Published online May 27, 2021. doi: 10.4254/wjh.v13.i5.543
Peer-review started: January 5, 2021
First decision: February 13, 2021
Revised: February 21, 2021
Accepted: March 31, 2021
Article in press: March 31, 2021
Published online: May 27, 2021
Processing time: 134 Days and 16.5 Hours
Cholestatic liver diseases are characterized by an accumulation of toxic bile acids (BA) in the liver, blood and other tissues which lead to progressive liver injury and poor prognosis in patients.
To discover and validate prognostic biomarkers of cholestatic liver diseases based on the urinary BA profile.
We analyzed urine samples by liquid chromatography-tandem mass spectrometry and investigated the use of the urinary BA profile to develop survival models that can predict the prognosis of hepatobiliary diseases. The urinary BA profile, a set of non-BA parameters, and the adverse events of liver transplant and/or death were monitored in 257 patients with cholestatic liver diseases for up to 7 years. The BA profile was characterized by calculating BA indices, which quantify the composition, metabolism, hydrophilicity, formation of secondary BA, and toxicity of the BA profile. We have developed and validated the bile-acid score (BAS) model (a survival model based on BA indices) to predict the prognosis of cholestatic liver diseases.
We have developed and validated a survival model based on BA (the BAS model) indices to predict the prognosis of cholestatic liver diseases. Our results demonstrate that the BAS model is more accurate and results in higher true-positive and true-negative prediction of death compared to both non-BAS and model for end-stage liver disease (MELD) models. Both 5- and 3-year survival probabilities markedly decreased as a function of BAS. Moreover, patients with high BAS had a 4-fold higher rate of death and lived for an average of 11 mo shorter than subjects with low BAS. The increased risk of death with high vs low BAS was also 2-4-fold higher and the shortening of lifespan was 6-7-mo lower compared to MELD or non-BAS. Similarly, we have shown the use of BAS to predict the survival of patients with and without liver transplant (LT). Therefore, BAS could be used to define the most seriously ill patients, who need earlier intervention such as LT. This will help provide guidance for timely care for liver patients.
The BAS model is more accurate than MELD and non-BAS models in predicting the prognosis of cholestatic liver diseases.
Core Tip: We have developed survival models based on bile acid (BA) indices to predict the prognosis of hepatobiliary diseases. Our BA models outperformed the model for end-stage liver disease and non-BA models in predicting the occurrence of the adverse events of death and/or liver transplant.
- Citation: Alamoudi JA, Li W, Gautam N, Olivera M, Meza J, Mukherjee S, Alnouti Y. Bile acid indices as biomarkers for liver diseases II: The bile acid score survival prognostic model. World J Hepatol 2021; 13(5): 543-556
- URL: https://www.wjgnet.com/1948-5182/full/v13/i5/543.htm
- DOI: https://dx.doi.org/10.4254/wjh.v13.i5.543
Cholestatic liver diseases are hepatobiliary diseases associated with a reducing in bile flow due to impairment in bile production or failure of bile flow into bile duct[1]. Chronic liver diseases account for greater than 41000 deaths in the United States in 2017, making it the 11th leading cause of mortality[2]. Most cholestatic diseases progress toward end stage liver failure, which likely requires liver transplantation. Even after liver transplantation, post-surgery complications are common, which may require liver re-transplantation[3].
Biomarkers currently used in the clinic for the diagnosis and prognosis of liver diseases are primarily serum liver enzymes such as aspartate transaminase (AST), alanine transaminase (ALT), and bilirubin. However, these markers have numerous shortfalls including the lack of specificity for liver or bile duct injuries as they can be elevated in hyperthyroidism, adrenal, heart, or muscle disorders. Also, severe cell injury has to occur before their levels increase[4,5]. Multifactorial models with multiple parameters based on these biomarkers are also frequently used and offer advantages compared to the use of their individual biomarker components such as the Child-Turcotte-Pugh score[2].
More recently, the model for end-stage liver disease (MELD) was developed to predict three-month mortality of patients with end-stage liver disease[5,6]. MELD is calculated based on serum creatinine, bilirubin, international normalized ratio (INR), and Na+, which are related to both liver and renal functions. MELD is currently used in many countries to classify patients awaiting transplantation to identify patients with the highest priority for liver transplant (LT)[6]. Since its implementation, MELD led to an intense reduction in the number of people waiting for liver transplant and decreased mortality on the waiting list without affecting post-transplant survival[7]. MELD is also an effective predictor of outcome in other conditions, such as patients have cirrhosis going for surgery and patients with alcoholic hepatitis or fulminant hepatic failure[7]. However, MELD is based on three objective laboratory variables, that are not necessarily liver specific. For example, serum bilirubin can be elevated in cases of hemolysis or sepsis. Serum creatinine can also be elevated from an underlying kidney disease that unrelated to hepatorenal syndrome and is a poor surrogate of renal function in cirrhotic patients[8]. In addition, patients may have an elevated INR which can be secondary to warfarin use. Any of these conditions can increase the MELD score and overestimate the liver disease severity[9]. Furthermore, several studies have shown that patients with cholestatic liver diseases may still have high mortality rates despite having low MELD scores[10,11].
Numerous clinical and preclinical studies have shown up to a 100-fold increase in BA concentrations in urine with various hepatobiliary diseases[12-16]. The impediment in bile flow associated with cholestatic liver diseases cause accumulation of toxic BA in the liver and blood, which can worsen the liver condition that lead to their accumulation and contribute to the unfavorable liver disease prognosis[17]. However, the potential use of BA as a marker for liver diseases have never translated into a widespread use in the clinic[18,19], due to major limitations including the major differences of the physiologic and pathologic effects of the various individual BA and the extremely high inter- and intra-individual variability of BA concentrations.
To this regard, we have developed the concept of “BA Indices”, which are ratios calculated from the absolute concentration of individual BA and their metabolites. BA indices offered numerous advantages over absolute BA concentrations including low intra- and inter-individual variability and resistance to the influence of food consumption, age, gender, body mass index (BMI), and moderate alcohol consumption[19-21]. In the 1st part of this study, we have demonstrated that BA indices outperformed serum liver enzymes as biomarkers for the diagnosis of cholestatic liver diseases. In this second part of the study, we have developed survival models based on BA indices to predict the prognosis of hepatobiliary diseases. Our BA models outperformed the non-BA and MELD models in predicting the occurrence of the adverse events of death and/or LT.
