Published online Nov 27, 2018. doi: 10.4254/wjh.v10.i11.807
Peer-review started: August 3, 2018
First decision: August 20, 2018
Revised: September 10, 2018
Accepted: October 23, 2018
Article in press: October 23, 2018
Published online: November 27, 2018
Processing time: 117 Days and 23.3 Hours
The severity of hepatic pathology and the response to treatment depend on the hepatitis virus genotype in the infected host. The objective of this review was to determine the distribution of hepatitis virus genotypes in West African countries. A systematic review of the literature in PubMed, Google Scholar and Science Direct was performed to identify 52 relevant articles reporting hepatitis A, B, C, D, E and G viruses genotypes. Hepatitis B virus (HBV) genotype E with a prevalence of 90.6% (95%CI: 0.891-0.920) found in this review, is characterized by low genetic diversity. Hepatitis C virus (HCV) genotypes 1 and 2 represented 96.4% of HCV infections in West African countries, while hepatitis delta virus, hepatitis A virus, hepatitis G virus genotypes 1 and HEV genotype 3 were reported in some studies in Ghana and Nigeria. HBV genotype E is characterized by high prevalence, low genetic diversity and wide geographical distribution. Further studies on the clinical implications of HBV genotype E and HCV genotypes 1 and 2 are needed for the development of an effective treatment against this viral hepatitis in West African countries. Surveillance of the distribution of different genotypes is also needed to reduce recombination rates and prevent the emergence of more virulent viral strains.
Core tip: The determination of hepatitis viruses genotypes is very important for the management and treatment of infected patients. Indeed, mutation development, disease progression and antiretroviral response are all dependent on the genotype of the infecting virus. Genotype determination is therefore very important to identify patients who are at increased risk of disease progression and to optimize treatment. The objective of this review was to determine the prevalence and distribution of different hepatitis viruses genotypes in 10 West African countries.
- Citation: Assih M, Ouattara AK, Diarra B, Yonli AT, Compaore TR, Obiri-Yeboah D, Djigma FW, Karou S, Simpore J. Genetic diversity of hepatitis viruses in West-African countries from 1996 to 2018. World J Hepatol 2018; 10(11): 807-821
- URL: https://www.wjgnet.com/1948-5182/full/v10/i11/807.htm
- DOI: https://dx.doi.org/10.4254/wjh.v10.i11.807
Viral hepatitis is an inflammation of the liver that may progress spontaneously to healing or lead to cirrhosis or hepatocellular carcinoma (HCC). Viral hepatitis caused 1.34 million deaths in 2015, a figure comparable to deaths from tuberculosis (TB) and human immunodeficiency virus (HIV). However, mortality attributable to TB and HIV is decreasing, while that due to hepatitis is constantly increasing[1]. There are five types of hepatitis viruses designated by the letters A, B, C, D and E. The most common forms of the disease are hepatitis A, B and C. Another human lymphotropic virus belonging to the Flaviviridae family and closely related to hepatitis C virus (HCV) was identified as hepatitis G virus (HGV) or GB virus C (GBV-C). However, several studies have shown that GBV-C/HGV infection is not clearly associated with any disease and may play a role in modulating HIV disease[2,3]. In a study conducted in Burkina Faso, a prevalence of 7.4% of HGV was reported in blood donors[4]. Viral hepatitis usually occurs as a result of parenteral contact with infected body fluids: Blood transfusions or contaminated blood products.
Hepatitis B virus (HBV) is ubiquitous, but the prevalence of infection varies across different regions of the world. According to the World Health Organization, about two billion people have been in contact with HBV worldwide, with more than 240 million cases of chronic infections[5]. About 80 to 120 million cases of chronic HBV infection occur in sub-Saharan Africa[6,7]. According to the carriage of hepatitis B surface antigen (HBsAg), there are zones of low endemicity (< 2%) such as Western Europe or North America; areas of average prevalence (2%-7%) such as North Africa or Eastern Europe and finally high endemic areas (> 8%) such as West Africa or Southeast Asia[8]. Indeed, hepatitis B is highly endemic in West Africa, with the highest prevalence in the world (> 8%). In sub-Saharan Africa, about 47% of HCC have been attributed to HBV[9]. Despite the availability of a vaccine, HBV remains a major public health problem, with approximately 686 thousand deaths per year worldwide due to the consequences (cirrhosis and HCC) of this infection[5].
More than 71 million people worldwide are chronically infected with HCV and may develop liver cirrhosis and/or HCC[10]. There is currently no effective vaccine against HCV; and about 399000 people die each year from hepatitis C, mainly cirrhosis and HCC. In North and West Africa, the prevalence of HCV infection ranges from 0.5% to 1.0%[10]. Hepatitis delta virus (HDV) is a small defective ribonucleic acid (RNA) virus that depends on HBV for the assembly of new virions and proliferation of infection to hepatocytes. HDV infection can be therefore prevented through vaccination or any strategy to eliminate HBV infection. Approximately 15 to 20 million people worldwide are co-infected with these two viruses, with a high risk of severe liver disease[11]. In a study of pregnant women in Benin, the prevalence of HDV was 11.4% in 15.5% of HBsAg-positive individuals[12].
There are several genotypes of hepatitis viruses with different clinical implications and distinct geographic distributions. HBV is classified into 10 genotypes (A-J) and about 40 subgenotypes with a correlation between genotypes and their modes of transmission[13]. In fact, HBV genotype A is found in North America, Europe, South-East Africa and India; genotypes B and C in Asia and Oceania while genotype D is the most common in North America, North Africa, Europe, the Middle-East and Oceania. HBV genotype E is hyperendemic in West Africa; genotype F is found in South America; and genotypes G and H are in Central and South America[13]. HCV has a high genetic diversity with a predominance of genotype 1 and 3 worldwide. The endemic strains of genotypes 1 and 2 are mainly found in West Africa, genotype 3 in Asia, genotypes 4 in Central Africa and the Middle East, while genotypes 5 and 6 are predominant in South Africa and South East Asia, respectively[14]. HCV genotype 2 is the most common genotype in West African countries, followed by genotype 1 and genotype 3[15]. HDV genotype I is more common in Europe and North America, genotype II is predominant in the Far East and Japan while genotype III is predominant in the Amazonian area and northern South America[16].
The severity of hepatic pathology and the response to treatment depend on the virus genotype in the infected host. For example, HBV genotype A infection tends to chronicity, whereas genotype D has a high frequency of mutation influencing response to treatment. Liver cirrhosis and progression to HCC are strongly associated with HBV genotypes C and D compared to other genotypes[17,18]. Furthermore, superinfection of chronic HBV patients by HDV leads to increased liver damage and more rapid progression of cirrhosis in 90% of cases[16]. HDV genotype III is thought to be associated with severe forms of liver disease, while a more moderate clinical evolution and a wide variety of clinical conditions are observed with genotypes II and I, respectively. The response to interferon treatments is more effective against HBV genotypes A and B compared to genotypes C, D and I. HBV genotype E seems to have the worst response to treatment[18]. Rapid progression of hepatic disease and HCC has also been associated with HBV genotype A1[13]. HCV subtype 1b is associated with a high risk of developing HCC compared to other genotypes[19]. Early generations of vaccines protected against subtype 1b, while genotype 3, which accounts for 30.1% of HCV global infections, is less likely to respond to first and second generation direct-acting antivirals currently used for HCV treatment[14,20].
Determination of the viral genotype is an essential element of the pre-therapeutic assessment, because it is one of the predictors of the response to treatment and determines the choice of molecules used with the new anti-HCV treatments. It has also been shown that HBV genotypes differ according to disease course, mutation development and response to antiviral therapy[21]. Indeed, genotype determination is important to identify patients who are at increased risk of disease progression and to optimize treatment[22]. Here, we review various publications on the genotypes of hepatitis viruses in West African countries [West African Economic and Monetary Union (WAEMU) countries, Ghana and Nigeria] in order to map the genotypes distribution and discuss the infections associated with the different viruses identified.
HBV belongs to the Hepadnaviridae family and the Orthohepadnavirus genus[23]. It is a double-stranded circular DNA enveloped virus of small circumference (1.6 million Dalton) associated with a DNA-dependent DNA polymerase that acts as a reverse transcriptase during replication. HBV is highly contagious, 100 times more contagious than HIV and can remain stable at 25 °C for seven days in dried blood. Sexual, parenteral (through the blood), mother-to-child or even close intrafamily non-sexual contact over a long period of time are the different modes of infection. The most common modes of spread of hepatitis B in endemic areas are mother-to-child transmission and exposure to infected blood. The appearance of a chronic infection is very common for infants infected by their mother before the age of 5 years.
Markers, such as the HBsAg, the HBs antibody (anti-HBs), the core antigen (HBcAg), the HBe antigen (HBeAg) and the HBe antibody (anti-HBe) make it possible to monitor the evolution of this virus. Despite the small size of the genome and the constraints imposed by its organization, HBV is highly variable.
HBV strains are divided into several genotypes. These genotypes are defined by a divergence of at least 8.0% of the whole genome nucleotide sequence and at least 4.1% in the S gene[24]. The main genotypes were divided into subgenotypes based on the divergence between 4.1% and 8.0% of their complete nucleotide sequence[25]. In the last decade, phylogenetic studies of sequences of different viral genomes have tentatively classified HBV into 10 genotypes (A-J)[26]. Genotypes and subgenotypes have a distinct geographic distribution[27]. Genotype A is the only predominant genotype in East Africa, where the prevalence of other genotypes is less than 5%[24,28]. The subgenotype A1 is predominant in Africa, while subgenotypes A3-A6 are found in Central and West Africa[29].
Subgenotype D1 is highly prevalent in East Africa[30]; D7 has been isolated in Tunisia[31]; and D8 has been characterized in Niger[32]. West Africa is the main focus of genotype E. Vaccination is the safest way to prevent HBV infection[33]. Major advances in the treatment of chronic HBV have been made with the development of nucleoside reverse transcriptase inhibitors with anti-HBV activity, such as L-nucleosides (lamivudine 3TC) or alkylphosphates (tenofovir disoproxil fumarate)[34].
