In addition, pretreatment genotyping is recommended if cirrhotic patients will be treated with SOF/VEL, as baseline RAS reduce SVR rates in HCV-3 cirrhotic patients treated with that regimen. Moreover, a recent study analyzing 539 HCV-3 infected patients showed that patients with baseline Y93H and/or A30K RAS had an SVR rate of 72.2%, while HCV-3 infected patients without NS5A RASs achieved an SVR rate of 95.7% (P = 0.002)[47]. Accordingly, a large meta-analysis that included more than 6500 subjects with chronic HCV infection reported reduced effectiveness of GLE/PIB in HCV-3 infected patients with baseline RAS like A30K, Y93H, and P53del, and recom
According to the American Association for the Study of Liver Diseases guidelines, pretreatment RAS testing is recommended in cirrhotic HCV-3 infected patients be
Even in the context of a low treatment failure rate (< 5%), the number of patients requiring retreatment is quite high because of the large number of patients with chronic HCV infection who are treated with DAA worldwide[22-24,29-30]. Currently, the main international treatment guidelines do not recommend massive testing of RAS before starting DAA treatment, although there are exceptions[49,50].
Treatment with SOF/VEL/VOX for 12 wk is one of the most promising pangenotypic regimens for rescuing patients who have failed treatment. Two phase III trials, POLARIS-1 and POLARIS-4, assessed the safety and efficacy of the SOF/VEL/VOX regimen for 12 wk in patients who failed treatment with NS3 and/or NS5A inhibitors[51]. In the POLARIS-1 study, which included 263 patients with NS5A inhibitor fai
The other available pangenotypic option for the treatment of patients with resistant variants is GLE/PIB. However, the combination did not have a suitable genetic barrier to achieve optimal SVR rates in patients failing previous DAA treatment[57]. In the MAGELLAN-1 Part 2 study, GLE/PIB was used for the retreatment of previous DAA failures. SVR12 was achieved by 89% and 91% of HCV-1 and HCV-4 infected patients who received 12 wk and 16 wk of treatment, respectively. Previous treatment with one inhibitor class (protease or NS5A) had no impact on SVR12, whereas past treatment with both classes of inhibitors was associated with lower SVR12 rates[57]. Another study adds support of the efficacy of the 16 wk regimen for retreatment of HCV-1 infected patients with a history of sofosbuvir/NS5A inhibitor treatment failure[58]. Consequently, treatment with GLE/PIB is recommended as an alternative regimen for the retreatment of patients who failed to a prior DAA regimen including a, NS5A or NS3 inhibitor. It is not recommended for patients who have failed treatment with the combination of both inhibitors[50]. Therefore, at present, the SOF/VEL/VOX com
Currently, the most challenging scenario is represented by patients who failed combinations containing the latest generation of pangenotypic DAA agents GLE/PIB and SOF/VEL/VOX. Thus, such patients who are very difficult to cure, the combinations of SOF/VEL/VOX or SOF/GLE/PIB with RBV for 12 wk, or without RBV for 16-24 wk, are the recommended options. In a previous study, 31 patients who failed GLE/PIB were retreated with SOF/VEL/VOX achieved an SVR of 94% despite the presence of NS5A RAS in 90% of the cases[59]. On the other hand, in the ongoing MAGELLAN-3 study, 23 patients who failed GLE/PIB and received treatment with SOF/GLE/PIB combined with RBV achieved an SVR of 96%, despite the presence of RAS in the NS5A region in 91% of them[60].
Recently, failure to SOF/VEL/VOX has been reported in 40 patients[61]. RAS testing after SOF/VEL/VOX failure showed that all HCV-1a had either NS3 or NS5A RAS. On the contrary, in HCV-1b, individual NS3 RAS were rather rare (11%), and the overall frequency of NS5A RAS was moderate (33%). Finally, for HCV-3, RAS in NS5A (56%) and in NS3 plus NS5A (28%) were relatively frequent. In 22 of the cases, rescue treatment with SOF/GLE/PIB, with or without RBV, for 12-24 wk achieved an SVR rate of 79%. Unfortunately, as all types of DAA drugs have been used in most de
The EASL currently recommends first line therapy regimens that do not require pretreatment RAS detection. The 2020 EASL Recommendations on Treatment of Hepatitis C state that in areas where the regimens are not available or not reimbursed, physicians who have access to reliable resistance tests can use the results to guide their decisions, according to[50]. Thus, the selected retreatment option depends on the availability of RAS testing, the actual access to the DAA agent indicated in the event of the failure, and the preference of the treating physician.