New and existing patients of the University of Nebraska Medical Center (UNMC) hepatology clinic, who were diagnosed with one or multi-hepatobiliary conditions due to chronic hepatitis C (n = 63) , hepatitis B (n = 14), alcoholic liver disease/alcoholic cirrhosis (n = 103), primary biliary cholangitis (n = 11), primary sclerosing cholangitis (n = 13), autoimmune hepatitis (n = 24), alpha-1-antitrypsin deficiency (n = 5), nonalcoholic fatty liver disease/nonalcoholic steatohepatitis (n = 51), carcinoma (n = 24), cryptogenic cirrhosis (n = 10), polycystic liver disease (n = 5), elevated liver function test (n = 18), and unknown etiology (n = 5), were enrolled in this study. Table 1 shows a summary of our patient population characteristics. A total of 257 patients (121 female and 136 male) between the ages of 19 and 83 years, who were treated for cholestatic liver diseases in UNMC, over the period from November of 2011 to December of 2018, were recruited into the study. All participants were followed up for up to 7 years by collecting urine samples for BA analysis and monitoring non-BA parameters and adverse events including liver transplant, and death from their medical records.
| Patients | Death | Liver transplant | |
| n | 257 | 27 | 25 |
| Gender | |||
| Male | 136 | 21 | 17 |
| Female | 121 | 6 | 8 |
| Age (yr) | |||
| mean ± SE | 52.2 ± 0.71 | 55.9 ± 1.88 | 52.9 ± 2.1 |
| Body mass index | |||
| mean ± SE | 30.7 ± 0.45 | 29.65 ± 1.19 | 29.11 ± 0.45 |
| Race | |||
| White | 217 | 26 | 24 |
| Black | 11 | 0 | 0 |
| Asian | 7 | 0 | 0 |
| Hispanic | 4 | 0 | 1 |
| Others | 18 | 1 | 0 |
| Non-BA parameters (mean ± SE) | |||
| Creatinine (mg/dL) | 1.02 ± 0.09 | ||
| Albumin (g/dL) | 3.53 ± 0.04 | ||
| INR | 1.19 ± 0.02 | ||
| Protime (s) | 12.01 ± 0.42 | ||
| AST (U/L) | 59.9 ± 4.07 | ||
| ALT (U/L) | 54.9 ± 4.26 | ||
| Bilirubin (mg/dL) | 1.75 ± 0.15 | ||
| AST/ALT | 1.28 ± 0.04 | ||
| MELD | 10.6 ± 0.34 | ||
| APRI | 1.15 ± 0.11 | ||
The study was approved by the Institutional Review Board at UNMC and written informed consents were provided for all participating subjects. The registry URL was (https://www.clinicaltrials.gov/ct2/show/NCT01200082?term=alnouti&draw=2&rank=1). The clinical trial number was NCT01200082. Thirty milliliters of urine samples were collected from patients on their first visit to the hepatology clinic. All urine samples were stored in -80 °C until analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS).
The performance of potential biomarkers from the urinary BA profile was also compared with and existing markers of liver function including ALT, AST, serum creatinine, albumin, protime, INR, bilirubin, AST/ALT ratio, and AST/platelet ratio index (APRI). These markers were monitored using the patients’ medical records. Bile acid quantification by liquid chromatography–tandem mass spectrometry
Urine samples were extracted using solid phase extraction as described previously[8,22,23]. BA concentrations were quantified by LC-MS/MS, as we described previously.
BA profile in urine was characterized using BA “indices”, which describe the composition, hydrophobicity, toxicity, and metabolism of total and individual BA as we have described previously[8,22,23].
All statistical analysis was performed using the Statistical Product and Service Solutions software, version 25 (IBM corporation, Armonk, NY, United States) and R software, version 3.6.3 (R Foundation for statistical Computing). A P value of 0.05 was considered significant for all the statistical tests described below.
Cox proportional hazards (PH) regression was used to develop survival models to predict the prognosis of hepatobiliary diseases in terms of progressing specifically into the end points/adverse events of death.
For the “death” models, the only endpoint/adverse event recorded was death at 3 and 5 years. We only had 7 and 17 deaths occurring within earlier time points including 1 and 2 years, respectively, which was not enough to develop survival models. Patients who underwent liver transplant (LT) were censored with the date of transplantation. Patients still alive at the end of each period (3 and 5 years) were considered as censored at that time. The term ‘‘censored’’ indicates that the patient was alive at that date and that was the end of the follow-up[22]. Patients dropped off, not due to the occurrence of adverse event, i.e. death, before the end of the follow-up period, were censored at the last day they were seen in the clinic.
In addition to the “death only” models above, we also constructed models to predict death and/or LT. We followed the same approach as the “death” models, with the exception that the endpoint was the occurrence of the adverse events of either death or LT. Patients whom did not have either of the adverse events at the end of each period (3 and 5 years) were censored at that time.
Individual BA and non-BA variables were analyzed as possible predictors of survival in a univariate Cox regression analysis. Values of these variables included in the statistical analysis were obtained at the time of patients’ first visits. Significant variables (P < 0.05), which were identified from the univariate analysis were included in the multivariate analysis. To build the multivariate model a backward elimination regression method was used to retain the most significant variables with retention criteria of P < 0.05.
Goodness of fit was performed by testing PH assumption for each covariate included in the final Cox model and for the global model as a whole. We used the bootstrapping for model validation.
Receiver operating characteristic (ROC) curve analyses was performed on the scores from the various multivariate Cox models to determine their cut-off values in differentiating patients with vs without the adverse event. The cut-off values with optimum specificity and sensitivity were selected and the areas under the ROC curve (AUC) values were calculated.
The average survival probability [S0 (t)] for a patient with an average score were calculated for different time points. To obtain the probability of survival for t years [S (t)], first the score e.g. bile-acid score (BAS) is calculated, and finally S (t) is calculated using this equation: Survival probability for t years: S (t) = S0(t)exp(BAS - BAS0).
Where, BAS0 is the average score from all patients in this study.
Kaplan-Meier plots were used to display survival curves. We have divided patients into two categories of high vs low risk and compared their survival with the Log-rank test and Breslow test[22]. We have tried the median cut-off values of the model scores to define high vs low risk.