HCV belongs to the family Flaviviridae and the genus hepacivirus[35]. HCV mainly infects hepatocytes but may also be present in blood mononuclear cells and dendritic cells[36]. HCV is a small single-stranded RNA virus of positive polarity, enveloped 55-65 nm in diameter. Parenteral route is the major mode of transmission of HCV. Transfusion and intravenous drug addiction are also routes of transmission. To this day, the main cause of HCV transmission in developed countries is drug abuse[37]. The HCV genome shows a high rate of mutations with considerable genetic heterogeneity of the virus in infected people worldwide. Phylogenetic approaches made it possible to classify HCV into 11 major genotypes (designated by the Arabic numerals from 1 to 11), with many subtypes (indicated by lower case letters a, b, c, etc.)[38].
Subgenotypes 1a, 1b, 2a, 2b and 3a are widely distributed worldwide[39], while 5a and 6a are common in South Africa and Southeast Asia[40,41]. Genotype 4 is predominant in Central Africa[42,43] and in North Africa[44]. In Africa, divergent HCV genotype 1 and 2 strains were found endemic in the West African subregion[45-47]. Recently, analysis of the epidemic history of HCV infections has traced modern HCV lines in West Africa to the 17th and 20th centuries[46]. The current standard treatment for chronic infection is the combination of pegylated interferon alpha (pegIFNα) and ribavirin (RBV)[48]. Currently, new therapeutic approaches using direct-acting antivirals have been developed for the treatment of chronic hepatitis C. These molecules inhibit certain stages of the viral cycle and prevent the production of viral particles by infected hepatocytes.
HDV infection: HDV is an infectious agent that can only infect patients previously or simultaneously infected with the HBV[49]. It is a single-stranded RNA negative polarity virus, 1700 nucleotides in size, which encodes a single structural protein, the hepatitis delta antigen (HDAg), and requires HBV to replicate. HDV infection can only occur with simultaneous coinfection with HBV or superinfection[1]. HDV transmission is predominantly parenteral, and the sexual route is less effective than HBV. Mother-to-child transmission is rare. Hepatitis D is a liver disease that can take the acute form and chronic form. There can be no hepatitis D in the absence of HBV. Co-infection with HBV or HDV superinfection results in more severe disease than HBV mono-infection[1]. HDV infection is diagnosed by high titers of immunoglobulin G (IgG) and immunoglobulin M (IgM) anti-HDV and confirmed by serum detection of HDV RNA by polymerase chain reaction[1]. HDV isolates in the world are divided into at least eight phylogenetically distinct genotypes[50].
Genotype 1 is the predominant form of HDV with worldwide distribution, while genotypes 2 and 4 are present in Japan and Taiwan and are often associated with a milder form of disease[50]. Genotype 3 has been reported in the Amazonian region[51]. Genotypes 5-8 were detected in the sera of patients of African origin[52]. In addition, genotype 8 infection was also detected in the state of Maranhão in northeastern Brazil[53]. Some studies have shown that genotypes 3 and 4 can be associated with particularly severe clinical forms of hepatitis (fulminant hepatitis)[51]. HDV infection is rarely studied in West Africa despite the high prevalence of HBV. There is currently no effective antiviral therapy for hepatitis D. The prevention of hepatitis D involves vaccination against hepatitis B. pegIFNα is the only effective anti-HDV drug; nucleoside analogues active against HBV have little or no effect on HDV replication[1].
HGV infection: The HGV, called GBV-C or HGV, is a flavivirus, such as HCV, that causes spontaneously resolving acute hepatitis or fulminant hepatitis. It can cause chronic infections. HGV is a single-stranded positive-strand RNA virus[54]. Its transmission is mainly parenteral. Maternal-fetal and sexual transmissions are higher than those seen with HCV. IFN is effective in normalizing hypertransaminasemia in infected patients, but relapse appears to be common when treatment is discontinued.
A systematic review of the literature was conducted to identify relevant articles reporting genotypes of hepatitis viruses in WAEMU countries including Ghana and Nigeria from 1996 to 2018. The research was conducted in French and/or English in three databases: PubMed, Google Scholar and Science Direct. The keywords used were “HBV and/or HBV and other viruses “+” the name of each of the 10 countries included in the study”. A filter limiting the search for keywords in the title and/or abstract of articles was used [PubMed: (tiab); Google Scholar: Allintitle and Science Direct: TITLE-ABSTR-KEY]. Searches with similar terms such as “hepatitis virus”, “hepatitis virus”, “hepatitis virus genotypes” or “hepatitis virus genotype” were also conducted.
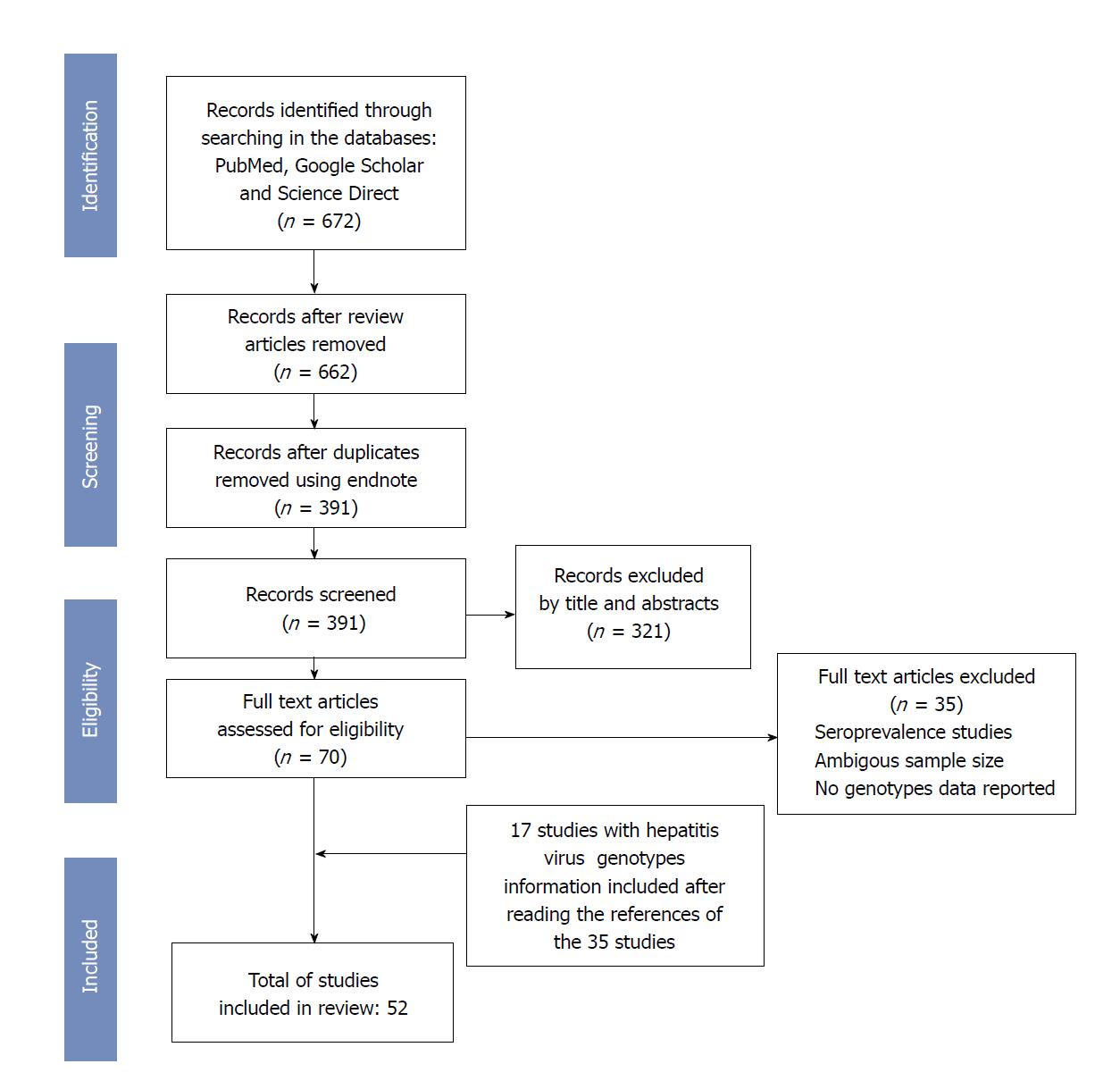
The studies were then selected on the basis of the following criteria: (1) data published in a peer-reviewed scientific journal; (2) only patients residing in one of the WAEMU countries, Ghana or Nigeria; and (3) patients from these countries infected with hepatitis viruses whose genotypes have been identified. All scientific publications (52) that reported data on genotypes of hepatitis viruses in populations from WAEMU countries, Ghana and Nigeria, between 1996 and 2018 and met the selection criteria were included in this systematic review (Figure 1). Eligible studies had to report the genotype of viral hepatitis in populations from included countries regardless of method used for viremia detection. Both risk groups or general population were eligible for inclusion.
HBV and HCV viremia detection were based on DNA/RNA amplification. Genotypes detection was performed using polymerase chain reaction or direct sequencing. HCV genotype classification was considered because in many studies, HCV cases were classified at the genotype level but not at the subtype level. Journal articles, publisher correspondence, news, letters, book chapters and studies whose data were ambiguous or could not be extracted were systematically excluded. The search and selection of the relevant articles in the three databases were carried out by two independent reviewers. The inclusion of a study by both reviewers was a requirement. In case of disagreement on the eligibility of a study, the problem was solved through a discussion and/or consensus with a third reviewer.
The genotyping data were extracted from the different studies carried out in Ghana, Nigeria and WAEMU countries. The data extracted from the various studies included in this review are: The first author, the year of the data publication, the study population, the type of study or data collection (prospective or retrospective), the country, the number of samples successfully genotyped and the results of identified genotypes.