In the current clinical setting, there is no need for baseline detection of RAS before DAA therapy initiation in naïve patients. The use of adequate pangenotypic regimes may overcome the effect of RAS in the first treatment. After treatment failure, RAS may be determined when available. Otherwise, SOF/VEL/VOX for 12 wk is the regimen of choice, as it has shown the highest SVR rates. GLE/PIB for 16 wk is an alternative regime and it may be used in patients who have failed NS5A or NS3 inhibitors, but not a combination of both. Failure to treatment with multiple DAA regimens may be the clearest clinical scenario for RAS detection. In such cases, rescue treatment can be guided based on the results. If after many failures, RAS detection is not available, treatment should be evaluated by multidisciplinary teams. SOF/VEL/ VOX or SOF/GLE/PIB with RBV for 12 wk or without RBV for 16-24 wk are the regimens of choice as they have shown effectiveness in curing these difficult-to-treat patients.
To Silvina Heisecke, from CEMIC-CONICET, for copyediting the manuscript.
| 1. | Pawlotsky JM. Mechanisms of antiviral treatment efficacy and failure in chronic hepatitis C. Antiviral Res. 2003;59:1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 110] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 2. | Feld JJ, Hoofnagle JH. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature. 2005;436:967-972. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 728] [Cited by in RCA: 740] [Article Influence: 35.2] [Reference Citation Analysis (0)] |
| 3. | Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, Goodman ZD, Koury K, Ling M, Albrecht JK. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958-965. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4736] [Cited by in RCA: 4559] [Article Influence: 182.4] [Reference Citation Analysis (4)] |
| 4. | Sánchez-Tapias JM, Diago M, Escartín P, Enríquez J, Romero-Gómez M, Bárcena R, Crespo J, Andrade R, Martínez-Bauer E, Pérez R, Testillano M, Planas R, Solá R, García-Bengoechea M, Garcia-Samaniego J, Muñoz-Sánchez M, Moreno-Otero R; TeraViC-4 Study Group. Peginterferon-alfa2a plus ribavirin for 48 versus 72 weeks in patients with detectable hepatitis C virus RNA at week 4 of treatment. Gastroenterology. 2006;131:451-460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 284] [Cited by in RCA: 264] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 5. | Mira JA, Rivero A, de Los Santos-Gil I, López-Cortés LF, Girón-González JA, Márquez M, Merino D, del Mar Viloria M, Téllez F, Ríos-Villegas MJ, Omar M, Rivero-Juárez A, Macías J, Pineda JA; Grupo HEPAVIR de la Sociedad Andaluza de Enfermedades Infecciosas (SAEI). Hepatitis C virus genotype 4 responds better to pegylated interferon with ribavirin than genotype 1 in HIV-infected patients. AIDS. 2012;26:1721-1724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 6. | Carrat F, Bani-Sadr F, Pol S, Rosenthal E, Lunel-Fabiani F, Benzekri A, Morand P, Goujard C, Pialoux G, Piroth L, Salmon-Céron D, Degott C, Cacoub P, Perronne C; ANRS HCO2 RIBAVIC Study Team. Pegylated interferon alfa-2b vs standard interferon alfa-2b, plus ribavirin, for chronic hepatitis C in HIV-infected patients: a randomized controlled trial. JAMA. 2004;292:2839-2848. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 652] [Cited by in RCA: 622] [Article Influence: 28.3] [Reference Citation Analysis (5)] |
| 7. | Soriano V, Miralles C, Berdún MA, Losada E, Aguirrebengoa K, Ocampo A, Arazo P, Cervantes M, de los Santos I, San Joaquín I, Echeverria S, Galindo MJ, Asensi V, Barreiro P, Sola J, Hernandez-Burruezo JJ, Guardiola J, Blanco F, Martin-Carbonero L, García-Samaniego J, Nuñez M; PRESCO Study Group. Premature treatment discontinuation in HIV/HCV-coinfected patients receiving pegylated interferon plus weight-based ribavirin. Antivir Ther. 2007;12:469-476. [PubMed] |