We have used multivariate cox regression analyses to build various models for the prediction of death. The performance of the different models in predicting the occurrence of death within 3- and 5-year periods were compared between the different models using the statistic outcomes from the Bootstrapping, Schoenfeld residuals, AUC, and Kaplan-Meier analyses.
Table 1 shows a summary of the characteristics of the patient population in our study. The demographic variables were (age, BMI, gender, and race). Subjects were divided into five race groups (White, Black, Asian, Hispanic, and others). During the 7-year follow-up period of 257 patients with cholestatic liver diseases, 27 patients (10.5%) died and 25 patients (9.7%) underwent liver transplantation.
We were interested in predicting the occurrence of adverse events of death within 3- and 5-year periods. During a 3-year follow-up period, 21 patients (8.2%) died and 19 patients (7.4%) underwent liver transplantation. While during a 5-year follow-up period, 25 patients (9.7%) died and 21 patients (8.2%) underwent liver transplantation.
Supplementary Table 1 shows the results of univariate Cox regression analyses for death prediction by BA Indices. Cox regression detects the risk of death associated with changes in BA indices. Positive regression coefficients imply that the risk of death increases with increasing the values of BA indices, while negative coefficients imply the risk of death increases with a decrease in the values of BA indices. We found correlation between the risk of death and many BA indices (P < 0.05).
The hazard ratio (HR) from Cox regressions analysis quantifies the magnitude of the risk of death per unit change in BA indices. Because BA concentrations and indices have different scales and units, we performed the same calculation per 10% and 20% of the mean value of each variable instead of per absolute unit. For example, for a 20% increase in the %CDCA, the risk of death increases 1.26-fold (HR: 1.26; P < 0.05).
We performed the same univariate cox regression analysis for demographics and non-BA parameters as well (Supplementary Table 2). Notably, the risk of death was significantly higher in males than females from this univariate analysis. Increasing levels of INR, protime, bilirubin, AST/ALT, APRI, and MELD also significantly increased the risk of death, whereas decreasing levels of albumin significantly increased the risk of death.
In multivariate analysis, a backward elimination regression was used to retain the most significant BA variables. The only BA variables retained in the multivariate model were %CDCA and %Tri-OH, which were independently predictive of survival (Table 2). For example, a 20% increase in the %CDCA and %Tri-OH increases the risk of death by 1.34-fold (HR: 1.34; P < 0.05) and 1.14-fold (HR: 1.14; P < 0.05), respectively. The BAS for individual patients can be calculated from this equation: BAS for death = 0.039 × %CDCA + 0.052 × %Tri-OH.
| BA indices (%) and non-BA parameters | B-value (regression coefficient) | Standard error | P value | Hazard ratio: Exp (B) | ||
| 1 unit change | 10% change | 20% change | ||||
| The BAS model | ||||||
| %CDCA | 0.039 | 0.010 | 0.000 | 1.040 | 1.159 | 1.344 |
| %Tri-OH | 0.052 | 0.016 | 0.001 | 1.053 | 1.069 | 1.142 |
| The non-BAS model | ||||||
| AST/ALT | 1.236 | 0.303 | 0.000 | 3.442 | 1.165 | 1.357 |
For example, for a patient with %CDCA of 20%, and a %Tri-OH of 50%, the BAS would be 3.38.
We performed the same multivariate Cox regression analysis for demographics and non-BA parameters as well. For demographic variables, gender was significant in univariate analysis, but did not retain in multivariate analysis when included in the BA model building. In contrast, gender retained in the multivariate analysis for the non-BA model, but with minimal improvement of model goodness of fit and validation (the Bootstrapping, Schoenfeld residuals, AUC, and Kaplan-Meier analyses). Therefore, we did not include gender in the multivariate Cox models and AST/ALT ratio was the only significant predictive variable of death (Table 2). For example, a 20% increase in the AST/ALT, increases the risk of death by 1.36-fold (HR: 1.36; P < 0.05). The non-BAS for individual patients can be calculated from this equation: non-BAS for death = 1.236 × AST/ALT.
In addition, we used the same methodology to develop other models including: (1) mixed BA and non-BA variables including demographics to test how the performance of a global BA- and non-BA mixed model compares to the BA-only and non-BA-only models; (2) MELD variables with coefficients from our data set to create a model with the original MELD variables, but with model coefficients derived from our data set; and (3) original MELD modified with BA and/or non-BA variables including demographics, to test if the performance of the original MELD can be improved by adding significant BA and non-BA parameters from the univariate analysis and vice versa (Supplementary Table 3). Overall, none of these strategies produced any statistically significant models neither they did improve the BA or non-BA-only model; therefore, were not further evaluated or validated.
Goodness of fit was performed by testing PH assumption for all the covariates of the final Cox model as well as for the global model as a whole, using a statistical test and a graphical diagnostic based on Schoenfeld residuals. A graphical diagnostic that shows a non-random pattern against time is evidence of violation of the PH assumption. The PH assumption is supported by a non-significant relationship between residuals and time. The Schoenfeld residual plots and P values supported the validity of the BA and non-BA models (Supplementary Figure 1).
We also used the bootstrapping validation. Bootstrapping validation results for the BA and non-BA models indicate that our regression coefficients were in the range of the 95%CI, P values were statistically significant for each covariate, bias values and standard error values were very small (Supplementary Table 4). We can conclude that the Bootstrapping validation results supported the validity of the BA and non-BA models.