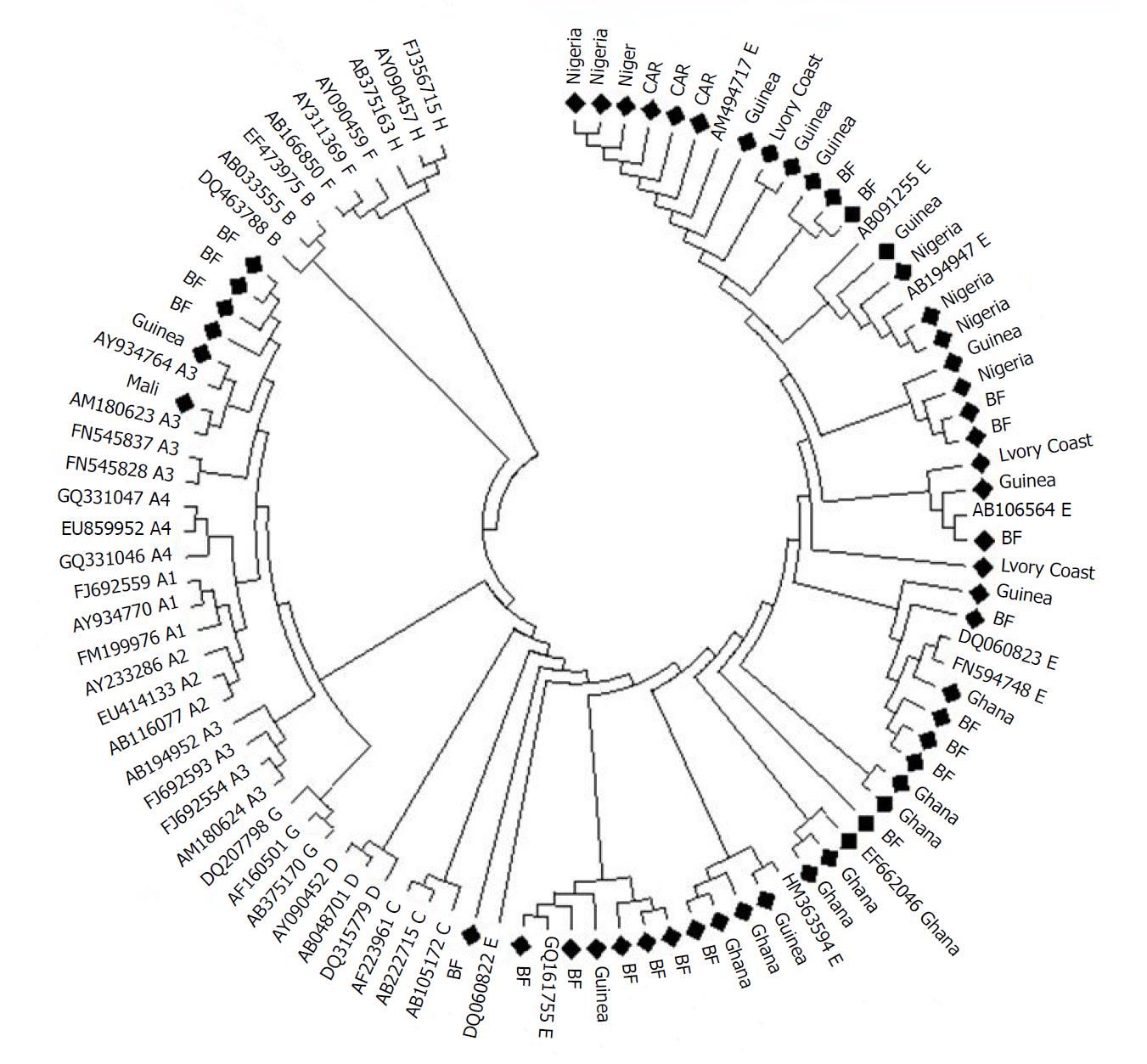
In multi-center studies, only data from the countries included in this mini-review were considered. Prevalence was determined by making the ratio of the genotype considered to the total number of samples tested for that genotype. Confidence intervals were calculated using the R software. Phylogenetic analysis was performed with 53 HBV sequences using the neighbor-joining algorithm based on the Kimura two-parameter distance estimation method. Only bootstrap values of > 80% are shown (1.000 replicates). The maps were made using genotyping data from each country (source: Dr. Ouattara AK).
The initial search in the three databases, according to the search strategy described in the methodology, found 391 articles after elimination of reviews and duplicates. Examination of titles and abstracts led to the elimination of 321 studies that did not meet the inclusion criteria of this review. Only 70 studies were considered eligible after full text review. This step allowed the exclusion of 35 articles presenting data reporting only seroprevalences or presenting ambiguous genotyping data. Finally, 52 studies, 35 of which were obtained after the full text examination and 17 after references examination of the 35 articles selected, were included in this systematic review.
Tables 1 and 2 present the characteristics of the different studies included in this systematic review. The majority of included studies used a prospective method of sample collection. A case-control study, two Cas reports, two multi-center studies and four cohort studies were included in the review, while the rest were cross-sectional or prospective studies. Twelve studies were performed in HIV-infected individuals compared to 11 in blood donors and seven in pregnant women, while populations and age groups were variable for the rest of the studies.
| Ref. | Year | Countries | Patients | Type of study | Samples | HBV genotypes (n) |
| Diarra et al[80] | 2018 | Burkina Faso | Occult HBV | Cross-sectional | 21 | E (17) and A3 (4) |
| Archampong et al[59] | 2017 | Ghana | HBV-HIV coinfected | Cross-sectional | 63 | E (58), A (4) and D (1) |
| Boyce et al[58] | 2017 | Ghana | HBV-HIV coinfected | Case reports | 3 | D/E (3) |
| Cella et al[63] | 2017 | Mali | Malian refugees | Cross-sectional | 16 | E (16) |
| Lawson-Ananissoh et al[81] | 2017 | Côte d’Ivoire | Chronic HBV | Prospective | 33 | E (27), A (6) |
| Dongdem et al[73] | 2016 | Ghana | Chronic Hepatitis B | Cross-sectional | 58 | E (47), A (8) and D (3) |
| Opaleye et al[82] | 2016 | Nigeria | HBV+ | Cross-sectional | 17 | E (17) |
| Compaore et al[62] | 2016 | Burkina Faso | HIV-1+ and HIV-1- | Case-Control | 120 | E (120) |
| Candotti et al[83] | 2016 | Burkina Faso | Blood donors | Prospective | 99 | E (71) A3QS (28) |
| Brah et al[84] | 2016 | Niger | HBV infected | Prospective | 23 | E (21), A3E (1) and D/E (1) |
| Boyd et al[85] | 2016 | Côte d’Ivoire | HBV-HIV coinfected | Prospective | 100 | E (98) and A (2) |
| Ampah et al[61] | 2016 | Ghana | Randomized volunteers | Prospective | 52 | E (52) |
| Traore et al[55] | 2015 | Mali | Adults volunteers | Cohort study | 90 | E (82), D/E (5), D (1) and A (2) |
| Faleye et al[86] | 2015 | Nigeria | Pregnant women | Cross-sectional | 6 | E (6) |
| Faleye et al[87] | 2015 | Nigeria | Asymptomatic individuals | Cross-sectional | 13 | E (13) |
| Maylin et al[88] | 2015 | Senegal | Chronic HBV | Cohort study | 87 | E (65), A (22) |
| Honge et al[76] | 2014 | Guinea-Bissau | HIV+ | Cross-sectional | 26 | E (25) and D (1) |
| De Paschale et al[12] | 2014 | Benin | Pregnant women | Prospective | 19 | E (19) |
| Forbi et al[56] | 2013 | West Africa1 | Pregnant women and HIV+ | Multicenter | 83 | E (74) and A (9) |
| Hübschen et al[89] | 2011 | Nigeria | Cohorts samples | Cohorts study | 163 | E (154) and A (9) |
| Geretti et al[90] | 2010 | Ghana | HIV+ | Cross-sectional | 86 | E (82) and A (4) |
| Forbi et al[69] | 2010 | Nigeria | Asymptomatic volunteers | Cross-sectional | 55 | E (53) and A3 (2) |
| Chekaraou et al[32] | 2010 | Niger | Blood donors | Cross-sectional | 24 | E (20), D/E (4) |
| Candotti et al[91] | 2007 | Ghana | Pregnant women | Cross-sectional | 70 | E (69) and A (1) |
| Vray et al[92] | 2006 | Senegal | Blood donors | Cross-sectional | 32 | E (23) and A (9) |
| Huy et al[60] | 2006 | Ghana | Blood donors | Cross-sectional | 12 | E (12) |
| Candotti et al[66] | 2006 | Ghana | Blood donors | Cross-sectional | 100 | E (87), A (10) and D (3) |
| Fujiwara et al[65] | 2005 | Benin | Blood donors | Cross-sectional | 21 | E (20) and A (1) |
| Mulders et al[64] | 2004 | West Africa2 | Measles or HIV+ | Multicenter | 79 | E (78), and A (1) |
| Suzuki et al[70] | 2003 | Côte d’Ivoire | HBV carriers | Cross-sectional | 48 | E (42), A (3) and D (3) |
| Ref. | Year | Countries | Patients | Type of study | Samples | Others hepatitis genotypes (n) |
| Abubakar et al[93] | 2017 | Nigeria | HCV+ | Prospective | 173 | HCV G1 (159) and G2 (14) |
| Ndiaye et al[94] | 2015 | Senegal | Drug users | Cohort study | 25 | HCV G1 (21), G2 (1), G3 (1) and G4 (2) |
| Henquell et al[95] | 2016 | Burkina Faso | woman | Case report | 1 | HCV G5 (1) |
| Opaleye [82] | 2016 | Nigeria | HBV+ | Cross-sectional | 14 | HDV G1 (14) |
| De Paschale et al[12] | 2014 | Benin | Pregnant women | Prospective | 6 | HCV G1 (1), G2 (5) |
| Honge et al[76] | 2014 | Guinea Bissau | HIV+ | Cross-sectional | 8 | HCV G2 (8) |
| Zeba et al[96] | 2014 | Burkina Faso | Blood donors | Cross-sectional | 36 | HCV G1 (4), G2 (22), G3 (8), G4 (2) |
| Forbi et al[56] | 2013 | Nigeria | Apparently healthy adult | Cross-sectional | 12 | HAV sub-G1A (12) |
| Diarra et al[97] | 2013 | Mali | Diabetic | Prospective | 25 | HCV G1 (7) and G2 (18) |
| Bouare et al[98] | 2013 | Mali | Old women | Prospective | 14 | HCV G1 (2) and G2 (12) |
| Forbi et al[47] | 2012 | Nigeria | Asymptomatic indigenes | Prospective | 60 | HCV G1 (51) and G2 (9) |
| Sombie et al[99] | 2011 | Burkina Faso | HCV+ | Prospective | 38 | HCV G1 (10), G2 (27) and G5 (1) |
| Bengue et al[100] | 2008 | Côte d’Ivoire | Blood donors | Prospective | 27 | HCV G1 (21), G2 (5) and G5 (1) |
| Plamondon et al[101] | 2007 | Guinea Bissau | Adult volunteers | Cross-sectional | 57 | HCV G1 (1) and G2 (56) |
| Simpore et al[102] | 2005 | Burkina Faso | Pregnant women | Prospective | 5 | HCV G1 (2) and G2 (3) |
| Rouet et al[103] | 2004 | Côte d’Ivoire | HIV+/Pregnant women | Cross-sectional | 6 | HCV G1 (3) and G2 (3) |
| Agwale et al[104] | 2004 | Nigeria | HIV+ under ART | Prospective | 12 | HCV G1 (9) and G2 (3) |
| Candotti et al[45] | 2003 | Ghana | Blood donors | Cross-sectional | 23 | HCV G1 (3) and G2 (20) |
| Buisson et al[105] | 2000 | Nigeria | Acute hepatitis | Cross-sectional | 7 | HEV G3 (7) |
| Saito et al[79] | 1999 | Ghana | HIV+ and HIV- | Cross-sectional | 9 | HGV G1 (9) |
| Wansbrough-Jones et al[106] | 1998 | Ghana | Blood donors | Cross-sectional | 7 | HCV G1 (2) and G2 (5) |
| Oni et al[107] | 1996 | Nigeria | blood donors | Cross-sectional | 5 | HCV G1 (2) and G4 (3) |
The frequency of the different genotypes was determined by dividing the number of samples presenting the genotype considered by the total number of successfully sequenced samples. In this systematic review, the largest number of successfully sequenced samples were recorded in Ghana (457/1620), followed by Nigeria (269/1620) and Côte d’Ivoire (251/1620). Genotyping studies of hepatitis viruses were rare in Guinea-Bissau and almost non-existent in Togo. The HBV genotype E was the predominantly isolated genotype in the various studies conducted in the WAEMU countries, Ghana and Nigeria (Table 1).