| 8. | Vispo E, Barreiro P, Pineda JA, Mira JA, Maida I, Martín-Carbonero L, Rodríguez-Nóvoa S, Santos I, López-Cortes LF, Merino D, Rivero A, Soriano V. Low response to pegylated interferon plus ribavirin in HIV-infected patients with chronic hepatitis C treated with abacavir. Antivir Ther. 2008;13:429-437. [PubMed] |
| 9. | Mira JA, López-Cortés LF, Barreiro P, Tural C, Torres-Tortosa M, de Los Santos Gil I, Martín-Rico P, Ríos-Villegas MJ, Hernández-Burruezo JJ, Merino D, López-Ruz MA, Rivero A, Muñoz L, González-Serrano M, Collado A, Macías J, Viciana P, Soriano V, Pineda JA. Efficacy of pegylated interferon plus ribavirin treatment in HIV/hepatitis C virus co-infected patients receiving abacavir plus lamivudine or tenofovir plus either lamivudine or emtricitabine as nucleoside analogue backbone. J Antimicrob Chemother. 2008;62:1365-1373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 47] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 10. | Holmes JA, Rutledge SM, Chung RT. Direct-acting antiviral treatment for hepatitis C. Lancet. 2019;393:1392-1394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 45] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 11. | Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Ogura Y, Izumi N, Marumo F, Sato C. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996;334:77-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 752] [Cited by in RCA: 735] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 12. | Pascu M, Martus P, Höhne M, Wiedenmann B, Hopf U, Schreier E, Berg T. Sustained virological response in hepatitis C virus type 1b infected patients is predicted by the number of mutations within the NS5A-ISDR: a meta-analysis focused on geographical differences. Gut. 2004;53:1345-1351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 110] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 13. | Okanoue T, Itoh Y, Hashimoto H, Yasui K, Minami M, Takehara T, Tanaka E, Onji M, Toyota J, Chayama K, Yoshioka K, Izumi N, Akuta N, Kumada H. Predictive values of amino acid sequences of the core and NS5A regions in antiviral therapy for hepatitis C: a Japanese multi-center study. J Gastroenterol. 2009;44:952-963. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 64] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 14. | Di Lello FA, Mira JA, Neukam K, Parra-Sánchez M, Guelfo JR, Cifuentes C, Macías J, Palomares JC, Gómez-Mateos J, Pineda JA, Real LM. Core amino acid variation at position 110 is associated with sustained virological response in Caucasian patients with chronic hepatitis C virus 1b infection. Arch Virol. 2014;159:3345-3351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 15. | Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ, Sulkowski M, McHutchison JG, Goldstein DB. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2776] [Cited by in RCA: 2734] [Article Influence: 160.8] [Reference Citation Analysis (1)] |
| 16. | Neukam K, Caruz A, Rivero-Juárez A, Barreiro P, Merino D, Real LM, Herrero R, Camacho A, Soriano V, Di Lello FA, Macías J, Rivero A, Pineda JA. Variations at multiple genes improve interleukin 28B genotype predictive capacity for response to therapy against hepatitis C infection. AIDS. 2013;27:2715-2724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 17. | Di Lello FA, Caruz A, Rallon NI, Rivero-Juarez A, Neukam K, Barreiro P, Camacho A, García-Rey S, Rivero A, Soriano V, Cifuentes C, Macias J, Pineda JA. Effects of the genetic pattern defined by low-density lipoprotein receptor and IL28B genotypes on the outcome of hepatitis C virus infection. Eur J Clin Microbiol Infect Dis. 2013;32:1427-1435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 18. | Pawlotsky JM. Hepatitis C Virus Resistance to Direct-Acting Antiviral Drugs in Interferon-Free Regimens. Gastroenterology. 2016;151:70-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 406] [Cited by in RCA: 425] [Article Influence: 42.5] [Reference Citation Analysis (1)] |