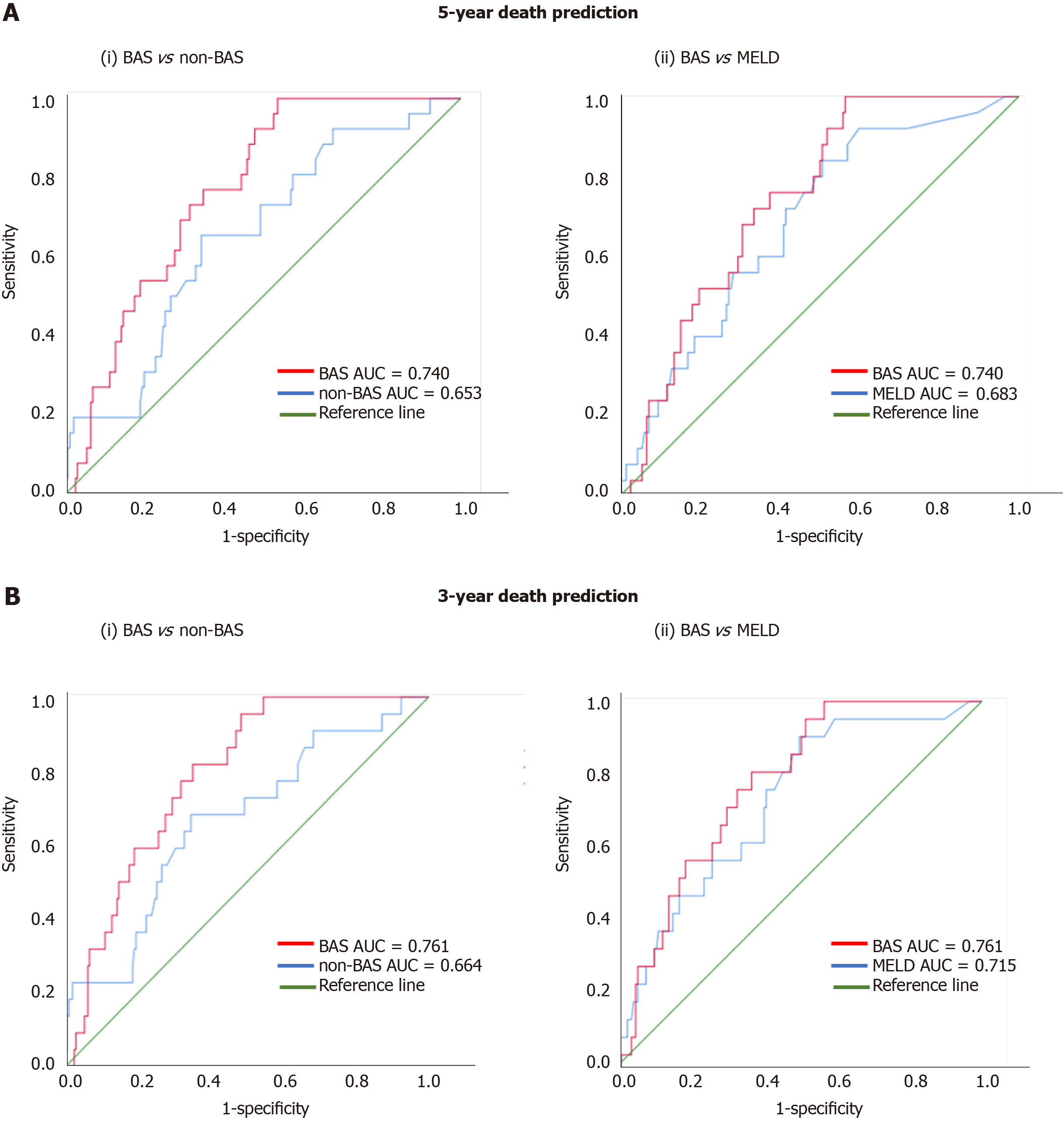
Figure 1 shows the ROC curves of the models for death prediction. For 5-year death prediction, the AUC for BAS, non-BAS, and MELD were 0.740, 0.653, and 0.683, respectively. For 3-year death prediction, the AUC for BAS, non-BAS, and MELD were 0.761, 0.664, and 0.715, respectively. Potential cut-off values selected based on the optimum sensitivity and specificity for different models. The ROC-optimum scores for BA, non-BA, and MELD models for death prediction were 2.71, 1.72, and 10, respectively (Table 3).
| Models | AUC (5-yr) | AUC (3-yr) | (Cutoff value; sensitivity, specificity) |
| BAS | 0.740 | 0.761 | (2.71; 74, 68) |
| non-BAS | 0.653 | 0.664 | (1.72; 67, 66) |
| MELD | 0.683 | 0.715 | (10; 62, 64) |
Table 4 presents the estimated survival probability [S0 (t)] for a patient with an average BAS0 of 2.24 (the average BAS from all 257 patients in this study) for different time points. To obtain the survival probability for t years [S (t)], first BAS is calculated, S0 (t) is identified from Table 4, and finally S (t) is calculated using this equation: Survival probability for t years: S (t) = S0(t)exp(BAS - BAS0).
| t (mo) | 5 | 7 | 14 | 24 | 36 | 60 | 76 |
| The BAS | |||||||
| S0 (t) | 0.993 | 0.985 | 0.971 | 0.948 | 0.934 | 0.916 | 0.901 |
| The non-BAS | |||||||
| S0 (t) | 0.989 | 0.978 | 0.958 | 0.924 | 0.902 | 0.876 | 0.855 |
Where, BAS0 is the average BAS from all patients in this study; namely 2.24, while BAS is the BAS for that particular patient. For the same example patient discussed above, the probability of surviving for at least 3 years is: Survival probability for (3) years = 0.934 exp (3.38 - 2.24) = 0.81 = 81%
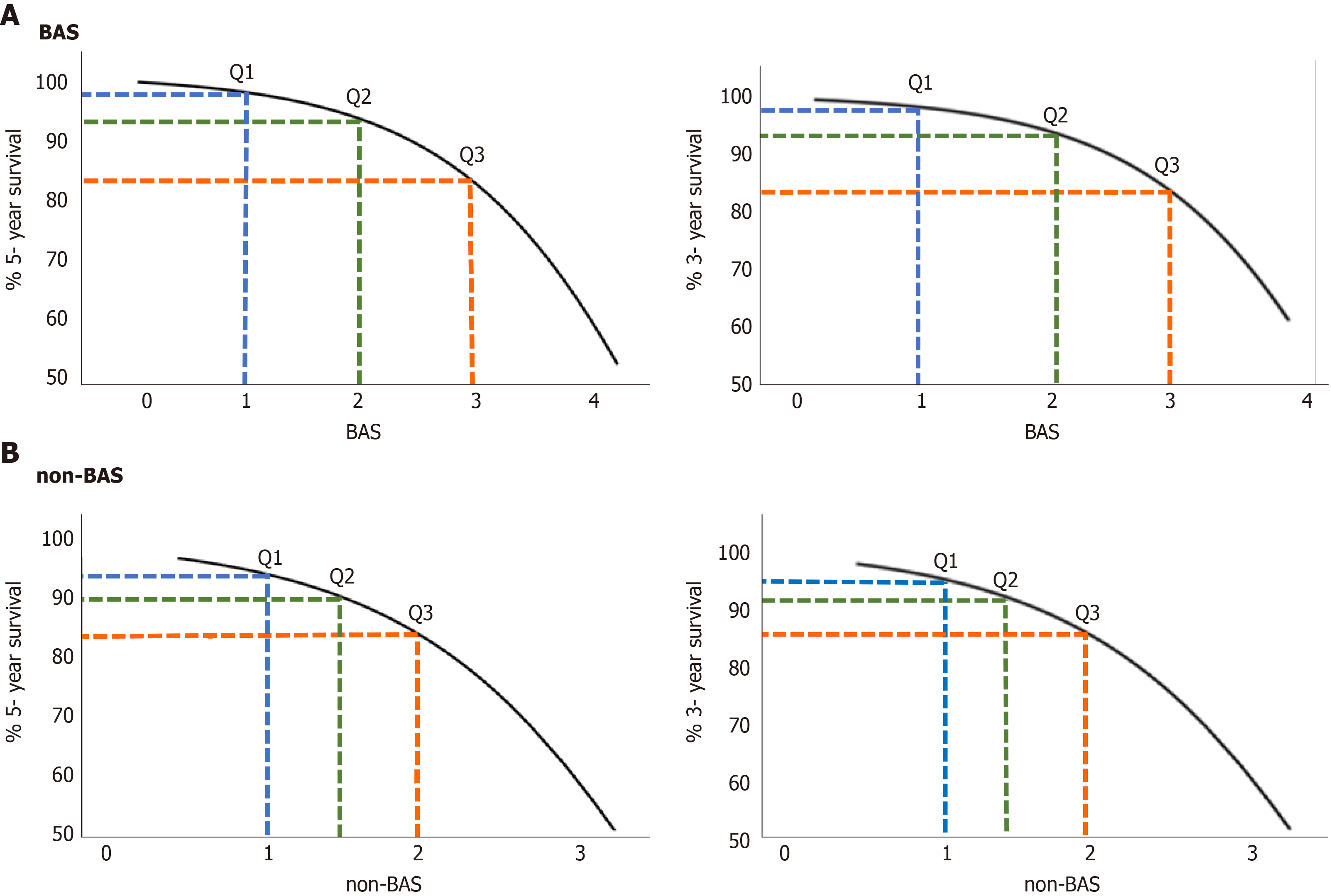