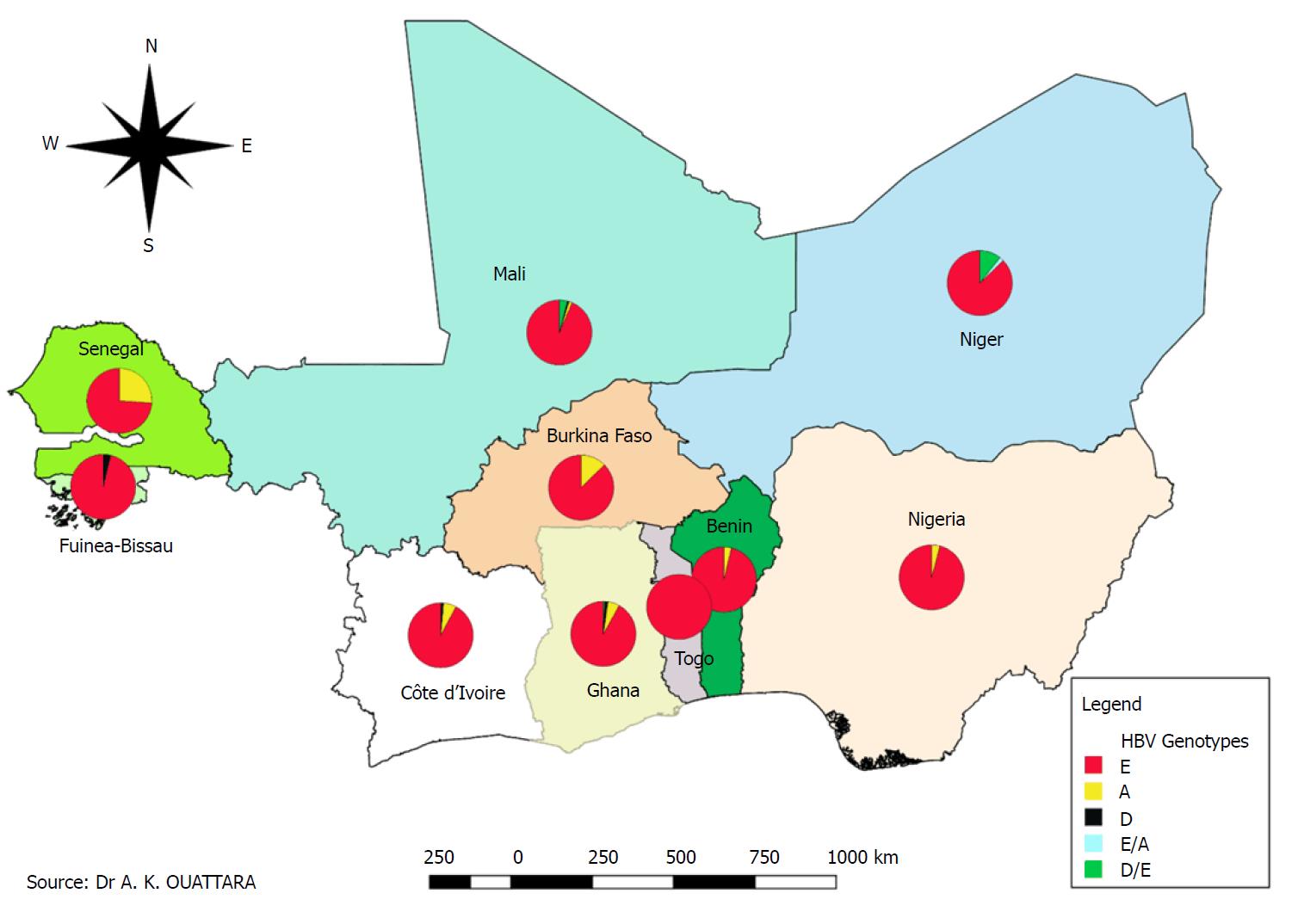
Indeed, out of a total of 1620 successfully sequenced HBV samples, E genotypes were individually isolated in 90.6% (1468/1620, 95%CI: 0.891-0.920) of HBV infection cases. In addition, its prevalence of recombination or coinfection with genotypes A and D was estimated at 0.86% (14/1620, 95%CI: 0.005-0.014) in our study area. HBV genotype E is characterized by low genetic diversity compared to other genotypes, including genotype A (Figure 2). The second HBV genotype reported in terms of frequency in the countries included in this review was genotype A with an individual prevalence of 7.8% (126/1620, 95%CI: 0.065-0.092), while a prevalence of 0.74% (12/1620, 95%CI: 0.004-0.013) of genotypes D was observed in the study area. A slight decrease in the overall frequency of HBV genotype E in West African countries was found between 2003 and 2010 (94.4%) compared to 2011-2018 (90.0%) with emergence of genotypes A and D. Some studies have focused on the genotyping of other hepatitis viruses with a predominance of HCV infections (528/570, Table 2). Figure 3 shows the geographical distribution of the different HBV genotypes in the countries included in this review.
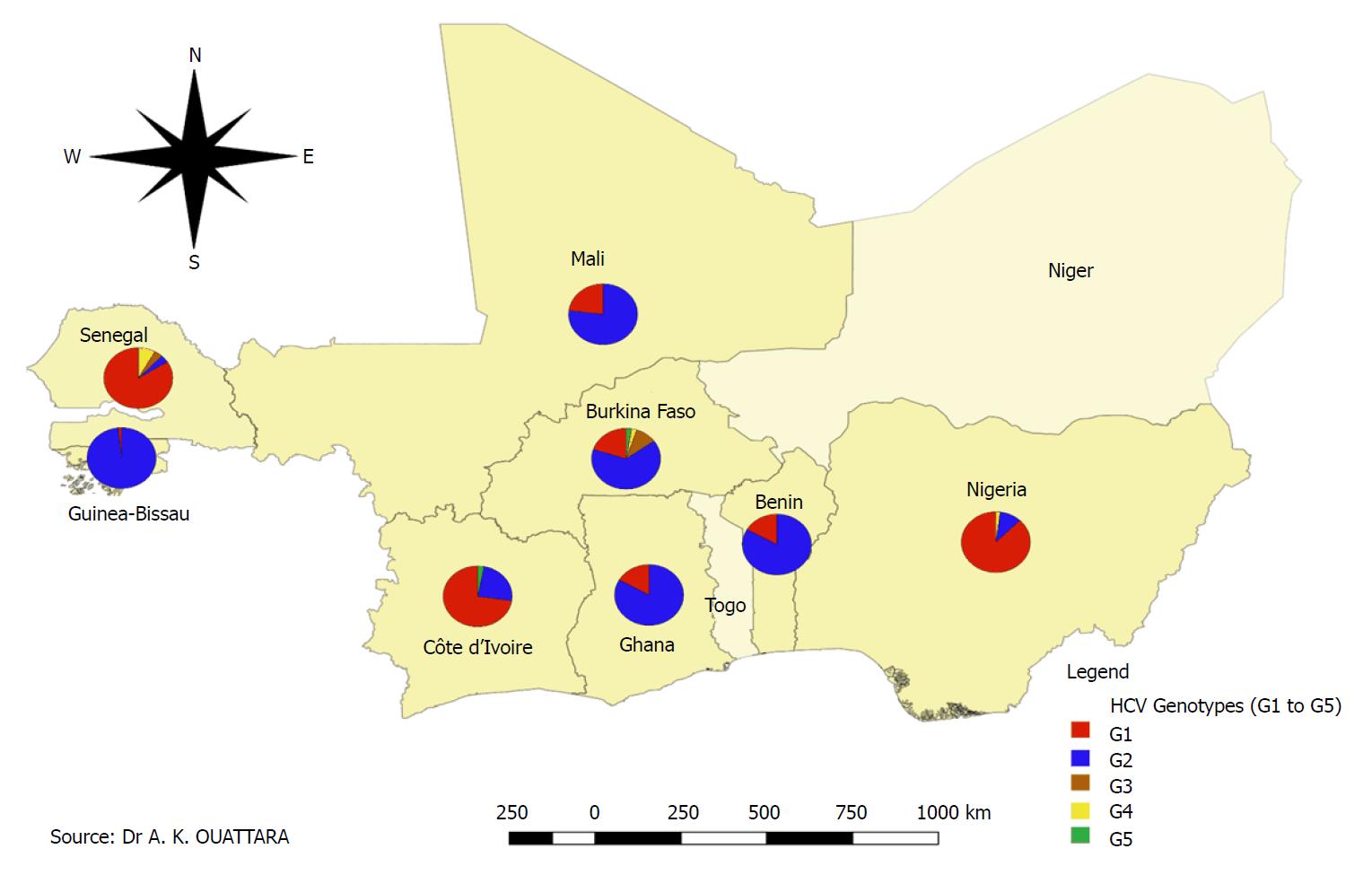
Of the 535 strains of HCV isolated and sequenced successfully, genotype 1 was found in the majority of cases of infections (56.4% or 298/528) against 40.0% (211/528) for genotype 2 while 3.6% (19/528) of the samples had genotypes 3 (9/19), 4 (7/19) and 5 (3/19) of HCV. HCV genotype 2 was most common in Benin, Burkina Faso, Ghana Guinea Bissau and Mali, while genotype 1 was predominant in Côte d’Ivoire, Senegal and Nigeria (Figure 4), with a high number of sequenced samples. Genotypes 1 of other hepatitis viruses such as HDV, a satellite virus still found in coinfection with HBV, HAV, HGV, and HEV genotype 3, have been reported in some studies in Ghana and Nigeria (Table 2).