| 19. | Neumann AU, Lam NP, Dahari H, Gretch DR, Wiley TE, Layden TJ, Perelson AS. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science. 1998;282:103-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1604] [Cited by in RCA: 1458] [Article Influence: 52.1] [Reference Citation Analysis (0)] |
| 20. | Domingo E, Sheldon J, Perales C. Viral quasispecies evolution. Microbiol Mol Biol Rev. 2012;76:159-216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 792] [Cited by in RCA: 733] [Article Influence: 52.4] [Reference Citation Analysis (0)] |
| 21. | Pearlman BL, Hinds AE. Review article: novel antivirals for hepatitis C-sofosbuvir/velpatasvir/voxilaprevir, glecaprevir/pibrentasvir. Aliment Pharmacol Ther. 2018;48:914-923. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 22. | Harrington PR, Komatsu TE, Deming DJ, Donaldson EF, O'Rear JJ, Naeger LK. Impact of hepatitis C virus polymorphisms on direct-acting antiviral treatment efficacy: Regulatory analyses and perspectives. Hepatology. 2018;67:2430-2448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 23. | Zeuzem S, Mizokami M, Pianko S, Mangia A, Han KH, Martin R, Svarovskaia E, Dvory-Sobol H, Doehle B, Hedskog C, Yun C, Brainard DM, Knox S, McHutchison JG, Miller MD, Mo H, Chuang WL, Jacobson I, Dore GJ, Sulkowski M. NS5A resistance-associated substitutions in patients with genotype 1 hepatitis C virus: Prevalence and effect on treatment outcome. J Hepatol. 2017;66:910-918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 178] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 24. | Pérez AB, Chueca N, Macías J, Pineda JA, Salmerón J, Rivero-Juárez A, Hidalgo-Tenorio C, Espinosa MD, Téllez F, Von-Wichmann MÁ, Omar M, Santos J, Hernández-Quero J, Antón JJ, Collado A, Lozano AB, García-Deltoro M, Casado M, Pascasio JM, Selfa A, Rosales JM, De la Iglesia A, Arenas JI, García-Bujalance S, Ríos MJ, Bernal E, Martínez O, García-Herola A, Vélez M, Rincón P, García F. Prevalence of resistance associated substitutions and efficacy of baseline resistance-guided chronic hepatitis C treatment in Spain from the GEHEP-004 cohort. PLoS One. 2019;14:e0221231. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 25. | Chen ZW, Li H, Ren H, Hu P. Global prevalence of pre-existing HCV variants resistant to direct-acting antiviral agents (DAAs): mining the GenBank HCV genome data. Sci Rep. 2016;6:20310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 77] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 26. | Martínez AP, Culasso ACA, Pérez PS, Romano V, Campos RH, Ridruejo E, García G, Di Lello FA. Polymorphisms associated with resistance to protease inhibitors in naïve patients infected with hepatitis C virus genotype 1 in Argentina: Low prevalence of Q80K. Virus Res. 2017;240:140-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 27. | Martínez AP, García G, Ridruejo E, Culasso AC, Pérez PS, Pereson MJ, Neukam K, Flichman D, Di Lello FA. Hepatitis C virus genotype 1 infection: Prevalence of NS5A and NS5B resistance-associated substitutions in naïve patients from Argentina. J Med Virol. 2019;91:1970-1978. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 28. | Bertoli A, Sorbo MC, Aragri M, Lenci I, Teti E, Polilli E, Di Maio VC, Gianserra L, Biliotti E, Masetti C, Magni CF, Babudieri S, Nicolini LA, Milana M, Cacciatore P, Sarmati L, Pellicelli A, Paolucci S, Craxì A, Morisco F, Palitti VP, Siciliano M, Coppola N, Iapadre N, Puoti M, Rizzardini G, Taliani G, Pasquazzi C, Andreoni M, Parruti G, Angelico M, Perno CF, Cento V, Ceccherini-Silberstein F; HCV Virology Italian Resistance Network (VIRONET-C). Prevalence of Single and Multiple Natural NS3, NS5A and NS5B Resistance-Associated Substitutions in Hepatitis C Virus Genotypes 1-4 in Italy. Sci Rep. 2018;8:8988. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 29. | Dietz J, Susser S, Vermehren J, Peiffer KH, Grammatikos G, Berger A, Ferenci P, Buti M, Müllhaupt B, Hunyady B, Hinrichsen H, Mauss S, Petersen J, Buggisch P, Felten G, Hüppe D, Knecht G, Lutz T, Schott E, Berg C, Spengler U, von Hahn T, Berg T, Zeuzem S, Sarrazin C; European HCV Resistance Study Group. Patterns of Resistance-Associated Substitutions in Patients With Chronic HCV Infection Following Treatment With Direct-Acting Antivirals. Gastroenterology. 2018;154:976-988.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 124] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 30. | Hezode C, Reau N, Svarovskaia ES, Doehle BP, Shanmugam R, Dvory-Sobol H, Hedskog C, McNally J, Osinusi A, Brainard DM, Miller MD, Mo H, Roberts SK, O'Leary JG, Shafran SD, Zeuzem S. Resistance analysis in patients with genotype 1-6 HCV infection treated with sofosbuvir/velpatasvir in the phase III studies. J Hepatol. 2018;68:895-903. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 78] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 31. | Fourati S, Pawlotsky JM. Virologic Tools for HCV Drug Resistance Testing. Viruses. 2015;7:6346-6359. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 38] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 32. | Verbinnen T, Van Marck H, Vandenbroucke I, Vijgen L, Claes M, Lin TI, Simmen K, Neyts J, Fanning G, Lenz O. Tracking the evolution of multiple in vitro hepatitis C virus replicon variants under protease inhibitor selection pressure by 454 deep sequencing. J Virol. 2010;84:11124-11133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 58] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 33. | Ikeda H, Watanabe T, Okuse C, Matsumoto N, Ishii T, Yamada N, Shigefuku R, Hattori N, Matsunaga K, Nakano H, Hiraishi T, Kobayashi M, Yasuda K, Yamamoto H, Yasuda H, Kurosaki M, Izumi N, Yotsuyanagi H, Suzuki M, Itoh F. Impact of resistance-associated variant dominancy on treatment in patients with HCV genotype 1b receiving daclatasvir/asunaprevir. J Med Virol. 2017;89:99-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 34. | Sharafi H, Alavian SM. Hepatitis C resistance to NS5A inhibitors: Is it going to be a problem? World J Hepatol. 2018;10:543-548. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 35. | Kjellin M, Kileng H, Akaberi D, Palanisamy N, Duberg AS, Danielsson A, Kristiansen MG, Nöjd J, Aleman S, Gutteberg T, Goll R, Lannergård A, Lennerstrand J. Effect of the baseline Y93H resistance-associated substitution in HCV genotype 3 for direct-acting antiviral treatment: real-life experience from a multicenter study in Sweden and Norway. Scand J Gastroenterol. 2019;54:1042-1050. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 36. | Neukam K, Martínez AP, Culasso ACA, Ridruejo E, García G, Di Lello FA. NS3 genomic sequencing and phylogenetic analysis as alternative to a commercially available assay to reliably determine hepatitis C virus subtypes 1a and 1b. PLoS One. 2017;12:e0182193. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 37. | Chevaliez S, Bouvier-Alias M, Brillet R, Pawlotsky JM. Hepatitis C virus (HCV) genotype 1 subtype identification in new HCV drug development and future clinical practice. PLoS One. 2009;4:e8209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 98] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 38. | Childs K, Davis C, Cannon M, Montague S, Filipe A, Tong L, Simmonds P, Smith D, Thomson EC, Dusheiko G, Agarwal K. Suboptimal SVR rates in African patients with atypical genotype 1 subtypes: Implications for global elimination of hepatitis C. J Hepatol. 2019;71:1099-1105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 64] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 39. | Wasitthankasem R, Vongpunsawad S, Siripon N, Suya C, Chulothok P, Chaiear K, Rujirojindakul P, Kanjana S, Theamboonlers A, Tangkijvanich P, Poovorawan Y. Genotypic distribution of hepatitis C virus in Thailand and Southeast Asia. PLoS One. 2015;10:e0126764. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 68] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 40. | Wei L, Lim SG, Xie Q, Văn KN, Piratvisuth T, Huang Y, Wu S, Xu M, Tang H, Cheng J, Le Manh H, Gao Y, Mou Z, Sobhonslidsuk A, Dou X, Thongsawat S, Nan Y, Tan CK, Ning Q, Tee HP, Mao Y, Stamm LM, Lu S, Dvory-Sobol H, Mo H, Brainard DM, Yang YF, Dao L, Wang GQ, Tanwandee T, Hu P, Tangkijvanich P, Zhang L, Gao ZL, Lin F, Le TTP, Shang J, Gong G, Li J, Su M, Duan Z, Mohamed R, Hou JL, Jia J. Sofosbuvir-velpatasvir for treatment of chronic hepatitis C virus infection in Asia: a single-arm, open-label, phase 3 trial. Lancet Gastroenterol Hepatol. 2019;4:127-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 90] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 41. | Pawlotsky JM. DAA failures in African patients with "unusual" HCV subtypes: Hey! Didn't you know there was another world? J Hepatol. 2019;71:1070-1072. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 42. | Messina JP, Humphreys I, Flaxman A, Brown A, Cooke GS, Pybus OG, Barnes E. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology. 2015;61:77-87. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1077] [Cited by in RCA: 1160] [Article Influence: 105.5] [Reference Citation Analysis (0)] |
| 43. | Sarrazin C, Dvory-Sobol H, Svarovskaia ES, Doehle BP, Pang PS, Chuang SM, Ma J, Ding X, Afdhal NH, Kowdley KV, Gane EJ, Lawitz E, Brainard DM, McHutchison JG, Miller MD, Mo H. Prevalence of Resistance-Associated Substitutions in HCV NS5A, NS5B, or NS3 and Outcomes of Treatment With Ledipasvir and Sofosbuvir. Gastroenterology. 2016;151:501-512.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 187] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 44. | Kwo P, Gane EJ, Peng CY, Pearlman B, Vierling JM, Serfaty L, Buti M, Shafran S, Stryszak P, Lin L, Gress J, Black S, Dutko FJ, Robertson M, Wahl J, Lupinacci L, Barr E, Haber B. Effectiveness of Elbasvir and Grazoprevir Combination, With or Without Ribavirin, for Treatment-Experienced Patients With Chronic Hepatitis C Infection. Gastroenterology. 2017;152:164-175.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 132] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 45. | Zeuzem S, Ghalib R, Reddy KR, Pockros PJ, Ben Ari Z, Zhao Y, Brown DD, Wan S, DiNubile MJ, Nguyen BY, Robertson MN, Wahl J, Barr E, Butterton JR. Grazoprevir-Elbasvir Combination Therapy for Treatment-Naive Cirrhotic and Noncirrhotic Patients With Chronic Hepatitis C Virus Genotype 1, 4, or 6 Infection: A Randomized Trial. Ann Intern Med. 2015;163:1-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 435] [Cited by in RCA: 428] [Article Influence: 38.9] [Reference Citation Analysis (0)] |