The relationship between estimated 5- and 3- year survival probability [S (t)] and the BAS in patients with liver disease are shown in Figure 2A. Survival probability decreases as a function of BAS. For example, the 5-year survival probability for patients with BAS of 1.2 (25th percentile of the population), 2.1 (50th percentile of the population i.e. median), and 3.1 (75th percentile of the population) are 97%, 93%, and 82%, respectively. Similarly, the 3-year survival probability for patients with the same BAS above, are 98%, 94%, and 85%, respectively.
Table 4 presents the estimated survival probability [S0 (t)] for a patient with an average non-BAS0 of 1.58 for different time points. The survival probability for (t) years is calculated using this equation: Survival probability for t years: S (t) = S0(t)exp(non-BAS - non-BAS0).
The relationship between estimated 5- and 3- year survival probability [S (t)] and the non-BAS in patients with liver disease are shown in Figure 2B. For example, the 5-year survival probability for patients with non-BAS of 1.1 (25th percentile of the population), 1.4 (50th percentile of the population), and 1.9 (75th percentile of the population) are 92%, 90%, and 83%, respectively. Similarly, the 3-year survival probability for patients with the same non-BAS above, are 95%, 91%, and 86%, respectively.
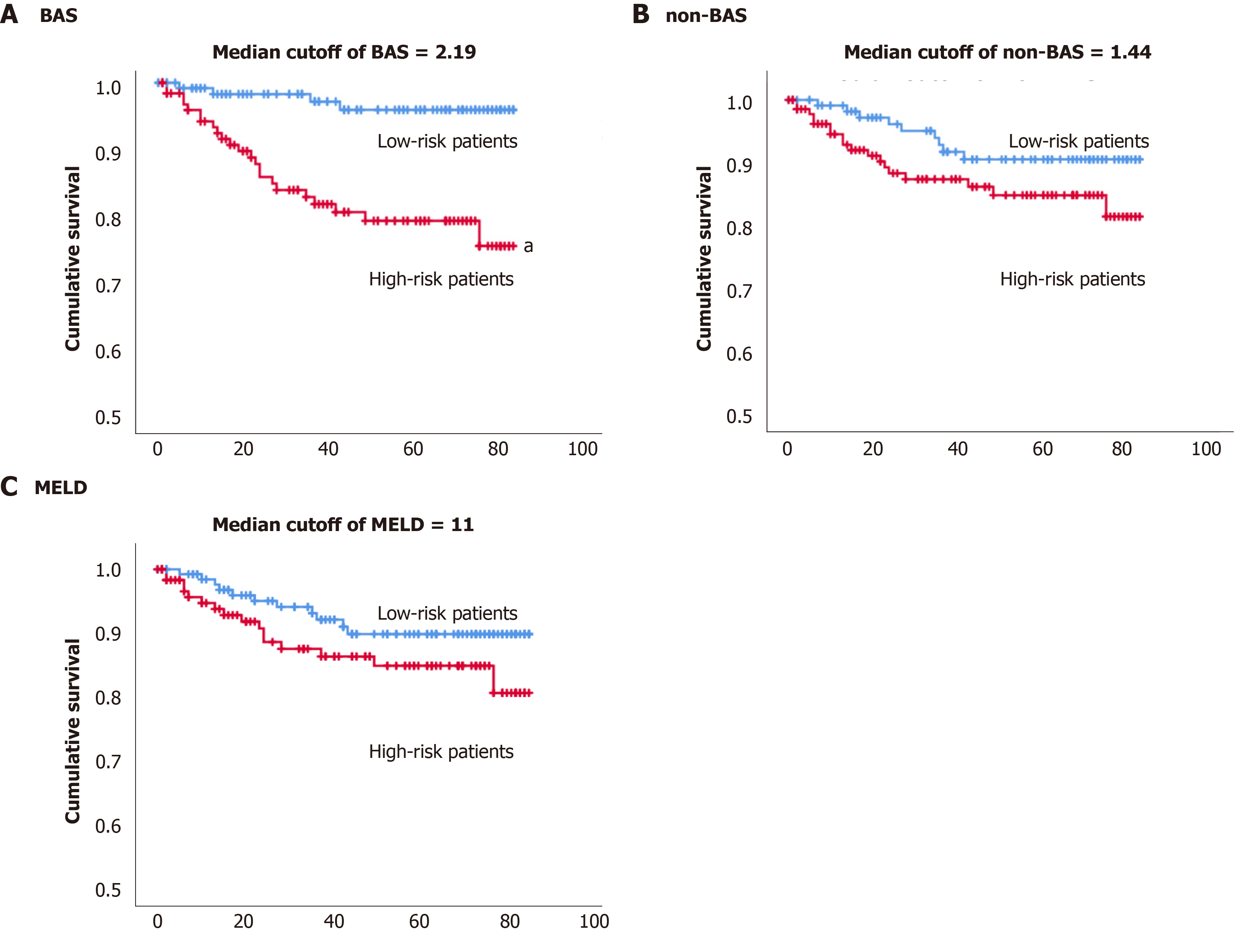
By the end of the study, up to 7 years monitoring of 257 patients with cholestatic liver diseases, 27 patients (10.5%) have died. The Kaplan-Meier estimator was used to estimate subjects’ survival free of adverse events over time. We have tried the median of the BAS of the population (2.19) cut-off value to define high vs low risk of death (Figure 3A). The estimated mean survival time was 71 mo (5.9 years) for the high-risk group and 82 mo (6.8 years) for the lower risk group based on the median BAS of 2.19 (Table 5). The P value of the log rank test and Breslow test were statistically significant (P value < 0.05), indicating the median cut-off of BAS, can differentiate low vs high risk of death.