The aim of this review was to map the genotypes of the different hepatitis viruses identified in WAEMU countries, Ghana and Nigeria. The systematic review in the PubMed, Google Scholar and Sciences Direct databases included 52 studies reporting genotypes of hepatitis A, B, C, D, E and G. The availability of genetic data varied across country due to the prevalence or clinical relevance of the virus or the difficulty of sequencing in a context of limited resources. Indeed, most of the genotyping studies (29/52) focused on HBV because of its endemicity in sub-Saharan Africa and its clinical implications[1]. In West Africa, chronic carriage of HBV in the general unvaccinated population is estimated to be between 10% and 18%[55]. Several studies (18/52) have also provided HCV genotype data, which is the second virus of clinical interest in this West African sub-region after HBV, while very little genetic data is available on HDV, a satellite of HBV. The genetic data on HAV come only from Nigeria where it is endemic[56], while the genotypes of hepatitis E and G viruses, very scarce in WAEMU countries[57], were reported respectively in Nigeria and Ghana.
Knowledge of hepatitis viruses genotypes is of great epidemiological and clinical interest. Indeed, genotypes are responsible for variable clinical manifestations with differences depending on the stage of the disease, mutations and response to treatments[58,59]. They are also an invaluable tool for mapping the molecular evolution and dynamics of infection transmission because the different genotypes have a distinct geographic distribution. The study of Archampong et al[59] demonstrated that the majority of HBV-positive and patients co-infected with lamivudine 3TC resistance were infected with HBV genotype E. This review confirms the endemicity of HBV genotype E, with a prevalence of 90.6% (1468/1620, 95%CI: 0.891-0.920) and a predominance of serotype awy4. Indeed, some studies conducted in Ghana[60,61], Burkina Faso[62] and Mali[63] exclusively reported the HBV genotype E in their study populations.
In addition to the presence of other genotypes, including HBV genotypes A and D, studies have reported a strong predominance of genotype E[64-66]. Similar observations have led several authors to support further the common presence of HBV genotype E in West African populations[24]. Indeed, the predominance and almost exclusive circulation of genotype E in sub-Saharan Africa certainly indicates its West African origin[67-69]. Its distribution is limited to West Africa, unlike other HBV genotypes, despite the migration of slaves from West Africa to North America[66]. This review also reports low genetic diversity of HBV genotype E in West Africa (Figure 2)[64,66]. The low genome diversity and large distribution of genotype E in West Africa suggests a recent introduction of this genotype in the human host[64,70]. It is possible that it has been introduced relatively recently into an animal reservoir (the chimpanzee) as well as for HIV or that variant of genotype D (the closest to genotype E) has acquired an evolutionary advantage[66]. HBV genotypes A and D were also reported in this review with respective prevalence of 7.8% (126/1620, 95%CI: 0.065-0.092) and 0.74% (12/1620, 95%CI: 0.004-0.013). HBV genotype A, which is also found in sub-Saharan Africa, has been reported in eight of the 10 countries included in this review. Indeed, genotype E is predominant in West Africa, while genotype A has a relatively high prevalence in East Africa[71].
In 2006, Candotti et al[66] reported a prevalence of 10% and 3%, respectively, for HBV genotypes A and D in blood donors in Ghana. Similar results have also highlighted the cocirculation of genotypes A and D in Ghana, Mali, Côte d’Ivoire and Nigeria[59]. The majority of genotypes A identified in Burkina Faso are quasi-A3 genotypes (A3Q) documented in West African populations[72]. Indeed, data from previous studies suggest a predominance of the A1 genotype in East Africa and the A3 genotype in West and Central Africa, while the A2 subgenotype has a high frequency in North Africa where the genotype D is predominant. Africa has a high diversity of HBV genotypes and subgenotypes displaying distinct geographical distributions. Genotype A is found mainly in south-eastern Africa, genotype E in western and central Africa and genotype D prevails in northern Africa. Genotype E is rarely found outside Africa, except in individuals of African descent.
Characterization of HBV genotypes allows clinicians to determine patients’ response to treatment and potential risks of complications[58,73]. Flink et al[74] have indeed reported that genotype A responds better to interferon alpha and pegIFNα than genotype D. Genotype recombination occurs in areas where multiple genotypes are in co-circulation, thus facilitating diversification between individuals within the general population. Our review reports a recombination prevalence of HBV genotype E with genotypes A and D of 0.87% (14/1599, 95%CI: 0.005-0.015). A/B, A/C, A/E, C/E, D/E and D/E/A recombination have been reported in West Africa[58]. Recombination requires co-infection with more than one genotype in the same patient. Appropriate treatment and elimination of risky behavior in people infected with the virus are therefore necessary for a considerable reduction in the spread of recombinant viruses.
The data in this review report five circulating HCV genotypes in the WAEMU countries, Ghana and Nigeria, with an overall predominance of genotype 1 (56.4%, 298/528). The prevalence of HCV genotype 1 has been reported by several authors in Nigeria, Senegal and Côte d’Ivoire, who record the most data presented in this review. Genotype 2 with a general frequency of 40.0% (211/528) was the main genotype found in Benin, Burkina Faso, Ghana, Guinea-Bissau and Mali (Figure 2). Indeed, several previous studies have reported a predominance of HCV genotype 2 in West Africa[75]. Most authors suggest a West African genotype 2 origin of HCV in the region, including The Gambia and Guinea-Bissau[46]. Indeed, HCV genotyping data in Guinea-Bissau report an almost exclusive predominance of genotype 2[76]. Candotti et al[45] reported 87.0% of genotype 2 was associated with chronic HCV infection with 13% of cases for genotype 1. In a study in Ghana in HCV/HIV coinfected patients, HCV sequences were phylogenetically assigned genotype 2 and subtypes 21 and 2r[77]. Although no published data on HCV genotypes were found in Togo, genotypes 2 and 1 were the most frequently isolated, with respective prevalence 73.2% and 17.1%, in a study conducted in 2014 in the Togolese general population.
Genotype 2 is, therefore, predominant in Togo, as it is in most parts of West Africa (unpublished data). In Martinique, where three quarters of the slaves sent in the 17th and 18th centuries came from West Africa, there is a great diversity of genotype 2[78]. The majority of molecular and epidemiological studies suggest that HCV genotype 2 has been present in West Africa for several centuries.
Data on the genotypes of other hepatitis viruses that are very infrequent or with relatively high frequencies in some areas have also been reported in this review. HAV genotype 1 and HEV and HDV were reported in Nigeria, while genotype 1 of HGV was found in Ghana. HAV, whose transmission is closely associated with lack of clean drinking water, unsuitable food, inadequate sanitation and poor personal hygiene, is prevalent in parts of Nigeria (World Health Organization). HDV is a satellite virus of HBV, because HDV only infects people with HBV. Limited data is available on circulating HDV genotypes. In a study in Togo, it was reported that 94.3% of the general population was infected with genotype 1, and 5.7% was infected with genotype 5. Studies on HGV are very limited[57,79]. The analysis of the 5% untranslated region nucleotide sequence of the genome of the HGV shows that the nine Ghanaian isolates of the HGV belong to genotype 1, the West-African type of the HGV[79].
The complexity of hepatitis virus genotypes often leads to a specificity of treatment associated with the genotype. The present review reporting a mapping of genotypes of hepatitis viruses A, B, C, D, E and G in the WAEMU, Ghana and Nigeria, reveals that the majority of studies conducted in Ghana and Nigeria have very little information on hepatitis D and G. In the WAEMU area including Ghana and Nigeria, HBV strains were classified as genotypes E, A, D with a predominance of genotype E and serotype ayw4. Genotype E is characterized by a high prevalence, low genetic diversity and wide geographical distribution.