| 46. | Zeuzem S, Serfaty L, Vierling J, Cheng W, George J, Sperl J, Strasser S, Kumada H, Hwang P, Robertson M, Wahl J, Barr E, Talwani R, Platt H. The safety and efficacy of elbasvir and grazoprevir in participants with hepatitis C virus genotype 1b infection. J Gastroenterol. 2018;53:679-688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 47. | Di Maio VC, Barbaliscia S, Teti E, Fiorentino G, Milana M, Paolucci S, Pollicino T, Morsica G, Starace M, Bruzzone B, Gennari W, Micheli V, Yu La Rosa K, Foroghi L, Calvaruso V, Lenci I, Polilli E, Babudieri S, Aghemo A, Raimondo G, Sarmati L, Coppola N, Pasquazzi C, Baldanti F, Parruti G, Perno CF, Angelico M, Craxì A, Andreoni M, Ceccherini-Silberstein F; HCV Virology Italian Resistance Network Group (Vironet C). Resistance analysis and treatment outcomes in hepatitis C virus genotype 3-infected patients within the Italian network VIRONET-C. Liver Int. 2021;41:1802-1814. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 48. | Zhang Y, Jiang X, Zhao Y, Xu Y. Effect of baseline resistance-associated substitutions on the efficiency of glecaprevir/pibrentasvir in chronic hepatitis C subjects: A meta-analysis. J Viral Hepat. 2021;28:177-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 49. | Ghany MG, Morgan TR; AASLD-IDSA Hepatitis C Guidance Panel. Hepatitis C Guidance 2019 Update: American Association for the Study of Liver Diseases-Infectious Diseases Society of America Recommendations for Testing, Managing, and Treating Hepatitis C Virus Infection. Hepatology. 2020;71:686-721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 560] [Article Influence: 93.3] [Reference Citation Analysis (0)] |
| 50. | European Association for the Study of the Liver; Clinical Practice Guidelines Panel: Chair; EASL Governing Board representative; Panel members. EASL recommendations on treatment of hepatitis C: Final update of the series☆. J Hepatol. 2020;73:1170-1218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 923] [Cited by in RCA: 866] [Article Influence: 144.3] [Reference Citation Analysis (0)] |
| 51. | Bourlière M, Gordon SC, Flamm SL, Cooper CL, Ramji A, Tong M, Ravendhran N, Vierling JM, Tran TT, Pianko S, Bansal MB, de Lédinghen V, Hyland RH, Stamm LM, Dvory-Sobol H, Svarovskaia E, Zhang J, Huang KC, Subramanian GM, Brainard DM, McHutchison JG, Verna EC, Buggisch P, Landis CS, Younes ZH, Curry MP, Strasser SI, Schiff ER, Reddy KR, Manns MP, Kowdley KV, Zeuzem S; POLARIS-1 and POLARIS-4 Investigators. Sofosbuvir, Velpatasvir, and Voxilaprevir for Previously Treated HCV Infection. N Engl J Med. 2017;376:2134-2146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 399] [Cited by in RCA: 431] [Article Influence: 47.9] [Reference Citation Analysis (0)] |
| 52. | Sarrazin C, Cooper CL, Manns MP, Reddy KR, Kowdley KV, Roberts SK, Dvory-Sobol H, Svarovskia E, Martin R, Camus G, Doehle BP, Stamm LM, Hyland RH, Brainard DM, Mo H, Gordon SC, Bourliere M, Zeuzem S, Flamm SL. No impact of resistance-associated substitutions on the efficacy of sofosbuvir, velpatasvir, and voxilaprevir for 12 wk in HCV DAA-experienced patients. J Hepatol. 2018;69:1221-1230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 51] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 53. | Da BL, Lourdusamy V, Kushner T, Dieterich D, Saberi B. Efficacy of sofosbuvir/velpatasvir/voxilaprevir in direct-acting antiviral experienced patients with hepatitis C virus. Eur J Gastroenterol Hepatol. 2021;33:859-861. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 54. | Belperio PS, Shahoumian TA, Loomis TP, Backus LI. Real-world effectiveness of sofosbuvir/velpatasvir/voxilaprevir in 573 direct-acting antiviral experienced hepatitis C patients. J Viral Hepat. 2019;26:980-990. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 55. | Degasperi E, Spinetti A, Lombardi A, Landonio S, Rossi MC, Pasulo L, Pozzoni P, Giorgini A, Fabris P, Romano A, Lomonaco L, Puoti M, Vinci M, Gatti F, Carolo G, Zoncada A, Bonfanti P, Russo FP, Aghemo A, Soria A, Centenaro R, Maggiolo F, Rovere P, Pasin F, Paon V, Faggiano G, Vario A, Grossi G, Soffredini R, Carriero C, Paolucci S, Noventa F, Alberti A, Lampertico P, Fagiuoli S; NAVIGATORE-Lombardia and Veneto Study Groups. Real-life effectiveness and safety of sofosbuvir/velpatasvir/voxilaprevir in hepatitis C patients with previous DAA failure. J Hepatol. 2019;71:1106-1115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 74] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 56. | Llaneras J, Riveiro-Barciela M, Lens S, Diago M, Cachero A, García-Samaniego J, Conde I, Arencibia A, Arenas J, Gea F, Torras X, Luis Calleja J, Antonio Carrión J, Fernández I, María Morillas R, Rosales JM, Carmona I, Fernández-Rodríguez C, Hernández-Guerra M, Llerena S, Bernal V, Turnes J, González-Santiago JM, Montoliu S, Figueruela B, Badia E, Delgado M, Fernández-Bermejo M, Iñarrairaegui M, Pascasio JM, Esteban R, Mariño Z, Buti M. Effectiveness and safety of sofosbuvir/velpatasvir/voxilaprevir in patients with chronic hepatitis C previously treated with DAAs. J Hepatol. 2019;71:666-672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 74] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 57. | Poordad F, Pol S, Asatryan A, Buti M, Shaw D, Hézode C, Felizarta F, Reindollar RW, Gordon SC, Pianko S, Fried MW, Bernstein DE, Gallant J, Lin CW, Lei Y, Ng TI, Krishnan P, Kopecky-Bromberg S, Kort J, Mensa FJ. Glecaprevir/Pibrentasvir in patients with hepatitis C virus genotype 1 or 4 and past direct-acting antiviral treatment failure. Hepatology. 2018;67:1253-1260. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 112] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 58. | Lok AS, Sulkowski MS, Kort JJ, Willner I, Reddy KR, Shiffman ML, Hassan MA, Pearlman BL, Hinestrosa F, Jacobson IM, Morelli G, Peter JA, Vainorius M, Michael LC, Fried MW, Wang GP, Lu W, Larsen L, Nelson DR. Efficacy of Glecaprevir and Pibrentasvir in Patients With Genotype 1 Hepatitis C Virus Infection With Treatment Failure After NS5A Inhibitor Plus Sofosbuvir Therapy. Gastroenterology. 2019;157:1506-1517.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 59. | Pearlman B, Perrys M, Hinds A. Sofosbuvir/Velpatasvir/Voxilaprevir for Previous Treatment Failures With Glecaprevir/Pibrentasvir in Chronic Hepatitis C Infection. Am J Gastroenterol. 2019;114:1550-1552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 60. | Wyles D, Weiland O, Yao B, Weilert F, Dufour JF, Gordon SC, Stoehr A, Brown A, Mauss S, Zhang Z, Pilot-Matias T, Rodrigues L Jr, Mensa FJ, Poordad F. Retreatment of patients who failed glecaprevir/pibrentasvir treatment for hepatitis C virus infection. J Hepatol. 2019;70:1019-1023. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 61. | Dietz J, Di Maio VC, de Salazar A, Merino D, Vermehren J, Paolucci S, Kremer AE, Lara M, Pardo MR, Zoller H, Degasperi E, Peiffer KH, Sighinolfi L, Téllez F, Graf C, Ghisetti V, Schreiber J, Fernández-Fuertes E, Boglione L, Muñoz-Medina L, Stauber R, Gennari W, Figueruela B, Santos J, Lampertico P, Zeuzem S, Ceccherini-Silberstein F, García F, Sarrazin C; HCV Virology Italian Resistance Network (VIRONET-C) collaborators; Spanish GEHEP-004 Collaborators; Members of the German HCV resistance study group. Failure on voxilaprevir, velpatasvir, sofosbuvir and efficacy of rescue therapy. J Hepatol. 2021;74:801-810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/Licenses/by-nc/4.0/
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Argentina
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Martin-Carbonero L S-Editor: Fan JR L-Editor: Filipodia P-Editor: Yuan YY