| Cutoff | Total n | n of events | Estimated mean (mo) | Standard error | 95%CI |
| BAS | |||||
| Median cutoff of 2.19 | |||||
| Low risk < 2.19 | 128 | 4 | 81.68 | 1.14 | 79.44-83.93 |
| High risk > 2.19 | 129 | 23 | 70.72 | 2.5 | 65.81-75.62 |
| Non-BAS | |||||
| Median cutoff of 1.44 | |||||
| Low risk < 1.44 | 118 | 9 | 78.68 | 1.70 | 75.34-82.02 |
| High risk > 1.44 | 139 | 18 | 73.97 | 2.21 | 69.64-78.29 |
| MELD | |||||
| Median cutoff of 11 | |||||
| Low risk < 11 | 133 | 11 | 78.06 | 1.71 | 74.71-81.42 |
| High risk > 11 | 124 | 16 | 73.91 | 2.35 | 69.29-78.52 |
Figure 3B shows the Kaplan Meier survival for the high vs low risk of death groups based on the median (1.44) for the non-BAS. The estimated mean survival time was 74 mo (6.2 years) for the high-risk group and 79 mo (6.6 years) for the lower risk group based on the median non-BAS of 1.44. The P value from the log rank test and Breslow test was insignificant (P value > 0.05), indicating the median of non-BAS (1.44) cannot differentiate low vs high risk of death (Table 5).
Figure 3C shows the Kaplan Meier survival for the high vs low risk of death groups based on the median (11) for the MELD model. The estimated mean survival time was 74 mo (6.2 years) for the high-risk group and 78 mo (6.5 years) for the lower risk group based on the median MELD of 11. The P value from the log rank test and Breslow test was insignificant (P value > 0.05), indicating the median of MELD (11) cannot differentiate low vs high risk of death (Table 5).
We have developed similar BAS and non-BAS multivariate cox models for the prediction of the adverse events of death and/or LT instead of death only (Supplementary Table 5). Both models were also validated using the same criteria (data not shown). For both 3 and 5-years prediction, AUC was > 0.74 for both models (Supplementary Figure 2 and Supplementary Table 6). Similar to the “death only” models, there were direct relationship between BAS and non-BAS and liver transplant-free survival (Supplementary Figure 3). The estimated mean liver transplant-survival time was 60 mo (4.9 years) for the high-risk group and 79 mo (6.6 years) for the lower risk group based on the median BAS (0.45), which were statistically different (Supplementary Figure 4 and Supplementary Table 7).
We developed a survival model based on BA indices to predict the prognosis of hepatobiliary diseases in terms of progressing into the end point/adverse event of death over a 3- and 5-year period of time. Using the multivariate Cox regression analysis, we have constructed these final models for death prediction: (1) The BAS model for death prediction: BAS for death = 0.039 × %CDCA + 0.052 × %Tri-OH; (2) The non-BAS model model for death prediction: non-BAS (non-BAS) for death = 1.236 × AST/ALT. BAS in this population ranged from 0-4, while the non-BAS ranged from 0.44-4.98.
Cholestatic diseases are associated with impaired bile flow to the intestine, which is expected to translate into reduced transformation of primary BA including CDCA and CA into secondary BA by intestinal bacteria. Therefore, accumulation of primary BA in the blood may indicate further impairment in bile flow and worsening of the liver diseases[8,22,23]. This is in agreement with the BAS model, where increased %CDCA and %Tri-OH BA (primarily consists of CA) were the most significant predictors of liver disease prognosis into death. Another interpretation for the accumulation of CDCA could be related to the fact that CDCA is the best substrate for bile salt export pump (BSEP), which is responsible for the efflux transport of BA across the canalicular membrane from hepatocytes into bile. Therefore, loss of BSEP function could be associated with the progression of the liver disease[8,22], which leads to CDCA accumulation in the liver and eventually into the systemic circulation.
Goodness of fit was performed by testing PH assumption using a statistical test and a graphical diagnostic based on Schoenfeld residuals. For death prediction, the PH assumption was met in both BA and non-BA models supporting their validity (Supplementary Figure 1). In addition, we used the bootstrapping method for model validation. Bootstrapping validation results supported the validity of both the BA and non-BA models for death prediction (Supplementary Table 4). Further validation efforts are also ongoing to build internal and eventually external data sets for more rigorous model validation.