The majority of HCV genotype data came from Nigeria, Senegal and Côte d’Ivoire were characterized by a predominance of genotype 1, while a high prevalence of genotype 2 was found in Benin, Burkina Faso, Ghana, in Guinea-Bissau and Mali. Further studies on the clinical implications of HBV genotype E are needed for the development of an effective treatment for HBV in West Africa. Monitoring the distribution of the different genotypes is also needed to reduce recombination levels and prevent the emergence of other viral strains. There is a diversity of genotypes and subtypes of hepatitis viruses with risks of recombination and emergence of even more virulent forms. Hepatitis viruses do not need a passport or visa to move from one country to another, and they have preceded us in WAEMU or ECOWAS. It is therefore appropriate for us to develop the adequate means to prevent, treat and even eradicate these viral infections using a vaccine covering all variants.
| 1. | World Health Organization. Global Hepatitis Report, 2017. Available from: http://www.who.int/hepatitis/publications/global-hepatitis-report2017/en/. |
| 2. | Halasz R, Weiland O, Sällberg M. GB virus C/hepatitis G virus. Scand J Infect Dis. 2001;33:572-580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 20] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 3. | Blackard JT, Ma G, Polen C, DuBois JC, Gast J, Radens CM, Sterling RK, Sherman KE. Recombination among GB virus C (GBV-C) isolates in the United States. J Gen Virol. 2016;97:1537-1544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 4. | Tao I, Bisseye C, Nagalo BM, Sanou M, Kiba A, Surat G, Compaoré TR, Traoré L, Nikiema JB, Pietra V. Screening of Hepatitis G and Epstein-Barr Viruses Among Voluntary non Remunerated Blood Donors (VNRBD) in Burkina Faso, West Africa. Mediterr J Hematol Infect Dis. 2013;5:e2013053. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 5. | World Health Organization. Global health sector strategy on viral hepatitis 2016-2021. Available from: http://www.who.int/hepatitis/strategy2016-2021/ghss-hep/en/. |
| 6. | Livinec B. Estimating the burden of hepatitis. Lancet. 2016;388:2738-2739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 7. | Stanaway JD, Flaxman AD, Naghavi M, Fitzmaurice C, Vos T, Abubakar I, Abu-Raddad LJ, Assadi R, Bhala N, Cowie B. The global burden of viral hepatitis from 1990 to 2013: findings from the Global Burden of Disease Study 2013. Lancet. 2016;388:1081-1088. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1042] [Cited by in RCA: 1014] [Article Influence: 101.4] [Reference Citation Analysis (0)] |
| 8. | Alter MJ. Epidemiology of hepatitis B in Europe and worldwide. J Hepatol. 2003;39 Suppl 1:S64-S69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 174] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 9. | Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol. 2006;45:529-538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1764] [Cited by in RCA: 1858] [Article Influence: 92.9] [Reference Citation Analysis (1)] |
| 10. | World Health Organization. Principaux repères sur l’hépatite C. Available from: http://www.who.int/fr/news-room/fact-sheets/detail/hepatitis-c. |
| 11. | Alfaiate D, Dény P, Durantel D. Hepatitis delta virus: From biological and medical aspects to current and investigational therapeutic options. Antiviral Res. 2015;122:112-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 12. | De Paschale M, Ceriani C, Cerulli T, Cagnin D, Cavallari S, Ndayaké J, Zaongo D, Priuli G, Viganò P, Clerici P. Prevalence of HBV, HDV, HCV, and HIV infection during pregnancy in northern Benin. J Med Virol. 2014;86:1281-1287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 13. | Rajoriya N, Combet C, Zoulim F, Janssen HLA. How viral genetic variants and genotypes influence disease and treatment outcome of chronic hepatitis B. Time for an individualised approach? J Hepatol. 2017;67:1281-1297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 127] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 14. | Messina JP, Humphreys I, Flaxman A, Brown A, Cooke GS, Pybus OG, Barnes E. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology. 2015;61:77-87. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1077] [Cited by in RCA: 1161] [Article Influence: 105.5] [Reference Citation Analysis (0)] |
| 15. | Petruzziello A, Marigliano S, Loquercio G, Cozzolino A, Cacciapuoti C. Global epidemiology of hepatitis C virus infection: An up-date of the distribution and circulation of hepatitis C virus genotypes. World J Gastroenterol. 2016;22:7824-7840. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 514] [Cited by in RCA: 585] [Article Influence: 58.5] [Reference Citation Analysis (16)] |
| 16. | Romeo R, Petruzziello A, Pecheur EI, Facchetti F, Perbellini R, Galmozzi E, Khan NU, Di Capua L, Sabatino R, Botti G. Hepatitis delta virus and hepatocellular carcinoma: an update. Epidemiol Infect. 2018;146:1612-1618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 17. | Petruzziello A. Epidemiology of Hepatitis B Virus (HBV) and Hepatitis C Virus (HCV) Related Hepatocellular Carcinoma. Open Virol J. 2018;12:26-32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 170] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 18. | Sunbul M. Hepatitis B virus genotypes: global distribution and clinical importance. World J Gastroenterol. 2014;20:5427-5434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 264] [Cited by in RCA: 304] [Article Influence: 25.3] [Reference Citation Analysis (8)] |
| 19. | Petruzziello A, Marigliano S, Loquercio G, Coppola N, Piccirillo M, Leongito M, Azzaro R, Izzo F, Botti GJIA, Cancer . Hepatitis C Virus (HCV) genotypes distribution among hepatocellular carcinoma patients in Southern Italy: a three year retrospective study. Infect Agents Cancer. 2017;12:52. [RCA] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 20. | Schietroma I, Scheri GC, Pinacchio C, Statzu M, Petruzziello A, Vullo V. Hepatitis C Virus and Hepatocellular Carcinoma: Pathogenetic Mechanisms and Impact of Direct-Acting Antivirals. Open Virol J. 2018;12:16-25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 21. | Shi YH. Correlation between hepatitis B virus genotypes and clinical outcomes. Jpn J Infect Dis. 2012;65:476-482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 58] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 22. | Tanwar S, Dusheiko G. Is there any value to hepatitis B virus genotype analysis? Curr Gastroenterol Rep. 2012;14:37-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 58] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 23. | Lazar C, Macovei A, Petrescu S, Branza-Nichita N. Activation of ERAD pathway by human hepatitis B virus modulates viral and subviral particle production. PLoS One. 2012;7:e34169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 71] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 24. | Norder H, Couroucé AM, Coursaget P, Echevarria JM, Lee SD, Mushahwar IK, Robertson BH, Locarnini S, Magnius LO. Genetic diversity of hepatitis B virus strains derived worldwide: genotypes, subgenotypes, and HBsAg subtypes. Intervirology. 2004;47:289-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 630] [Cited by in RCA: 651] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 25. | Schaefer S. Hepatitis B virus: significance of genotypes. J Viral Hepat. 2005;12:111-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 167] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 26. | Tatematsu K, Tanaka Y, Kurbanov F, Sugauchi F, Mano S, Maeshiro T, Nakayoshi T, Wakuta M, Miyakawa Y, Mizokami M. A genetic variant of hepatitis B virus divergent from known human and ape genotypes isolated from a Japanese patient and provisionally assigned to new genotype J. J Virol. 2009;83:10538-10547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 338] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 27. | Schaefer S. Hepatitis B virus taxonomy and hepatitis B virus genotypes. World J Gastroenterol. 2007;13:14-21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 243] [Cited by in RCA: 247] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 28. | Kurbanov F, Tanaka Y, Mizokami M. Geographical and genetic diversity of the human hepatitis B virus. Hepatol Res. 2010;40:14-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 158] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 29. | Kramvis A, Kew MC. Epidemiology of hepatitis B virus in Africa, its genotypes and clinical associations of genotypes. Hepatol Res. 2007;37:S9-S19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 233] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 30. | Banerjee A, Kurbanov F, Datta S, Chandra PK, Tanaka Y, Mizokami M, Chakravarty R. Phylogenetic relatedness and genetic diversity of hepatitis B virus isolates in Eastern India. J Med Virol. 2006;78:1164-1174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 77] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 31. | Meldal BH, Moula NM, Barnes IH, Boukef K, Allain JP. A novel hepatitis B virus subgenotype, D7, in Tunisian blood donors. J Gen Virol. 2009;90:1622-1628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 71] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 32. | Abdou Chekaraou M, Brichler S, Mansour W, Le Gal F, Garba A, Dény P, Gordien E. A novel hepatitis B virus (HBV) subgenotype D (D8) strain, resulting from recombination between genotypes D and E, is circulating in Niger along with HBV/E strains. J Gen Virol. 2010;91:1609-1620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 63] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 33. | World Health Organization. Guidelines for the prevention, care and treatment of persons with chronic hepatitis B infection. Available from: http://www.who.int/hiv/pub/hepatitis/hepatitis-b-guidelines/en/. |
| 34. | Zoulim F. Hepatitis B virus resistance to antiviral drugs: where are we going? Liver Int. 2011;31 Suppl 1:111-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 103] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 35. | Lundin M, Monné M, Widell A, Von Heijne G, Persson MA. Topology of the membrane-associated hepatitis C virus protein NS4B. J Virol. 2003;77:5428-5438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 152] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 36. | Navas MC, Fuchs A, Schvoerer E, Bohbot A, Aubertin AM, Stoll-Keller F. Dendritic cell susceptibility to hepatitis C virus genotype 1 infection. J Med Virol. 2002;67:152-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 65] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 37. | Murray KF, Richardson LP, Morishima C, Owens JW, Gretch DR. Prevalence of hepatitis C virus infection and risk factors in an incarcerated juvenile population: a pilot study. Pediatrics. 2003;111:153-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 37] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 38. | Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology. 2014;59:318-327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 965] [Cited by in RCA: 991] [Article Influence: 82.6] [Reference Citation Analysis (2)] |
| 39. | Ramia S, Eid-Fares J. Distribution of hepatitis C virus genotypes in the Middle East. Int J Infect Dis. 2006;10:272-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 71] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 40. | Pybus OG, Barnes E, Taggart R, Lemey P, Markov PV, Rasachak B, Syhavong B, Phetsouvanah R, Sheridan I, Humphreys IS. Genetic history of hepatitis C virus in East Asia. J Virol. 2009;83:1071-1082. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 173] [Cited by in RCA: 167] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 41. | Prabdial-Sing N, Puren AJ, Bowyer SM. Sequence-based in silico analysis of well studied hepatitis C virus epitopes and their variants in other genotypes (particularly genotype 5a) against South African human leukocyte antigen backgrounds. BMC Immunol. 2012;13:67. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 42. | Ndong-Atome GR, Makuwa M, Ouwe-Missi-Oukem-Boyer O, Pybus OG, Branger M, Le Hello S, Boye-Cheik SB, Brun-Vezinet F, Kazanji M, Roques P. High prevalence of hepatitis C virus infection and predominance of genotype 4 in rural Gabon. J Med Virol. 2008;80:1581-1587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 24] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 43. | Cantaloube JF, Gallian P, Laperche S, Elghouzzi MH, Piquet Y, Bouchardeau F, Jordier F, Biagini P, Attoui H, de Micco P. Molecular characterization of genotype 2 and 4 hepatitis C virus isolates in French blood donors. J Med Virol. 2008;80:1732-1739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 44. | Antaki N, Craxi A, Kamal S, Moucari R, Van der Merwe S, Haffar S, Gadano A, Zein N, Lai CL, Pawlotsky JM. The neglected hepatitis C virus genotypes 4, 5 and 6: an international consensus report. Liver Int. 2010;30:342-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 120] [Article Influence: 7.5] [Reference Citation Analysis (2)] |
| 45. | Candotti D, Temple J, Sarkodie F, Allain JP. Frequent recovery and broad genotype 2 diversity characterize hepatitis C virus infection in Ghana, West Africa. J Virol. 2003;77:7914-7923. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 111] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 46. | Markov PV, Pepin J, Frost E, Deslandes S, Labbé AC, Pybus OG. Phylogeography and molecular epidemiology of hepatitis C virus genotype 2 in Africa. J Gen Virol. 2009;90:2086-2096. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 75] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 47. | Forbi JC, Purdy MA, Campo DS, Vaughan G, Dimitrova ZE, Ganova-Raeva LM, Xia GL, Khudyakov YE. Epidemic history of hepatitis C virus infection in two remote communities in Nigeria, West Africa. J Gen Virol. 2012;93:1410-1421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 48. | NIH Consensus Statement on Management of Hepatitis C: 2002. NIH Consens State Sci Statements. 2002;19:1-46. [PubMed] |
| 49. | Madejón A, Romero M, Hernández Á, García-Sánchez A, Sánchez-Carrillo M, Olveira A, García-Samaniego J. Hepatitis B and D viruses replication interference: Influence of hepatitis B genotype. World J Gastroenterol. 2016;22:3165-3174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 9] [Cited by in RCA: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 50. | Alvarado-Mora MV, Locarnini S, Rizzetto M, Pinho JR. An update on HDV: virology, pathogenesis and treatment. Antivir Ther. 2013;18:541-548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 62] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 51. | Gomes-Gouvêa MS, Soares MC, Bensabath G, de Carvalho-Mello IM, Brito EM, Souza OS, Queiroz AT, Carrilho FJ, Pinho JR. Hepatitis B virus and hepatitis delta virus genotypes in outbreaks of fulminant hepatitis (Labrea black fever) in the western Brazilian Amazon region. J Gen Virol. 2009;90:2638-2643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 73] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 52. | Dény P. Hepatitis delta virus genetic variability: from genotypes I, II, III to eight major clades? Curr Top Microbiol Immunol. 2006;307:151-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 69] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 53. | Barros LM, Gomes-Gouvêa MS, Pinho JR, Alvarado-Mora MV, Dos Santos A, Mendes-Corrêa MC, Caldas AJ, Sousa MT, Santos MD, Ferreira AS. Hepatitis Delta virus genotype 8 infection in Northeast Brazil: inheritance from African slaves? Virus Res. 2011;160:333-339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 38] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 54. | Stapleton JT, Foung S, Muerhoff AS, Bukh J, Simmonds P. The GB viruses: a review and proposed classification of GBV-A, GBV-C (HGV), and GBV-D in genus Pegivirus within the family Flaviviridae. J Gen Virol. 2011;92:233-246. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 204] [Cited by in RCA: 231] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 55. | Traoré F, Gormally E, Villar S, Friesen MD, Groopman JD, Vernet G, Diallo S, Hainaut P, Maiga MY. Molecular characteristics of Hepatitis B and chronic liver disease in a cohort of HB carriers from Bamako, Mali. BMC Infect Dis. 2015;15:180. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 56. | Forbi JC, Esona MD, Agwale SM. Molecular characterization of hepatitis A virus isolates from Nigeria. Intervirology. 2013;56:22-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 57. | Tao I, Compaoré TR, Diarra B, Djigma F, Zohoncon TM, Assih M, Ouermi D, Pietra V, Karou SD, Simpore J. Seroepidemiology of hepatitis B and C viruses in the general population of burkina faso. Hepat Res Treat. 2014;2014:781843. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 58. | Boyce CL, Ganova-Raeva L, Archampong TNA, Lartey M, Sagoe KW, Obo-Akwa A, Kenu E, Kwara A, Blackard JT. Identification and comparative analysis of hepatitis B virus genotype D/E recombinants in Africa. Virus Genes. 2017;53:538-547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 9] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 59. | Archampong TN, Boyce CL, Lartey M, Sagoe KW, Obo-Akwa A, Kenu E, Blackard JT, Kwara A. HBV genotypes and drug resistance mutations in antiretroviral treatment-naive and treatment-experienced HBV-HIV-coinfected patients. Antivir Ther. 2017;22:13-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 60. | Huy TT, Ishikawa K, Ampofo W, Izumi T, Nakajima A, Ansah J, Tetteh JO, Nii-Trebi N, Aidoo S, Ofori-Adjei D. Characteristics of hepatitis B virus in Ghana: full length genome sequences indicate the endemicity of genotype E in West Africa. J Med Virol. 2006;78:178-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 61. | Ampah KA, Pinho-Nascimento CA, Kerber S, Asare P, De-Graft D, Adu-Nti F, Paixão IC, Niel C, Yeboah-Manu D, Pluschke G. Limited Genetic Diversity of Hepatitis B Virus in the General Population of the Offin River Valley in Ghana. PLoS One. 2016;11:e0156864. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 62. | Compaore TR, Diarra B, Assih M, Obiri-Yeboah D, Soubeiga ST, Ouattara AK, Tchelougou D, Bisseye C, Bakouan DR, Compaore IP. HBV/HIV co-infection and APOBEC3G polymorphisms in a population from Burkina Faso. BMC Infect Dis. 2016;16:336. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 63. | Cella E, Ceccarelli G, Vita S, Lai A, Presti AL, Blasi A, Palco ML, Guarino MP, Zehender G, Angeletti S. First epidemiological and phylogenetic analysis of Hepatitis B virus infection in migrants from Mali. J Med Virol. 2017;89:639-646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 64. | Mulders MN, Venard V, Njayou M, Edorh AP, Bola Oyefolu AO, Kehinde MO, Muyembe Tamfum JJ, Nebie YK, Maiga I, Ammerlaan W. Low genetic diversity despite hyperendemicity of hepatitis B virus genotype E throughout West Africa. J Infect Dis. 2004;190:400-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 141] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 65. | Fujiwara K, Tanaka Y, Orito E, Ohno T, Kato T, Sugihara K, Hasegawa I, Sakurai M, Ito K, Ozasa A. Distribution of HBV genotypes among HBV carriers in Benin:phylogenetic analysis and virological characteristics of HBV genotype E. World J Gastroenterol. 2005;11:6410-6415. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 24] [Cited by in RCA: 27] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 66. | Candotti D, Opare-Sem O, Rezvan H, Sarkodie F, Allain JP. Molecular and serological characterization of hepatitis B virus in deferred Ghanaian blood donors with and without elevated alanine aminotransferase. J Viral Hepat. 2006;13:715-724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 64] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 67. | Hübschen JM, Andernach IE, Muller CP. Hepatitis B virus genotype E variability in Africa. J Clin Virol. 2008;43:376-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 32] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 68. | Andernach IE, Nolte C, Pape JW, Muller CP. Slave trade and hepatitis B virus genotypes and subgenotypes in Haiti and Africa. Emerg Infect Dis. 2009;15:1222-1228. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 98] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 69. | Forbi JC, Vaughan G, Purdy MA, Campo DS, Xia GL, Ganova-Raeva LM, Ramachandran S, Thai H, Khudyakov YE. Epidemic history and evolutionary dynamics of hepatitis B virus infection in two remote communities in rural Nigeria. PLoS One. 2010;5:e11615. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 83] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 70. | Suzuki S, Sugauchi F, Orito E, Kato H, Usuda S, Siransy L, Arita I, Sakamoto Y, Yoshihara N, El-Gohary A. Distribution of hepatitis B virus (HBV) genotypes among HBV carriers in the Cote d’Ivoire: complete genome sequence and phylogenetic relatedness of HBV genotype E. J Med Virol. 2003;69:459-465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 59] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 71. | Bekondi C, Olinger CM, Boua N, Talarmin A, Venard V, Muller CP, Le Faou A. [Characterization of hepatitis B virus strains from the Central African Republic: preliminary results]. Pathol Biol (Paris). 2008;56:310-313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 72. | Olinger CM, Venard V, Njayou M, Oyefolu AO, Maïga I, Kemp AJ, Omilabu SA, le Faou A, Muller CP. Phylogenetic analysis of the precore/core gene of hepatitis B virus genotypes E and A in West Africa: new subtypes, mixed infections and recombinations. J Gen Virol. 