We used ROC analysis to compare the accuracy of our prognostic models. The higher the AUC under the ROC curve, the greater the overall accuracy of the marker in distinguishing between groups. For prognostic models, AUC of 0.9 or greater is rarely seen, AUC between 0.8 and 0.9 indicates excellent diagnostic accuracy, and any AUC over 0.7 may be considered clinically useful[23,24]. ROC curves are also used to determine cut-off values which quantify the normal ranges of biomarkers. The selection of optimum cut-off values is a tradeoff between sensitivity and specificity. Accordingly, scores for the BA, non-BA, and MELD models for death prediction of 2.71, 1.72, and 10, respectively, were identified as cut-off values with optimum sensitivity vs specificity (Table 3).
For 5-year death prediction, the AUC for BAS was 0.74 compared to 0.65 for non-BAS and 0.68 for MELD models (Figure 1A). Similarly, for 3-year death prediction, the AUC for BAS was 0.76 compared to 0.66 for non-BAS and 0.71 for MELD models (Figure 1B). In addition, BAS sensitivity in death prediction (74% vs 67% and 62%) was 7% and 12% higher than non-BAS and MELD, respectively. BAS specificity was also higher than non-BAS and MELD (68% vs 66% and 64%). Therefore, ROC analysis show that BAS is more accurate and results in higher true-positive and true-negative prediction of death compared to both non-BAS and MELD.
The Cox survival model can be used to predict the survival probability at any time point. The survival probability for t years [S (t)] was calculated for every subject using both BAS and non-BAS models, as: Survival probability for (t) years: S (t) = S0 (t) exp (BAS -2.24), survival probability for (t) years: S (t) = S0 (t) exp (non-BAS -1.58).
Where S0 (t) presents the estimated survival probability for a patient with an average BAS of 2.24 or non-BAS of 1.58 for different time points (Table 4).
As shown in Figure 2, both 5- and 3-year survival probabilities decrease as a function of both BA and non-BAS. For example, the 3-year survival probability for patients with BAS of 1.2 (25th percentile of the population), 2.1 (50th percentile of the population i.e. median), and 3.1 (75th percentile of the population) are 98%, 94%, and 85%, respectively. While, the 3-year survival probability for patients with equivalent non-BAS (25th, 50th, and 75th population percentiles) are 95%, 91%, and 86%, respectively.
The Kaplan-Meier estimator was used to estimate subjects’ survival free of adverse event over time. Median cut-off for BAS (2.19) was able to differentiate low vs high risk of death. While the median cut-offs for non-BAS and MELD were not able to differentiate low vs high risk of death (Figure 3 and Table 5).
Twenty-three patients with high BAS (> the median BAS of 2.19) died vs four patients with low BAS (< the median BAS of 2.19) for the entire study. Therefore, 19 more patients died with high compared to low BAS. In contrast, nine and five more subjects with high non-BAS and high MELD have died compared to low non-BAS and low MELD, respectively. Also, patients with low BAS lived for an average of 82 mo, while patients with high BAS lived for an average of 71 mo since their diagnosis with the liver diseases. Therefore, patients with low BAS lived 11 mo longer than patients with high BAS. On the other hand, patients with low non-BAS or low MELD (< median score), lived, in average, for only five or four months longer, compared to the high non-BAS or high MELD (high score), respectively (Table 5). Consequently, the shortening of lifespan between patients with high vs low BAS was 6-7 mo more compared to high non-BAS or high MELD. Also, the number of deaths with high BAS is 2-4-fold higher than that with high non-BAS or high MELD. Therefore, it can be concluded that in this patient population, patients with high BAS are at a much higher risk of death compared to patients with high MELD or high non-BAS.
Similar conclusions can be made regarding the death and/or LT prediction models. Patients with high BAS lived without need for LT 2-5 mo less than patients with high non-BAS or high MELD. Therefore, patients with high BAS are at a higher risk of death and/or LT compared to patients with high MELD or high non-BAS (Supplementary Figures 2-4) and (Supplementary Tables 5-7).
In summary, we have developed and validated a survival model based on BA (the BAS model) indices to predict the prognosis of cholestatic liver diseases. Our results demonstrate that the BAS model is more accurate and results in higher true-positive and true-negative prediction of death compared to both non-BAS and MELD models. Both 5- and 3-year survival probabilities markedly decreased as a function of BAS. Moreover, patients with high BAS had a 4-fold higher rate of death and lived for an average of 11 mo shorter than subjects with low BAS. The increased risk of death with high vs low BAS was also 2-4-fold higher and the shortening of lifespan was 6-7-mo lower compared to MELD or non-BAS. Similarly, we have shown the use of BAS to predict the survival of patients with and without LT. Therefore, BAS could be used to define the most seriously ill patients, who need earlier intervention such as LT. This will help provide guidance for timely care for liver patients.
Most cholestatic diseases progress toward end stage liver failure, which likely requires liver transplantation. Numerous clinical and preclinical studies have shown up to a 100-fold increase in bile acids (BA) concentrations in urine with various hepatobiliary diseases. However, due to their high inter-and intra-individual variability, BA has not been used in clinic as markers for the diagnosis and prognosis of liver diseases. To this end, we have developed the concept of BA indices and utilized it to build a survival model to predict the prognosis of liver diseases.
Biomarkers currently used in the clinic for the diagnosis and prognosis of liver diseases are primarily serum liver enzymes. Model for end-stage liver disease (MELD) was developed to predict three-month mortality of patients with end-stage liver disease. MELD is based on three objective laboratory variables that are not necessarily liver specific. The potential use of BA as a marker for liver diseases has never translated into a widespread use in the clinic. To this end, we have developed the concept of BA indices and utilized it to build a survival model to predict the prognosis of liver diseases.
The objective of this project was to discover and validate prognostic biomarkers of cholestatic liver diseases based on the urinary BA profile. We investigated the use of the urinary BA profile to develop survival models to predict the prognosis of hepatobiliary diseases. One application for BAS could be to define the most seriously ill liver patients, who may need earlier intervention such as liver transplantation.
Sample analysis: Liquid chromatography-tandem mass spectrometry. Statistical analysis: univariate and multivariate Cox proportional hazards regression, testing proportional hazards assumption, receiver operating characteristic curve, survival probability, and Kaplan-Meier plots.
The bile-acid score (BAS) model (a survival model based on BA indices) was more accurate and results in higher true-positive and true-negative prediction of death compared to both non-BAS and MELD models. Both 3- and 5-year survival probabilities markedly decreased as a function of BAS. Patients with high BAS had a 4-fold higher rate of death and lived for an average of 11 mo shorter than subjects with low BAS. The increased risk of death with high vs low BAS was also 2-4-fold greater and the shortening of lifespan was 6-7-mo lower compared to MELD or non-BAS.
We have developed and validated a survival model (the BAS model) based on BA indices to predict the prognosis of cholestatic liver diseases.
BAS could be used to define the most seriously ill patients, who need earlier intervention such as liver transplant. This will help provide guidance for timely care for liver patients.
The authors wish to thank the nurses of the CRC (Mary Ann Martin, Cindy Cowarden, Caroline Peterson, Claire Haier, and Mary Phillips) and the staff for their valuable contributions to managing the health control arm of the study, recruiting subjects, and collecting samples.
| 1. | Hofmann AF. The continuing importance of bile acids in liver and intestinal disease. Arch Intern Med. 1999;159:2647-2658. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 637] [Cited by in RCA: 725] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 2. | Monte MJ, Marin JJ, Antelo A, Vazquez-Tato J. Bile acids: chemistry, physiology, and pathophysiology. World J Gastroenterol. 2009;15:804-816. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 364] [Cited by in RCA: 413] [Article Influence: 24.3] [Reference Citation Analysis (9)] |
| 3. | Khurana S, Raufman JP, Pallone TL. Bile acids regulate cardiovascular function. Clin Transl Sci. 2011;4:210-218. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 124] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 4. | Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov. 2008;7:678-693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 869] [Cited by in RCA: 1083] [Article Influence: 60.2] [Reference Citation Analysis (0)] |
| 5. | Maillette de Buy Wenniger L, Beuers U. Bile salts and cholestasis. Dig Liver Dis. 2010;42:409-418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 134] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 6. | Pauli-Magnus C, Meier PJ. Hepatocellular transporters and cholestasis. J Clin Gastroenterol. 2005;39:S103-S110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 65] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 7. | Palmer RH. Bile acids, liver injury, and liver disease. Arch Intern Med. 1972;130:606-617. [PubMed] |
| 8. | Bathena SP, Thakare R, Gautam N, Mukherjee S, Olivera M, Meza J, Alnouti Y. Urinary bile acids as biomarkers for liver diseases II. Signature profiles in patients. Toxicol Sci. 2015;143:308-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 9. | Makino I, Hashimoto H, Shinozaki K, Yoshino K, Nakagawa S. Sulfated and nonsulfated bile acids in urine, serum, and bile of patients with hepatobiliary diseases. Gastroenterology. 1975;68:545-553. [PubMed] |
| 10. | Summerfield JA, Cullen J, Barnes S, Billing BH. Evidence for renal control of urinary excretion of bile acids and bile acid sulphates in the cholestatic syndrome. Clin Sci Mol Med. 1977;52:51-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 11. | Takikawa H, Beppu T, Seyama Y. Urinary concentrations of bile acid glucuronides and sulfates in hepatobiliary diseases. Gastroenterol Jpn. 1984;19:104-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 38] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 12. | van Berge Henegouwen GP, Brandt KH, Eyssen H, Parmentier G. Sulphated and unsulphated bile acids in serum, bile, and urine of patients with cholestasis. Gut. 1976;17:861-869. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 132] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 13. | LaRusso NF, Shneider BL, Black D, Gores GJ, James SP, Doo E, Hoofnagle JH. Primary sclerosing cholangitis: summary of a workshop. Hepatology. 2006;44:746-764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 168] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 14. | Dueland S, Reichen J, Everson GT, Davis RA. Regulation of cholesterol and bile acid homoeostasis in bile-obstructed rats. Biochem J. 1991;280 (Pt 2):373-377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 86] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 15. | Kawai H, Kudo N, Kawashima Y, Mitsumoto A. Efficacy of urine bile acid as a non-invasive indicator of liver damage in rats. J Toxicol Sci. 2009;34:27-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 16. | Geuken E, Visser D, Kuipers F, Blokzijl H, Leuvenink HG, de Jong KP, Peeters PM, Jansen PL, Slooff MJ, Gouw AS, Porte RJ. Rapid increase of bile salt secretion is associated with bile duct injury after human liver transplantation. J Hepatol. 2004;41:1017-1025. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 84] [Article Influence: 3.8] [Reference Citation Analysis (1)] |
| 17. | Buis CI, Geuken E, Visser DS, Kuipers F, Haagsma EB, Verkade HJ, Porte RJ. Altered bile composition after liver transplantation is associated with the development of nonanastomotic biliary strictures. J Hepatol. 2009;50:69-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 78] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 18. | Ramaiah SK. A toxicologist guide to the diagnostic interpretation of hepatic biochemical parameters. Food Chem Toxicol. 2007;45:1551-1557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 313] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 19. | Ozer J, Ratner M, Shaw M, Bailey W, Schomaker S. The current state of serum biomarkers of hepatotoxicity. Toxicology. 2008;245:194-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 736] [Cited by in RCA: 852] [Article Influence: 44.8] [Reference Citation Analysis (0)] |
| 20. | Huang WM, Seubert DE, Donnelly JG, Liu M, Javitt NB. Intrahepatic cholestasis of pregnancy: detection with urinary bile acid assays. J Perinat Med. 2007;35:486-491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 21. | Muraji T, Harada T, Miki K, Moriuchi T, Obatake M, Tsugawa C. Urinary sulfated bile acid concentrations in infants with biliary atresia and breast-feeding jaundice. Pediatr Int. 2003;45:281-283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 16] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 22. | Bathena SP, Thakare R, Gautam N, Mukherjee S, Olivera M, Meza J, Alnouti Y. Urinary bile acids as biomarkers for liver diseases I. Stability of the baseline profile in healthy subjects. Toxicol Sci. 2015;143:296-307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 47] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 23. | Bathena SP, Mukherjee S, Olivera M, Alnouti Y. The profile of bile acids and their sulfate metabolites in human urine and serum. J Chromatogr B Analyt Technol Biomed Life Sci. 2013;942-943:53-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 107] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 24. | Pagano M, Gauvreau K. Principles of Biostatistics. 2nd ed. Brooks/Cole: Duxbury, 2000: 259-331. |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/Licenses/by-nc/4.0/
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Sticova E, Wu ZQ S-Editor: Zhang H L-Editor: A P-Editor: Xing YX