2006;87:1163-1173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 123] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 73. | Dongdem AZ, Dzodzomenyo M, Asmah RH, Nyarko KM, Nortey P, Agyei A, Adjei DN, Kenu E, Adjei AA. Hepatitis B virus genotypes among chronic hepatitis B patients reporting at Korle-Bu teaching hospital, Accra, Ghana. Pan Afr Med J. 2016;25:5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 74. | Flink HJ, van Zonneveld M, Hansen BE, de Man RA, Schalm SW, Janssen HL; HBV 99-01 Study Group. Treatment with Peg-interferon alpha-2b for HBeAg-positive chronic hepatitis B: HBsAg loss is associated with HBV genotype. Am J Gastroenterol. 2006;101:297-303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 203] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 75. | Purdy MA, Forbi JC, Sue A, Layden JE, Switzer WM, Opare-Sem OK, Phillips RO, Khudyakov YE. A re-evaluation of the origin of hepatitis C virus genotype 2 in West Africa. J Gen Virol. 2015;96:2157-2164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 76. | Hønge BL, Jespersen S, Medina C, Té Dda S, da Silva ZJ, Lewin S, Østergaard L, Erikstrup C, Wejse C, Laursen AL. Hepatitis B and Delta virus are prevalent but often subclinical co-infections among HIV infected patients in Guinea-Bissau, West Africa: a cross-sectional study. PLoS One. 2014;9:e99971. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 77. | Geretti AM, King S, Adjei-Asante K, Appiah LT, Owusu DO, Sarfo FS, Chadwick D, Phillips RO, Beloukas A. Hepatitis C Virus (HCV) RNA screening and sequencing using dry plasma spots. J Clin Virol. 2017;97:18-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 78. | Martial J, Morice Y, Abel S, Cabié A, Rat C, Lombard F, Edouard A, Pierre-Louis S, Garsaud P, Béra O. Hepatitis C virus (HCV) genotypes in the Caribbean island of Martinique: evidence for a large radiation of HCV-2 and for a recent introduction from Europe of HCV-4. J Clin Microbiol. 2004;42:784-791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 36] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 79. | Saito T, Ishikawa K-i, Osei-Kwasi M, Kaneko T, Brandful JA, Nuvor V, Aidoo S, Ampofo W, Apeagyei FA, Ansah JE, Adu-Sarkodie Y, Nkrumah FK, Abea K. Prevalence of hepatitis G virus and characterization of viral genome in Ghana. Hepatol Res. 1999;13:221-231. [DOI] [Full Text] |
| 80. | Diarra B, Yonli AT, Sorgho PA, Compaore TR, Ouattara AK, Zongo WA, Tao I, Traore L, Soubeiga ST, Djigma FW. Occult Hepatitis B Virus Infection and Associated Genotypes among HBsAg-negative Subjects in Burkina Faso. Mediterr J Hematol Infect Dis. 2018;10:e2018007. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 81. | Lawson-Ananissoh L, Attia K, Diallo D, Doffou S, Kissi Y, Bangoura D, Kouame D, Mahassadi K, Yao-Bathaix F, Yoman T. Distribution and Clinical Implications of the Genotypes of the Hepatitis B Virus in 33 Chronic Carriers of Hepatitis B Virus in Cote-d’Ivoire. J Afr Hepato Gastroentero. 2017;11:1-5. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 82. | Opaleye OO, Japhet OM, Adewumi OM, Omoruyi EC, Akanbi OA, Oluremi AS, Wang B, Tong Hv, Velavan TP, Bock CT. Molecular epidemiology of hepatitis D virus circulating in Southwestern Nigeria. Virol J. 2016;13:61. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 83. | Candotti D, Diarra B, Bisseye C, Tao I, Pham Quang K, Sanou M, Laperche S, Sanogo R, Allain JP, Simpore J. Molecular characterization of hepatitis B virus in blood donors from Burkina Faso: Prevalence of quasi-subgenotype A3, genotype E, and mixed infections. J Med Virol. 2016;88:2145-2156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 84. | Brah S, Moussa S, Inoua A, Alhousseini DM, Daou M, Madougou B, Romera MH, Hamadou A, Adehossi E, Parola P. Molecular characterization of hepatitis B virus from chronically-infected patients in Niamey, Niger. Int J Infect Dis. 2016;45:18-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 85. | Boyd A, Maylin S, Moh R, Mahjoub N, Gabillard D, Eholié SP, Danel C, Anglaret X, Zoulim F, Girard PM. Hepatitis B surface antigen quantification as a predictor of seroclearance during treatment in HIV-hepatitis B virus coinfected patients from Sub-Saharan Africa. J Gastroenterol Hepatol. 2016;31:634-644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 86. | Faleye TO, Adewumi MO, Ifeorah IM, Omoruyi EC, Bakarey SA, Akere A, Awokunle F, Ajibola HO, Makanjuola DO, Adeniji JA. Detection of hepatitis B virus isolates with mutations associated with immune escape mutants among pregnant women in Ibadan, southwestern Nigeria. Springerplus. 2015;4:43. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 87. | Faleye TO, Adewumi OM, Ifeorah IM, Akere A, Bakarey AS, Omoruyi EC, Oketunde K, Awonusi OB, Ajayi MR, Adeniji JA. Detection and circulation of hepatitis B virus immune escape mutants among asymptomatic community dwellers in Ibadan, southwestern Nigeria. Int J Infect Dis. 2015;39:102-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 88. | Maylin S, Sire JM, Mbaye PS, Simon F, Sarr A, Evra ML, Fall F, Daveiga J, Diallo A, Debonne JM. Short-term spontaneous fluctuations of HBV DNA levels in a Senegalese population with chronic hepatitis B. BMC Infect Dis. 2015;15:154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 89. | Hübschen JM, Mbah PO, Forbi JC, Otegbayo JA, Olinger CM, Charpentier E, Muller CP. Detection of a new subgenotype of hepatitis B virus genotype A in Cameroon but not in neighbouring Nigeria. Clin Microbiol Infect. 2011;17:88-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 37] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 90. | Geretti AM, Patel M, Sarfo FS, Chadwick D, Verheyen J, Fraune M, Garcia A, Phillips RO. Detection of highly prevalent hepatitis B virus coinfection among HIV-seropositive persons in Ghana. J Clin Microbiol. 2010;48:3223-3230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 77] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 91. | Candotti D, Danso K, Allain JP. Maternofetal transmission of hepatitis B virus genotype E in Ghana, west Africa. J Gen Virol. 2007;88:2686-2695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 88] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 92. | Vray M, Debonne JM, Sire JM, Tran N, Chevalier B, Plantier JC, Fall F, Vernet G, Simon F, Mb PS. Molecular epidemiology of hepatitis B virus in Dakar, Sénégal. J Med Virol. 2006;78:329-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 44] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 93. | Abubakar UM, Yahaya M, Maishanu SH, Ibrahim I, Ishaq AR, Nnaemeka AM, Ahmad AS, Yahaya M. Molecular Epidemiology of HCV Genotype in Relation to Viral Load of Infected Individuals in Northwestern Nigeria. GJMS. 2017;. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 94. | Ndiaye O, Gozlan J, Diop-Ndiaye H, Sall AS, Chapelain S, Leprêtre A, Maynart M, Gueye M, Lo G, Thiam M. Usefulness of Dried Blood Spots (DBS) to perform hepatitis C virus genotyping in drug users in Senegal. J Med Virol. 2017;89:484-488. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 95. | Henquell C, Yameogo S, Sangaré L. First genome characterization of a novel hepatitis C virus genotype 5 variant. Infect Genet Evol. 2016;39:173-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 96. | Zeba MT, Sanou M, Bisseye C, Kiba A, Nagalo BM, Djigma FW, Compaoré TR, Nebié YK, Kienou K, Sagna T. Characterisation of hepatitis C virus genotype among blood donors at the regional blood transfusion centre of Ouagadougou, Burkina Faso. Blood Transfus. 2014;12 Suppl 1:s54-s57. [PubMed] |
| 97. | Diarra M, Konaté A, Diakité Y, Samaké KD, Coulibaly HS, Kassambra Y, Tounkara M, Kaya AS, Kallé A, Sidibé AT. Hepatitis C virus infection among diabetics in CHU Gabriel Touré and Bamako Center of diabetes control (Mali). J Afr Hepato Gastroenterol. 2013;188. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 98. | Bouare N, Gothot A, Delwaide J, Bontems S, Vaira D, Seidel L, Gerard P, Gerard C. Epidemiological profiles of human immunodeficiency virus and hepatitis C virus infections in Malian women: Risk factors and relevance of disparities. World J Hepatol. 2013;5:196-205. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 1.0] [Reference Citation Analysis (6)] |
| 99. | Sombie R, Bougouma A, Somda S, Sangare L, Lompo O, Kabore Z, Tieno H, Drabo J, Ilboudo D. Chronic hepatitis C: epidemiology, diagnosis and treatment in Yalgado-Ouedraogo teaching hospital in Ouagadougou. J Afr Hepato Gastroenterol. 2011;1:6-13. [RCA] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 100. | Bengue AK-M, Kouacou MJL, Ekaza E, Siransy-Bogui L, Nrsquo DC, Labonté P, Dosso M. Hepatitis C virus infection in Abidjan Cote d Ivoire: heterogeneity of genotypes. Sci Res Essays. 2008;139. |
| 101. | Plamondon M, Labbé AC, Frost E, Deslandes S, Alves AC, Bastien N, Pepin J. Hepatitis C virus infection in Guinea-Bissau: a sexually transmitted genotype 2 with parenteral amplification? PLoS One. 2007;2:e372. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 102. | Simpore J, Ilboudo D, Samandoulougou A, Guardo P, Castronovo P, Musumeci S. HCV and HIV co-infection in pregnant women attending St. Camille Medical Centre in Ouagadougou (Burkina Faso). J Med Virol. 2005;75:209-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 103. | Rouet F, Chaix ML, Inwoley A, Msellati P, Viho I, Combe P, Leroy V, Dabis F, Rouzioux C; ANRS 1236 DITRAME-B&C Study Group. HBV and HCV prevalence and viraemia in HIV-positive and HIV-negative pregnant women in Abidjan, Côte d’Ivoire: the ANRS 1236 study. J Med Virol. 2004;74:34-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 103] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 104. | Agwale SM, Tanimoto L, Womack C, Odama L, Leung K, Duey D, Negedu-Momoh R, Audu I, Mohammed SB, Inyang U. Prevalence of HCV coinfection in HIV-infected individuals in Nigeria and characterization of HCV genotypes. J Clin Virol. 2004;31 Suppl 1:S3-S6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 32] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 105. | Buisson Y, Grandadam M, Nicand E, Cheval P, van Cuyck-Gandre H, Innis B, Rehel P, Coursaget P, Teyssou R, Tsarev S. Identification of a novel hepatitis E virus in Nigeria. J Gen Virol. 2000;81:903-909. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 79] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 106. | Wansbrough-Jones MH, Frimpong E, Cant B, Harris K, Evans MR, Teo CG. Prevalence and genotype of hepatitis C virus infection in pregnant women and blood donors in Ghana. Trans R Soc Trop Med Hyg. 1998;92:496-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 42] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 107. | Oni AO, Harrison TJ. Genotypes of hepatitis C virus in Nigeria. J Med Virol. 1996;49:178-186. [DOI] [Full Text] |
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Burkina Faso
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C, C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Arriagada GL, Chen CJ, Petruzziello A S- Editor: Ji FF L- Editor: Filipodia E- Editor: Tan WW